Significance
Globular proteins perform many of the chemical reactions required for life. The prevailing model of globular protein structure, which is based on studies in dilute solutions, emphasizes the requirement for a well-packed hydrophobic interior, but minimizes the importance of the exterior, provided it is hydrophilic. We demonstrate that the exterior plays a significant role when a globular protein is studied under physiologically relevant conditions. By changing a surface residue we show that attractive interactions between the protein surface and the cytosol modulate the stability of the protein, even though the change has a negligible effect in dilute solution. Recognizing and quantifying such intracellular interactions will aid in understanding and manipulating the biological role of proteins.
Keywords: H/D exchange, quinary interactions, protein NMR, protein thermodynamics
Abstract
Protein quinary interactions organize the cellular interior and its metabolism. Although the interactions stabilizing secondary, tertiary, and quaternary protein structure are well defined, details about the protein–matrix contacts that comprise quinary structure remain elusive. This gap exists because proteins function in the crowded cellular environment, but are traditionally studied in simple buffered solutions. We use NMR-detected H/D exchange to quantify quinary interactions between the B1 domain of protein G and the cytosol of Escherichia coli. We demonstrate that a surface mutation in this protein is 10-fold more destabilizing in cells than in buffer, a surprising result that firmly establishes the significance of quinary interactions. Remarkably, the energy involved in these interactions can be as large as the energies that stabilize specific protein complexes. These results will drive the critical task of implementing quinary structure into models for understanding the proteome.
The inside of cells is packed with macromolecules whose concentrations reach 300–400 g/L (1). Compared with the ideal (dilute) environments conventionally used to study proteins, crowding inside cells can significantly alter the biophysical landscape of proteins, including their equilibrium thermodynamic stability (2–6). Experimental and computational efforts establish that crowding effects arise from a combination of short-range (steric) repulsions and longer-range (often called soft) interactions between macromolecules (7–13). Despite a growing number of in-cell protein studies (2–6), there is no quantitative information about the energetics of quinary interactions.
Amide proton exchange experiments have been used for more than 50 y to measure equilibrium protein stability, defined as the Gibbs free energy required to open the protein and expose individual backbone amide protons to solvent, (14). For the B1 domain of protein G (GB1), equals −RTln(kobs/kuns), where R is the gas constant, T is the absolute temperature, kobs is the observed rate of exchange, and kuns is the rate in an unstructured peptide (6). We know that the cytoplasm does not affect kuns (15). Most importantly, we know that for exchange under these conditions approximates the free energy required to denature the protein, (6). Therefore, these experiments provide a thermodynamically rigorous measure of equilibrium global protein stability. Using this information, we quantified the stability of GB1 at the residue level in Escherichia coli (6) via NMR-detected backbone amide hydrogen/deuterium exchange (16).
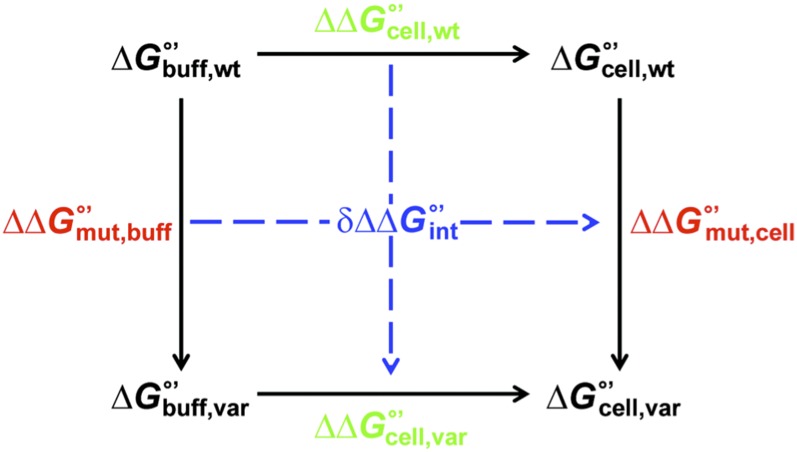
Thermodynamic cycles (17) can be used to quantify the energetics of interactions between proteins in specific protein complexes (17, 18) and between side chains in globular proteins (19, 20). Briefly, the individual effects of two single-site amino acid changes are compared with the combined effect of both mutations. If the sites interact, the sum of the contributions from the single-site changes will not equal the contribution from the double mutant. The difference between the two values measures the strength of the interaction.
We realized that transferring a variant (“var”) from buffer (“buff”) to cells (“cell”) is analogous to making a second mutation to the protein (Fig. 1 and SI Appendix, Fig. S1). Discrepancies in the horizontal (and vertical) sides of Fig. 1 define the free energy () associated with quinary interactions. Differences in the residue-level stability change caused by the mutation () in cells and in buffer are used to calculate :
A negative value of indicates the introduction of an attractive interaction (relative to WT) upon transferring the mutant from buffer to cells.
Fig. 1.
Thermodynamic cycle used to quantify quinary interactions.
Results
Innocuous Mutation in Buffer Is Destabilizing in Cells.
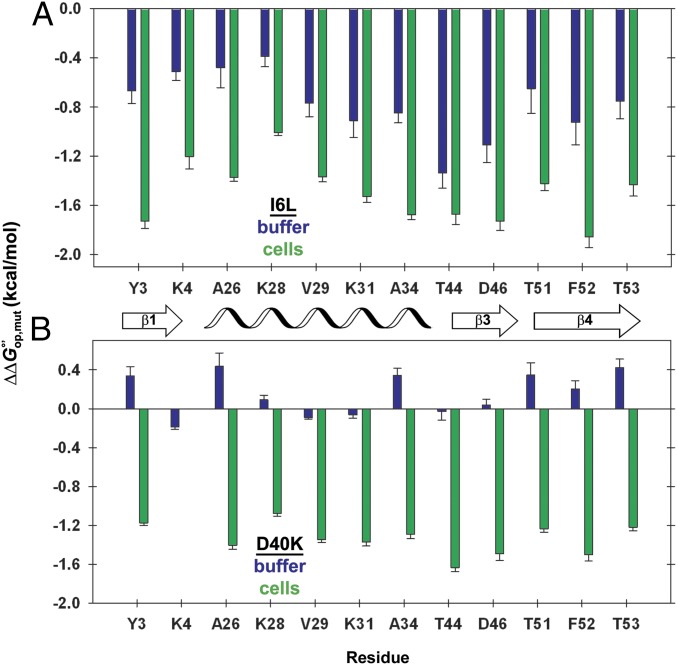
We measured the stability of WT GB1 and four variants (I6L, D40K, D40N, and D40A) in buffer and in cells (6) (SI Appendix, Tables S1–S11). Constant values of across the primary structure might be expected because approximates under our conditions, yet small variations are observed (Fig. 2). The variations arise from two effects. First, there is an inherent uncertainty in kuns, because the values come from model peptides, not GB1. Second, the variation reflects subtle differences in the free energy required to expose each proton. At present, these factors cannot be separated. Therefore, we focus on average values of and (Table 1). The I6L mutation has a modest effect on stability in buffer and in cells (Fig. 2A). The D40K mutation gives dramatically different results (Fig. 2B): The average effect in cells is 10-fold larger than that in buffer (Table 1). Thus, a mutation with an innocuous effect in buffer experiences new and significant quinary interactions that alter its stability in cells.
Fig. 2.
Stability change from charge reversal is small in buffer, but large in cells. (A and B) Changes in stability () caused by the (A) I6L and (B) D40K mutations in cells (green) and in buffer (blue) for quantifiable residues. The number of residues is limited by overlap in NMR spectra and by rates of exchange that are too large to quantify. Error bars represent the uncertainty propagated from triplicate measurements. Complete datasets for D40K and D40N are given in SI Appendix. The datasets for the WT protein and the I6L variant in cells have been published (6).
Table 1.
Average and (kcal/mol, 37 °C)
| Free energy | I6L | D40A | D40N | D40K |
| ∆∆G°′op,mut,buff | −0.8 ± 0.2 | 0.15 ± 0.09 | 0.30 ± 0.08 | 0.14 ± 0.1 |
| ∆∆G°′op,mut,cell | −1.50 ± 0.08 | −1.0 ± 0.1 | −1.0 ± 0.1 | −1.34 ± 0.07 |
| δ∆∆G°′op,int | −0.7 ± 0.2 | −1.1 ± 0.2 | −1.3 ± 0.1 | −1.5 ± 0.1 |
Strength of Quinary Interactions.
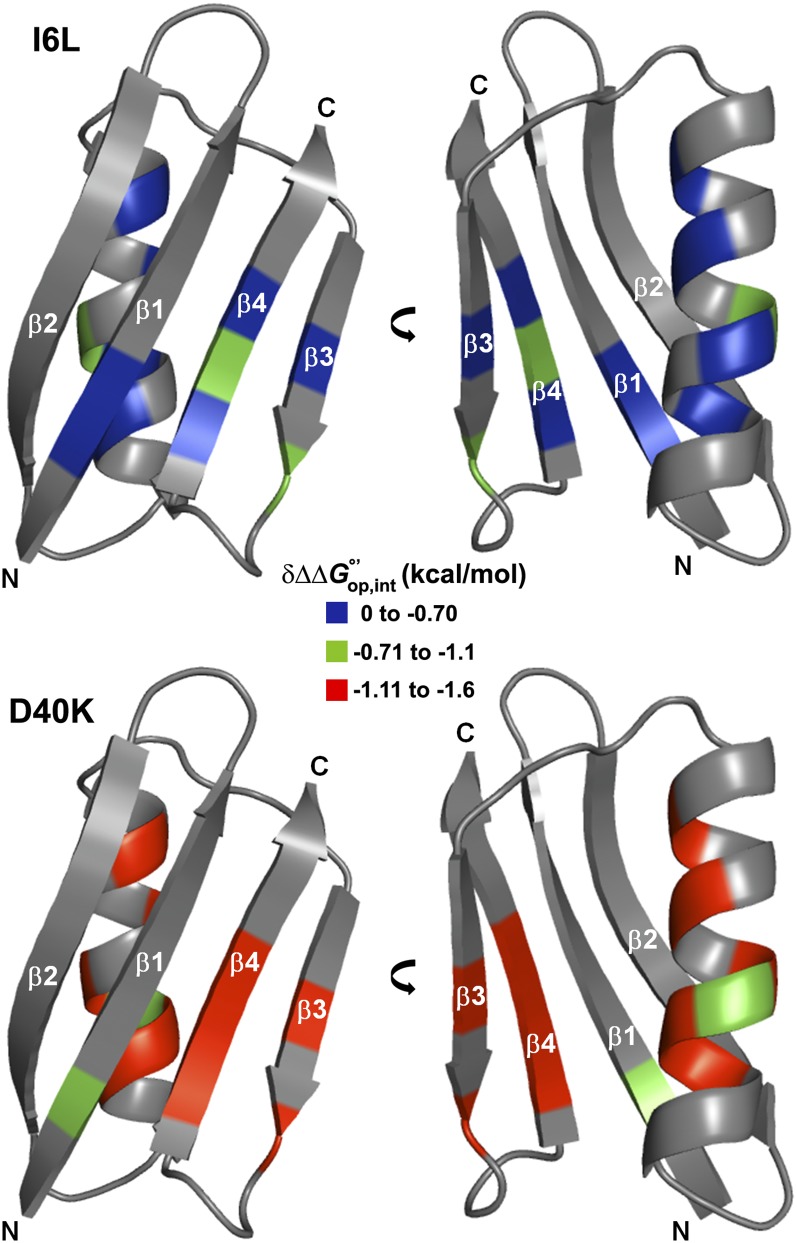
We calculated interaction free energies between the protein and the cytoplasm, using the cycle shown in Fig. 1. Our original implementation (6) of this cycle suggested the presence of quinary interactions between the cytosol and the I6L variant of GB1 that are absent in buffer. The strength of these interactions for I6L is modest (Table 1; see also Fig. 4) and reasonable considering that the substitution shifts only the position of a surface-exposed, nonpolar (methyl) group (21), such that interactions with the cytoplasm are expected to be similar to those experienced in WT GB1. This result led us to test the hypothesis with a variant predicted to produce a larger effect.
Fig. 4.
Structure of WT GB1 (Protein Data Bank ID code 1pgb) colored by the strength of quinary interactions () in the variants. The coupled effect of mutating GB1 in cells is significantly more destabilizing to D40K, where the mutation involves a charged surface residue. Gray residues yield no data.
Whereas the steric repulsions associated with crowding stabilize proteins (8), the associated longer-range attractive interactions can be destabilizing (9–13). We also know that both GB1 and most of the proteins in E. coli are negatively charged (22). We reckoned that an appropriate surface charge reversal might increase attractive interactions, decreasing the stability in cells, while maintaining WT stability in buffer. We targeted D40 because it is the only acidic residue in a surface-exposed loop and it lacks intraprotein side chain hydrogen bonds (23). We chose D40K as the mutation because it changes the net charge of the protein by +2.
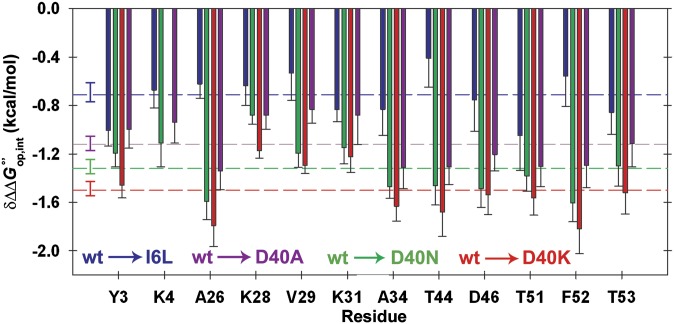
Most importantly, the interaction energies of the D40K and the I6L mutants are strikingly different (Table 1 and Figs. 3 and 4). The average interaction free energy caused by the D40K mutation is twice that caused by the I6L change. To further test our ideas, we studied the D40N and D40A variants, which change the charge by +1. As expected, the average interaction free energy decreases as negative charge and polar contacts are removed (Table 1 and Fig. 3).
Fig. 3.
Interaction free energies () with the cytosol are large for charge-changing mutations. Values for I6L, D40A, D40N, and D40K variants are shown in blue, green, and red, respectively. Error bars represent the uncertainty propagated from triplicate measurements. Dashed lines and associated error bars are the average values and their SDs of the mean. K4 crosspeak volume was insufficient for quantification in the D40K variant.
We attempted to quantify quinary interactions involving two other surface-exposed charge reversals, E19K and E56K, but their stabilities in cells and buffer were too low for quantification. The most likely reason for the decreased stability is their known participation in intramolecular hydrogen bonds (24). It would also be interesting to examine the effects of charge-enhancing mutations (e.g., K to D or E), which we predict would increase GB1 stability in cells relative to wild type. The difficulty again lies in finding a residue, like D40, that lacks regular secondary structure and H bonding. There are no suitable lysine residues in GB1, and there are no arginine residues. These observations highlight the necessity of choosing side chains, such as that of D40, that are not involved in intraprotein interactions when quantifying quinary interactions.
Structural Changes Are Small.
The most important assumption in our analysis is that the structures of the native state and denatured state ensemble of the two proteins are the same in buffer and in cells. This assumption appears to be valid because patterns of H/D exchange data across the sequence in buffer and in cells are similar (i.e., protection factors are consistent) and the in-cell heteronuclear single quantum coherence (HSQC) spectra of GB1 and the variants studied can be overlaid (Fig. 5). We do not expect that molecular chaperones confound our results because chaperones are required for only a small portion (≤5%) of the proteome (25), and small, rapid folders like GB1 most likely reach their native states without these catalysts.
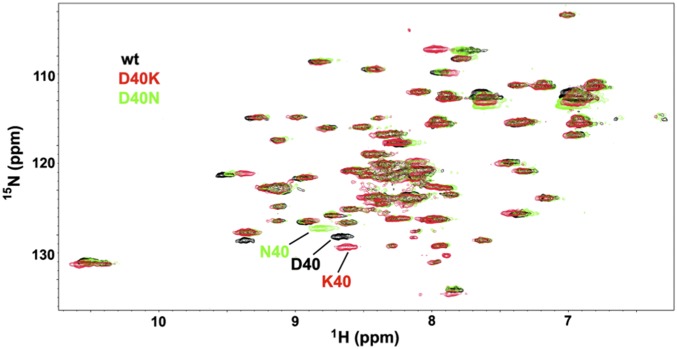
Fig. 5.
Overlaid in-cell 15N-1H HSQC spectra of WT, D40K, and D40N GB1. Expression of I6L GB1 is too low to observe crosspeaks. The in-cell samples were gently harvested after acquisition and the supernatants analyzed. Lack of crosspeaks in all supernatant spectra indicate the proteins do not leak during the course of the experiments.
Discussion
The importance of electrostatic interactions to protein stability was dismissed over 70 y ago, despite several contrary observations (26–28). This situation led to Kauzmann’s recognition of the hydrophobic effect (29) as the dominant stabilizing factor (30, 31). Richards’ subsequent demonstration that the core of a globular protein is as well packed as crystals of small organic compounds (32) shifted the attention to buried intramolecular interactions and led to the synthesis of these ideas by Lattman and Rose (33), who concluded that changes in the core alter the stability of a protein, but not its fold. Although the focus of protein folding studies moved primarily to core packing and hydrophobicity, McConkey (34) noted that protein isoelectric points are conserved despite the evolutionary driving force of random mutagenesis. Recognizing the key role of protein surface charge, he coined the term “quinary structure” (34) to describe the interactions that organize the inside of cells, and Srere coined the term “metabolon” to describe this organization (35).
The ability to quantify protein stability in cells now leads us to reassess the role of surface charge–charge interactions. Both large and small stability changes have been observed for intramolecular ion-pair interactions in buffer (36, 37), but our findings demonstrate that these observations are not necessarily applicable in cells, where intermolecular interactions abound between macromolecules in the crowded cellular interior. The effect is large: The D40K interaction free energy (∼1 kcal/mol) accounts for nearly one-fifth of the protein’s total stability (∼7 kcal/mol) and is in the range of interaction free energies observed in specific protein–protein complexes (18). The implications for larger proteins and for the charge-altering posttranslational modifications required for signal transduction (e.g., phosphorylation, acetylation, myristoylization, and sulfation) cannot be ignored. We suggest that surface residues can be as important as core residues to folding and stability when proteins are studied under physiologically relevant conditions. We are not, however, challenging the crucial role of core packing because the structure of the protein appears to be the same in buffer and in cells.
Anfinsen’s thermodynamic hypothesis states, “the native conformation is determined by the totality of interatomic interactions … in a given environment” and that a protein can be properly understood only “under conditions similar to those for which it was selected—the so-called physiological state” (ref. 38, p. 223). The emphasis on the physiological state has, until recently, been ignored because nearly all studies have been conducted with purified proteins in simple buffered solutions. This void is being filled by studies showing differences between folding in buffer and folding in cells (2–6). Here, we have taken the next step by demonstrating that nonspecific, quinary interactions with the cytoplasmic milieu can be modulated in a way that alters protein stability. This observation reveals a role for surface residues that will aid in understanding native protein function. Although Anfinsen precisely stated the existence of such interactions (38), and McConkey (34) and Srere (35) recognized their biological significance, their energetic contribution went unrecognized until now because we were looking in the wrong place: buffer, instead of cells.
Materials and Methods
Vector.
The pET11a plasmid containing the T2Q GB1 (WT) gene and its I6L variant have been described (6). The D40K variant was produced by site-directed mutagenesis (QuikChange; Agilent) with the following primers: forward 5′ C GAC AAC GGT GTT AAA GGT GAA TGG ACC 3′ and reverse 5′ GGT CCA TTC ACC TTT AAC ACC GTT GTC G 3′ (mutation underlined). The D40N variant made use of the following primers: forward 5′ C GGT GTT AAC GGT GAA TGG ACC 3′ and reverse 5′ C ACC GTT AAC ACC GTT GTC G 3′. The D40A variant made use of the following primers: forward 5′ GGT GTT GCG GGT GAA TGG ACC 3′ and reverse 5′ CGC AAC ACC GTT GTC GTT AGC 3′.
Protein Purification.
Isolation and purification of 15N-enriched WT GB1 and its variants have been described (6). However, the D40K variant does not bind to the ion exchange column owing to a less negative charge (Znet = −2 compared with Znet = −4 for WT and I6L GB1). Fractions containing D40K GB1 were collected in the wash step of anion exchange chromatography and further purified via size exclusion chromatography.
In-Cell H/D Exchange.
The protocol for measuring H/D exchange in E. coli with quenched cell lysates has been reported (6). For WT GB1, aliquots were removed ∼1 h, 2 h, 3 h, 5 h, 8 h, 13 h, and 22 h after initiating exchange. Four samples prepared between 45 min and 3 h were sufficient to capture decay profiles of the destabilized I6L, D40A, D40N, and D40K variants. Data were acquired at 37 °C. Consistent with the observations of Waudby et al., we find that intracellular pH decreases with time (39). The internal pH was measured by comparing the shift of the K10 amide proton to the shift of the W43 amide proton by using a titration curve of the protein in buffer (SI Appendix, Fig. S2). The pH dropped to ∼5.8 after 2 h. Exchange rates were converted to equilibrium unfolding free energies by using intrinsic rates from SPHERE (pH 5.8, 37 °C, alanine as oligopeptide basis) (40). Tables of rates and free energies are found in SI Appendix, Tables S1–S11 and ref. 6.
In Vitro H/D Exchange.
Wild-type GB1 exchange rates measured using a quench step with discrete samples (to mimic the in-cell protocol) yield the same values as those measured with the traditional method involving serial HSQC acquisitions on a single exchange sample. Consequently, the serial method was used here. Experimental details and data analysis have been described (6). The in vitro H/D exchange data were acquired at pH 5.8 and 37 °C in 20 mM citrate buffer and analyzed using the intrinsic rates described above.
Supplementary Material
Acknowledgments
We thank Leonard Spicer for providing the plasmid encoding T2Q GB1, Peter Crowley and Ciara Kyne for aiding in GB1 chemical shift assignments, Ashutosh Tripathy for assistance with calorimetry, Marc ter Horst for spectrometer maintenance, members of the G.J.P. laboratory for encouragement, and Elizabeth Pielak for comments on the manuscript. The National Science Foundation (MCB-1410854 and MCB-1410854) supported this work.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1417415112/-/DCSupplemental.
References
- 1.Zimmerman SB, Trach SO. Estimation of macromolecule concentrations and excluded volume effects for the cytoplasm of Escherichia coli. J Mol Biol. 1991;222(3):599–620. doi: 10.1016/0022-2836(91)90499-v. [DOI] [PubMed] [Google Scholar]
- 2.Dhar A, et al. Protein stability and folding kinetics in the nucleus and endoplasmic reticulum of eucaryotic cells. Biophys J. 2011;101(2):421–430. doi: 10.1016/j.bpj.2011.05.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ebbinghaus S, Dhar A, McDonald JD, Gruebele M. Protein folding stability and dynamics imaged in a living cell. Nat Methods. 2010;7(4):319–323. doi: 10.1038/nmeth.1435. [DOI] [PubMed] [Google Scholar]
- 4.Guzman I, Gelman H, Tai J, Gruebele M. The extracellular protein VlsE is destabilized inside cells. J Mol Biol. 2014;426(1):11–20. doi: 10.1016/j.jmb.2013.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ignatova Z, et al. From the test tube to the cell: Exploring the folding and aggregation of a β-clam protein. Biopolymers. 2007;88(2):157–163. doi: 10.1002/bip.20665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Monteith WB, Pielak GJ. Residue level quantification of protein stability in living cells. Proc Natl Acad Sci USA. 2014;111(31):11335–11340. doi: 10.1073/pnas.1406845111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crowley PB, Chow E, Papkovskaia T. Protein interactions in the Escherichia coli cytosol: An impediment to in-cell NMR spectroscopy. ChemBioChem. 2011;12(7):1043–1048. doi: 10.1002/cbic.201100063. [DOI] [PubMed] [Google Scholar]
- 8.Zhou HX, Rivas G, Minton AP. Macromolecular crowding and confinement: Biochemical, biophysical, and potential physiological consequences. Annu Rev Biophys. 2008;37:375–397. doi: 10.1146/annurev.biophys.37.032807.125817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McGuffee SR, Elcock AH. Diffusion, crowding & protein stability in a dynamic molecular model of the bacterial cytoplasm. PLoS Comput Biol. 2010;6(3):e1000694. doi: 10.1371/journal.pcbi.1000694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harada R, Tochio N, Kigawa T, Sugita Y, Feig M. Reduced native state stability in crowded cellular environment due to protein-protein interactions. J Am Chem Soc. 2013;135(9):3696–3701. doi: 10.1021/ja3126992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sarkar M, Smith AE, Pielak GJ. Impact of reconstituted cytosol on protein stability. Proc Natl Acad Sci USA. 2013;110(48):19342–19347. doi: 10.1073/pnas.1312678110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sarkar M, Li C, Pielak GJ. Soft interactions and crowding. Biophys Rev. 2013;5(2):187–194. doi: 10.1007/s12551-013-0104-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sarkar M, Lu J, Pielak GJ. Protein crowder charge and protein stability. Biochemistry. 2014;53(10):1601–1606. doi: 10.1021/bi4016346. [DOI] [PubMed] [Google Scholar]
- 14.Linderstrøm-Lang KU. 1958. Deuterium exchange and protein structure. Symposium on Protein Structure, ed Neuberger A (Methuen, London), pp 23–34.
- 15.Smith AE, Sarkar M, Young GB, Pielak GJ. Amide proton exchange of a dynamic loop in cell extracts. Protein Sci. 2013;22(10):1313–1319. doi: 10.1002/pro.2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Englander SW, Kallenbach NR. Hydrogen exchange and structural dynamics of proteins and nucleic acids. Q Rev Biophys. 1983;16(4):521–655. doi: 10.1017/s0033583500005217. [DOI] [PubMed] [Google Scholar]
- 17.Carter PJ, Winter G, Wilkinson AJ, Fersht AR. The use of double mutants to detect structural changes in the active site of the tyrosyl-tRNA synthetase (Bacillus stearothermophilus) Cell. 1984;38(3):835–840. doi: 10.1016/0092-8674(84)90278-2. [DOI] [PubMed] [Google Scholar]
- 18.Schreiber G, Fersht AR. Energetics of protein-protein interactions: Analysis of the barnase-barstar interface by single mutations and double mutant cycles. J Mol Biol. 1995;248(2):478–486. doi: 10.1016/s0022-2836(95)80064-6. [DOI] [PubMed] [Google Scholar]
- 19.Horovitz A, Fersht AR. Co-operative interactions during protein folding. J Mol Biol. 1992;224(3):733–740. doi: 10.1016/0022-2836(92)90557-z. [DOI] [PubMed] [Google Scholar]
- 20.Green SM, Shortle D. Patterns of nonadditivity between pairs of stability mutations in staphylococcal nuclease. Biochemistry. 1993;32(38):10131–10139. doi: 10.1021/bi00089a032. [DOI] [PubMed] [Google Scholar]
- 21.Smith CK, Withka JM, Regan L. A thermodynamic scale for the β-sheet forming tendencies of the amino acids. Biochemistry. 1994;33(18):5510–5517. doi: 10.1021/bi00184a020. [DOI] [PubMed] [Google Scholar]
- 22.Spitzer J, Poolman B. The role of biomacromolecular crowding, ionic strength, and physicochemical gradients in the complexities of life’s emergence. Microbiol Mol Biol Rev. 2009;73(2):371–388. doi: 10.1128/MMBR.00010-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gronenborn AM, et al. A novel, highly stable fold of the immunoglobulin binding domain of streptococcal protein G. Science. 1991;253(5020):657–661. doi: 10.1126/science.1871600. [DOI] [PubMed] [Google Scholar]
- 24.Gallagher T, Alexander P, Bryan P, Gilliland GL. Two crystal structures of the B1 immunoglobulin-binding domain of streptococcal protein G and comparison with NMR. Biochemistry. 1994;33(15):4721–4729. [PubMed] [Google Scholar]
- 25.Lorimer GH. A quantitative assessment of the role of the chaperonin proteins in protein folding in vivo. FASEB J. 1996;10(1):5–9. doi: 10.1096/fasebj.10.1.8566548. [DOI] [PubMed] [Google Scholar]
- 26.Mirsky AE, Pauling L. On the structure of native, denatured, and coagulated proteins. Proc Natl Acad Sci USA. 1936;22(7):439–447. doi: 10.1073/pnas.22.7.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eyring H, Stearn A. The application of the theory of absolute reaction rates to proteins. Chem Rev. 1939;24:253–270. [Google Scholar]
- 28.Jacobsen CF, Linderstrøm-Lang K. Salt linkages in proteins. Nature. 1949;164(4166):411–412. doi: 10.1038/164411a0. [DOI] [PubMed] [Google Scholar]
- 29.Kauzmann W. Some factors in the interpretation of protein denaturation. Adv Protein Chem. 1959;14:1–63. doi: 10.1016/s0065-3233(08)60608-7. [DOI] [PubMed] [Google Scholar]
- 30.Dill KA. Dominant forces in protein folding. Biochemistry. 1990;29(31):7133–7155. doi: 10.1021/bi00483a001. [DOI] [PubMed] [Google Scholar]
- 31.Baldwin RL. Dynamic hydration shell restores Kauzmann’s 1959 explanation of how the hydrophobic factor drives protein folding. Proc Natl Acad Sci USA. 2014;111(36):13052–13056. doi: 10.1073/pnas.1414556111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Richards FM. Areas, volumes, packing and protein structure. Annu Rev Biophys Bioeng. 1977;6:151–176. doi: 10.1146/annurev.bb.06.060177.001055. [DOI] [PubMed] [Google Scholar]
- 33.Lattman EE, Rose GD. Protein folding—What’s the question? Proc Natl Acad Sci USA. 1993;90(2):439–441. doi: 10.1073/pnas.90.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McConkey EH. Molecular evolution, intracellular organization, and the quinary structure of proteins. Proc Natl Acad Sci USA. 1982;79(10):3236–3240. doi: 10.1073/pnas.79.10.3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Srere PA. The metabolon. Trends Biochem Sci. 1985;10(3):109–110. [Google Scholar]
- 36.Pace CN, Alston RW, Shaw KL. Charge-charge interactions influence the denatured state ensemble and contribute to protein stability. Protein Sci. 2000;9(7):1395–1398. doi: 10.1110/ps.9.7.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spector S, et al. Rational modification of protein stability by the mutation of charged surface residues. Biochemistry. 2000;39(5):872–879. doi: 10.1021/bi992091m. [DOI] [PubMed] [Google Scholar]
- 38.Anfinsen CB. Principles that govern the folding of protein chains. Science. 1973;181(4096):223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 39.Waudby CA, et al. In-cell NMR characterization of the secondary structure populations of a disordered conformation of α-synuclein within E. coli cells. PLoS ONE. 2013;8(8):e72286. doi: 10.1371/journal.pone.0072286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y-Z. 1995 Protein and peptide structure and interactions studied by hydrogen exchange and NMR. PhD thesis (University of Pennsylvania, Philadelphia). Available at www.fcc.edu/research/labs/roder/sphere/. Accessed December 9, 2014.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.