Abstract
Cells of the monocyte/macrophage lineage are an important target for HIV-1 infection. They are often at anatomical sites linked to HIV-1 transmission and are an important vehicle for disseminating HIV-1 throughout the body, including the central nervous system. Monocytes do not support extensive productive HIV-1 replication, but they become more susceptible to HIV-1 infection as they differentiate into macrophages. The mechanisms guiding susceptibility of HIV-1 replication in monocytes versus macrophages are not entirely clear. We determined whether endogenous activity of β-catenin signaling impacts differential susceptibility of monocytes and monocyte-derived macrophages (MDMs) to productive HIV-1 replication. We show that monocytes have an approximately 4-fold higher activity of β-catenin signaling than MDMs. Inducing β-catenin in MDMs suppressed HIV-1 replication by 5-fold while inhibiting endogenous β-catenin signaling in monocytes by transfecting with a dominant negative mutant for the downstream effector of β-catenin (TCF-4) promoted productive HIV-1 replication by 6-fold. These findings indicate that β-catenin/TCF-4 is an important pathway for restricted HIV-1 replication in monocytes and plays a significant role in potentiating HIV-1 replication as monocytes differentiate into macrophages. Targeting this pathway may provide a novel strategy to purge the latent reservoir from monocytes/macrophages, especially in sanctuary sites for HIV-1 such as the central nervous system.
INTRODUCTION
It has been three decades since HIV-1 was identified as the etiologic agent of AIDS [1-4]. Considerable progress has been made in antiretroviral therapy, which has pushed HIV-1 to become a chronic infection. Given this, concerted scientific efforts are being made towards a functional cure. Eradication of HIV-1 is especially challenging because the virus remains latent in a number of reservoirs, evading the action of current antiretroviral therapy which requires active replication to interrupt different stages of the HIV-1 life cycle, such as entry, reverse transcription, integration, and assembly.
In response to danger signals and/or microenvironment triggers, monocytes differentiate into multiple cell lineages (dendritic cells, microglia, Kupffer cells, and osteoblasts). With this capability, monocytes resemble a Trojan horse in disseminating HIV-1 to various organs, including the central nervous system [5, 6]. Monocytes differentiate into macrophages in response to immune stimulation [7]. TLR2s or GM-CSF stimulation drives monocytes to differentiate into M-1-like macrophages, while IL-4, IL-13, or M-CSF drives monocytes to become M2-like macrophages. However, this is not a strict polarization paradigm for macrophages. Macrophages are highly plastic cells that can further respond to their specific tissue microenvironment, largely driven by surrounding cytokines, to differentiate into macrophages that do not necessarily fall under a classical M1 or M2 macrophage phenotype [8, 9].
Monocytes restrict HIV-1 productive replication and become susceptible to HIV-1 replication once they differentiate into macrophages [10-12] [13]. Although macrophages support HIV-1 replication, the degree of HIV-1 replication within macrophage subsets varies and maybe context-specific and phenotype specific[11]. HIV-1 enters monocytes by classical binding and fusion using CD4 receptor and CCR5 co-receptor. HIV-1 undergoes reverse transcription, but robust virion release does not occur in monocytes [16-18]. As monocytes differentiate to macrophages, they become more susceptible to productive HIV-1 replication through mechanism(s) that are not entirely clear or well integrated within each other. A number of mechanisms have been proposed to explain the refractory nature of monocytes to productive HIV-1 replication [13] [10, 14, 15] [16] [17, 18] [19-22]. Initially, lower expression of CCR5 was thought to restrict HIV-1 entry into monocytes [15, 23]. However, bypassing the CD4/co-receptor (CCR5/CXCR4) entry requirement of HIV-1 by pseudotyping with Vesicular Stomatitis Virus (VSV)-G envelope still did not circumvent the restriction to productive HIV-1 replication [19, 20]. These studies point to post-entry mechanisms that hinder HIV-1 replication in monocytes. Proposed post-entry mechanisms of HIV-1 blockade in monocytes include slow reverse transcription kinetics, delayed nuclear import and integration and low or absent expression of key host factors for HIV-1 transcription such as deoxythymidine trisphosphate (dTTP), cyclin T1 and NFAT5 [21, 24-26]. dTTP along with thymidine phosphorylase convert thymine to thymidine. Supplementing monocytes with D-thymidine to increase dTTP did not relieve restricted HIV-1 replication in monocytes[19]. Similarly, cyclin T1, which along with CDK9 forms the positive transcription elongation factor b (P-TEFb) that is critical for Tat-mediated activation of the HIV-1 LTR promoter, is low in monocytes. Low level of cyclin T1 is due to high endogenous expression of microRNA 198, which suppresses cyclin T1 expression[24]. Nonetheless, transfection of monocytes with cyclin T1 expression vector also did not render them susceptible to productive HIV-1 replication [26]. Given that removing known restrictive mechanisms does not render monocytes permissive to HIV-1 replication suggests that there are additional prominent mechanisms for the HIV-1 blockade in monocytes.
Our laboratory has identified the Wnt/β-catenin pathway as a potent inhibitor of HIV-1 replication [27-30]. We showed that Wnt/β-catenin signaling is inversely associated with HIV-1 replication in peripheral blood lymphocytes and in astrocytes. Given the importance of monocytes/macrophages in the pathogenesis of HIV-1-1 and their contribution to the persistence and progression of the disease, we evaluated the role of the Wnt/β-catenin pathway in regulating HIV-1 permissiveness in monocytes and macrophages.
The Wnt/β-catenin signaling pathway is highly conserved among species and is involved in a complex signal transduction pathway which regulate the transcriptional activity of hundreds of genes impacting diverse physiologic functions, including survival, differentiation, and proliferation [31, 32]. Wnt/β-catenin signaling is initiated by binding of a Wnt protein to a seven transmembrane frizzled receptor. In humans there are 19 identified soluble secreted Wnts and at least 10 frizzled receptors. Binding of Wnt to frizzled often requires the recruitment of low-density lipoprotein receptor-related protein (LRP) 5/6 inducing an intracellular signal cascade that leads to the stabilization of β-catenin as a hypophosphorylated protein. β-catenin is the central mediator of this pathway and acts as a transcriptional co-factor through interacting with members of the T-cell factor/lymphoid enhancer (TCF/LEF) transcription factors which lead to target gene transcription regulation. β-catenin also associates with E-cadherins to provide structural support for adhesion.
Because β-catenin is emerging as a host factor that regulates HIV-1 replication in multiple compartments [33], we evaluated whether differential expression of β-catenin in monocytes and MDMs may contribute to restricted HIV-1 replication in monocytes. A better understanding of pathways involved in restricting HIV-1 replication in monocytes can inform new modes of therapy to target this latent reservoir of HIV-1 and/or engage this pathway to eliminate active HIV-1 replication, especially among patients that fail HAART or do not reach undetectable viral load.
MATERIALS AND METHODS
Cell culture
Blood was collected from healthy laboratory donors in accordance with institutional guidelines on conduct of human research. Peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll-Hypaque density gradient centrifugation. Monocytes were obtained by adherence to tissue culture treated flasks for four hours at 37°C in serum-free RPMI 1640 medium (Biowhittaker; Walkersville, MD). Cells were then washed and cultured in complete media (RPMI 1640 medium supplemented with 10% heat-inactivated fetal bovine serum (Sigma, St Louis, MO), 1% penicillin-streptomycin (Gibco-BRL, Grand Island, NY), and 2mM L-glutamine (Gibco-BRL, Grand Island, NY)). Monocytes (1×106 cells/ml) were differentiated into monocyte-derived macrophages (MDMs) by treatment with 100ng/ml granulocyte-macrophage colony stimulating factor (GM-CSF) or macrophage colony stimulating factor (M-CSF) (Sigma, St Louis, MO) and incubation at 37°C in 5% CO2 for seven days. U1 cells were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH from Dr. Thomas Folks and propagated as recommended. U1 cells is a monocytic cell line latently infected with HIV-1, it has two proviral copies of HIV-1.
HIV-1 infection and detection by flow cytometry and p24 ELISA
Monocytes and MDMs were infected with HIV-1 Bal (AIDS Research and Reference Reagent Program, Germantown, MD) at 10 ng HIV-1 p24/1×106 cells for four hours at 37°C. Cells were then washed three times to remove unbound virus and incubated at 37°C in 5% CO2. MDM medium also contained GM-CSF (100 ng/ml) or M-CSF (100ng/ml). Supernatant was harvested at day 7, unless otherwise indicated, to measure HIV-1 p24 by conventional enzyme-linked immunosorbent assay (ELISA) (AIDS vaccine program, Frederick, MD). Alternatively, cells were analyzed by flow cytometry on a BD FACSVerse (BD Biosciences, San Jose, CA) for presence of HIV-1 Core Antigen-1 (Beckman Coulter, Brea California),
DNA constructs and nucleofection
The TCF-4 dominant-negative (DN) mutant construct has a DNA binding domain but lacks the N-terminus required for β-catenin binding [34] and was a gift from James O’Kelly (University of California, Los Angeles). This construct blocks β-catenin signaling [34-36]. TOPflash and FOPflash constructs are widely used to evaluate β-catenin dependent signaling events that drive the expression of Wnt/β-catenin target genes. TOPflash is a TCF/LEF reporter plasmid containing two sets of three copies of wild type TCF/LEF binding sites upstream of a Thymidine Kinase (TK) minimal promoter and a luciferase reporter gene. FOPflash contains mutated TCF/LEF binding sites upstream of the same TK promoter and luciferase open reading frame as TOPflash. FOPflash is used as a negative control for TOPflash activity. Both TOPflash and FOPflash were purchased from Millipore (Billerica, MA). The Renilla luciferase internal control vector is driven by cytomegalovirus (CMV) immediate early promoter region and was purchased from Promega (Madison, WI). The vector encoding green fluorescent protein (GFP) is also driven by the CMV immediate early promoter region and was purchased from Lonza Inc. (Portsmouth, NH). Nucleofection of monocytes and monocyte-derived macrophages (MDMs) was performed using the Amaxa monocyte or macrophage nucleofector kit, as recommended by the manufacturer (Lonza; Gaithersburg, MD). Briefly, 3×106 to 7×106 monocytes, or 7×105 macrophages were nucleofected with TCF-4 DN construct at 2μg of plasmid/1×106 cells using program Y-001 or Y-010 for monocytes and macrophages, respectively. Cells were left to rest for 24 hours in complete medium at 37°C in 5% CO2 prior to further use, as dictated by each experiment. 3×106 U1 monocytic cells were nucleofected using the same amaxa Y-001 program with 1μg of pCDNA or DNTCF4, cells were incubated in complete medium at 37°C in 5% CO2 for 3 days and harvested for cellular RNA.
Quantitative real-time RT-PCR
RNA was isolated using TRIzol reagent (Invitrogen, CA), according to the manufacturer’s recommendations. Subsequently, cDNA was synthesized using Quantitect reverse transcription kit (Qiagen, Maryland). Real-time PCR was performed using a Quantitect SYBR green PCR kit (Qiagen, Maryland) in a 7500 Real Time PCR System (Applied Biosystems, Foster City, CA) using 7500 software v2.0.1. Melting curve analysis was performed to ensure that the primers amplified the desired amplicon and that primer-dimers were absent. Primers used for mRNAs were: HIV-1-GAG forward-5-AGAGAAGGCTTTCAGCCCAGAAGT-3 and reverse-5-TGCACTGGATGCACTCTATCCCAT-3; HIV-1-ENV forward-5-ACGAGGATTGTGGAACTTCTGGGA-3 and reverse-5-TGGCATTGAGCAAGCTAACAGCAC-3; HIV-1-REV forward-5-TCCTTGGCACTTATCTGGGACGAT-3 and reverse-5-TCCCAGAAGTTCCACAATCCTCGT-3; and GAPDH forward-5-CTTCAACGACCACTTTGT-3 and reverse-5-TGGTCCAGGGGTCTTACT-3. Fold change in mRNA expression was calculated by relative quantification using the comparative CT method using GAPDH as endogenous control.
Flow cytometry
Freshly isolated monocytes and adhered monocytes with or without GM-CSF (100ng/ml) or M-CSF (100ng/ml) treatment were immunostained with appropriate antibodies using conventional surface and/or intracellular staining methods. Cells were stained intracellularly using the BD Cytofix/Cytoperm Buffer kit (BD Biosciences, San Jose CA) according to manufacturer’s instructions. Intracellular active β-catenin was measured using a primary monoclonal β-catenin antibody that detects the active form of β-catenin that is hypophosphorylated on serine 37 and threonine 41 (US Biological, MA) and a secondary goat F (ab’2) anti-mouse (CALTAG, CA) antibody. Intracellular HIV-1 Core Antigen was detected using a monoclonal antibody (Beckman Coulter Brea, CA). CD3, CD4, CD14, CD16, CD80, CD86 and HLA-DR surface stains were detected using antibodies from BD Biosciences (Franklin Lakes, NJ). LSR II flow cytometer with FACSDiva software (BD) was used for data collection in Figure 2. BDFACSVerse flow cytometer with BD FACSuite software (BD) was used for data collection in figures 1 and 5. FlowJo software (Tree Star; Ashland, OR) was used for analysis. Side scatter area (SSC-A) versus Aqua Live/Dead staining and forward scatter area (FSC-A) versus forward scatter height (FSC-H) were employed to exclude dead cells and doublets, respectively.
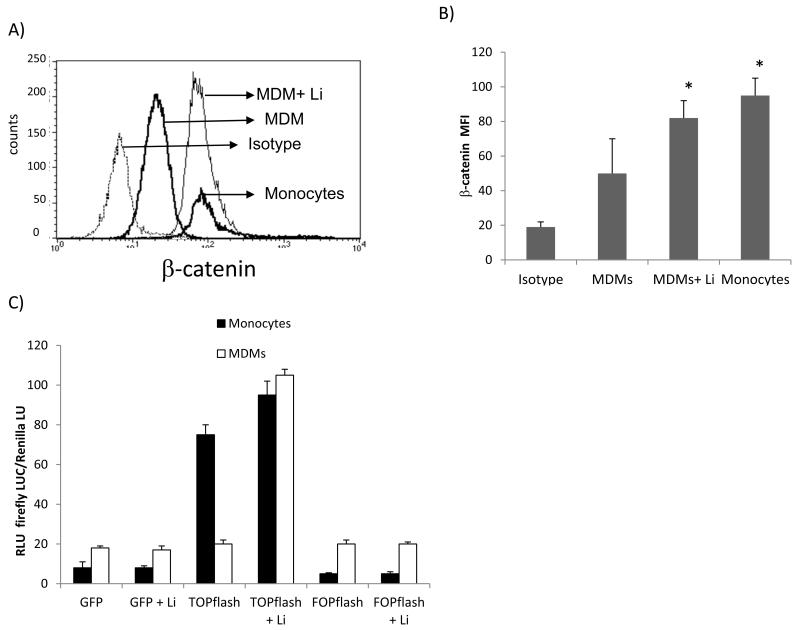
Figure 2. Endogenous β-catenin signaling is higher in monocytes than MDMs.
Freshly isolated monocytes (4 hr adherence) or GM-CSF MDMs were evaluated for endogenous and inducible active β-catenin protein. LiCl at 1mM was used to activate β-catenin. A representative histogram of active β-catenin level measured by conventional intracellular flow cytometry (A). Cumulative data of at least three independent experiments measuring active β-catenin level by flow cytometry (B). Monocytes and GM-CSF MDMs were transfected with green fluorescent protein construct (GFP), TOPflash construct, or FOPflash construct. The cells were then left untreated or treated with lithium (1mM). All cultures were co-transfected with an internal transfection control (Renilla). Approximately 12 hrs later, firefly luciferase and Renilla activities were measured using a dual luciferase assay (C). Data are presented as firefly luciferase over Renilla luciferase relative light units (RLU). Asterisks in (B) denote p<0.05 in comparison to MDMs alone.
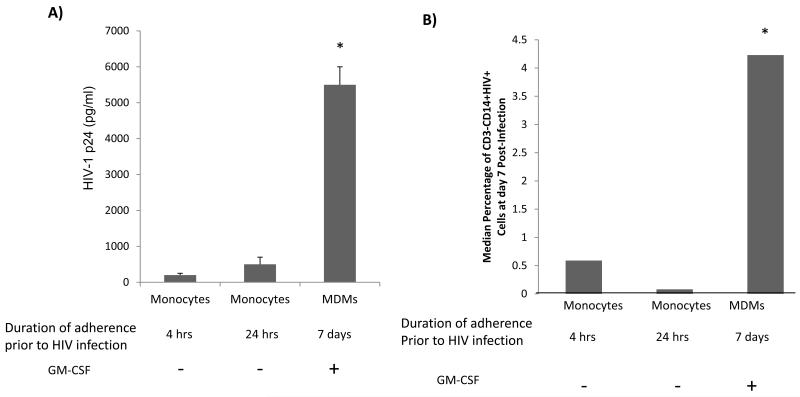
Figure 1. Higher level of HIV-1 replication in MDMs than monocytes, regardless of duration of monocyte adherence.
Monocytes were collected and adhered to tissue culture treated flasks for four hours or 24 hours prior to infection with HIV-1 Bal (10 ng p24/1×106 cells). Monocytes were also isolated by adherence for 7 days and maintained in the presence of GM-CSF (100 ng/ml) for seven days prior to HIV-1 infection. Seven days post-HIV-1 infection, HIV-1 p24 was measured by conventional ELISA (A) or intracellular flow cytometry for CD3, CD4, CD14, active β- catenin, and HIV-1 p24 (B). The median percentage of live, CD3-CD14+HIV-1 p24+ cells was determined (B). Data are based on at least three different donors. Asterisk denotes p<0.05 in comparison to first two columns.
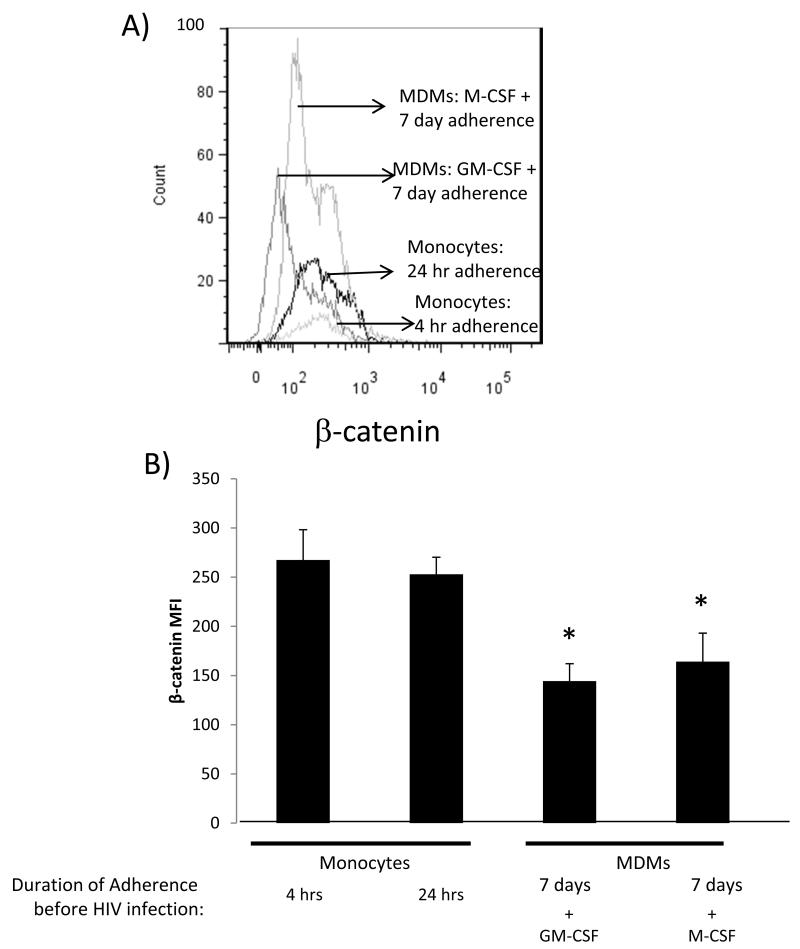
Figure 5. M-CSF MDMs down regulate β-catenin expression.
Monocytes were adhered for 4 hours or 24 hours or treated with GM-CSF or M-CSF for 7 days and active β-catenin expression measured by intracellular flow cytometry. A representative histogram of active β-catenin level measured by conventional intracellular flow cytometry is shown in (A) and cumulative data of mean active β-catenin mean fluorescence intensity (MFI) based on at least two independent experiments is shown in (B). Asterisk denote p<0.05 in comparison to first two columns.
Statistical analysis
Groups were compared using the Student’s t-Test when the data were normally distributed. When the data were not normally distributed, groups were compared using the non-parametric Mann-Whitney test. All tests were two-tailed with a p-value < 0.05 considered significant. Prism software (GraphPad Prism, San Diego, CA) was used for data analysis.
RESULTS
Differential susceptibility to HIV-1 infection between monocytes and MDMs
Several reports have demonstrated that monocytes are not permissive to productive HIV-1 infection, although they can harbor HIV-1 DNA [11, 12]. Few studies reported that extended in vitro culturing of monocytes promoted HIV-1 replication but did not fully differentiate the monocytes to MDMs [46, 47]. To investigate the impact of extended adherence of monocytes on level of HIV-1 replication, monocytes were adhered to plastic for four or 24 hours prior to infection with HIV-1 Bal or differentiated to MDMs by treatment with GM-CSF for seven days prior to HIV-1 infection. Regardless of the duration of monocyte adherence, monocytes did not support robust level of HIV-1 replication in comparison to MDMs (Fig.1). Specifically, HIV-1 replication was enhanced by 5-fold in MDMs in comparison to monocytes, as evaluated by HIV-1-1 p24 released into the supernatant (Fig.1a) and by 8-fold as measured by HIV-1 p24 intracellular staining by flow cytometry within MDMs defined as CD3-CD14+ cells (Fig.1b). The detection of HIV-1 p24 using flow cytometry validates the p24 ELISA data and rules out the contribution of minor contaminating T cells in higher susceptibility of MDMs to HIV-1 replication. Specifically, HIV-1 p24 was present in approximately 0.6% of CD3-CD14+ monocytes cultured for 4 hours prior to HIV-1 infection but in 4% of CD3-CD14+ GM-CSF treated MDMs (Fig. 1b). These data confirm the well established observation that monocytes are restricted to productive HIV-1 replication and that as monocytes differentiate to MDMs they become increasingly supportive of HIV-1 productive replication. Further, the restriction of monocytes to productive HIV-1 replication is not overcome by extended attachment of monocytes in the absence of GM-CSF.
Monocytes express higher level of endogenous β-catenin signaling than MDMs
β-catenin signaling is a host pathway that restricts HIV-1 replication in a number of cell types, including lymphocytes and astrocytes [27, 37-39]. Although both monocytes and MDMs are CD4+, only MDMs support robust productive HIV-1 replication (Fig.1). We therefore evaluated whether endogenous expression of β-catenin signaling may also dictate resistance or susceptibility to productive HIV-1 replication in monocytes Vs. macrophages. First, we evaluated endogenous expression of β-catenin in monocytes and MDMs. Monocytes were isolated from PBMCs of healthy adult blood donors, used fresh or differentiated to MDMs by treatment with GM-CSF. Active β-catenin protein expression was evaluated by intracellular flow cytometry using an antibody that recognizes the active form of β-catenin that is hypophosphorylated at ser37 and Thr41, which are sites that target β-catenin for ubiquitination and degradation (47). Monocytes expressed significantly higher level of endogenous active β-catenin protein than MDMs (Fig.2a-b). Expression of endogenous active β-catenin in monocytes was approximately one log higher in monocytes than MDMs (Fig. 2a). β-catenin expression can be increased in MDMs by treatment with lithium chloride (LiCl; 1mM), which induces β-catenin by inhibiting GSK3β (37). Inducible β-catenin in MDMs by LiCl resulted in the up-regulation of β-catenin to levels comparable to that measured in monocytes (Fig.2a-b).
Given that β-catenin can either function as a transcriptional co-activator or bind to cadherins, we measured β-catenin-mediated transcriptional activity using TOP or FOP-flash reporter constructs. These constructs are conventionally used to monitor endogenous and inducible β-catenin-mediated signaling. TOPflash is a DNA construct containing two sets of three β-catenin pathway (TCF) while FOPflash contains mutated TCF/LEF sites; both are linked to the firefly luciferase reporter. Monocytes and MDMs were transfected with TOP, FOP, or green fluorescence protein (GFP) with Renilla luciferase as internal control and left untreated or treated with LiCl to induce β-catenin activity. Monocytes expressed the highest level of endogenous β-catenin-mediated transcriptional activity, which was further induced by LiCl treatment (Fig.2c). MDMs, conversely, expressed a 4-fold lower endogenous level of β-catenin signaling in comparison to monocytes. β-catenin expression can be induced in both monocytes and MDMs post treatment with LiCl. However, the degree of β-catenin signaling induction was much greater in MDMs than monocytes (Fig.2c), reflecting higher endogenous activity in monocytes. Collectively, these data indicate that monocytes, which are resistant to productive HIV-1 replication, express higher levels of endogenous β-catenin, which is down-regulated as monocytes differentiate into macrophages and become more susceptible to productive HIV-1 replication.
Inhibiting β-catenin renders monocytes susceptible to productive HIV-1 replication
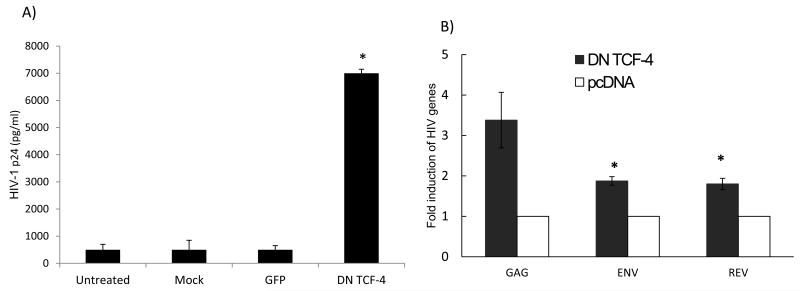
Given that monocytes express high levels of endogenous active β-catenin, we evaluated whether β-catenin restricts HIV-1 replication in monocytes. We performed loss of function studies by transfecting monocytes with a TCF-4 dominant-negative mutant plasmid. TCF-4 is a downstream effector of β-catenin signaling, demonstrated to engage β-catenin to repress HIV-1 transcription [28, 30]. The DN TCF-4 plasmid lacks the N-terminus required for β-catenin binding. Monocytes were nucleofected with TCF-4 DN mutant construct, green fluorescent protein (GFP), or nucleofected without a plasmid (mock nucleofection), or the cells were left without nucleofection. The cells were rested for 24 hours prior to infection with HIV-1 Bal and HIV-1 p24 level was measured by ELISA at day 7 post-infection. Monocytes that were nucleofected with TCF-4 DN mutant construct supported robust HIV-1 replication that was approximately 6-fold higher than monocytes nucleofected with GFP, mock nucleofected, or not nucleofected (Fig. 3a) (p< 0.05). To assess whether inhibition of TCF-4 can also induce HIV-1 from latently infected monocytes, U1/HIV-1 cells, which are latently infected and harbor two copies of provirus, were transfected with DN TCF-4 or backbone vector. Inhibiting TCF-4 significantly enhanced HIV-1 transcripts (gag, pol, env) from U1 cells by 2-4 fold (Fig.3b). These data indicate that inhibition of TCF-4 leads to HIV-1 activation from latently infected monocytes. Together, these data demonstrate that β-catenin/TCF-4 signaling is an endogenous inhibitor of HIV-1 replication in monocytes.
Figure 3. Inhibiting β-catenin-mediated signaling in monocytes renders them susceptible to HIV-1 replication and leads to re-activation of latent HIV-1 from promonocytic cells.
a) Freshly isolated monocytes were nucleofected with a dominant negative (DN) TCF-4 mutant construct (2 g/1×106 cells), green fluorescent protein (GFP) construct, nucleofected without a construct (mock), or left without nucleofection. The cells were rested for 24 hours post nucleofection and then infected with HIV-1 Bal. HIV-1 p24 concentration was measured by ELISA on day 7 post-infection (A). U1 cells which harbor 2 HIV-1 proviral copies were nucleofected with DN TCF-4 or backbone vector (pcDNA) plasmids and on day 3 post nucleofection qRT-PCR was performed to amplify for HIV-1 gag, env, and rev transcripts and data normalized to GAPDH gene expression (B). Data are based on three independent experiments. Asterisks denote p<0.05 in comparison to untreated cultures (A) or paired pcDNA treatments (B).
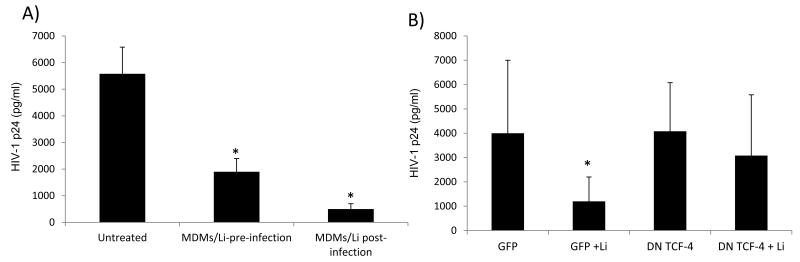
Activating β-catenin signaling in MDMs suppresses HIV-1 replication
Unlike monocytes, MDMs express low levels of β-catenin. We therefore evaluated whether induction of β-catenin in MDMs could be associated with reduction in HIV-1 replication. GM-CSF-differentiated MDMs were either treated with LiCl for 24 hours before they were infected with HIV-1 Bal at 10 ng p24/1×106 cells, or after infection. Treating the cells with 1 mM LiCl for only 24 hours prior to HIV-1 infection reduced HIV-1 replication by 3-fold (p<0.0001), while treating the cells with LiCl after the infection reduced HIV-1 p24 by 5-fold (p<0.0001) (Fig. 4a). While LiCl is a well described inducer of β-catenin, it may have effects that are independent of β-catenin. To investigate whether the ability of LiCl to inhibit HIV-1 replication in MDMs is mediated by its effect on β-catenin, MDMs were transfected with TCF-4 DN plasmid, GFP, or mock transfected. Approximately 24 hours post-transfection, cells were infected with HIV-1 Bal (10 ng p24/1×106 cells). After infection, the cells were treated with 1 mM LiCl or left untreated. HIV-1 p24 level was measured by ELISA on day 7. The ability of LiCl to inhibit HIV-1 replication was abrogated by interfering with β-catenin signaling (Fig. 4b). These data demonstrate that LiCl exerts its inhibitory effect on HIV-1 replication in MDMs through activation of β-catenin-mediated signaling.
Figure 4. Inhibition of β-catenin mediated signaling enhances HIV-1 replication in MDMs.
MDMs were pre-treated with LiCl (1mM) for 24 hrs or left untreated then infected with HIV-1 Bal. Post-infection the cells were left untreated or treated with LiCl. HIV-1 p24 was measured by ELISA on day 7 post-infection (A). MDMs were nucleofected with GFP or TCF-4 DN mutant constructs, rested for 24 hrs, then infected with HIV-1 Bal and left untreated or treated with LiCl. HIV-1 p24 was measured on day 7 by ELISA. Data are representative of at least three experiments (B). Asterisks denote p< 0.05.
Differentiation of monocytes by M-CSF also leads to down-regulation of β-catenin expression
Ex vivo, monocytes can be differentiated to macrophages by GM-CSF or M-CSF treatments to generate M1- and M2- like macrophages, respectively [40]. HIV-1 infection drives macrophages toward an M-1 like phenotype (54-55). For this reason we used GM-CSF to differentiate monocytes to MDMs. We assessed whether β-catenin is differentially expressed in GM-CSF MDM in comparison to M-CSF MDM. Monocytes adhered for 4 or 24 hrs had robust expression of endogenous β-catenin. Treating the cells with GM-CSF or M-CSF diminished β-catenin expression within MDMs to a similar level in comparison to monocytes (Fig.5). Interestingly, although not reaching statistical significance, we observed a trend towards a higher level of β-catenin expression within M-CSF MDMs in comparison to GM-CSF MDMs (Fig.5a). This difference may play a role in differential permissiveness of M-CSF MDMs vs. GM-CSF MDMs to HIV-1 productive replication within these subsets. Nonetheless, these data indicate that differentiation of monocytes, regardless of GM-CSF or M-CSF skewing, lead to down regulation of β-catenin expression. Taken together, our studies indicate that down regulation of β-catenin as monocytes differentiate to macrophages promotes susceptibility of these cells to productive HIV-1 replication, while robust expression of β-catenin is a host restriction factor for HIV-1 replication in monocytes
DISCUSSION
We demonstrate here that the β-catenin pathway is active to a higher extent in monocytes than MDMs. Inhibiting β-catenin activity in monocytes rendered them permissive to HIV-1 replication to the same degree as observed in differentiated monocytes (MDMs). Inversely, enhancing β-catenin activity in MDMs potently suppressed HIV-1 replication. A number of host factors have been linked to restricted HIV-1 replication in monocytes, including APOBEC3 A and G [41] and SAM domain- and HD domain containing protein 1 (SAMHD1)[42]. Our data demonstrate that β-catenin signaling is also restrictive factor for HIV-1 replication in monocytes. Whether there is cross talk between β-catenin signaling and these additional host restriction factors is still not clear. At least for SMADH-1, its expression is not modulated by monocyte treatment with GM-CSF or M-CSF, whereas we show here that monocyte treatment with GM-CSF or M-CSF lead to a profound down regulation of β-catenin signaling.
β-catenin signaling impacts hundreds of genes which in turn can have an effect on HIV-1 replication. However, the ability of β-catenin signaling to repress HIV-1 replication is a direct process. We have made significant progress in understanding how β-catenin signaling represses HIV-1 replication. Those studies have defined TCF-4 binding sites on the HIV-1 LTR promoter, particularly at the β-148 nt site from the initiation of transcription, whereby TCF-4, β-catenin, and SMAR-1 tether to pull the HIV-1 DNA into the nuclear matrix and away from the transcriptional machinery leading to repression of HIV-1 transcription [30]. β-catenin signaling has been linked to repression of antiviral innate immune response [43], which along with direct inhibition of HIV-1 transcription may contribute to refractoriness of monocytes to productive HIV replication. Additionally, the interaction between β-catenin and E-cadherins at the tight junction of the cell membrane may inhibit HIV-1 release [39, 44]. Vpu, an HIV-1 auxiliary protein that is important for HIV-1 replication in macrophages, overcomes this restriction by disrupting the association between β-catenin and E-cadherin [39]. Vpu also downregulates CD4 by recruiting β-TrCP, a member of the skpI-Cdc53-F-box E3 ubiquitin ligase for the degradation of CD4. In recruiting β-TrCP, Vpu sequesters it away from other cellular substrates including β-catenin and the NF-κB inhibitor IκB, which may result in the accumulation and stabilization of these substrates [42-44]. Therefore β-catenin stabilization and NFκB inhibition may both contribute to HIV-1 restriction in monocytes. Indeed, some studies have indicated that active Wnt/β-catenin signaling may antagonize NFκB activity [45]. Lastly, HIV-1 has a number of viral proteins that overcome restriction factors. For example, Vpr overcomes restriction to viral release by destabilizing tetherin while Vif degrades APOBEC3 to overcome its lethal deamination of viral DNA[46]. We recently demonstrated that Tat overcomes β-catenin mediated repression by inhibiting this signaling pathway [30]. Therefore in monocytes, where Tat levels may not be sufficiently reached, β-catenin restricts robust level of HIV-1 replication, However, as Tat threshold is reached in permissive cells such as macrophages, this pathway will still limit the extent of HIV-1 replication, but Tat will drive higher level of HIV-1 transcription/replication. It is thus a balance between Tat level within a cell and β-catenin signal integrity that at least in part regulates HIV-1 replication in macrophages.
Monocytes are also likely to secrete a factor that inhibits HIV-1 replication in trans. In heterokeryons between HIV-1 permissive T4 HeLa cells and monocytes, the heterokeryons were restricted to HIV-1 replication [22] This finding suggests that monocytes secrete a dominant factor that actively suppresses HIV-1 replication in otherwise HIV-1 permissive T4 HeLa cells. While the identity of this factor is not known, the fact that the β-catenin pathway can be activated by soluble Wnt ligands suggests that monocytes may secrete Wnt ligands that suppress HIV-1 replication in adjacent cells. Further, within the monocyte and macrophage population are various subsets that are phenotypically and functionally diverse [47-50]. Based on transcriptome analyses, CD16+ monocytes are more differentiated than CD16-monocytes, although they are distinctly different than macrophages[49, 50]. CD16+ monocytes also harbor higher levels of HIV-1 in vivo and are more permissive to HIV-1 replication in vitro than CD16-monocytes[51]. We have not been able to detect differences in β-catenin level between these two monocyte populations. However, endogenous β-catenin expression may be different among various macrophage subsets that can in turn regulate extent of HIV-1 replication. We observed a trend, in some donors, of higher β-catenin expression in M-CSF vs. GM-CSF-derived macrophages. At least in vitro, M-CSF MDMs are more susceptible to HIV-1 replication than GM-CSF MDMs [52], which is consistent with our overall observation that β-catenin expression is inversely correlated with HIV-1 productive replication [33]. In vivo, it is the microenvironment consisting of multiple/converging signals that will dictate the degree of macrophage permissiveness to HIV-1 replication and in some cases these signals may trigger HIV-1 latency within macrophages, as reported in the CNS.
β-catenin activity can be regulated by specific cytokines. We previously demonstrated that IFN inhibits β-catenin[37] and we show here that GM-CSF and M-CSF also inhibit β-catenin. The ability of cytokines to modulate β-catenin expression may in turn regulate the susceptibility of tissue macrophages to HIV-1. Macrophages from cervical/vaginal explants support HIV-1 replication and are likely to play an important role in HIV-1 sexual transmission [53, 54], but intestinal jejunum and alveolar macrophages are not highly permissive to HIV-1 [53, 55]. This differential susceptibility of tissue macrophages to productive HIV-1 replication may, in part, be dependent on cytokine-regulation of β-catenin at various tissue sites. Understanding the mechanisms of restricted HIV-1 replication mediated by β-catenin in monocytes can highlight a new approach for anti-viral therapy and enhance our understanding of HIV-1 pathogenesis in monocytes/macrophages.
Acknowledgment
This work was supported by NIH grants R01 NS060632 (LA); R01 NIMH100628 (LA), PO1 AI082971, Project III (LA), and F32 F32NS080657 (MHR). The studies were also supported by the Chicago Developmental Center for AIDS Research (D-CFAR, P30 AI 082151), an NIH funded program supported by NIAID, NCI, NIMH, NIDA, NICHD, NHLBI, and NCCAM.
Footnotes
Authorship: LA designed the research. YA, MHR, MSS, and SN performed the experiments. All analyzed the data and all authors contributed to the writing and editing of the manuscript.
None of authors have a conflict of interest to declare.
The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: HIV-1 Bal from Dr. Suzanne Gallo.
LITERATURE CITED
- 1.Gallo RC, Salahuddin SZ, Popovic M, Shearer GM, Kaplan M, Haynes BF, Palker TJ, Redfield R, Oleske J, Safai B, et al. Frequent detection and isolation of cytopathic retroviruses (HTLV-III) from patients with AIDS and at risk for AIDS. Science. 1984;224:500–503. doi: 10.1126/science.6200936. [DOI] [PubMed] [Google Scholar]
- 2.Popovic M, Sarngadharan MG, Read E, Gallo RC. Detection, isolation, and continuous production of cytopathic retroviruses (HTLV-III) from patients with AIDS and pre-AIDS. Science. 1984;224:497–500. doi: 10.1126/science.6200935. [DOI] [PubMed] [Google Scholar]
- 3.Gallo RC, Sarin PS, Gelmann EP, Robert-Guroff M, Richardson E, Kalyanaraman VS, Mann D, Sidhu GD, Stahl RE, Zolla-Pazner S, Leibowitch J, Popovic M. Isolation of human T-cell leukemia virus in acquired immune deficiency syndrome (AIDS) Science. 1983;220:865–867. doi: 10.1126/science.6601823. [DOI] [PubMed] [Google Scholar]
- 4.Barre-Sinoussi F, Chermann JC, Rey F, Nugeyre MT, Chamaret S, Gruest J, Dauguet C, Axler-Blin C, Vezinet-Brun F, Rouzioux C, Rozenbaum W, Montagnier L. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS) Science. 1983;220:868–871. doi: 10.1126/science.6189183. [DOI] [PubMed] [Google Scholar]
- 5.Alexaki A, Liu Y, Wigdahl B. Cellular reservoirs of HIV-1 and their role in viral persistence. Curr HIV Res. 2008;6:388–400. doi: 10.2174/157016208785861195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fischer-Smith T, Bell C, Croul S, Lewis M, Rappaport J. Monocyte/macrophage trafficking in acquired immunodeficiency syndrome encephalitis: lessons from human and nonhuman primate studies. J Neurovirol. 2008;14:318–326. doi: 10.1080/13550280802132857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008;8:958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coma G, Pena R, Blanco J, Rosell A, Borras FE, Este JA, Clotet B, Ruiz L, Parkhouse RM, Bofill M. Treatment of monocytes with interleukin (IL)-12 plus IL-18 stimulates survival, differentiation and the production of CXC chemokine ligands (CXCL)8, CXCL9 and CXCL10. Clin Exp Immunol. 2006;145:535–544. doi: 10.1111/j.1365-2249.2006.03145.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darwich L, Coma G, Pena R, Bellido R, Blanco EJ, Este JA, Borras FE, Clotet B, Ruiz L, Rosell A, Andreo F, Parkhouse RM, Bofill M. Secretion of interferon-gamma by human macrophages demonstrated at the single-cell level after costimulation with interleukin (IL)-12 plus IL-18. Immunology. 2009;126:386–393. doi: 10.1111/j.1365-2567.2008.02905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bergamaschi A, Pancino G. Host hindrance to HIV-1 replication in monocytes and macrophages. Retrovirology. 2010;7:31. doi: 10.1186/1742-4690-7-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cassol E, Alfano M, Biswas P, Poli G. Monocyte-derived macrophages and myeloid cell lines as targets of HIV-1 replication and persistence. J Leukoc Biol. 2006;80:1018–1030. doi: 10.1189/jlb.0306150. [DOI] [PubMed] [Google Scholar]
- 12.Kedzierska K, Crowe SM. The role of monocytes and macrophages in the pathogenesis of HIV-1 infection. Curr Med Chem. 2002;9:1893–1903. doi: 10.2174/0929867023368935. [DOI] [PubMed] [Google Scholar]
- 13.Rich EA, Chen IS, Zack JA, Leonard ML, O’Brien WA. Increased susceptibility of differentiated mononuclear phagocytes to productive infection with human immunodeficiency virus-1 (HIV-1) J Clin Invest. 1992;89:176–183. doi: 10.1172/JCI115559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sonza S, Maerz A, Uren S, Violo A, Hunter S, Boyle W, Crowe S. Susceptibility of human monocytes to HIV type 1 infection in vitro is not dependent on their level of CD4 expression. AIDS Res Hum Retroviruses. 1995;11:769–776. doi: 10.1089/aid.1995.11.769. [DOI] [PubMed] [Google Scholar]
- 15.Tuttle DL, Harrison JK, Anders C, Sleasman JW, Goodenow MM. Expression of CCR5 increases during monocyte differentiation and directly mediates macrophage susceptibility to infection by human immunodeficiency virus type 1. J Virol. 1998;72:4962–4969. doi: 10.1128/jvi.72.6.4962-4969.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong C, Kwas C, Wu L. Transcriptional restriction of human immunodeficiency virus type 1 gene expression in undifferentiated primary monocytes. J Virol. 2009;83:3518–3527. doi: 10.1128/JVI.02665-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pion M, Granelli-Piperno A, Mangeat B, Stalder R, Correa R, Steinman RM, Piguet V. APOBEC3G/3F mediates intrinsic resistance of monocyte-derived dendritic cells to HIV-1 infection. J Exp Med. 2006;203:2887–2893. doi: 10.1084/jem.20061519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palmieri C, Trimboli F, Puca A, Fiume G, Scala G, Quinto I. Inhibition of HIV-1 replication in primary human monocytes by the IkappaB-alphaS32/36A repressor of NF-kappaB. Retrovirology. 2004;1:45. doi: 10.1186/1742-4690-1-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Triques K, Stevenson M. Characterization of restrictions to human immunodeficiency virus type 1 infection of monocytes. J Virol. 2004;78:5523–5527. doi: 10.1128/JVI.78.10.5523-5527.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neil S, Martin F, Ikeda Y, Collins M. Postentry restriction to human immunodeficiency virus-based vector transduction in human monocytes. J Virol. 2001;75:5448–5456. doi: 10.1128/JVI.75.12.5448-5456.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henderson AJ, Zou X, Calame KL. C/EBP proteins activate transcription from the human immunodeficiency virus type 1 long terminal repeat in macrophages/monocytes. J Virol. 1995;69:5337–5344. doi: 10.1128/jvi.69.9.5337-5344.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaushik R, Zhu X, Stranska R, Wu Y, Stevenson M. A cellular restriction dictates the permissivity of nondividing monocytes/macrophages to lentivirus and gammaretrovirus infection. Cell Host Microbe. 2009;6:68–80. doi: 10.1016/j.chom.2009.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Naif HM, Li S, Alali M, Sloane A, Wu L, Kelly M, Lynch G, Lloyd A, Cunningham AL. CCR5 expression correlates with susceptibility of maturing monocytes to human immunodeficiency virus type 1 infection. J Virol. 1998;72:830–836. doi: 10.1128/jvi.72.1.830-836.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sung TL, Rice AP. miR-198 inhibits HIV-1 gene expression and replication in monocytes and its mechanism of action appears to involve repression of cyclin T1. PLoS Pathog. 2009;5:e1000263. doi: 10.1371/journal.ppat.1000263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arfi V, Riviere L, Jarrosson-Wuilleme L, Goujon C, Rigal D, Darlix JL, Cimarelli A. Characterization of the early steps of infection of primary blood monocytes by human immunodeficiency virus type 1. J Virol. 2008;82:6557–6565. doi: 10.1128/JVI.02321-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Michel N, Goffinet C, Ganter K, Allespach I, Kewalramani VN, Saifuddin M, Littman DR, Greene WC, Goldsmith MA, Keppler OT. Human cyclin T1 expression ameliorates a T-cell-specific transcriptional limitation for HIV in transgenic rats, but is not sufficient for a spreading infection of prototypic R5 HIV-1 strains ex vivo. Retrovirology. 2009;6:2. doi: 10.1186/1742-4690-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar A, Zloza A, Moon RT, Watts J, Tenorio AR, Al-Harthi L. Active {beta}-catenin signaling is an inhibitory pathway of HIV replication in peripheral blood mononuclear cells. J Virol. 2008;82:2813–2820. doi: 10.1128/JVI.02498-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Narasipura SD, Henderson LJ, Fu SW, Chen L, Kashanchi F, Al-Harthi L. Role of beta-catenin and TCF/LEF family members in transcriptional activity of HIV in astrocytes. J Virol. 2012;86:1911–1921. doi: 10.1128/JVI.06266-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li W, Henderson LJ, Major EO, Al-Harthi L. IFN-{gamma} Mediates Enhancement of HIV Replication in Astrocytes by Inducing an Antagonist of the {beta}-Catenin Pathway (DKK1) in a STAT 3-Dependent Manner. J Immunol. 2011;186:6771–6778. doi: 10.4049/jimmunol.1100099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henderson LJ, Narasipura SD, Adarichev V, Kashanchi F, Al-Harthi L. Identification of novel T cell factor 4 (TCF-4) binding sites on the HIV long terminal repeat which associate with TCF-4, beta-catenin, and SMAR1 to repress HIV transcription. J Virol. 2012;86:9495–9503. doi: 10.1128/JVI.00486-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moon RT, Brown JD, Torres M. WNTs modulate cell fate and behavior during vertebrate development. Trends Genet. 1997;13:157–162. doi: 10.1016/s0168-9525(97)01093-7. [DOI] [PubMed] [Google Scholar]
- 32.Miller JR, Moon RT. Signal transduction through beta-catenin and specification of cell fate during embryogenesis. Genes Dev. 1996;10:2527–2539. doi: 10.1101/gad.10.20.2527. [DOI] [PubMed] [Google Scholar]
- 33.Al-Harthi L. Interplay between Wnt/beta-catenin signaling and HIV: virologic and biologic consequences in the CNS. J Neuroimmune Pharmacol. 2012;7:731–739. doi: 10.1007/s11481-012-9411-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xie D, Yin D, Tong X, O’Kelly J, Mori A, Miller C, Black K, Gui D, Said JW, Koeffler HP. Cyr61 is overexpressed in gliomas and involved in integrin-linked kinase-mediated Akt and beta-catenin-TCF/Lef signaling pathways. Cancer Res. 2004;64:1987–1996. doi: 10.1158/0008-5472.can-03-0666. [DOI] [PubMed] [Google Scholar]
- 35.Chung EJ, Hwang SG, Nguyen P, Lee S, Kim JS, Kim JW, Henkart PA, Bottaro DP, Soon L, Bonvini P, Lee SJ, Karp JE, Oh HJ, Rubin JS, Trepel JB. Regulation of leukemic cell adhesion, proliferation, and survival by beta-catenin. Blood. 2002;100:982–990. doi: 10.1182/blood.v100.3.982. [DOI] [PubMed] [Google Scholar]
- 36.Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler KW, Vogelstein B, Clevers H. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC−/− colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 37.Carroll-Anzinger D, Al-Harthi L. Gamma interferon primes productive human immunodeficiency virus infection in astrocytes. J Virol. 2006;80:541–544. doi: 10.1128/JVI.80.1.541-544.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wortman B, Darbinian N, Sawaya BE, Khalili K, Amini S. Evidence for regulation of long terminal repeat transcription by Wnt transcription factor TCF-4 in human astrocytic cells. J Virol. 2002;76:11159–11165. doi: 10.1128/JVI.76.21.11159-11165.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salim A, Ratner L. Modulation of beta-catenin and E-cadherin interaction by Vpu increases human immunodeficiency virus type 1 particle release. J Virol. 2008;82:3932–3938. doi: 10.1128/JVI.00430-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mantovani A, Sica A, Sozzani S, Allavena P, Vecchi A, Locati M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004;25:677–686. doi: 10.1016/j.it.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 41.Bergamaschi A, Pancino G. Host hindrance to HIV-1 replication in monocytes and macrophages. Retrovirology. 7:31. doi: 10.1186/1742-4690-7-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laguette N, Sobhian B, Casartelli N, Ringeard M, Chable-Bessia C, Segeral E, Yatim A, Emiliani S, Schwartz O, Benkirane M. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature. 2011;474:654–657. doi: 10.1038/nature10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baril M, Es-Saad S, Chatel-Chaix L, Fink K, Pham T, Raymond VA, Audette K, Guenier AS, Duchaine J, Servant M, Bilodeau M, Cohen E, Grandvaux N, Lamarre D. Genome-wide RNAi screen reveals a new role of a WNT/CTNNB1 signaling pathway as negative regulator of virus-induced innate immune responses. PLoS Pathog. 2013;9:e1003416. doi: 10.1371/journal.ppat.1003416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Estrabaud E, Le Rouzic E, Lopez-Verges S, Morel M, Belaidouni N, Benarous R, Transy C, Berlioz-Torrent C, Margottin-Goguet F. Regulated degradation of the HIV-1 Vpu protein through a betaTrCP-independent pathway limits the release of viral particles. PLoS Pathog. 2007;3:e104. doi: 10.1371/journal.ppat.0030104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hoeflich KP, Luo J, Rubie EA, Tsao MS, Jin O, Woodgett JR. Requirement for glycogen synthase kinase-3beta in cell survival and NF-kappaB activation. Nature. 2000;406:86–90. doi: 10.1038/35017574. [DOI] [PubMed] [Google Scholar]
- 46.Harris RS, Hultquist JF, Evans DT. The restriction factors of human immunodeficiency virus. J Biol Chem. 2012;287:40875–40883. doi: 10.1074/jbc.R112.416925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fischer-Smith T, Croul S, Adeniyi A, Rybicka K, Morgello S, Khalili K, Rappaport J. Macrophage/microglial accumulation and proliferating cell nuclear antigen expression in the central nervous system in human immunodeficiency virus encephalopathy. Am J Pathol. 2004;164:2089–2099. doi: 10.1016/S0002-9440(10)63767-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fischer-Smith T, Croul S, Sverstiuk AE, Capini C, L’Heureux D, Regulier EG, Richardson MW, Amini S, Morgello S, Khalili K, Rappaport J. CNS invasion by CD14+/CD16+ peripheral blood-derived monocytes in HIV dementia: perivascular accumulation and reservoir of HIV infection. J Neurovirol. 2001;7:528–541. doi: 10.1080/135502801753248114. [DOI] [PubMed] [Google Scholar]
- 49.Ancuta P, Liu KY, Misra V, Wacleche VS, Gosselin A, Zhou X, Gabuzda D. Transcriptional profiling reveals developmental relationship and distinct biological functions of CD16+ and CD16-monocyte subsets. BMC Genomics. 2009;10:403. doi: 10.1186/1471-2164-10-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martinez FO. The transcriptome of human monocyte subsets begins to emerge. J Biol. 2009;8:99. doi: 10.1186/jbiol206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ellery PJ, Tippett E, Chiu YL, Paukovics G, Cameron PU, Solomon A, Lewin SR, Gorry PR, Jaworowski A, Greene WC, Sonza S, Crowe SM. The CD16+ monocyte subset is more permissive to infection and preferentially harbors HIV-1 in vivo. J Immunol. 2007;178:6581–6589. doi: 10.4049/jimmunol.178.10.6581. [DOI] [PubMed] [Google Scholar]
- 52.Komuro I, Yokota Y, Yasuda S, Iwamoto A, Kagawa KS. CSF-induced and HIV-1-mediated distinct regulation of Hck and C/EBPbeta represent a heterogeneous susceptibility of monocyte-derived macrophages to M-tropic HIV-1 infection. J Exp Med. 2003;198:443–453. doi: 10.1084/jem.20022018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shen R, Richter HE, Clements RH, Novak L, Huff K, Bimczok D, Sankaran-Walters S, Dandekar S, Clapham PR, Smythies LE, Smith PD. Macrophages in vaginal but not intestinal mucosa are monocyte-like and permissive to human immunodeficiency virus type 1 infection. J Virol. 2009;83:3258–3267. doi: 10.1128/JVI.01796-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Greenhead P, Hayes P, Watts PS, Laing KG, Griffin GE, Shattock RJ. Parameters of human immunodeficiency virus infection of human cervical tissue and inhibition by vaginal virucides. J Virol. 2000;74:5577–5586. doi: 10.1128/jvi.74.12.5577-5586.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nakata K, Weiden M, Harkin T, Ho D, Rom WN. Low copy number and limited variability of proviral DNA in alveolar macrophages from HIV-1-infected patients: evidence for genetic differences in HIV-1 between lung and blood macrophage populations. Mol Med. 1995;1:744–757. [PMC free article] [PubMed] [Google Scholar]