Significance
Hypothalamic agouti-related peptide (AGRP) neurons control food intake and body weight. G protein-coupled receptor 17 (GPR17) was recently shown to be expressed in these neurons and controls their activity, thereby reducing body weight and food intake in mice. In the current study, we demonstrate that Gpr17-deficient mice have normal hypothalamic and circulating AGRP levels. Body weight, food intake, and glucose homeostasis appear normal in the GPR17-deficient mice. The current data do not validate GPR17 as a therapeutic target for obesity or type 2 diabetes.
Keywords: GPR17, AGRP, diabetes, body weight, food intake
Abstract
G protein-coupled receptor 17 (GPR17) was recently reported to be a Foxo1 target in agouti-related peptide (AGRP) neurons. Intracerebroventricular injection of GPR17 agonists induced food intake, whereas administration of an antagonist to the receptor reduced feeding. These data lead to the conclusion that pharmacological modulation of GPR17 has therapeutic potential to treat obesity. Here we report that mice deficient in Gpr17 (Gpr17−/−) have similar food intake and body weight compared with their wild-type littermates. Gpr17−/− mice have normal hypothalamic Agrp mRNA expression, AGRP plasma levels, and metabolic rate. GPR17 deficiency in mice did not affect glucose homeostasis or prevent fat-induced insulin resistance. These data do not support a role for GPR17 in the control of food intake, body weight, or glycemic control.
Hypothalamic neurons expressing agouti-related peptide (AGRP) play an important role in feeding (1–3). AGRP acts as an inverse agonist on melanocortin 4 receptors (4, 5). Leptin and insulin are anorectic hormones, which partly regulate food intake and energy homeostasis by inhibiting AGRP neurons (6–8). The transcription factor Foxo1 is expressed in AGRP neurons and represents a shared mediator of the pathways activated by insulin and leptin to suppress food intake (9, 10). Accordingly, Foxo1 expression is reduced by insulin and leptin and these hormones’ effect on feeding is inhibited following hypothalamic Foxo1 overexpression (9). Moreover, activation of Foxo1 in the hypothalamus increases food intake and body weight, whereas inhibition of Foxo1 decreases both (9). Finally, ablation of Foxo1 in AGRP neurons results in reduced food intake, leanness, and improved glycemic control (11).
The G protein-coupled receptor, GPR17, is a prominent Foxo1 target and highly expressed in AGRP neurons (11). GPR17 is phylogenetically related to nucleotide P2Y and cysteinyl-leukotriene receptors (12). It has been reported that uridyl-diphosphate glucose (UDP glucose) and leukotriene D4 (LTD4) activate GPR17, whereas the P2Y12 antagonist, Cangrelor, suppresses receptor activity (13). Consistent with these findings, intracerebroventricular injection of UDP glucose and LTD4 in mice stimulated food intake, whereas administration of Cangrelor suppressed it (11). However, other studies have failed to demonstrate activation of GPR17 with UDP glucose and LTD4 (14, 15). In this study, we generated and fed Gpr17-deficient (Gpr17−/−) mice a normal and a high-fat diet (HFD). Under these experimental conditions, we did not observe an effect of GPR17 on food intake, body weight, or glycemic control. These data call into question the therapeutic potential of inhibiting GPR17 to treat obesity and related metabolic disorders.
Results
Reporter Gene Expression from the Gpr17 Locus and AGRP Plasma Levels in Gpr17−/− and Wild-Type Mice.
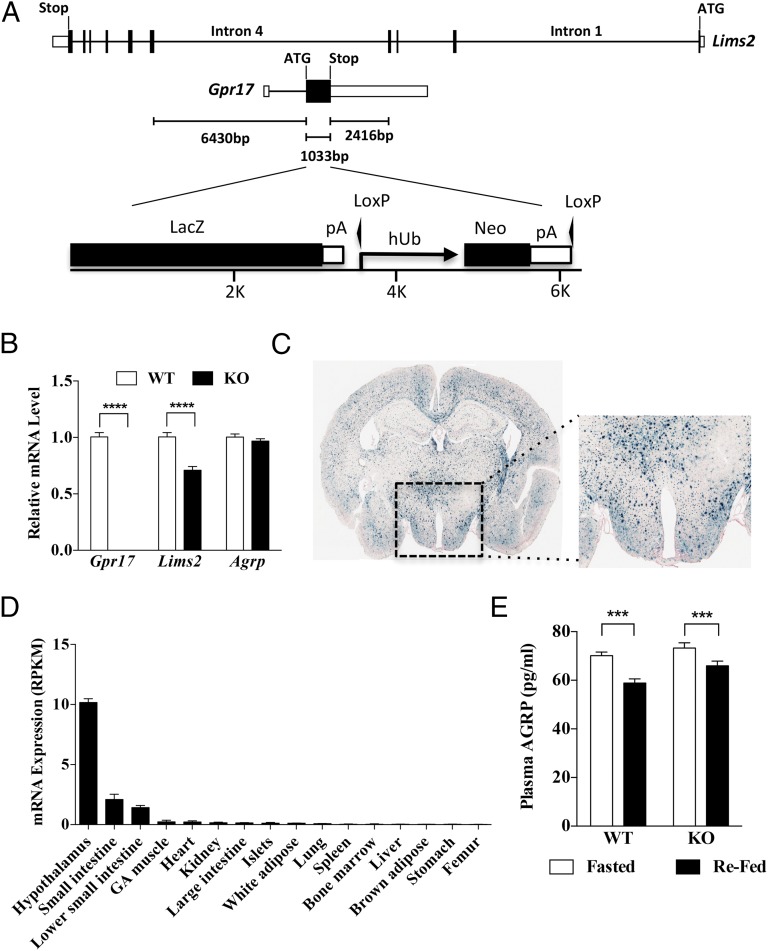
Gpr17−/− mice were developed by homologous recombination using Regeneron’s VelociGene technology. The murine Gpr17 gene [National Center for Biotechnology Information (NCBI) Reference Sequence: NM_001025381.2] is a single exon contained entirely within the coding region of the Lims2 gene (NCBI Reference Sequence: NM_144862.3), albeit on the opposite strand. We deleted a 1,032-bp segment with the lacZ fused in frame just after Gpr17’s ATG without deleting any exons from Lims2 (Fig. 1A). No Gpr17 mRNA was detected in the hypothalamus of knockout (KO) animals (Fig. 1B). Lims2 expression was reduced by 29% in Gpr17−/− mice (Fig. 1B). Gpr17−/− mice were born in the expected Mendelian ratios. Reporter expression was found in multiple areas of the brain and the hypothalamus (Fig. 1C), consistent with a previous report (16). Gpr17 is expressed in few peripheral tissues at lower levels than in the hypothalamus (Fig. 1D). Gpr17−/− and wild-type mice had similar levels of hypothalamic Agrp mRNA and circulating AGRP in the fasted state (Fig. 1 B and E). Refeeding of Gpr17−/− and control mice produced an equivalent decrement in the AGRP plasma levels (Fig. 1E).
Fig. 1.
Locus organization, brain lacZ reporter gene expression, and AGRP plasma levels of Gpr17−/− mice. (A) Gpr17 is nested inside Lims2 in the reverse strand such that its single coding exon is fully contained in the long intron 4 (9,879 bp) of Lims2. Coding exons are shown as black boxes, UTR as white boxes, introns as straight line. Deletion of the VG12229 allele is well contained inside the Lims2 intron, far away from splicing signals (distances shown). LacZ-floxed-Neo cassette inserted in frame after Gpr17's ATG. Drug resistance coding sequence was driven by human ubiquitin promoter. (B) Gpr17, Lims2, and Agrp mRNA levels in hypothalamus from Gpr17−/− (KO) and wild-type (WT) mice. Data expressed in reads per kilobase per million (RPKM). (C) LacZ reporter staining in the endogenous Gpr17 loci in whole brain section. Inset shows region of the hypothalamus at 5× magnification. (D) Gpr17 expression using RNAseq in 16 mouse tissues of C57BL/6 mice on chow diet (n = 6 per group). (E) AGRP plasma levels in fasted (4 h) and refed Gpr17−/− (KO) and wild-type (WT) mice. All groups had seven animals if not otherwise indicated. Values are mean ± SEM. ***P < 0.001; ****P < 0.0001.
Gpr17−/− Mice Have Normal Food Intake, Body Weight, and Metabolic Rate.
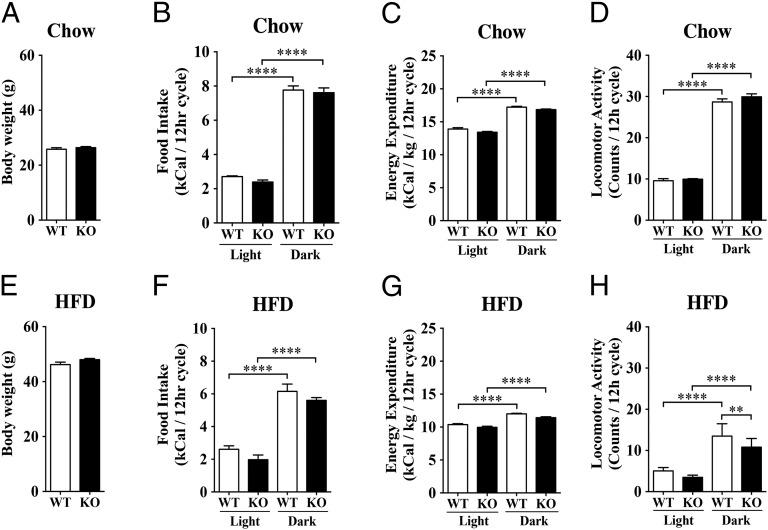
Mice lacking GPR17 had normal body weight compared with their wild-type littermates (Fig. 2A). Consistent with this data, there were no differences in the light and dark phases of the light cycle in food intake (Fig. 2B) or energy expenditure (Fig. 2C). Data for oxygen consumption (VO2), carbon dioxide production (VCO2), respiratory quotient (RER), energy expenditure, and food intake in the light and dark phases of the light cycle are shown in Fig. S1 A–E. Locomotor activity was similar between both groups of mice (Fig. 2D and Fig. S1F). Gpr17−/− and wild-type mice fed a HFD for 10 wk had a 70% increase in body weight, but showed no difference in food intake or energy expenditure (Fig. 2 E–G and Fig. S1 G–K). There were no significant differences in body weight between the KO mice and controls at any time point over the first 9 wk of high-fat feeding. Locomotor activity was slightly reduced in HFD mice (Fig. 2H and Fig. S1L). No genotypic differences in body composition were observed in mice on standard chow or HFD, except the Gpr17−/− mice on the chow diet tended to have higher body fat content (Table 1). Hypothalamic Gpr17 and Agrp mRNA expression did not change in response to high-fat feeding (Fig. S2A). AGRP plasma levels were similar between Gpr17−/− and wild-type mice fed a HFD (Fig. S2B). These data suggest that ablation of GPR17 does not affect food intake or body weight in mice.
Fig. 2.
Body weight, food intake, energy expenditure, and locomotor activity in Gpr17−/− and wild-type mice. (A and E) Body weight was measured in male wild-type (WT) and Gpr17−/− (KO) mice on chow diet (13 wk of age) and high-fat feeding (HFD; 24 wk of age). Mice were on HFD from weeks 14–24. (B and F) Food intake during dark and light phases of the light cycle in wild-type (WT) and Gpr17−/− (KO) mice on chow diet or HFD. (C and G) Energy expenditure during dark and light phases of the light cycle in wild-type (WT) and Gpr17−/− (KO) mice on chow diet or HFD. (D and H) Locomotor activity during dark and light phases of the light cycle in wild-type (WT) and Gpr17−/− (KO) mice on chow diet or HFD. All groups had eight animals. Values are mean ± SEM. **P < 0.01; ****P < 0.0001.
Table 1.
Body composition measurements in Gpr17−/− and wild-type mice
| Chow | High-fat diet | |||
| WT | KO | WT | KO | |
| Body weight, g | 25.9 ± 0.5 | 26.4 ± 0.4 | 44.1 ± 0.9 | 45.9 ± 0.6 |
| Lean volume, cm3 | 16.9 ± 0.4 | 16.7 ± 0.3 | 18.1 ± 0.5 | 18.6 ± 0.5 |
| Fat volume, cm3 | 3.02 ± 0.15 | 3.89 ± 0.24* | 20.0 ± 0.78 | 21.6 ± 0.36 |
| Bone volume, cm3 | 1.48 ± 0.03 | 1.44 ± 0.02 | 1.48 ± 0.03 | 1.47 ± 0.02 |
| Bone density, mgHA/cm3 | 304 ± 3.1 | 300 ± 3.7 | 306 ± 1.6 | 305 ± 1.6 |
| Bone mineral content, mgHA | 449 ± 13.2 | 433 ± 8.6 | 454 ± 10.7 | 448 ± 7.7 |
| Body fat, % | 11.0 ± 0.6 | 14.0 ± 1.0* | 41.7 ± 1.4 | 43.2 ± 0.7 |
Body composition was measured using μCT in Gpr17−/− (KO) and wild-type (WT) mice on chow diet (13 wk of age) and following 10 wk of high-fat feeding. All groups had eight animals. Values are mean ± SEM. *P < 0.05.
Gpr17−/− Mice Have Normal Glucose Homeostasis and Insulin Sensitivity.
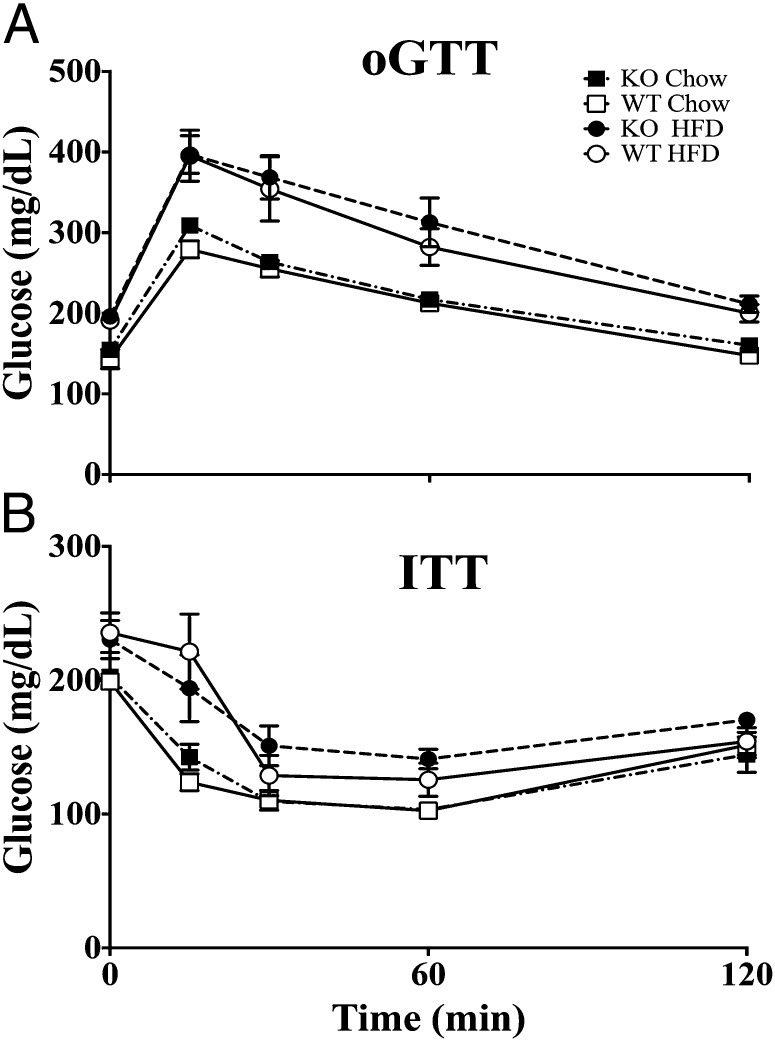
Gpr17−/− mice had normal glycemic control as revealed by an oral glucose tolerance test (oGTT) (Fig. 3A) or insulin tolerance test (ITT) (Fig. 3B) compared with wild-type mice. Feeding Gpr17−/− and wild-type mice a HFD for 10 wk impaired glucose tolerance (Fig. 3A) and insulin sensitivity (Fig. 3B). No genotypic differences in glucose or insulin tolerance were observed in mice on normal chow or HFD. These data show that GPR17 does not control glucose homeostasis or protect from glucose intolerance in response to a HFD.
Fig. 3.
Oral glucose and insulin tolerance tests in Gpr17−/− and wild-type mice. (A) Glucose tolerance test in male wild-type (WT) and Gpr17−/− (KO) mice on chow diet (11 wk of age) and high-fat feeding (HFD; 23 wk of age). Mice were on HFD from weeks 14 to 24. (B) Insulin tolerance test in wild-type (WT) and Gpr17−/− (KO) mice on chow diet and HFD for 11 wk. All groups had eight animals. Values are mean ± SEM.
Discussion
The main findings of the present study are that (i) ablation of GPR17 does not regulate circulating AGRP levels, food intake, metabolic rate, and body weight and (ii) GPR17 does not control glucose homeostasis or prevent high-fat–induced insulin resistance.
GPR17 is abundantly expressed in AGRP neurons and was recently suggested to control feeding using pharmacological modulators of the receptor and intracerebroventricular administration in mice (11). However, these compounds have subsequently been questioned as pharmacological modulators of GPR17 (14, 15). In this study, we confirm that Gpr17 is highly expressed in the hypothalamus of mice. However, wild-type and Gpr17−/− mice show no differences in food intake or metabolic rate and can mount a significant and similar increase in body weight in response to high-fat feeding. Gpr17−/− mice have normal Agrp hypothalamic mRNA expression and circulating AGRP levels.
The Gpr17 gene is contained entirely within the coding region of the Lims2 gene, but on the opposite strand. LIMS2 belongs to the family of focal adhesion proteins and functions as a component of the integrin signaling pathway (17). The small but significant reduction in Lims2 mRNA expression is unlikely to affect the metabolism of Gpr17−/− mice because Lims2-deficient mice do not have an apparent phenotype (18) and Agrp expression was similar in Gpr17−/− and wild-type mice.
β-Galactosidase (lacZ) staining of GPR17 in the brain showed widespread expression, suggesting that the receptor has a more general function rather than a specific role in AGRP neurons. Previous studies have shown that GPR17 is expressed in oligodendrocyte precursors (19) and negatively regulated by the oligodendrocyte maturation transcription factor, Olig1 (20). Whereas ablation of GPR17 in mice caused only a slight advance in central nervous system (CNS) myelination, overexpression of GPR17 inhibited myelinogenesis within the CNS of mice (16), suggesting it plays a role not just in development of the CNS, but could also negatively impact the adult nervous system following traumatic injury. Consistent with this data, knockdown of Gpr17 mRNA levels in a focal ischemia rat model attenuated short-term neuron loss, brain atrophy, and microglial activation after reperfusion (21). Thus, GPR17 seems to control oligodendrocyte function rather than AGRP neuronal activity.
The Agrp-Foxo1−/− mice (which have low Gpr17 expression) exhibit reduced hepatic glucose production and improved glucose homeostasis (11). Here we demonstrate a lack of involvement of GPR17 in the regulation of glucose control. Moreover, Gpr17−/− mice are not protected from HFD-induced insulin resistance. The nature of the Foxo1 target gene(s) mediating the beneficial effects on food intake and glycemic control in the Agrp-Foxo1−/− mice remains unknown.
In conclusion, the present data do not support a role for GPR17 in the regulation of food intake or glucose homeostasis. Thus, GPR17 does not appear to be a viable therapeutic target for the treatment of obesity and type 2 diabetes.
Materials and Methods
Animals.
Gpr17−/− mice (100% C57BL/6NTac background) were generated by homologous recombination using Regeneron’s VelociGene technology (22). VelociGene allele identification number is VG12229. Male mice were used throughout this study and housed four to five per cage in a controlled environment (12-h light/dark cycle, 23 ± 1 °C, 60–70% humidity) and fed ad libitum with standard chow (Purina Laboratory Rodent Diet 5001, LabDiet). Some mice were fed a high-fat diet (Research Diets, D12492; 60% fat by calories). In the food intake, body weight, metabolic rate, and glucose studies, comparisons were made between wild-type and knockout littermates. C57BL/6 mice (males; Taconic) were used for Gpr17 expression studies. All animal procedures were conducted in compliance with protocols approved by the Regeneron Pharmaceuticals Institutional Animal Care and Use Committee.
Glucose Homeostasis Measurements.
For the insulin tolerance test, mice (male; 10 wk of age) were fasted for 4 h before testing. The fast started at 9:00 AM, 2 h after initiation of the light cycle. Fasting glucose was measured via tail bleed using the Accu-Chek Compact Plus (Roche Diagnostics). Insulin stock was prepared from Humulin R (Lilly). Mice on normal chow were injected i.p. with 0.75 units insulin/kg body weight. Mice on the HFD (male; 23 wk of age; subjected to high-fat feeding for 10 wk) were injected i.p. with 2.0 units insulin/kg body weight. Blood glucose was measured with the glucometer via tail bleed at 0, 15, 30, 60, and 120 min postinjection. For the oral glucose tolerance test, mice were fasted overnight (16 h) followed by oral gavage of glucose at 2 g/kg body weight. The fast started at 5:00 PM, 2 h before initiation of the dark cycle. Blood glucose level was evaluated at 0, 15, 30, 60, and 120 min postinjection.
Body Composition Assessments.
MicroCT (μCT) measurements were performed using the Quantum FX micro CT preclinical in vivo imaging system (Caliper Life Sciences). Mice (male) fed the chow diet were 13 wk of age at time of μCT; mice (male) fed the HFD were 24 wk of age at time of μCT and on the HFD for the last 11 wk. Mice were anesthetized using Forane (Baxter). Scans were performed using field of view setting = 60 and scan technique = Dyn17Sec2. Data conversion was performed using the Caliper analysis software (Caliper Life Sciences).
Metabolic Rate and Food Intake Measurements.
Metabolic cage data were generated using the Oxymax Lab Animal Monitoring System: CLAMS (Columbus Instruments). Mice (male; 9 wk of age) were individually monitored in cages with center feeds for 96 h. Food intake was measured continuously and divided into calories consumed per light and dark phase of the light cycle. VO2 and VCO2 were measured in 17-min intervals over a 4-d span and plotted over time in hours. Energy expenditure was calculated as a function of the respiratory quotient and the oxygen consumption, normalized to body weight.
RNA Preparation and RNA Sequencing Read Mapping.
Total RNA was purified using MagMAX-96 for Microarrays Total RNA Isolation kit (Ambion), according to manufacturer specifications. Genomic DNA was removed using MagMAXTurboDNase buffer and TURBO DNase from the MagMAX kit listed above. mRNA was purified from total RNA using Dynabeads mRNA Purification kit (Invitrogen). Strand-specific RNA sequencing (RNA-seq) libraries were prepared using ScriptSeq mRNA-Seq Library Preparation kit (Epicentre). Twelve-cycle PCR was performed to amplify libraries. Sequencing was performed on Illumina HiSeq2000 by a multiplexed single-read run with 33 cycles. Raw sequence data (BCL files) were converted to FASTQ format via Illumina Casava 1.8.2. Reads were decoded based on their barcodes and read quality was evaluated with FastQC (www.bioinformatics.babraham.ac.uk/projects/fastqc/). Reads were mapped to the mouse transcriptome (NCBI Build37.2) using Bowtie (bowtie-bio.sourceforge.net/index.shtml) allowing two mismatches. Reads mapped to the sense-strand exons of a gene were summed at the gene level.
Serum Chemistry.
Serum chemistry analysis was performed using the ADVIA 1800 Clinical Chemistry System (Siemens Healthcare Diagnostics). Whole mouse blood was collected by retroperitoneal bleed under anesthesia into a BD Microtainer serum separator tube and stored at −20 °C.
Hypothalamic Gene Expression.
Hypothalamus was dissected from whole brain using an acrylic mouse brain block (Harvard Apparatus), placed in RNAlater (Qiagen), and homogenized in a Precellys mill using the Precellys Ceramic kit 1.4/2.8 mm (Peqlab). Total RNA was isolated with the RNeasy Fibrous Tissue Mini kit (Qiagen), including DNase I digestion and quantified with the Quant-iT RiboGreen RNA assay (Life Technologies). Reverse transcription to cDNA was done using the BioRad iScript cDNA synthesis kit. Gpr17, Agrp, and Lims2 cDNA were quantified with the digital droplet PCR method (23) using the ddPCR Supermix for probes (no UDP) kit on a QX-200 system (BioRad). TaqMan primer-probe sets are available upon request.
LacZ Staining.
LacZ histochemistry was performed as described earlier (24). Slides were scanned on Aperio ScanScope AT Turbo (Leica) and annotated with Aperio 5ImageScope.
Plasma AGRP Measurements.
The concentration of circulating AGRP was measured in mouse EDTA plasma using the mouse AGRP fluorescent immunoassay kit from Phoenix Pharmaceuticals, according to the manufacturer’s instructions. Signal calibration was done based on a standard curve obtained with known AGRP concentrations between 10 and 1,000 pg/mL.
Data Analysis.
All data are expressed as mean ± SEM. Two-way ANOVA with Bonferroni’s post hoc analysis was performed on data collected for hypothalamic gene expression, ITT, oGTT, and metabolic cage studies. Unpaired Student’s t test was used to analyze data collected from the μCT and serum chemistry studies. All analysis was performed using GraphPad PRISM 6.0e (GraphPad Software).
Supplementary Material
Acknowledgments
We thank Esther Latres, Claire Kammermeier, Martin Stephan, Pierre Wenski, Jörn Wandschneider, Uwe Butty, and Alejo Mujica for advice and excellent technical assistance.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1424968112/-/DCSupplemental.
References
- 1.Ollmann MM, et al. Antagonism of central melanocortin receptors in vitro and in vivo by agouti-related protein. Science. 1997;278(5335):135–138. doi: 10.1126/science.278.5335.135. [DOI] [PubMed] [Google Scholar]
- 2.Wortley KE, et al. Agouti-related protein-deficient mice display an age-related lean phenotype. Cell Metab. 2005;2(6):421–427. doi: 10.1016/j.cmet.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 3.Gropp E, et al. Agouti-related peptide-expressing neurons are mandatory for feeding. Nat Neurosci. 2005;8(10):1289–1291. doi: 10.1038/nn1548. [DOI] [PubMed] [Google Scholar]
- 4.Haskell-Luevano C, Monck EK. Agouti-related protein functions as an inverse agonist at a constitutively active brain melanocortin-4 receptor. Regul Pept. 2001;99(1):1–7. doi: 10.1016/s0167-0115(01)00234-8. [DOI] [PubMed] [Google Scholar]
- 5.Chai BX, et al. Inverse agonist activity of agouti and agouti-related protein. Peptides. 2003;24(4):603–609. doi: 10.1016/s0196-9781(03)00104-9. [DOI] [PubMed] [Google Scholar]
- 6.Breen TL, Conwell IM, Wardlaw SL. Effects of fasting, leptin, and insulin on AGRP and POMC peptide release in the hypothalamus. Brain Res. 2005;1032(1-2):141–148. doi: 10.1016/j.brainres.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 7.Takahashi KA, Cone RD. Fasting induces a large, leptin-dependent increase in the intrinsic action potential frequency of orexigenic arcuate nucleus neuropeptide Y/Agouti-related protein neurons. Endocrinology. 2005;146(3):1043–1047. doi: 10.1210/en.2004-1397. [DOI] [PubMed] [Google Scholar]
- 8.Könner AC, et al. Insulin action in AgRP-expressing neurons is required for suppression of hepatic glucose production. Cell Metab. 2007;5(6):438–449. doi: 10.1016/j.cmet.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 9.Kim MS, et al. Role of hypothalamic Foxo1 in the regulation of food intake and energy homeostasis. Nat Neurosci. 2006;9(7):901–906. doi: 10.1038/nn1731. [DOI] [PubMed] [Google Scholar]
- 10.Kitamura T, et al. Forkhead protein FoxO1 mediates Agrp-dependent effects of leptin on food intake. Nat Med. 2006;12(5):534–540. doi: 10.1038/nm1392. [DOI] [PubMed] [Google Scholar]
- 11.Ren H, et al. FoxO1 target Gpr17 activates AgRP neurons to regulate food intake. Cell. 2012;149(6):1314–1326. doi: 10.1016/j.cell.2012.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parravicini C, Ranghino G, Abbracchio MP, Fantucci P. GPR17: Molecular modeling and dynamics studies of the 3-D structure and purinergic ligand binding features in comparison with P2Y receptors. BMC Bioinformatics. 2008;9:263. doi: 10.1186/1471-2105-9-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ciana P, et al. The orphan receptor GPR17 identified as a new dual uracil nucleotides/cysteinyl-leukotrienes receptor. EMBO J. 2006;25(19):4615–4627. doi: 10.1038/sj.emboj.7601341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qi AD, Harden TK, Nicholas RA. Is GPR17 a P2Y/leukotriene receptor? Examination of uracil nucleotides, nucleotide sugars, and cysteinyl leukotrienes as agonists of GPR17. J Pharmacol Exp Ther. 2013;347(1):38–46. doi: 10.1124/jpet.113.207647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hennen S, et al. Decoding signaling and function of the orphan G protein-coupled receptor GPR17 with a small-molecule agonist. Sci Signal. 2013;6(298):ra93. doi: 10.1126/scisignal.2004350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Y, et al. The oligodendrocyte-specific G protein-coupled receptor GPR17 is a cell-intrinsic timer of myelination. Nat Neurosci. 2009;12(11):1398–1406. doi: 10.1038/nn.2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y, Chen K, Guo L, Wu C. Characterization of PINCH-2, a new focal adhesion protein that regulates the PINCH-1-ILK interaction, cell spreading, and migration. J Biol Chem. 2002;277(41):38328–38338. doi: 10.1074/jbc.M205576200. [DOI] [PubMed] [Google Scholar]
- 18.Stanchi F, et al. Consequences of loss of PINCH2 expression in mice. J Cell Sci. 2005;118(Pt 24):5899–5910. doi: 10.1242/jcs.02686. [DOI] [PubMed] [Google Scholar]
- 19.Fratangeli A, et al. The regulated expression, intracellular trafficking, and membrane recycling of the P2Y-like receptor GPR17 in Oli-neu oligodendroglial cells. J Biol Chem. 2013;288(7):5241–5256. doi: 10.1074/jbc.M112.404996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xin M, et al. Myelinogenesis and axonal recognition by oligodendrocytes in brain are uncoupled in Olig1-null mice. J Neurosci. 2005;25(6):1354–1365. doi: 10.1523/JNEUROSCI.3034-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao B, et al. The new P2Y-like receptor G protein-coupled receptor 17 mediates acute neuronal injury and late microgliosis after focal cerebral ischemia in rats. Neuroscience. 2012;202:42–57. doi: 10.1016/j.neuroscience.2011.11.066. [DOI] [PubMed] [Google Scholar]
- 22.Valenzuela DM, et al. High-throughput engineering of the mouse genome coupled with high-resolution expression analysis. Nat Biotechnol. 2003;21(6):652–659. doi: 10.1038/nbt822. [DOI] [PubMed] [Google Scholar]
- 23.Hindson BJ, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem. 2011;83(22):8604–8610. doi: 10.1021/ac202028g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adams N, Gale N. High resolution gene expression analysis in mice using genetically inserted reporter genes. In: Lois SPC, editor. Mammalian and Avian Transgenesis—New Approaches. Springer; Heidelberg: 2006. pp. 131–172. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.