Abstract
Eukaryotic replication origins are defined by the ORC-dependent loading of the Mcm2-7 helicase complex onto chromatin in G1. Paradoxically, there is a vast excess of Mcm2-7 relative to ORC assembled onto chromatin in G1. These excess Mcm2-7 complexes exhibit little co-localization with ORC or replication foci and can function as dormant origins. We dissected the mechanisms regulating the assembly and distribution of the Mcm2-7 complex in the Drosophila genome. We found that in the absence of cyclin E/Cdk2 activity, there was a 10-fold decrease in chromatin-associated Mcm2-7 relative to the levels found at the G1/S transition. The minimal amounts of Mcm2-7 loaded in the absence of cyclin E/Cdk2 activity were strictly localized to ORC binding sites. In contrast, cyclin E/Cdk2 activity was required for maximal loading of Mcm2-7 and a dramatic genome-wide reorganization of the distribution of Mcm2-7 that is shaped by active transcription. Thus, increasing cyclin E/Cdk2 activity over the course of G1 is not only critical for Mcm2-7 loading, but also for the distribution of the Mcm2-7 helicase prior to S-phase entry.
Keywords: cell cycle, chromatin, DNA replication, Mcm2-7
Introduction
The duplication of a eukaryotic genome within the confines of S-phase is a remarkable event. First, the sheer scale of the process—tens to thousands of million base pairs of DNA—needs to be precisely copied once and only once within just a few hours. In addition, DNA replication forks must initiate within and progress through diverse local chromatin environments including accessible euchromatin and repressive heterochromatin. And finally, the process has to be dynamic and capable of responding to environmental and developmental cues. Thus, with every cell cycle, thousands of DNA replication start sites (origins) must be selected and activated in a regulated and coordinated manner to ensure that the entire genome is faithfully duplicated (reviewed in Masai et al, 2010).
Each potential origin of replication is marked by the origin recognition complex (ORC) (Bell & Stillman, 1992; Rao & Stillman, 1995). In G1, the Mcm2-7 complex, the replicative helicase, is loaded as a double hexamer at ORC binding sites in an ORC-, Cdc6-, and Cdt1-dependent manner to form the pre-replicative complex (pre-RC) (Evrin et al, 2009; Remus et al, 2009; Gambus et al, 2011). The assembly of the pre-RC in G1 “licenses” the origin for potential activation in the subsequent S-phase. As a cell enters S-phase, cyclin-dependent kinase (CDK) and Dbf4-dependent kinase (DDK) activate the Mcm2-7 helicase by the recruitment of Cdc45 and the GINS complex to form the CMG (Cdc45, Mcm2-7, and GINS) holo-helicase complex (reviewed in Tanaka & Araki, 2013). After initiation of DNA replication, the Mcm2-7 helicase, as part of the CMG complex, travels with and unwinds the DNA ahead of the replication fork (Aparicio et al, 1997; Labib, 2000; Pacek & Walter, 2004; Pacek et al, 2006; Sekedat et al, 2010).
In theory, a double hexamer of Mcm2-7 loaded at each origin should be sufficient to replicate the genome, with a single hexamer of Mcm2-7 traveling with each bidirectional DNA replication fork. However, nuclear Mcm2-7 protein levels are in vast excess relative to ORC or the number of replication origins (Burkhart et al, 1995; Lei et al, 1996; Donovan et al, 1997; Mahbubani et al, 1997; Edwards et al, 2002). In vitro, multiple double hexamers of Mcm2-7 are able to be loaded at ORC binding sites by reiterative rounds of ATP hydrolysis (Bowers et al, 2004; Evrin et al, 2013). Electron microscopy revealed that these double hexamers are distributed throughout the DNA template (Evrin et al, 2009; Remus et al, 2009), suggesting the ability of the Mcm2-7 complex to passively translocate away from ORC binding sites. Similarly, in vivo immunofluorescence and chromatin association studies have revealed that Mcm2-7 are broadly distributed throughout the nucleus and exhibit little co-localization with ORC (Madine et al, 1995; Krude et al, 1996; Romanowski et al, 1996b; Ritzi et al, 1998; Edwards et al, 2002; Harvey & Newport, 2003). Together, these seemingly paradoxical observations regarding the location and quantity of Mcm2-7 have been termed the “MCM Paradox” (Takahashi et al, 2005).
Although the mechanisms regulating the loading of multiple Mcm2-7 complexes and their distribution throughout the genome are unclear, increasing data suggest that the excess Mcm2-7 are important for maintaining genome stability during replicative stress. Mcm2-7 levels can be depleted by more than 90% with little, if any, impact on progression through S-phase (Woodward et al, 2006; Crevel et al, 2007; Ge et al, 2007; Ibarra et al, 2008). However, in the absence of the full complement of Mcm2-7, there is a marked reduction in the utilization of dormant origins and an increase in cell death when cells encounter replicative stress during S-phase (Crevel & Cotterill, 1991; Woodward et al, 2006; Ge et al, 2007; Ibarra et al, 2008). Thus, the full complement of Mcm2-7 is critical for the activation of dormant replication origins and function to preserve genome integrity during replicative stress.
We set out to examine this “MCM Paradox” from a biochemical and genome-wide perspective by using chromatin association assays and genome-wide chromatin immunoprecipitation (ChIP) to quantify the loading and distribution of Mcm2-7 at different points in the cell cycle of Drosophila Kc cells. Specific questions we set out to address included: (i) How does Mcm2-7 loading onto chromatin progress during G1? (ii) Do Mcm2-7 complexes load adjacent to ORC? (iii) Where are Mcm2-7 distributed throughout the genome? and (iv) Do other chromosomal features (e.g. transcription units) impact the distribution of Mcm2-7?
Results
Mcm2-7 loading is regulated by cyclin E/Cdk2 activity
Mcm2-7 chromatin association is dynamic throughout the cell cycle. Mcm2-7 are loaded onto chromatin in G1, travel ahead of the replication fork in S-phase, and are removed from the chromatin as S-phase progresses (reviewed in Masai et al, 2010). In order to examine the dynamics of Mcm2-7 chromatin association, we developed approaches to arrest Drosophila Kc cells at specific points in the cell cycle when Mcm2-7 would presumably be associated or not associated with ORC at potential origins of replication. We were particularly interested in identifying a defined cell cycle point immediately before Mcm2-7 helicase activation and its movement with the DNA replication fork.
We reasoned that inhibition of cyclin E/Cdk2 kinase activity would be sufficient to arrest cells in G1 immediately prior to origin activation. We first depleted cyclin E and Cdk2 separately using RNAi in Drosophila Kc cells. In Drosophila, cyclin E is the regulatory subunit of the cyclin-dependent kinase 2 (Cdk2), both of which, like their mammalian homologs, function together to drive cells out of G1 and into S-phase (Dulic et al, 1992; Ohtsubo & Roberts, 1993; Knoblich et al, 1994). We also overexpressed Dacapo, a p27 homolog and potent inhibitor of cyclin E/Cdk2 kinase activity (Lane et al, 1996), from a copper-responsive metallothionein promoter. As controls, cells were arrested in early G1 by RNAi depletion of Dup/Cdt1, a key replication licensing factor required for Mcm2-7 loading (Whittaker et al, 2000) and at the G1/S transition by treatment with 1 mM hydroxyurea (HU). Dup/Cdt1 RNAi, cyclin E RNAi, Cdk2 RNAi, Dacapo overexpression (+Dacapo), and HU treatment all resulted in a cell cycle arrest with the majority (70–85%) of cells having a 2C DNA content by flow cytometry (Fig1A). In contrast, only 30–40% of the cells from an asynchronous population of Drosophila cells exhibit 2C DNA content (Supplementary Fig S1D).
Figure 1.
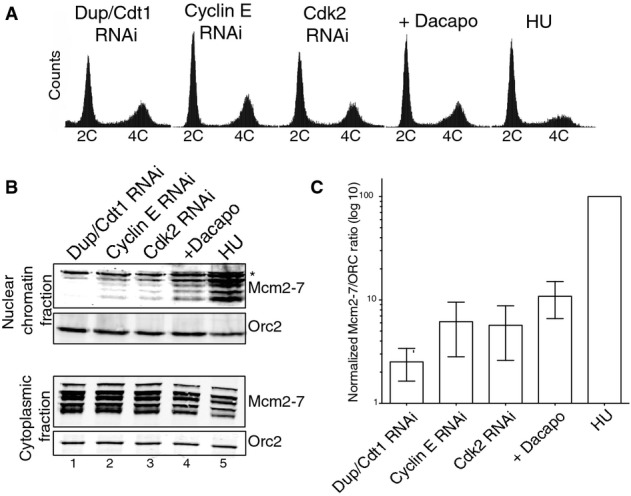
- FACS profiles of DNA content for cells arrested at different points in the cell cycle by Dup/Cdt1 RNAi, cyclin E RNAi, Cdk2 RNAi, Dacapo overexpression (+Dacapo), and 1 mM HU.
- Analysis of Mcm2-7 nuclear chromatin association at different points in the cell cycle. Nuclear chromatin fractions and cytoplasmic extracts were assayed for Orc2 and Mcm2-7 by Western blot. A non-specific band is indicated by an asterisk (*).
- Quantification of the ratio of chromatin-bound Mcm2-7 relative to Orc2 (log10 scale) for a minimum of 11 replicates (mean ± SD).
We next assessed the relative amounts of nuclear chromatin-associated Mcm2-7 in each of these conditions by chromatin fractionation (Fig1B). A polyclonal antibody specific to Drosophila Orc2 (Austin et al, 1999) and a monoclonal antibody (AS1.1) that recognizes all six Mcm2-7 subunits (Chen et al, 2007) were used to assess the chromatin-associated and whole cell extract levels of ORC and Mcm2-7. As expected, cells lacking the licensing factor, Dup/Cdt1, exhibited no detectable chromatin-associated Mcm2-7 (Fig1B, lane 1). The background levels of Mcm2-7 chromatin association in the Dup/Cdt1-depleted cells were similar to the levels observed in G2-arrested cells where the presence of geminin prevents pre-RC formation (McGarry & Kirschner, 1998) (Supplementary Fig S1B). In contrast, cells arrested at the G1/S transition by HU treatment had a robust accumulation of nuclear Mcm2-7 (Fig1B, lane 5). Interestingly, cyclin E and Cdk2 RNAi cells exhibited an intermediate phenotype (Fig1B, lanes 2 and 3) with considerably less nuclear chromatin-associated Mcm2-7 than HU-arrested cells. We observed a slight increase in the amount of chromatin-associated Mcm2-7 in +Dacapo cells (Fig1B, lane 4) over both cyclin E and Cdk2 RNAi cells. However, the amount of chromatin-bound Mcm2-7 in +Dacapo cells was still markedly reduced compared to levels seen in HU-treated cells. In contrast to the cell cycle fluctuations in chromatin-associated Mcm2-7 levels, ORC remained constant for each condition surveyed. Importantly, the cell cycle differences observed in Mcm2-7 chromatin association were not simply due to changes in protein levels as the levels of ORC and Mcm2-7 remained constant in the non-nuclear cytoplasmic fraction. The chromatin-associated Mcm2-7/ORC ratio was calculated for each condition, normalized to the level in HU to obtain a fold difference, and plotted on a log10 scale (Fig1C). A 40-fold difference in chromatin-associated Mcm2-7 was observed between Dup/Cdt1 RNAi and HU and 9-fold to 15-fold differences in loading were observed between HU and cells with impaired cyclin E/Cdk2 kinase activity (cyclin E RNAi, Cdk2 RNAi, and +Dacapo). Together, our results suggest that while minimal loading of the Mcm2-7 complex can occur in the absence of cyclin E/Cdk2 activity, the loading of the full complement of Mcm2-7 requires cyclin E/Cdk2 kinase activity.
Maximal loading of Mcm2-7 coincides with entry into S-phase
Our results suggest that Mcm2-7 levels continue to increase during G1 concomitant with increasing Cdk2 kinase activity prior to entry into S-phase. However, it remained possible that the maximal Mcm2-7 loading we observed was perhaps a consequence of replicative stress resulting from HU treatment and the activation of dormant replication origins (Anglana et al, 2003). Thus, we wanted to assess Mcm2-7 loading without perturbing S-phase or activating the intra-S-phase checkpoint.
To monitor pre-RC assembly as cells synchronously enter S-phase from G1, we induced overexpression of Dacapo to block cyclin E/Cdk2 kinase activity and prevent the full complement of Mcm2-7 from being assembled on the chromatin in G1. We then released the cells from the Dacapo arrest by removing the inducer (Cu2+) of Dacapo expression. Entry into the next cell cycle was prevented by the addition of the mitotic inhibitor, colcemid, to the medium. As cells progressed into S-phase, we monitored DNA content, Dacapo protein levels, and the chromatin-associated levels of Mcm2-7 and ORC (Fig2). We found that most cells entered S-phase by 3 h and had completed DNA replication by 9 h. Entry into S-phase at 3 h coincided with a decrease in Dacapo expression and a marked increase (∽9-fold) in Mcm2-7 association with the chromatin. The increase in Mcm2-7 chromatin association at the onset of S-phase entry was transient as Mcm2-7 were likely removed from chromatin with passage of the DNA replication fork (Kuipers et al, 2011). Together, these results demonstrate that the cyclin E/Cdk2 kinase-dependent loading of the full complement of Mcm2-7 is a regulated process that occurs during a normal cell cycle.
Figure 2.
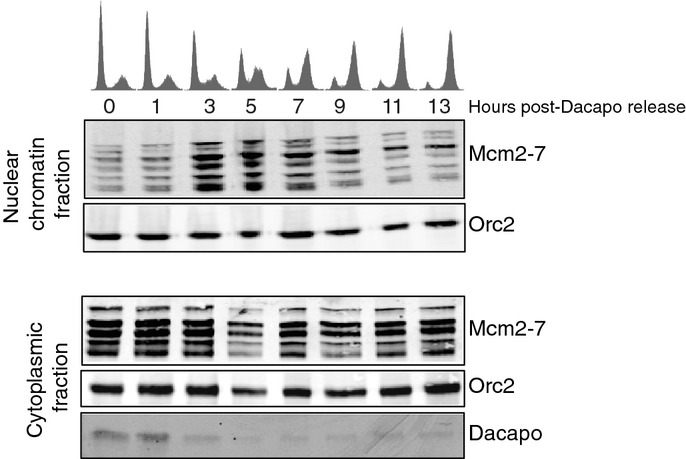
The full complement of Mcm2-7 is loaded during an unperturbed G1/S transition
Cell cycle analysis by FACS of cells arrested in G1 by Dacapo overexpression followed by release back into the cell cycle for 13 h (top). Western blot analysis of Mcm2-7 and Orc2 for the nuclear chromatin and cytoplasmic fractions of Mcm2-7, Orc2, and Dacapo.
We also examined Mcm2-7 nuclear localization by immunofluorescence in an asynchronous cell population (Supplementary Fig S2). In order to visualize chromatin-associated Mcm2-7 in the nucleus, it is necessary to remove the cytoplasmic and non-chromatin-associated Mcm2-7 population (which are the majority) by permeabilizing and washing the cells prior to fixation (Claycomb et al, 2002). Unfortunately, the permeabilization prior to fixation makes it impossible to monitor non-chromatin-associated factors such as cyclin E or Cdk2 levels. In the absence of a suitable cell cycle marker for cells in G1 or early S-phase, we instead used mono-methylation of lysine 20 on histone H4 (H4K20me1) as a marker of cells in late S-phase or G2/M. H4K20me1 is a well-characterized cell cycle-regulated chromatin mark that is generated by PR-Set7/Set8 in very late S-phase and persists on the chromatin up until mitosis (Abbas et al, 2010; Oda et al, 2010) (Supplementary Fig S2A). Thus, cells in late S-phase and G2/M will be identifiable by elevated levels of H4K20me1 and, in contrast, cells in G1 and early to mid-S-phase will be marked by very low levels of H4K20me1. We found that H4K20me1 and Mcm2-7 staining cell populations were mutually exclusive with very little overlap consistent with Mcm2-7 being removed from chromatin by the end of S-phase (Supplementary Fig S2B). The remaining H4K20me1-negative cells displayed a range of Mcm2-7 staining from barely detectable to a robust signal, presumably representing cells in early G1 (low cyclin E/Cdk2 activity) and cells in late G1/early S-phase (high cyclin E/Cdk2 activity), respectively (Supplementary Fig S2C). Thus, even in a dividing population of cells, we can clearly detect multiple stages of Mcm2-7 loading in G1 and early S-phase cells. These findings also parallel an increase in chromatin-associated Mcm2-7 from late M phase into and peaking at the G1/S transition (Kuipers et al, 2011; Symeonidou et al, 2013).
The cyclin E/Cdk2 kinase-dependent phase of Mcm2-7 loading requires the canonical pre-RC assembly pathway
It is well documented that ORC, Cdc6, and Cdt1 are able to load multiple Mcm2-7 double hexamers onto DNA templates in vitro (Bowers et al, 2004; Evrin et al, 2009; Remus et al, 2009). In vivo, the situation is more complex with ORC, Cdc6, and Cdt1 dynamically associating with chromatin (McNairn et al, 2005; Xouri et al, 2007; Sonneville et al, 2012) and ATP hydrolysis driving both the loading and release of the Mcm2-7 helicase complex (Frigola et al, 2013). It was evident from our biochemical studies that Dup/Cdt1 was required to load minimal amounts of Mcm2-7 in a cyclin E-independent manner (Fig1). However, it remained unclear whether the full complement of Mcm2-7 loading we observed, which is stimulated by cyclin E/Cdk2 kinase activity, was dependent on the canonical pre-RC assembly pathway (e.g. Cdc6 and Dup/Cdt1).
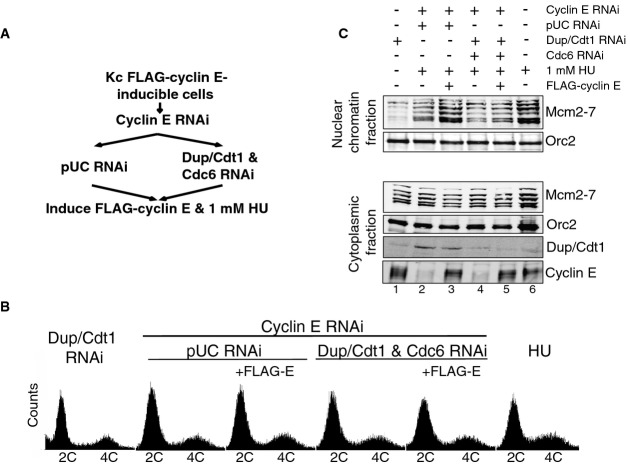
To determine whether the full complement of Mcm2-7 loading we observed in HU-arrested cells is dependent on the established pre-RC assembly pathway, we depleted cells of Cdc6 and Dup/Cdt1 immediately after the cyclin E/Cdk2-independent pre-RC assembly step (Fig3A). Specifically, we arrested cells in early G1 by RNAi depletion of cyclin E for 24 h followed by treatment with either control (pUC) or Cdc6 and Dup/Cdt1 dsRNA for 24 h. Cyclin E was subsequently restored by overexpression of RNAi-resistant FLAG-cyclin E. To assess Mcm2-7 loading at a static point in the cell cycle immediately after entry into S-phase, the cells were released into medium containing 1 mM HU. As expected, the majority of cells from each condition arrested with a 2C DNA content (Fig3B).
Figure 3.
- Schematic of the experiment.
- FACS profiles of DNA content for each cell population assayed.
- Western blot analysis for Orc2 and Mcm2-7 in the nuclear chromatin fraction and Orc2, Mcm2-7, Dup/Cdt1, and cyclin E in the cytoplasmic fraction for each condition assayed.
We found that cells depleted of Dup/Cdt1 and Cdc6 were unable to recruit additional Mcm2-7 when cyclin E expression and progression through G1 were restored by FLAG-cyclin E overexpression (Fig3C, lanes 4 and 5). In contrast, control RNAi (pUC) cells were able to load additional Mcm2-7 when driven through G1 by FLAG-cyclin E overexpression (Fig3C, lane 3). The increase of chromatin-associated Mcm2-7 was not due to a difference in FLAG-cyclin E overexpression. These results indicate that Dup/Cdt1 and Cdc6 are needed to load the full complement of Mcm2-7 onto chromatin throughout G1 in a regulated manner.
Genome-wide distribution of Mcm2-7 is determined by cyclin E/Cdk2 activity and local transcription
Earlier immunofluorescence-based studies noted that Mcm2-7 do not strictly co-localize with ORC in the nucleus during S-phase (Madine et al, 1995; Krude et al, 1996; Romanowski et al, 1996b). The dichotomy in Mcm2-7 chromatin association that we observed between cells arrested by cyclin E RNAi and cells arrested at the G1/S transition by HU treatment prompted us to investigate the genome-wide distribution of Mcm2-7 relative to ORC. Specifically, we used chromatin immunoprecipitation to address where Mcm2-7 was localized in relation to ORC in cyclin E- and HU-arrested cells.
We used genome-wide mapping experiments to localize ORC in asynchronous cells (MacAlpine et al, 2010; Roy et al, 2010) (Fig4A) and Mcm2-7 in cyclin E RNAi (Fig4B) treated cells. We identified 5,135 ORC peaks and 3,792 Mcm2-7 peaks throughout the Drosophila Kc genome. The concordance between ORC and Mcm2-7 in cyclin E RNAi treated cells was > 93%, consistent with the critical role of ORC in pre-RC assembly (Fig4C). Not surprisingly, we found that ORC and Mcm2-7 were specifically enriched at early activating origins of DNA replication mapped by the incorporation of a nucleotide analog, BrdU, during an HU arrest (Supplementary Fig S3).
Figure 4.
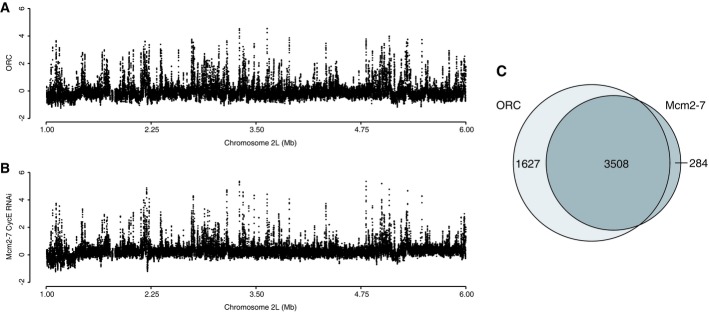
- Genome-wide analysis of ORC localization by ChIP-chip. ORC enrichment from asynchronous cells is depicted for a 5-Mb section of chromosome 2L.
- Genome-wide analysis of Mcm2-7 localization in early G1 by ChIP-chip. Mcm2-7 enrichment from cyclin E RNAi-depleted cells is depicted for a 5-Mb section of chromosome 2L.
- Venn diagram depicting the overlap between ORC and Mcm2-7 peaks.
Strikingly, we found a very different pattern of Mcm2-7 localization at the G1/S transition during an HU arrest (Fig5A). In contrast to the tight co-localization with ORC observed in the cyclin E-depleted cells, we observed a “binary” pattern of Mcm2-7 localization across the genome. Specifically, we observed broad chromosomal regions containing Mcm2-7 signal punctuated by the absence of Mcm2-7 localization. The dramatic change in Mcm2-7 was not due to a global change in chromatin configuration in the HU-arrested cells because ORC binding remained indistinguishable between the asynchronous and HU-arrested cells (Supplementary Fig S4). Together with our biochemical results, these data suggest that the full complement of Mcm2-7 we observed at the G1/S transition has re-distributed from ORC binding sites. We do not believe that Mcm2-7 are completely coating the DNA, but rather that we are observing the likelihood of detecting Mcm2-7 signal within specific genomic features (see below).
Figure 5.
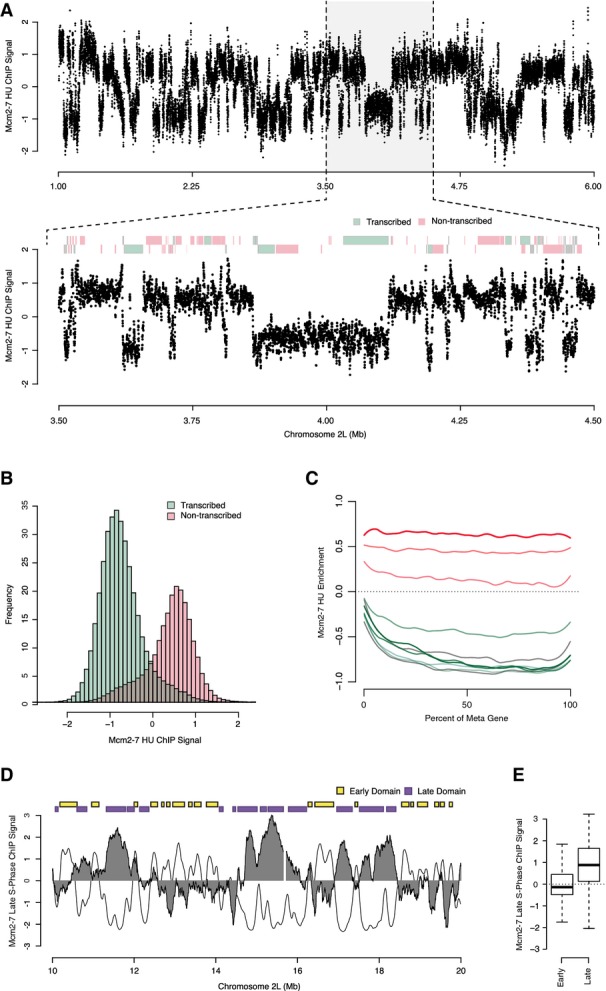
- Genome-wide analysis of Mcm2-7 localization at the G1/S transition by ChIP-chip. Mcm2-7 enrichment from HU-arrested cells is depicted for a 5-Mb section of chromosome 2L. Inset: transcribed (green) and non-transcribed (red) genes are indicated above with genes on the positive strand on the top and those on the negative on the bottom.
- Bimodal distribution of Mcm2-7 enrichment over transcribed and non-transcribed genes. Histogram showing the distribution of probe scores found within transcribed (green) and non-transcribed (red) genes.
- “Meta”-gene analysis of Mcm2-7 enrichment for different deciles of gene expression and their aggregated probe intensities.
- Mcm2-7 is displaced from chromatin by DNA replication. Genome-wide analysis of Mcm2-7 localization in late S-phase (6 h post HU release) by ChIP-chip. Mcm2-7 enrichment is depicted for a 10-Mb section of chromosome 2L (filled gray), replication timing profile (black line) and early (yellow) and late (purple) replication timing domains.
- Box-plots representing late S-phase Mcm2-7 ChIP signal found within early (246) or late (167) replication domains.
The binary distribution of Mcm2-7 across the genome prompted us to investigate genomic features that may be associated with the broad regions of high or low Mcm2-7 levels along the chromosome. The Mcm2-7 localization pattern relative to annotated genomic features suggested that Mcm2-7 may be displaced by actively transcribed genes (Fig5A). To quantitatively assess the Mcm2-7 distribution relative to transcription units, we generated histograms of Mcm2-7 enrichment for transcribed and non-transcribed genes (Fig5B) and found a bimodal pattern of Mcm2-7 enrichment. Specifically, active genes had no or very little Mcm2-7 signal, whereas inactive or non-transcribed genes exhibited an elevated Mcm2-7 signal (P < 1.02 × 10−257; t = 40.16). We also considered that the bimodal distribution of Mcm2-7 enrichment between active and inactive genes might be due to the activation of early origins that are enriched near actively transcribed genes. However, we found that the bimodal distribution for Mcm2-7 was not dependent on early origin activity, but instead was a specific feature of annotated transcripts (Supplementary Fig S5).
The enrichment of Mcm2-7 signal at non-transcribed genes was indistinguishable from intergenic levels (Supplementary Fig S6), suggesting that active transcription was responsible for displacing Mcm2-7 from chromatin. We reasoned that if active transcription was sufficient to displace Mcm2-7 from chromatin, then in the absence of additional Mcm2-7 loading, we should expect that both low and high transcription levels should be able to displace Mcm2-7 from chromatin. To address this, we binned the 14,594 genes into deciles based on their relative gene expression levels and plotted the Mcm2-7 enrichment relative to a “meta”-gene body (Fig5C). We found that Mcm2-7 levels were depleted over the entire transcription unit for actively transcribed genes. The seven most expressed deciles (green and gray lines) exhibited the same degree of Mcm2-7 depletion, suggesting that any amount of transcription is sufficient to displace Mcm2-7 from the chromatin.
We sought to directly test the requirement for active transcription in the displacement of the Mcm2-7 complex from gene bodies. As we only observe differences in Mcm2-7 distribution during an HU arrest when further pre-RC assembly is blocked, we would need to specifically alter transcription during the progression from G1 to S, which would likely interfere with the transcription of key cell cycle-regulated genes (e.g. E2F-regulated genes). As an alternative approach, we examined the genome-wide localization of Mcm2-7 in two different Drosophila cell lines (Kc167 and S2) treated with HU. Although much of the transcriptome is the same between these cell lines (Cherbas et al, 2011), we identified approximately 100 genes that were significantly transcribed in one cell line and off in the other. As before, we found that the Mcm2-7 complex was excluded from actively transcribed genes in both cell lines (Supplementary Fig S7). However, when we specifically examined those genes that were only transcribed in one cell line, we observed a significant (Mann–Whitney–Wilcoxon test; P < 7 × 10−5) decrease in Mcm2-7 occupancy that was specific for the transcribed cell line. We conclude that transcription is required to displace the Mcm2-7 complexes from gene bodies. Together, these results suggest that individual Mcm2-7 complexes can be displaced by active transcription and that after the G1/S transition, they cannot be re-established or translocate into these regions.
The Mcm2-7 helicase complex is inherently dynamic throughout the cell cycle; it is loaded onto chromatin, travels with the replication fork, and is rapidly removed from the chromatin by passage of the replication fork (Madine et al, 1995; Krude et al, 1996; Romanowski et al, 1996a; Kuipers et al, 2011). Our biochemical and genomic data indicate that the full complement of Mcm2-7 is loaded onto chromatin in late G1 and redistributes throughout non-transcribed genes and intergenic sequences by the G1/S transition. If this broad distribution of chromatin-associated Mcm2-7 at the G1/S transition represents the true biological distribution of the Mcm2-7 complex, then we would expect that these Mcm2-7 complexes would be displaced by replication during S-phase. To test this hypothesis, we arrested cells at the G1/S transition by treatment with 1 mM HU and then released them synchronously into S-phase. We surveyed the genome-wide distribution of Mcm2-7 near the end of S-phase (6 h after HU release) (Fig5D, dark gray). We found that Mcm2-7 had been displaced from early replicating regions of the genome and only remained at those sequences copied in late S-phase (Fig5D, black line and purple segments). To quantify these results, we used our prior segmentation of the genome into early (yellow) and late (purple) replicating domains (Lubelsky et al, 2014) and compared it to the distribution of Mcm2-7 at the end of S-phase. We found a significant enrichment of Mcm2-7 associated with late replicating domains relative to early domains (Fig5E, P < 2.37 × 10−68, t = 18.22). Together, these data argue that the broad distribution of Mcm2-7 we observe at the G1/S transition represents the full and functional complement of Mcm2-7 on the chromatin.
Discussion
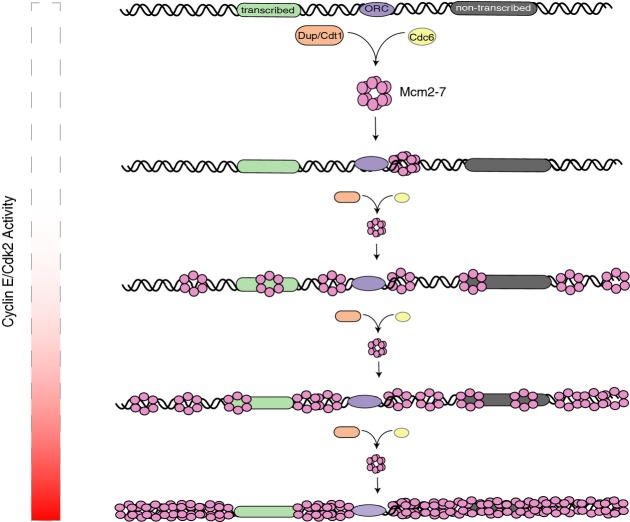
In order to ensure the fidelity of the DNA replication program and genome integrity, there are many more Mcm2-7 helicase complexes loaded onto DNA than potential start sites marked by ORC. We have examined this “MCM Paradox” using both genomic and biochemical approaches to understand the mechanisms by which the full complement of excess Mcm2-7 are loaded and distributed throughout the genome to preserve genomic integrity. We found that in early G1, minimal levels of Mcm2-7 were loaded onto chromatin at ORC binding sites independent of cyclin E/Cdk2 activity (Fig6). Concurrent with and dependent on increasing cyclin E/Cdk2 activity during G1, we observed an increase in Mcm2-7 loading that culminated in a 10-fold higher level of chromatin-associated Mcm2-7 at the G1/S transition. Both the cyclin E/Cdk2 activity-independent Mcm2-7 loading and cyclin E/Cdk2 activity-dependent Mcm2-7 loading were dependent on the canonical pre-RC assembly pathway. Strikingly, the full complement of Mcm2-7 when assembled onto chromatin was not restricted to sequences immediately adjacent to ORC binding sites, but rather distributed throughout the genome and shaped by active transcription.
Figure 6.
Model of pre-RC assembly and redistribution of Mcm2-7 in G1
Mcm2-7 is loaded at ORC binding sites immediately after entry into G1. As G1 progresses, increasing cyclin E/Cdk2 activity promotes the loading of additional Mcm2-7 complexes resulting in the full complement of Mcm2-7 being loaded by the end of G1. All Mcm2-7 loading is dependent on Cdc6 and Cdt1. Prior to or coinciding with the entry into S-phase, the full complement of Mcm2-7 redistributes along the chromosomes and is displaced from transcribed genes.
Precise regulation of cyclin E/Cdk2 kinase activity is critical for pre-RC assembly and genomic stability
Pre-RC assembly begins in telophase (Dimitrova et al, 1999), continues through G1, and culminates just prior to entry into S-phase (Symeonidou et al, 2013). The precipitous drop in CDK activity as cells exit mitosis likely leads to the limited amounts of Mcm2-7 being assembled onto chromatin that we detect specifically at ORC binding sites. As the cells progress into G1, increasing levels of Cdk2 activity contribute to cyclin E/Cdk2-dependent Mcm2-7 loading. Indeed, a gradual increase of Cdk2 activity occurs following exit from mitosis in cycling human cells (Spencer et al, 2013). Conversely, at some point, increasing CDK activity leads to the inhibition of pre-RC assembly and ultimately origin activation (reviewed in Masai et al, 2010). In S. cerevisiae, CDK activity targeted toward pre-RC components directly impairs pre-RC assembly (Drury et al, 1997; Elsasser et al, 1999; Nguyen et al, 2001); however, in higher eukaryotes, the evidence for a direct role is less clear. In human cell culture, phosphorylation of Cdt1 by CDK actively promotes its degradation by the SCF-Skp2 E3 ligase (Liu et al, 2004; Nishitani et al, 2006). However, this appears to be a minor pathway as the bulk of Cdt1 is degraded in S-phase by PCNA-coupled Cul4-Ddb1 destruction (Arias & Walter, 2006; Jin et al, 2006; Senga et al, 2006).
How does cyclin E/Cdk2 activity promote the loading of the full Mcm2-7 complement? Prior experiments performed on quiescent mammalian cells re-entering the cell cycle suggested that cyclin E/Cdk2 kinase activity stabilizes Cdc6 and Cdc7, two factors critical for pre-RC assembly and initiation, respectively (Mailand & Diffley, 2005; Chuang et al, 2009). In contrast, cycling cells become competent to assemble the pre-RC immediately after the metaphase to anaphase transition (Clijsters et al, 2013). Following exit from anaphase, Cdc6 is degraded by APC/C-Cdh1; however, there is a short window between late G1 and S where Cdc6 protein may accumulate before being degraded in S-phase by a PIP-box and Crl4-Cdt2 mediated mechanism (Clijsters & Wolthuis, 2014). This second wave of Cdc6 expression in late G1 is likely dependent on cyclin E/Cdk2 activity and E2f-regulated gene expression and may serve to promote loading the full complement of Mcm2-7 on the chromatin. Alternatively, the Mcm2-7 complexes, which are loaded proximal to the ORC binding sites in a cyclin E- and Cdk2-independent manner, may serve to promote additional Mcm2-7 loading upon exposure to increasing cyclin E/Cdk2 kinase activity prior to S-phase by a direct Mcm2-7–cyclin E–Cdt1 interaction (Geng et al, 2007). The transition from a cyclin E/Cdk2 kinase-independent mode to cyclin E/Cdk2 kinase-dependent mode of Mcm2-7 loading would facilitate a feed forward mechanism to amplify Mcm2-7 loading as G1 progresses.
The precise regulation of cyclin E/Cdk2 kinase activity during G1 is critical for pre-RC assembly and genome stability. Cyclin E/Cdk2 kinase activity is required to load the full Mcm2-7 complement, yet deregulation of cyclin E/Cdk2 kinase activity results in a shortening of G1, a lengthening of S-phase, and genomic instability (Ohtsubo & Roberts, 1993; Resnitzky et al, 1994; Wimmel et al, 1994; Spruck et al, 1999). The genomic instability and lengthening of S-phase appear to be due to a defect in pre-RC assembly (Ekholm-Reed et al, 2004). We speculate that deregulation of cyclin E/Cdk2 kinase activity would result in the cells transitioning from mitosis being immediately exposed to high levels of cyclin E/Cdk2 kinase activity instead of the gradual increase that normally occurs over the course of G1 (Spencer et al, 2013). Thus, insufficient pre-RCs would be assembled on chromatin as the cyclin E-deregulated cells rapidly enter S-phase and may not properly redistribute throughout the genome. In vivo, mutations that impact Mcm2-7 levels and pre-RC assembly have been linked to genome instability and tumorigenesis (Pruitt et al, 2007; Shima et al, 2007; Chuang et al, 2010).
Genome-wide redistribution of Mcm2-7
Our chromatin immunoprecipitation studies of Mcm2-7 localization provide the first genome-wide view of the Mcm2-7 distribution in a higher eukaryote and also reveal a dramatic reorganization of Mcm2-7 during late G1. In the absence of cyclin E/Cdk2 kinase activity (early G1), we find that there is near perfect concordance between ORC and Mcm2-7 peaks. A similar distribution of overlapping ORC and Mcm2-7 peaks is also observed in S. cerevisiae (Wyrick et al, 2001; Eaton et al, 2010). Not surprisingly, these genome-wide datasets from S. cerevisiae reinforce the essential role of ORC in pre-RC assembly and suggest that Mcm2-7 loading is a local phenomenon restricted to the adjacent sequences. In contrast, in Drosophila, we see a dramatic change in the distribution of Mcm2-7 that occurs by the G1/S transition. Not only are more Mcm2-7 loaded onto the chromatin, but they are also broadly enriched along the entire chromosome and not restricted to ORC binding sites.
The distribution of the full Mcm2-7 complement along the genome is shaped by active transcription. Mcm2-7 signal is enriched in both intergenic and non-transcribed sequences, but depleted from actively transcribed transcripts. The displacement of Mcm2-7 is not localized to exons, but rather occurs over the entirety of the transcript suggesting that local sequence bias (e.g. increased GC content in exons) is not a contributing factor. We do not envision that the full complement of Mcm2-7 is coating these intergenic and non-transcribed regions, but rather represent an increased probability of finding an Mcm2-7 complex in static regions of the genome relative to those that are actively undergoing transcription.
We propose that Mcm2-7 residing in transcribed regions are displaced by the passage of RNA Pol II during transcription. The displacement of Mcm2-7 during transcription is analogous to the removal of Mcm2-7 from inactive origins during passage of the replication fork in S-phase (Madine et al, 1995; Krude et al, 1996; Romanowski et al, 1996a; Kuipers et al, 2011). The bimodal distribution of Mcm2-7 at intergenic and non-transcribed genes relative to active genes may only be established after the transition into S-phase. Prior to S-phase and the cessation of pre-RC assembly, Mcm2-7 are likely in a cycle of loading, translocation, and subsequent eviction in transcribed regions; however, once pre-RC assembly is inhibited they are no longer able to re-occupy transcribed regions following eviction by active transcription.
The mechanism(s) by which the Mcm2-7 enrichment transitions from ORC-specific localization to a broad and distributed pattern along the chromosome is not clear. We envision at least two possibilities—translocation along the DNA away from ORC or the loading of Mcm2-7 at distal sites by chromosomal looping. In vitro reconstitution of Mcm2-7 loading on circular DNA templates revealed Mcm2-7 double hexamers distributed randomly on the template irrespective of the location of the ACS or ORC binding site (Evrin et al, 2009; Remus et al, 2009). Thus, in vitro, Mcm2-7 double hexamers can be loaded and are free to translocate along the dsDNA template. However, it is very difficult to imagine how, in vivo, Mcm2-7 complexes could translocate along the DNA given the chromatin obstacles such as nucleosomes, DNA binding proteins, and active transcription. Nucleosomes would have to be displaced and re-assembled throughout the genome requiring ATP-dependent chromatin remodeling activities and histone chaperones (Alabert & Groth, 2012). Alternatively, the loading of Mcm2-7 at sites distal to ORC may be achieved by chromatin looping mediated by cohesin complexes that are close to ORC in Drosophila (MacAlpine et al, 2010). In mammalian cells, cohesin has been shown to be required for looping chromatin at replication factories (Guillou et al, 2010).
Defining an origin of replication
The search for sequence-based replicators, similar to the ARS element in S. cerevisiae, has been a holy grail for the mammalian replication field. In vitro, any sequence can be replicated in Xenopus extracts and plasmid-based assays have also exhibited very promiscuous replication (Krysan & Calos, 1991). Recently, the analysis of mammalian replication intermediates by next-generation sequencing identified G4-quadruplex sequences as potential replicators (Cayrou et al, 2011; Besnard et al, 2012; Valton et al, 2014). Despite the identification of G4 quadruplex structures as origins of replication, there is little concordance between datasets, indicating that these degenerate structures, which occur every few kilobases, are not sufficient for origin specification. Work from the Hamlin group using two-dimensional gel electrophoresis demonstrated that many genomic fragments have the potential to harbor inefficient replication origins (Mesner et al, 2006). We propose that the broad distribution of the full complement of Mcm2-7 we observe contributes to the apparent promiscuity of metazoan origin selection. In vitro experiments in Xenopus have shown that ORC is not required for initiation after pre-RC assembly (Hua & Newport, 1998; Rowles et al, 1999). Thus, those Mcm2-7 helicases located distally from ORC would still have the potential to be activated and could likely function as dormant origins in the presence of replicative stress.
In our prior studies, we have demonstrated that early origins of replication, which are resistant to HU, are enriched for ORC binding sites (MacAlpine et al, 2010). These ORC binding sites also serve as focal points for the cyclin E-independent Mcm2-7 loading. Together, these data suggest that Mcm2-7 loaded proximal to ORC binding sites have an increased likelihood of activation during early S-phase. In S. cerevisiae, Cdc45 specifically associates with early activating origins in G1 of the cell cycle (Aparicio et al, 1999). Thus, during early G1 in Drosophila, Cdc45 may specifically associate with the few Mcm2-7 complexes loaded proximal to ORC binding sites, increasing the likelihood of activation of this subset of pre-RCs during entry into S-phase.
Materials and Methods
Drosophila cell culture
Cells were cultured in 150-mm plates at an approximate density of 1 × 106 cells/ml in Schneider's Insect Cell Medium (Invitrogen) supplemented with 10% heat-inactivated FBS (Hyclone) and 1% penicillin/streptomycin/glutamine (Invitrogen). Dacapo was overexpressed for 48 h in the presence of 500 μM copper sulfate. FLAG-tagged cyclin E was overexpressed in the presence of 500 μM copper sulfate in the medium. To arrest at G1/S, cells were incubated with 1 mM HU for 24 h (unless otherwise noted). G2-arrested cells were treated with 3% DMSO and assayed after 48 h.
dsRNA synthesis and RNAi
Primers were designed with a 5′ T7 sequence (TTAATACGACTCACTATAGGGAGA) to amplify a 500–900 bp region in the gene of interest. The region was amplified using standard PCR followed by gel extraction (Qiagen Qiaquick gel extraction) and the PCR product was subsequently used as a template for dsRNA production (Promega T7 Ribomax express large scale expression). The dsRNA was purified by phenol/chloroform extraction followed by ethanol precipitation. Primers used for each target: Dup/Cdt1 dsRNA, F: 5′ctatcagtatcaagaacaggcg, R: 5′tgctttccaccagactg; cyclin E dsRNA, F: 5′gccatccgtcacataagca, R: 5′atcgtggaagcaagcagac; Cdk2 dsRNA, F: 5′tgtggccctcaaaaagattc, R: 5′gaagtaagcgtgctgcagtg; Cdc6 dsRNA, F: 5′gccacagcacctgatcagtccttcgcg, R: 5′gcagtttacaagaaactatgcacc; pUC dsRNA, F: 5′agctcactcaaaggcggtaa, R: 5′gcctacatacctcgctctgc.
Cells were washed with serum-free Schneider's insect medium (Invitrogen), resuspended to 1 × 106 cells/ml in serum-free insect medium, and then replated. dsRNA (15 μg/1 × 106 cells/ml) was added to the medium and incubated for 1 h at 25°C. Medium with 2× serum was added to make the total medium 1× in respect to serum. Cyclin E, Cdk2 and pUC RNAi cells were assessed 48 h after RNAi, Dup/Cdt1 and Cdc6 RNAi cells were assessed after 24 h.
FACS preparation and analysis
Cells were harvested by centrifugation, washed with ice-cold 1× PBS, resuspended in residual 1× PBS, and fixed overnight at 4°C with ice-cold 100% ethanol. Next, the cells were centrifuged, the supernatant removed, and washed with 1× PBS—1% fetal bovine serum. The cells were resuspended in a final volume of 800 μl 1× PBS—1% fetal bovine serum with 50 mg/ml of propidium iodide and 0.1 mg/ml RNaseA. The cells were incubated at 37°C for 1 h. Equal numbers of cells per condition were assayed using a FACSCanto machine running BD FACSDiva software and R bioconductor flowCore package to generate histograms.
Chromatin fractionation
Fractionation of the chromatin was optimized for Drosophila cell lines from established protocols (Hancock, 1974; Ritzi et al, 1998). Cells were harvested and then washed with ice-cold 1× PBS twice. Cells were resuspended in hypotonic buffer [10 mM HEPES-KOH (pH 7.5), 20 mM KCl, 0.25 mM EDTA, and a Roche protease mini tab], kept on ice for 4 min then centrifuged for 4 min at 700 g at 4°C. The pellet was resuspended in lysis buffer [10 mM HEPES-KOH (pH 7.5), 70 mM NaCl, 20 mM KCl, 5 mM MgCl2, 2 mM CaCl2, 0.5% NP-40, and a Roche protease inhibitor tab] and underlayed with 1 volume of 30% sucrose. The lysate was centrifuged for 3 min at 3,000 g at 4°C. The supernatant was set aside as the whole cell lysate sample. The pellet was resuspended in lysis buffer and centrifuged with a sucrose cushion. The pellet was washed with low salt buffer [20 mM HEPES/KOH (pH 7.5), 0.5 mM MgCl2, 0.3 M sucrose, KCl, and Roche protease inhibitor tab] and centrifuged at 4°C for 3 min at 3,000 g. The pellet was washed again in low salt buffer and resuspended in 2× SDS + β-ME loading buffer. To verify that the proteins associated with the nuclear pellet were soluble, they were washed in high salt buffer (480 mM HEPES/KOH (pH 7.5), 0.5 mM MgCl2, 0.3 M sucrose, KCl, and Roche protease inhibitor tab) (Supplementary Fig S8A). Alternatively, to solubilize the chromatin-bound fraction, pellets were resuspended in low salt buffer supplemented with 5 mM CaCl2 and treated with 45 U of micrococcal nuclease for 10 min prior to quenching with 5 mM EGTA (Supplementary Fig S8B).
Immunofluorescence
For double labeling of Mcm2-7 and H4K20me1, cells were treated with 0.5% Triton X-100 in PBS for 1 min, washed, fixed with 4% paraformaldehyde in PBS for 10 min, and blocked with 3% BSA in PBS-T (0.1% Triton in PBS). Cells were stained with monoclonal Mcm2-7 antibody (AS1.1, 1:100) and anti-H4K20me1 antibody (ab9051, 1:1,000), followed by secondary detection with Alexa Fluor 568 goat anti-mouse and Alexa Fluor 633 goat anti-rabbit antibodies (1:500). Images were acquired with Zeiss Axio Imager wide field fluorescence microscope, and the median intensities of individual cells (n = 277) from each channel were measured by ImageJ. The plot of H4K20me1 signal versus Mcm2-7 signal was generated in R.
Western blot analysis
Mcm2-7 monoclonal mouse antibody (AS1.1) was used at 1:100 (Chen et al, 2007), Orc2 rabbit polyclonal antibody was used at 1:3,000 (Austin et al, 1999), Dacapo (NP-1) used at 1:1,000 (Iowa Hybridoma Bank), Dup/Cdt1 guinea pig raised antibody was used at 1:5,000, and cyclin E antibody raised in rabbits was used at 1:500 (d-300 Santa Cruz sc-33748). Secondary antibodies: goat anti-mouse IRDye 800CW IgG (LiCor), goat anti-rabbit Alexa Fluor 680 IgG (Invitrogen), and goat anti-guinea pig IRDye 800 (Rockland Immunochemicals), were all used at 1:10,000. Westerns were visualized and quantified using LiCor infrared technology and gray scaled in ImageJ.
ChIP-chip sample preparation
Samples were prepared as described previously (MacAlpine et al, 2010). Mcm2-7 (AS1.1) antibody was used at a 1/25 dilution. All experiments were performed in duplicate.
ChIP-Chip and ChIP-Seq analysis
Within-replicate probe intensities were determined and between-slide intensities normalized via the R (R Development Core Team, 2008) package limma (Smyth, 2004). Replicated probe intensities were determined via MA2C (Song et al, 2007). Subsequently, all analyses were done in R using the combined replicate probe intensities. Gene bodies were obtained from (Graveley et al, 2011). Mcm2-7 ChIP-seq and corresponding input samples were performed on HU-arrested Kc and S2 cells. log2 RPKM ratios of Mcm ChIP-seq to input sample were calculated over all Drosophila gene models from the 5.12 annotation release, excluding any genes overlapping a Kc or S2 early origin. Genes were then subsequently subsetted into untranscribed (< 1 RPKM) and transcribed (> 4 RPKM) groups based on expression data from Cherbas et al (2011). log2 ratios of Mcm2-7 ChIP-seq to input samples were then compared between the cell lines for four gene classes: untranscribed genes in both Kc and S2 (4,593 genes), transcribed genes in both Kc and S2 (4,804 genes), untranscribed in Kc and transcribed in S2 (91 genes), and transcribed in Kc and untranscribed in S2 (128 genes). The Mann–Whitney–Wilcoxon Test was used to evaluate differences in the log2 ratio distributions between Kc and S2.
FLAG-cyclin E stable transfection
SLIC cloning (Li & Elledge, 2012) was used to introduce a FLAG fusion tag in frame at either the N-terminus or C-terminus of cyclin E cDNA (clone LD22682) in the pMK-CTAP plasmid (pMK-CTAP was a gift from Artavanis laboratory). Briefly, the plasmid was digested with a single restriction site enzyme, N term = XhoI, C term = SpeI. The 5′ ends were treated with T4 DNA polymerase (NEB) for 45 min at room temperature. Approximately 150 ng of vector in a 1:1 ratio was annealed to FLAG primers in 1× ligation mix (Invitrogen) at 37°C for 30 min. The reactions were transformed into max efficiency DH5α cells (Invitrogen). Effectene transfection reagent kit (Qiagen) protocol was followed for suspension cells in 100-mm dish, with the only exception being that 150 μl Effectene reagent was added. Cells were plated at 1 × 106 cells/ml and incubated at 25°C for 2 days. Hygromycin B (Sigma) was then added to 0.125 μg/ml.
Data access
All genomic data are publicly available at NCBI GEO and SRA data repositories with the following accessions numbers: GSE17282, GSE17283, GSE41349, GSE63915, SRP050273.
Acknowledgments
We thank Bob Duronio, Don Fox, and MacAlpine laboratory members for critical comments on the manuscript. Don Rio for the Dacapo construct, Steve Bell for Mcm2-7 antibody, and the Artavanis laboratory for pMK plasmid. This work was supported by an American Cancer Society Research Scholar Grant (120222-RSG-11-048-01-DMC) and the National Institutes of Health (1R01GM104097-01A1).
Author contributions
SKP and DMM designed the experiments and project. SKP performed the biochemical experiments, HKM performed the genome-wide experiments, JAP and JAB executed the bioinformatic analysis, and YL performed the immunofluorescence experiments. SKP, HKM, and DMM wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Supplementary Figures S1–S8
Review Process File
References
- Abbas T, Shibata E, Park J, Jha S, Karnani N, Dutta A. CRL4(Cdt2) regulates cell proliferation and histone gene expression by targeting PR-Set7/Set8 for degradation. Mol Cell. 2010;40:9–21. doi: 10.1016/j.molcel.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alabert C, Groth A. Chromatin replication and epigenome maintenance. Nat Rev Mol Cell Biol. 2012;13:153–167. doi: 10.1038/nrm3288. [DOI] [PubMed] [Google Scholar]
- Anglana M, Apiou F, Bensimon A, Debatisse M. Dynamics of DNA replication in mammalian somatic cells: nucleotide pool modulates origin choice and interorigin spacing. Cell. 2003;114:385–394. doi: 10.1016/s0092-8674(03)00569-5. [DOI] [PubMed] [Google Scholar]
- Aparicio OM, Weinstein DM, Bell SP. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell. 1997;91:59–69. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- Aparicio OM, Stout AM, Bell SP. Differential assembly of Cdc45p and DNA polymerases at early and late origins of DNA replication. Proc Natl Acad Sci USA. 1999;96:9130–9135. doi: 10.1073/pnas.96.16.9130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias EE, Walter JC. PCNA functions as a molecular platform to trigger Cdt1 destruction and prevent re-replication. Nat Cell Biol. 2006;8:84–90. doi: 10.1038/ncb1346. [DOI] [PubMed] [Google Scholar]
- Austin RJ, Orr-Weaver TL, Bell SP. Drosophila ORC specifically binds to ACE3, an origin of DNA replication control element. Genes Dev. 1999;13:2639–2649. doi: 10.1101/gad.13.20.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell SP, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- Besnard E, Babled A, Lapasset L, Milhavet O, Parrinello H, Dantec C, Marin JM, Lemaitre JM. Unraveling cell type-specific and reprogrammable human replication origin signatures associated with G-quadruplex consensus motifs. Nat Struct Mol Biol. 2012;19:837–844. doi: 10.1038/nsmb.2339. [DOI] [PubMed] [Google Scholar]
- Bowers JL, Randell JC, Chen S, Bell SP. ATP hydrolysis by ORC catalyzes reiterative Mcm2-7 assembly at a defined origin of replication. Mol Cell. 2004;16:967–978. doi: 10.1016/j.molcel.2004.11.038. [DOI] [PubMed] [Google Scholar]
- Burkhart R, Schulte D, Hu D, Musahl C, Gohring F, Knippers R. Interactions of human nuclear proteins P1Mcm3 and P1Cdc46. Eur J Biochem. 1995;228:431–438. [PubMed] [Google Scholar]
- Cayrou C, Coulombe P, Vigneron A, Stanojcic S, Ganier O, Peiffer I, Rivals E, Puy A, Laurent-Chabalier S, Desprat R, Mechali M. Genome-scale analysis of metazoan replication origins reveals their organization in specific but flexible sites defined by conserved features. Genome Res. 2011;21:1438–1449. doi: 10.1101/gr.121830.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, de Vries MA, Bell SP. Orc6 is required for dynamic recruitment of Cdt1 during repeated Mcm2-7 loading. Genes Dev. 2007;21:2897–2907. doi: 10.1101/gad.1596807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherbas L, Willingham A, Zhang D, Yang L, Zou Y, Eads BD, Carlson JW, Landolin JM, Kapranov P, Dumais J, Samsonova A, Choi JH, Roberts J, Davis CA, Tang H, van Baren MJ, Ghosh S, Dobin A, Bell K, Lin W, et al. The transcriptional diversity of 25 Drosophila cell lines. Genome Res. 2011;21:301–314. doi: 10.1101/gr.112961.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang LC, Teixeira LK, Wohlschlegel JA, Henze M, Yates JR, Mendez J, Reed SI. Phosphorylation of Mcm2 by Cdc7 promotes pre-replication complex assembly during cell-cycle re-entry. Mol Cell. 2009;35:206–216. doi: 10.1016/j.molcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang CH, Wallace MD, Abratte C, Southard T, Schimenti JC. Incremental genetic perturbations to MCM2-7 expression and subcellular distribution reveal exquisite sensitivity of mice to DNA replication stress. PLoS Genet. 2010;6:e1001110. doi: 10.1371/journal.pgen.1001110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claycomb JM, MacAlpine DM, Evans JG, Bell SP, Orr-Weaver TL. Visualization of replication initiation and elongation in Drosophila. J Cell Biol. 2002;159:225–236. doi: 10.1083/jcb.200207046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clijsters L, Ogink J, Wolthuis R. The spindle checkpoint, APC/C(Cdc20), and APC/C(Cdh1) play distinct roles in connecting mitosis to S phase. J Cell Biol. 2013;201:1013–1026. doi: 10.1083/jcb.201211019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clijsters L, Wolthuis R. PIP-box-mediated degradation prohibits re-accumulation of Cdc6 during S phase. J Cell Sci. 2014;127:1336–1345. doi: 10.1242/jcs.145862. [DOI] [PubMed] [Google Scholar]
- Crevel G, Cotterill S. DNA replication in cell-free extracts from Drosophila melanogaster. EMBO J. 1991;10:4361–4369. doi: 10.1002/j.1460-2075.1991.tb05014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crevel G, Hashimoto R, Vass S, Sherkow J, Yamaguchi M, Heck MM, Cotterill S. Differential requirements for MCM proteins in DNA replication in Drosophila S2 cells. PLoS ONE. 2007;2:e833. doi: 10.1371/journal.pone.0000833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova DS, Todorov IT, Melendy T, Gilbert DM. Mcm2, but not RPA, is a component of the mammalian early G1-phase prereplication complex. J Cell Biol. 1999;146:709–722. doi: 10.1083/jcb.146.4.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan S, Harwood J, Drury LS, Diffley JF. Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc Natl Acad Sci USA. 1997;94:5611–5616. doi: 10.1073/pnas.94.11.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drury LS, Perkins G, Diffley JF. The Cdc4/34/53 pathway targets Cdc6p for proteolysis in budding yeast. EMBO J. 1997;16:5966–5976. doi: 10.1093/emboj/16.19.5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulic V, Lees E, Reed SI. Association of human cyclin E with a periodic G1-S phase protein kinase. Science. 1992;257:1958–1961. doi: 10.1126/science.1329201. [DOI] [PubMed] [Google Scholar]
- Eaton ML, Galani K, Kang S, Bell SP, MacAlpine DM. Conserved nucleosome positioning defines replication origins. Genes Dev. 2010;24:748–753. doi: 10.1101/gad.1913210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards MC, Tutter AV, Cvetic C, Gilbert CH, Prokhorova TA, Walter JC. MCM2-7 complexes bind chromatin in a distributed pattern surrounding the origin recognition complex in Xenopus egg extracts. J Biol Chem. 2002;277:33049–33057. doi: 10.1074/jbc.M204438200. [DOI] [PubMed] [Google Scholar]
- Ekholm-Reed S, Spruck CH, Sangfelt O, van Drogen F, Mueller-Holzner E, Widschwendter M, Zetterberg A, Reed SI. Mutation of hCDC4 leads to cell cycle deregulation of cyclin E in cancer. Cancer Res. 2004;64:795–800. doi: 10.1158/0008-5472.can-03-3417. [DOI] [PubMed] [Google Scholar]
- Elsasser S, Chi Y, Yang P, Campbell JL. Phosphorylation controls timing of Cdc6p destruction: a biochemical analysis. Mol Biol Cell. 1999;10:3263–3277. doi: 10.1091/mbc.10.10.3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrin C, Clarke P, Zech J, Lurz R, Sun J, Uhle S, Li H, Stillman B, Speck C. A double-hexameric MCM2-7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc Natl Acad Sci USA. 2009;106:20240–20245. doi: 10.1073/pnas.0911500106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrin C, Fernandez-Cid A, Zech J, Herrera MC, Riera A, Clarke P, Brill S, Lurz R, Speck C. In the absence of ATPase activity, pre-RC formation is blocked prior to MCM2-7 hexamer dimerization. Nucleic Acids Res. 2013;41:3162–3172. doi: 10.1093/nar/gkt043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigola J, Remus D, Mehanna A, Diffley JF. ATPase-dependent quality control of DNA replication origin licensing. Nature. 2013;495:339–343. doi: 10.1038/nature11920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambus A, Khoudoli GA, Jones RC, Blow JJ. MCM2-7 form double hexamers at licensed origins in Xenopus egg extract. J Biol Chem. 2011;286:11855–11864. doi: 10.1074/jbc.M110.199521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge XQ, Jackson DA, Blow JJ. Dormant origins licensed by excess Mcm2-7 are required for human cells to survive replicative stress. Genes Dev. 2007;21:3331–3341. doi: 10.1101/gad.457807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng Y, Lee YM, Welcker M, Swanger J, Zagozdzon A, Winer JD, Roberts JM, Kaldis P, Clurman BE, Sicinski P. Kinase-independent function of cyclin E. Mol Cell. 2007;25:127–139. doi: 10.1016/j.molcel.2006.11.029. [DOI] [PubMed] [Google Scholar]
- Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, Artieri CG, van Baren MJ, Boley N, Booth BW, Brown JB, Cherbas L, Davis CA, Dobin A, Li R, Lin W, Malone JH, Mattiuzzo NR, Miller D, Sturgill D, et al. The developmental transcriptome of Drosophila melanogaster. Nature. 2011;471:473–479. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillou E, Ibarra A, Coulon V, Casado-Vela J, Rico D, Casal I, Schwob E, Losada A, Mendez J. Cohesin organizes chromatin loops at DNA replication factories. Genes Dev. 2010;24:2812–2822. doi: 10.1101/gad.608210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock R. Interphase chromosomal deoxyribonucleoprotein isolated as a discrete structure from cultured cells. J Mol Biol. 1974;86:649–663. doi: 10.1016/0022-2836(74)90187-9. [DOI] [PubMed] [Google Scholar]
- Harvey KJ, Newport J. CpG methylation of DNA restricts prereplication complex assembly in xenopus egg extracts. Mol Cell Biol. 2003;23:6769–6779. doi: 10.1128/MCB.23.19.6769-6779.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua XH, Newport J. Identification of a preinitiation step in DNA replication that is independent of origin recognition complex and cdc6, but dependent on cdk2. J Cell Biol. 1998;140:271–281. doi: 10.1083/jcb.140.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra A, Schwob E, Mendez J. Excess MCM proteins protect human cells from replicative stress by licensing backup origins of replication. Proc Natl Acad Sci USA. 2008;105:8956–8961. doi: 10.1073/pnas.0803978105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Arias EE, Chen J, Harper JW, Walter JC. A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol Cell. 2006;23:709–721. doi: 10.1016/j.molcel.2006.08.010. [DOI] [PubMed] [Google Scholar]
- Knoblich JA, Sauer K, Jones L, Richardson H, Saint R, Lehner CF. Cyclin E controls S phase progression and its down-regulation during Drosophila embryogenesis is required for the arrest of cell proliferation. Cell. 1994;77:107–120. doi: 10.1016/0092-8674(94)90239-9. [DOI] [PubMed] [Google Scholar]
- Krude T, Musahl C, Laskey RA, Knippers R. Human replication proteins hCdc21, hCdc46 and P1Mcm3 bind chromatin uniformly before S-phase and are displaced locally during DNA replication. J Cell Sci. 1996;109:309–318. doi: 10.1242/jcs.109.2.309. [DOI] [PubMed] [Google Scholar]
- Krysan PJ, Calos MP. Replication initiates at multiple locations on an autonomously replicating plasmid in human cells. Mol Cell Biol. 1991;11:1464–1472. doi: 10.1128/mcb.11.3.1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuipers MA, Stasevich TJ, Sasaki T, Wilson KA, Hazelwood KL, McNally JG, Davidson MW, Gilbert DM. Highly stable loading of Mcm proteins onto chromatin in living cells requires replication to unload. J Cell Biol. 2011;192:29–41. doi: 10.1083/jcb.201007111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib K. Uninterrupted MCM2-7 function required for DNA replication fork progression. Science. 2000;288:1643–1647. doi: 10.1126/science.288.5471.1643. [DOI] [PubMed] [Google Scholar]
- Lane ME, Sauer K, Wallace K, Jan YN, Lehner CF, Vaessin H. Dacapo, a cyclin-dependent kinase inhibitor, stops cell proliferation during Drosophila development. Cell. 1996;87:1225–1235. doi: 10.1016/s0092-8674(00)81818-8. [DOI] [PubMed] [Google Scholar]
- Lei M, Kawasaki Y, Tye BK. Physical interactions among Mcm proteins and effects of Mcm dosage on DNA replication in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:5081–5090. doi: 10.1128/mcb.16.9.5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MZ, Elledge SJ. SLIC: a method for sequence- and ligation-independent cloning. Methods Mol Biol. 2012;852:51–59. doi: 10.1007/978-1-61779-564-0_5. [DOI] [PubMed] [Google Scholar]
- Liu E, Li X, Yan F, Zhao Q, Wu X. Cyclin-dependent kinases phosphorylate human Cdt1 and induce its degradation. J Biol Chem. 2004;279:17283–17288. doi: 10.1074/jbc.C300549200. [DOI] [PubMed] [Google Scholar]
- Lubelsky Y, Prinz JA, DeNapoli L, Li Y, Belsky JA, MacAlpine DM. DNA replication and transcription programs respond to the same chromatin cues. Genome Res. 2014;24:1102–1114. doi: 10.1101/gr.160010.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlpine HK, Gordan R, Powell SK, Hartemink AJ, MacAlpine DM. Drosophila ORC localizes to open chromatin and marks sites of cohesin complex loading. Genome Res. 2010;20:201–211. doi: 10.1101/gr.097873.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madine MA, Khoo CY, Mills AD, Musahl C, Laskey RA. The nuclear envelope prevents reinitiation of replication by regulating the binding of MCM3 to chromatin in Xenopus egg extracts. Curr Biol. 1995;5:1270–1279. doi: 10.1016/s0960-9822(95)00253-3. [DOI] [PubMed] [Google Scholar]
- Mahbubani HM, Chong JP, Chevalier S, Thommes P, Blow JJ. Cell cycle regulation of the replication licensing system: involvement of a Cdk-dependent inhibitor. J Cell Biol. 1997;136:125–135. doi: 10.1083/jcb.136.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailand N, Diffley JF. CDKs promote DNA replication origin licensing in human cells by protecting Cdc6 from APC/C-dependent proteolysis. Cell. 2005;122:915–926. doi: 10.1016/j.cell.2005.08.013. [DOI] [PubMed] [Google Scholar]
- Masai H, Matsumoto S, You Z, Yoshizawa-Sugata N, Oda M. Eukaryotic chromosome DNA replication: where, when, and how? Annu Rev Biochem. 2010;79:89–130. doi: 10.1146/annurev.biochem.052308.103205. [DOI] [PubMed] [Google Scholar]
- McGarry TJ, Kirschner MW. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell. 1998;93:1043–1053. doi: 10.1016/s0092-8674(00)81209-x. [DOI] [PubMed] [Google Scholar]
- McNairn AJ, Okuno Y, Misteli T, Gilbert DM. Chinese hamster ORC subunits dynamically associate with chromatin throughout the cell-cycle. Exp Cell Res. 2005;308:345–356. doi: 10.1016/j.yexcr.2005.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesner LD, Crawford EL, Hamlin JL. Isolating apparently pure libraries of replication origins from complex genomes. Mol Cell. 2006;21:719–726. doi: 10.1016/j.molcel.2006.01.015. [DOI] [PubMed] [Google Scholar]
- Nguyen VQ, Co C, Li JJ. Cyclin-dependent kinases prevent DNA re-replication through multiple mechanisms. Nature. 2001;411:1068–1073. doi: 10.1038/35082600. [DOI] [PubMed] [Google Scholar]
- Nishitani H, Sugimoto N, Roukos V, Nakanishi Y, Saijo M, Obuse C, Tsurimoto T, Nakayama KI, Nakayama K, Fujita M, Lygerou Z, Nishimoto T. Two E3 ubiquitin ligases, SCF-Skp2 and DDB1-Cul4, target human Cdt1 for proteolysis. EMBO J. 2006;25:1126–1136. doi: 10.1038/sj.emboj.7601002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda H, Hubner MR, Beck DB, Vermeulen M, Hurwitz J, Spector DL, Reinberg D. Regulation of the histone H4 monomethylase PR-Set7 by CRL4(Cdt2)-mediated PCNA-dependent degradation during DNA damage. Mol Cell. 2010;40:364–376. doi: 10.1016/j.molcel.2010.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsubo M, Roberts JM. Cyclin-dependent regulation of G1 in mammalian fibroblasts. Science. 1993;259:1908–1912. doi: 10.1126/science.8384376. [DOI] [PubMed] [Google Scholar]
- Pacek M, Walter JC. A requirement for MCM7 and Cdc45 in chromosome unwinding during eukaryotic DNA replication. EMBO J. 2004;23:3667–3676. doi: 10.1038/sj.emboj.7600369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacek M, Tutter AV, Kubota Y, Takisawa H, Walter JC. Localization of MCM2-7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol Cell. 2006;21:581–587. doi: 10.1016/j.molcel.2006.01.030. [DOI] [PubMed] [Google Scholar]
- Pruitt SC, Bailey KJ, Freeland A. Reduced Mcm2 expression results in severe stem/progenitor cell deficiency and cancer. Stem Cells. 2007;25:3121–3132. doi: 10.1634/stemcells.2007-0483. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2008. . ISBN 3-900051-07-0, URL http://www.R-project.org. [Google Scholar]
- Rao H, Stillman B. The origin recognition complex interacts with a bipartite DNA binding site within yeast replicators. Proc Natl Acad Sci USA. 1995;92:2224–2228. doi: 10.1073/pnas.92.6.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remus D, Beuron F, Tolun G, Griffith JD, Morris EP, Diffley JF. Concerted loading of Mcm2-7 double hexamers around DNA during DNA replication origin licensing. Cell. 2009;139:719–730. doi: 10.1016/j.cell.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnitzky D, Gossen M, Bujard H, Reed SI. Acceleration of the G1/S phase transition by expression of cyclins D1 and E with an inducible system. Mol Cell Biol. 1994;14:1669–1679. doi: 10.1128/mcb.14.3.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritzi M, Baack M, Musahl C, Romanowski P, Laskey RA, Knippers R. Human minichromosome maintenance proteins and human origin recognition complex 2 protein on chromatin. J Biol Chem. 1998;273:24543–24549. doi: 10.1074/jbc.273.38.24543. [DOI] [PubMed] [Google Scholar]
- Romanowski P, Madine MA, Laskey RA. XMCM7, a novel member of the Xenopus MCM family, interacts with XMCM3 and colocalizes with it throughout replication. Proc Natl Acad Sci USA. 1996a;93:10189–10194. doi: 10.1073/pnas.93.19.10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanowski P, Madine MA, Rowles A, Blow JJ, Laskey RA. The Xenopus origin recognition complex is essential for DNA replication and MCM binding to chromatin. Curr Biol. 1996b;6:1416–1425. doi: 10.1016/s0960-9822(96)00746-4. [DOI] [PubMed] [Google Scholar]
- Rowles A, Tada S, Blow JJ. Changes in association of the Xenopus origin recognition complex with chromatin on licensing of replication origins. J Cell Sci. 1999;112:2011–2018. doi: 10.1242/jcs.112.12.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Ernst J, Kharchenko PV, Kheradpour P, Negre N, Eaton ML, Landolin JM, Bristow CA, Ma L, Lin MF, Washietl S, Arshinoff BI, Ay F, Meyer PE, Robine N, Washington NL, Di Stefano L, Berezikov E, Brown CD, Candeias R, et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330:1787–1797. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekedat MD, Fenyo D, Rogers RS, Tackett AJ, Aitchison JD, Chait BT. GINS motion reveals replication fork progression is remarkably uniform throughout the yeast genome. Mol Syst Biol. 2010;6:353. doi: 10.1038/msb.2010.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senga T, Sivaprasad U, Zhu W, Park JH, Arias EE, Walter JC, Dutta A. PCNA is a cofactor for Cdt1 degradation by CUL4/DDB1-mediated N-terminal ubiquitination. J Biol Chem. 2006;281:6246–6252. doi: 10.1074/jbc.M512705200. [DOI] [PubMed] [Google Scholar]
- Shima N, Alcaraz A, Liachko I, Buske TR, Andrews CA, Munroe RJ, Hartford SA, Tye BK, Schimenti JC. A viable allele of Mcm4 causes chromosome instability and mammary adenocarcinomas in mice. Nat Genet. 2007;39:93–98. doi: 10.1038/ng1936. [DOI] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Song JS, Johnson WE, Zhu X, Zhang X, Li W, Manrai AK, Liu JS, Chen R, Liu XS. Model-based analysis of two-color arrays (MA2C) Genome Biol. 2007;8:R178. doi: 10.1186/gb-2007-8-8-r178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonneville R, Querenet M, Craig A, Gartner A, Blow JJ. The dynamics of replication licensing in live Caenorhabditis elegans embryos. J Cell Biol. 2012;196:233–246. doi: 10.1083/jcb.201110080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer SL, Cappell SD, Tsai FC, Overton KW, Wang CL, Meyer T. The proliferation-quiescence decision is controlled by a bifurcation in CDK2 activity at mitotic exit. Cell. 2013;155:369–383. doi: 10.1016/j.cell.2013.08.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spruck CH, Won KA, Reed SI. Deregulated cyclin E induces chromosome instability. Nature. 1999;401:297–300. doi: 10.1038/45836. [DOI] [PubMed] [Google Scholar]
- Symeonidou IE, Kotsantis P, Roukos V, Rapsomaniki MA, Grecco HE, Bastiaens P, Taraviras S, Lygerou Z. Multi-step loading of human minichromosome maintenance proteins in live human cells. J Biol Chem. 2013;288:35852–35867. doi: 10.1074/jbc.M113.474825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi TS, Wigley DB, Walter JC. Pumps, paradoxes and ploughshares: mechanism of the MCM2-7 DNA helicase. Trends Biochem Sci. 2005;30:437–444. doi: 10.1016/j.tibs.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Araki H. Helicase activation and establishment of replication forks at chromosomal origins of replication. Cold Spring Harb Perspect Biol. 2013;5:a010371. doi: 10.1101/cshperspect.a010371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valton AL, Hassan-Zadeh V, Lema I, Boggetto N, Alberti P, Saintome C, Riou JF, Prioleau MN. G4 motifs affect origin positioning and efficiency in two vertebrate replicators. EMBO J. 2014;33:732–746. doi: 10.1002/embj.201387506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker AJ, Royzman I, Orr-Weaver TL. Drosophila double parked: a conserved, essential replication protein that colocalizes with the origin recognition complex and links DNA replication with mitosis and the down-regulation of S phase transcripts. Genes Dev. 2000;14:1765–1776. [PMC free article] [PubMed] [Google Scholar]
- Wimmel A, Lucibello FC, Sewing A, Adolph S, Muller R. Inducible acceleration of G1 progression through tetracycline-regulated expression of human cyclin E. Oncogene. 1994;9:995–997. [PubMed] [Google Scholar]
- Woodward AM, Gohler T, Luciani MG, Oehlmann M, Ge X, Gartner A, Jackson DA, Blow JJ. Excess Mcm2-7 license dormant origins of replication that can be used under conditions of replicative stress. J Cell Biol. 2006;173:673–683. doi: 10.1083/jcb.200602108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyrick JJ, Aparicio JG, Chen T, Barnett JD, Jennings EG, Young RA, Bell SP, Aparicio OM. Genome-wide distribution of ORC and MCM proteins in S. cerevisiae: high-resolution mapping of replication origins. Science. 2001;294:2357–2360. doi: 10.1126/science.1066101. [DOI] [PubMed] [Google Scholar]
- Xouri G, Squire A, Dimaki M, Geverts B, Verveer PJ, Taraviras S, Nishitani H, Houtsmuller AB, Bastiaens PI, Lygerou Z. Cdt1 associates dynamically with chromatin throughout G1 and recruits Geminin onto chromatin. EMBO J. 2007;26:1303–1314. doi: 10.1038/sj.emboj.7601597. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1–S8
Review Process File