Abstract
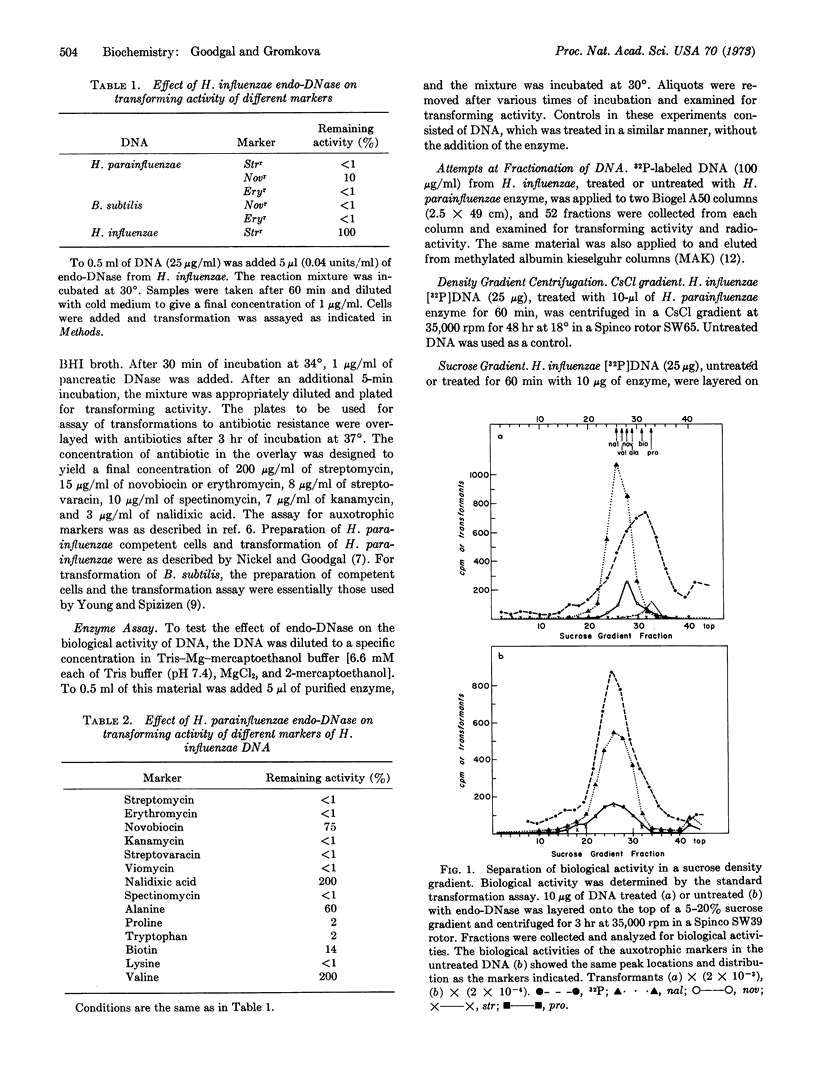
Hemophilus parainfluenzae endodeoxyribonuclease was used to degrade the DNA of H. influenzae and to follow the biological activity of 14 markers associated with this DNA. It was found that some H. influenzae markers were completely inactivated by endodeoxyribonuclease treatment, while others appeared to retain all or almost all of their original activity. The bulk of the H. influenzae DNA was reduced to double-stranded pieces of the order of 8 × 105 to 1 × 106 daltons. Velocity sedimentation of the DNA in sucrose gradients disclosed that markers that retained biological activity were present in DNA particles that were of the order of 1 × 106 daltons or larger, and indicated a close correlation between the size of the DNA fragment and the amount of biological activity retained. These data suggest that H. parainfluenzae endodeoxyribonuclease breaks DNA at specific sites. The nalr marker was shown to have twice as much biological activity after treatment with endodeoxyribonuclease when assayed at saturating DNA concentrations. In the linear portion of the DNA dose-response curve, the biological activity of this marker was reduced 3- to 10-fold compared to untreated DNA (in accord with the reduced size of its DNA). These data demonstrate a specific enrichment of the nalr marker by about 6- to 20-fold, and suggest a technique for the separation and purification of specific segments of DNA.
Keywords: bacterial transformation, Hemophilus species, separating markers, unique DNA segments
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- ALEXANDER H. E., LEIDY G. Induction of streptomycin resistance in sensitive Hemophilus influenzae by extracts containing desoxyribonucleic acid from resistant Hemophilus influenzae. J Exp Med. 1953 Jan;97(1):17–31. doi: 10.1084/jem.97.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BARNHART B. J., HERRIOTT R. M. PENETRATION OF DEOXYRIBONUCLEIC ACID INTO HEMOPHILUS INFLUENZAE. Biochim Biophys Acta. 1963 Sep 17;76:25–39. [PubMed] [Google Scholar]
- Catlin B. W., Bendler J. W., 3rd, Goodgal S. H. The type b capsulation locus of Haemophilus influenzae: map location and size. J Gen Microbiol. 1972 May;70(3):411–422. doi: 10.1099/00221287-70-3-411. [DOI] [PubMed] [Google Scholar]
- Danna K., Nathans D. Specific cleavage of simian virus 40 DNA by restriction endonuclease of Hemophilus influenzae. Proc Natl Acad Sci U S A. 1971 Dec;68(12):2913–2917. doi: 10.1073/pnas.68.12.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgell M. H., Hutchison C. A., 3rd, Sclair M. Specific endonuclease R fragments of bacteriophage phiX174 deoxyribonucleic acid. J Virol. 1972 Apr;9(4):574–582. doi: 10.1128/jvi.9.4.574-582.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GOODGAL S. H., HERRIOTT R. M. Studies on transformations of Hemophilus influenzae. I. Competence. J Gen Physiol. 1961 Jul;44:1201–1227. doi: 10.1085/jgp.44.6.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromkova R., Goodgal S. H. Action of haemophilus endodeoxyribonuclease on biologically active deoxyribonucleic acid. J Bacteriol. 1972 Mar;109(3):987–992. doi: 10.1128/jb.109.3.987-992.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly T. J., Jr, Smith H. O. A restriction enzyme from Hemophilus influenzae. II. J Mol Biol. 1970 Jul 28;51(2):393–409. doi: 10.1016/0022-2836(70)90150-6. [DOI] [PubMed] [Google Scholar]
- Litt M., Marmur J., Ephrussi-Taylor H., Doty P. THE DEPENDENCE OF PNEUMOCOCCAL TRANSFORMATION ON THE MOLECULAR WEIGHT OF DEOXYRIBOSE NUCLEIC ACID. Proc Natl Acad Sci U S A. 1958 Feb;44(2):144–152. doi: 10.1073/pnas.44.2.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meselson M., Yuan R. DNA restriction enzyme from E. coli. Nature. 1968 Mar 23;217(5134):1110–1114. doi: 10.1038/2171110a0. [DOI] [PubMed] [Google Scholar]
- Michalka J., Goodgal S. H. Genetic and physical map of the chromosome of Hemophilus influenzae. J Mol Biol. 1969 Oct 28;45(2):407–421. doi: 10.1016/0022-2836(69)90115-6. [DOI] [PubMed] [Google Scholar]
- NICKEL L., GOODGAL S. H. EFFECT OF INTERSPECIFIC TRANSFORMATION ON LINKAGE RELATIONSHIPS OF MARKERS IN HAEMOPHILUS INFLUENZAE AND HAEMOPHILUS PARAINFLUENZAE. J Bacteriol. 1964 Dec;88:1538–1544. doi: 10.1128/jb.88.6.1538-1544.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randolph M. L., Setlow J. K. Mechanism of inactivation of Haemophilus influenzae transforming deoxyribonucleic acid by sonic radiation. J Bacteriol. 1972 Jul;111(1):186–191. doi: 10.1128/jb.111.1.186-191.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SUEOKA N., CHENG T. Y. Fractionation of nucleic acids with the methylated albumin column. J Mol Biol. 1962 Mar;4:161–172. doi: 10.1016/s0022-2836(62)80048-5. [DOI] [PubMed] [Google Scholar]
- Smith H. O., Wilcox K. W. A restriction enzyme from Hemophilus influenzae. I. Purification and general properties. J Mol Biol. 1970 Jul 28;51(2):379–391. doi: 10.1016/0022-2836(70)90149-x. [DOI] [PubMed] [Google Scholar]
- YOUNG F. E., SPIZIZEN J. Physiological and genetic factors affecting transformation of Bacillus subtilis. J Bacteriol. 1961 May;81:823–829. doi: 10.1128/jb.81.5.823-829.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]


