Abstract
Osteogenesis Imperfecta (OI) type V is characterized by increased bone fragility, long bone deformities, hyperplastic callus formation and calcification of interosseous membranes. It is caused by a recurrent mutation in the 5’ UTR of the IFITM5 gene (c.−14C>T). This mutation introduces an alternative start codon, adding 5 amino acid residues to the N-terminus of the protein. The mechanism whereby this novel IFITM5 protein causes OI type V is yet to be defined. To address this, we created transgenic mice expressing either the wild type or the OI type V mutant IFITM5 under the control of an osteoblast-specific Col1a1 2.3kb promoter. These mutant IFITM5 transgenic mice exhibited perinatal lethality, whereas wild-type IFITM5 transgenic mice showed normal growth and development. Skeletal preparations and radiographs performed on E15.5 and E18.5 OI type V transgenic embryos revealed delayed/abnormal mineralization and skeletal defects including abnormal rib cage formation, long bone deformities and fractures. Primary osteoblast cultures, derived from mutant mice calvaria at E18.5, showed decreased mineralization by Alizarin-Red staining and RNA isolated from calvaria showed reduced expression of osteoblast differentiation markers such as Osteocalcin, as compared to non-transgenic littermates and wild-type mice calvaria, consistent with the in vivo phenotype. Importantly, overexpression of wild-type Ifitm5 did not manifest a significant bone phenotype. Collectively, our results suggest that expression of mutant IFITM5 causes abnormal skeletal development, low bone mass, and abnormal osteoblast differentiation. Given that neither overexpression of the wild type Ifitm5, as shown in our model, nor knock-out of Ifitm5, as previously published, showed significant bone abnormalities, we conclude that the IFITM5 mutation in OI type V acts in a neomorphic fashion.
Keywords: genetic animal models, osteoblasts, osteogenesis imperfecta, bone μCT, matrix mineralization
Introduction
Osteogenesis imperfecta (OI) is the most common osteodysplasia and is characterized by low bone mass and bone fragility. The majority of cases are caused by dominant mutations in the type I collagen genes (COL1A1, COL1A2); however, recessive mutations in collagen modifying genes, chaperones, and signaling molecules have been identified.(1-17) Recently, dominant mutations in osteogenesis imperfecta type V have been described.(18-22) Specifically, OI type V is characterized with clinical features including large, hypertrophic callus formation as well as calcification of the interosseous membrane.(23) Patients with OI type V exhibit significant phenotypic variability, but several groups have recently reported that all patients have the same mutation (c.−14C>T) in the interferon inducible transmembrane protein family 5 (IFITM5, also known as BRIL) gene.(18-22) This mutation introduces a novel in frame start codon in the 5’ untranslated region of the gene and adds 5 amino acid residues to the amino terminus of the protein.(24) The molecular consequences of this mutation are unknown.
IFITM5 is a transmembrane protein that is highly conserved and first appeared in bony fish, and exhibits a bone specific expression pattern in mammals.(25, 26) Within bone, its expression has been shown to be osteoblast specific.(27, 28) It has two transmembrane domains, and recent studies suggest that the amino terminus resides intracellularly, while the carboxy-terminus is extracellular.(27, 29) These domains are likely to interact with other proteins, and it is of note that the patient mutations result in a change to the N-terminus. In vitro studies suggest that there is a dose-dependent effect of Ifitm5 levels on mineralization. Higher levels increase mineralization and vice versa.(27) Furthermore, it was shown that IFITM5 competes for interaction of CD9 with an immune complex consisting of FK506 binding protein 11 (FKBP11), CD81 and prostaglandin F2 receptor negative regulator (FPRP). Binding of IFITM5 to this complex increases the expression of interferon-induced genes in osteoblasts, also suggesting a role of IFITM5 in the immune interactions within bone.(28, 30)
In vivo studies of Ifitm5 deficient mice have revealed only minor effects on bone.(28) In these studies, newborn knockout mice exhibited reduced bone length and bone deformities (bowing of long bones); however, this did not persist further in the postnatal period as adults showed no deformities nor changes in bone morphometric parameters as evidenced by micro-computed tomography (μCT) (28). Therefore, mutations in IFITM5 leading to OI type V are inconsistent with the loss of function mouse model. Here, we describe the phenotype of the transgenic mice expressing the novel start codon of Ifitm5 corresponding to the recurrent human OI type V mutation.
Materials and Methods
Generation of transgenic mice
Murine Ifitm5 cDNA was placed under the Col1a1 2.3 kb promoter. Mutagenesis was performed to obtain the c.−14C>T mutation. The tyrosinase minigene was included in these constructs to facilitate identification of transgenics. Constructs with wild type or mutant Ifitm5 were injected in-house. Mice were maintained on a FVB/N background. Genotyping was based upon identification of the WPRE cassette in addition to visualization of black eyes resulting from the presence of the tyrosinase cassette.(31) Mice were housed in the Baylor College of Medicine vivarium. These studies were approved by the Baylor College of Medicine Institutional Animal Care and Use Committee as well as Center for Comparative Medicine.
Radiographic Analysis/ Skeletal Preparations
Radiographs of transgenics and their respective non-transgenic littermates were taken (identical views) with the Kubtec XPERT80 (Kubtec X-ray, Milford, CT). E18.5 or E15.5 mutant transgenics and their non-transgenic littermates, or wild type controls, were sacrificed and fixed with 95% Ethanol, then stained overnight in Alcian blue (containing 0.015% Alcian blue 8GX, 20% Acetic acid, and 80% Ethanol 95%), and soaked in 2% KOH solution for 24 hours to remove remnants of soft tissue. Skeletal preps were then stained overnight in Alizarin red solution (containing 0.005% Alizarin red S in 1% KOH), cleared in 1% KOH/20% glycerol solution and stored in glycerol/95% Ethanol 1:1 solution.
Histology
E18.5 and E15.5 hindlimbs and E15.5 forelimbs were collected then fixed in 4% paraformaldehyde overnight, at room temperature. They were stored in 70% ethanol and then embedded in paraffin. Hindlimbs were sagittally sectioned at 7μm. They were then stained with hematoxylin and eosin, picrosirius red, alcian blue with nuclear fast red, and von kossa according to standard methods. These were imaged using a Zeiss Axioplan 2 microscope.
Calvarial cultures
Calvaria were dissected from E18.5 mutant transgenics, their non-transgenic littermates, and wild type controls. Each calvarium was further dissected and incubated in digestion medium, containing alpha MEM with 0.05% Trypsin-EDTA and 0.1mg/ml Collagenase P, with frequent shaking and mixing (by pipetting) for a total of 2.5 hours. Cultures were then incubated in alpha MEM containing 15% FBS, 1% glutamine and 1% penicillin/streptomycin, in 6 well plates for 5 days, until confluency was achieved. On the 5th day, cells were reseeded for further analysis, and stained for alkaline phosphatase activity and with Alizarin red, as specified below.
Mineralization assessment and staining for alkaline phosphatase activity
Primary calvaria cells derived from either mutant transgenics, wild type transgenics or their non-transgenic littermates, were seeded into 6-well plates and cultured for 7 days, then fixed in 4% paraformaldehyde in PBS. Fixed cultures were incubated for 20 minutes in staining solution, containing Naphthol AS-MX phosphate (0.1 mg/ml), N,N dimethyl formamide (0.5%), MgCl2 (2mM), Fast blue BB salt (0.6 mg/ml) and Tris-HCl pH 8.5 (0.1M). Stained cultures were washed in PBS, dried and scanned. For Alizarin red staining, cells were grown for 18-20 days in culture prior to fixation. Fixed cells were incubated for 20 minutes in Alizarin red (40mM solution), then washed in water, dried and scanned. Following scanning of cultures, Alizarin red staining was dissolved in 10% cetylpyridinium chloride solution for 2 hours before quantification (measurement of absorbance at 550nm). Intensity of alkaline phosphatase staining was quantified by ImageJ software to compare between samples.
Quantitative real-time PCR
Total RNA was extracted from snap frozen (liquid nitrogen) E18.5 mouse calvaria using Trizol reagent (Invitrogen). The Superscript III RT system (Invitrogen) was used to synthesize cDNA from total RNA according to the manufacturer's protocol. Quantitative RT-PCR was performed on a LightCycler.v 1.5 (Roche) using gene-specific primers and SYBR Green I reagent (Roche). β2-Microglobulin and/or Gapdh were used as reference genes.
Mass spectrometry
Collagen was prepared from minced E18.5 calvaria. Type I α-chains were extracted by heat denaturation (90C) in SDS-PAGE sample buffer, resolved on 6% SDS-PAGE gels,(32) cut from gels and digested with trypsin in-gel.(33)
Calvarial tissue was also digested with bacterial collagenase as described.(34) Collagenase-generated peptides were separated by reverse phase HPLC (C8, Brownlee Aquapore RP-300, 4.6 mm × 25 cm) with a linear gradient of acetonitrile:n-propanol (3:1 v/v) in aqueous 0.1% (v/v) trifluoroacetic acid.(35) Individual fractions were analyzed by LC-MS.
Peptides were analyzed by electrospray LC/MS using an LTQ XL ion-trap mass spectrometer, (Thermo Scientific) equipped with in-line liquid chromatography on a C4 5um capillary column (300 um × 150 mm; Higgins Analytical RS-15M3-W045) and eluted at 4.5 μl/min. The LC mobile phase consisted of buffer A (0.1% formic acid in MilliQ water) and buffer B (0.1% formic acid in 3:1 acetonitrile:n-propanol v/v). An electrospray ionization source (ESI) introduced the LC sample stream into the mass spectrometer with a spray voltage of 3kV. Proteome Discoverer search software (Thermo Scientific) was used for peptide identification using the NCBI protein database.
Micro-computed tomography
Micro-CT was performed as previously described.(36) Briefly, spine and femur samples were placed into 16mm tubes filled with 70% ethanol and scanned at 16 micron resolution using a ScanCo μct40 scanner. Trabecular and cortical analysis was performed using the ScanCo software. The trabecular region of the L4 vertebra was manually selected (contoured) every 5 slices, and then the remaining slices were morphed to include the region of interest, for a total of 100 slices. The region was thresholded at 210 with a gauss setting of 0. Cortical analysis was performed on 50 slices of the femoral midshaft, using the same thresholding.
Results
Generation of a transgenic mouse model for OI type V
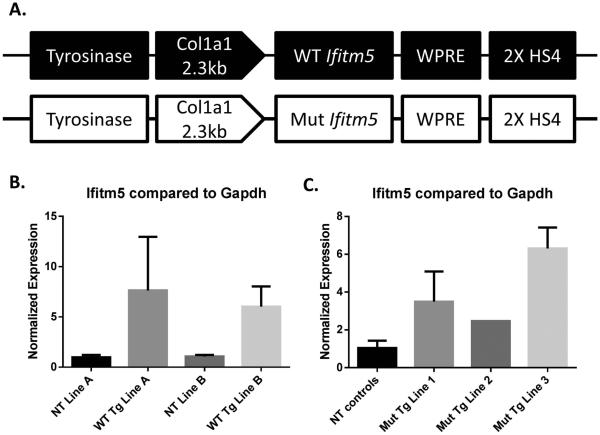
We generated transgenic mice containing the murine Ifitm5 cDNA with and without the variant corresponding to the human mutation c.−14C>T found in OI type V patients. The transgene contains a tyrosinase minigene for visual genotyping, and the Ifitm5 cDNA is expressed under the Col1a1 2.3kb promoter (Figure 1A). Two lines overexpressing wild type Ifitm5 (WT Tg) had overexpression levels between 6 and 7- fold, by qPCR, in P0 calvaria compared to non-transgenic littermates (NT) at the R3 generation (Figure 1B). WT Tg line A was used for subsequent studies. Three founders overexpressing mutant Ifitm5 (Mut Tg) were established with overexpression levels between 2- and 7-fold in E18.5 calvaria compared to non-transgenic littermates (Figure 1C). Mutant transgenic line 1 was used for subsequent studies.
Figure 1. A transgenic mouse model of OI type V.
A transgenic construct of mutant Ifitm5 (c.−14C>T) and wild type Ifitm5 (control) under the Col1a1 2.3 kb promoter expresses in osteoblasts. (A). Quantitative real time PCR indicates overexpression of Ifitm5 in wild type transgenic lines (WT Tg) compared to non-transgenic (NT) littermates (B). Quantitative real time PCR shows overexpression of Ifitm5 in mutant transgenic lines (Mut Tg) compared to non-transgenic (NT) littermates (C). Data in (B, C) are means ± SD.
Overexpression of mutant Ifitm5 results in severe skeletal defects
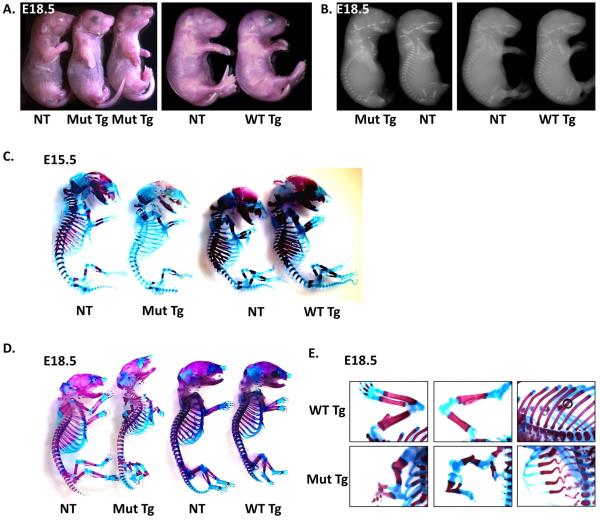
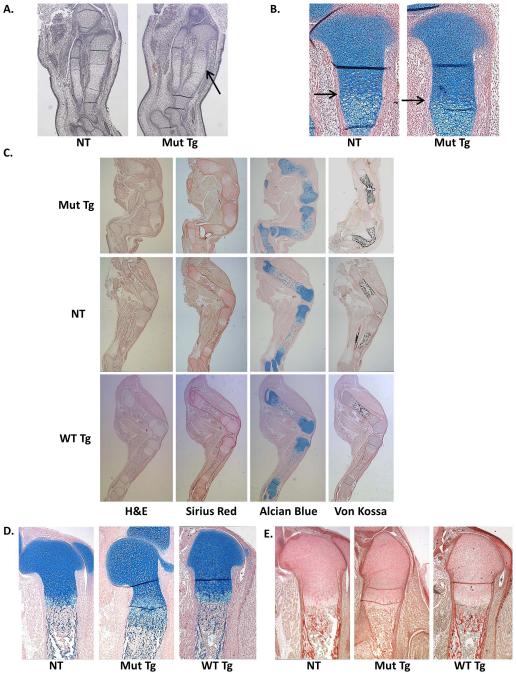
Mice expressing the c.−14C>T Ifitm5 transgene exhibited severe skeletal defects as well as lethality at birth. These mice have downward facing limbs at birth, but do not otherwise show growth defects (Figure 2A). Radiographs taken of E18.5 mutant transgenic mice show a reduction in mineralization, not seen in wild type transgenic mice (Figure 2B). After observing these phenotypes, we proceeded to perform skeletal preparations at E15.5 and E18.5. At E15.5, a delay in mineralization was apparent in the mutant transgenic mice (Figure 2C). This defect persisted at E18.5, when severe defects of the limbs including in utero fractures were also identified (Figure 2D-E). In utero fractures of the forelimbs and hindlimbs and severe ribcage deformity were observed in the mutant transgenic mice. These defects were not seen in the wild type transgenic mice (Figure 2E). At E15.5 tibia dysplasia is already present, although fractures are not evident (Figure 3A). Furthermore, histological analysis of the hindlimbs of the mutant transgenic mice at E18.5 showed fractures of both the femur and tibia. Picrosirius red staining failed to reveal any major changes in collagen distribution within these limbs; von kossa staining showed some residual mineralization in the hindlimbs. Although von kossa staining appears to be increased in mutant samples, this is likely due to the extreme bent bone and seems to be an artifact of the histology preparation. Interestingly, alcian blue staining for proteoglycans showed a diffuse staining pattern throughout the limbs, unlike in non-transgenic littermates where staining was primarily restricted to the growth plates (Figure 3C). The growth plates of these mice appear to be extended, and the cellular morphology within the bones was more chondrocyte-like than in non-transgenic littermates (Figure 3D, E). This extended growth plate appears by E15.5 with altered cellular morphology (Figure 3B). To confirm that these skeletal phenotype is not due to insertional mutagenesis, we also studied skeletal preparations from an additional mutant transgenic founder, skeletal preparations from limbs of mutant transgenic line 2, and histology of mutant transgenic line 3 (Supplementary Figure 1). The phenotypic features were consistent across all transgenic lines studied.
Figure 2. Mutant transgenic mice have severe skeletal defects and perinatal lethality.
Transgenic mice overexpressing the mutant Ifitm5 show limb deformities compared to non-transgenic littermates and wild type transgenics at E18.5 (A). Radiographs of E18.5 mutant transgenics show reduced mineralization, while wild type transgenics appear normal (B). Skeletal preparations of mutant transgenics at E15.5 (C) and E18.5 (D) show reduced alizarin red staining and fractures which are not evident in wild type transgenic or non-transgenic littermates. Skeletal preparations of E18.5 mutant transgenic mice compared to wild type transgenic controls show abnormalities of the forelimbs, hindlimbs and ribs (E).
Figure 3. Mutant transgenic mice have in utero fractures and morphological defects of the hindlimbs.
Histology of the E15.5 tibia of non-transgenic controls and mutant transgenic mice stained with H&E with arrow indicating limb dysplasia (A), and with alcian blue and nuclear fast red with arrow indicating growth plate expansion (B). Histology of E18.5 non-transgenic controls, mutant transgenic littermates, and wild-type transgenic control mice by H&E, picrosirius red, alcian blue with nuclear fast red, and von kossa staining of the hindlimbs (C). The growth plate is extended in mutant transgenics, as indicated by alcian blue staining of the proximal tibia (D). Bone morphology is altered in the proximal tibia, as shown in picrosirius red staining (E).
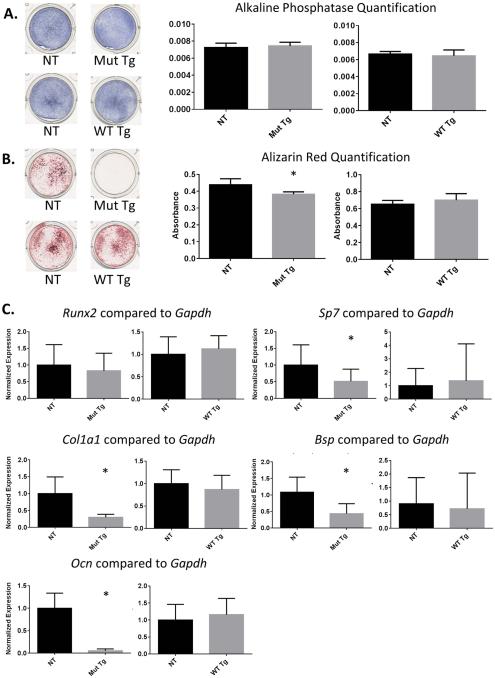
Because of the delay in mineralization evidenced by the skeletal preparations, as well as previous reports implicating Ifitm5 in mineralization, we isolated calvarial osteoblasts from the transgenic mice and control non-transgenic littermates. Alkaline phosphatase staining of these cells showed no changes after 7 days in culture (Figure 4A). Alizarin red staining at 18-21 days of culture was reduced in cells derived from mutant transgenic mice, but not in wild type transgenic cultures, as compared to their respective non-transgenic controls (Figure 4B). Quantitative real-time PCR of calvarial bone cDNA showed reduced levels of osterix (Sp7), collagen type I alpha I (Col1a1), bone sialoprotein (Bsp) and osteocalcin (Ocn), but no change was detected in runt related transcription factor 2 (Runx2) expression (Figure 4C).
Figure 4. Calvarial osteoblasts from mutant transgenics have mineralization defects.
Alkaline phosphatase staining of mutant and wild type transgenic calvarial osteoblasts shows no changes (A). Alizarin red staining of calvarial osteoblasts shows a mineralization defect in mutant transgenics but not in wild type transgenics as compared to non-transgenic littermates (n=5-6 biological replicates plus 3 technical replicates of each) (B). Quantitative real-time PCR shows no change in Runx2 levels in calvarial bone samples, but a reduction in Sp7, Col1a1, Bsp and Ocn levels (C). Results from a t-test in (A, B, and C) are means ± SD, *p < .05.
Other mouse models of osteogenesis imperfecta exhibit posttranslationally overmodified type I bone collagen, including abnormalities in crosslinking.(3, 4, 37-41) Although Ifitm5 likely plays a role in cell signaling, we wanted to evaluate whether alterations in type I collagen modifications could contribute to the bone phenotype. Gel electrophoresis of the α1(I) and α2(I) collagen chains showed no changes in their migration pattern, consistent with absence of gross post-translational over-modification (Figure 5A). Furthermore, there were no changes in α1(I) C-telopeptide lysine hydroxylation by mass spectrometric analysis, indicating no effect on bone collagen crosslinking as is observed in forms of OI caused by mutations of the collagen chaperone Fkbp10.(38, 40, 41) There were also no changes in the hydroxylation of the P986 residue of the α1(I) chain nor in the P707 residue of the α2(I) chain, which are both altered in other types of OI with mutations in the 3-prolyl-hydroxyation complex (Figure 5B-D). (3, 4, 37)
Figure 5. Mutant transgenics have no alterations in collagen post-translational modification in calvarial bone.
Type I collagen isolated from calvaria of E18.5 mutant transgenics shows no changes in electrophoretic migration pattern (A). There are no changes in 3-hydroxylation of the P986 residue of α1(I) (B), the P707 residue of α2(I) (C), or in hydroxylation of the C-telopeptide lysine of α1(I) (D). Results indicate absolute values.
Overexpression of wild type Ifitm5 does not alter normal bone parameters
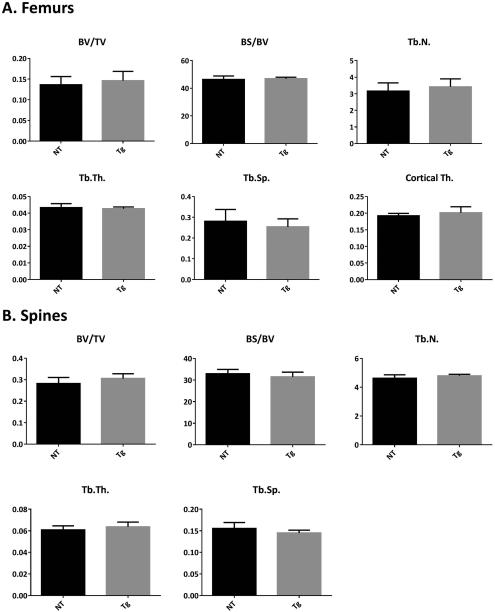
Another possibility is that mutation in IFITM5 could lead to an excess of normal IFITM5 function. To assess the consequences of overexpression of wild type Ifitm5, we performed μCT analysis of 3 month old femurs and spines from the R3 generation of both WT IFITM5 transgenic mouse lines. No significant changes were observed in the trabecular or cortical parameters of femurs or spines in either line in males (Figure 6, Supplementary Figure 3 ). Minor increases in BV/TV and Tb.N. were observed in female femurs of line A, consistent with previous reports of Ifitm5 overexpression increasing mineralization (Supplementary Figure 2). In summary, the overexpression of wild type Ifitm5 did not result in a phenotype suggesting brittle bone disease in neither the neonate nor adult mouse bone.
Figure 6. Wild type transgenic mice have no changes in adult architectural parameters.
μCT analyses of 3 month femurs (A) and spines (B) from wild type transgenic mice have no changes in trabecular or cortical parameters compared to non-transgenic littermates. Analysis is on line A male mice. Results are displayed in mean ± SD.
Discussion
Osteogenesis imperfecta is genetically heterogeneous, and an increasing number of causative gene mutations have been identified. This has resulted in the identification of new genes important in bone homeostasis and development. In OI type V, a single recurrent mutation (c.−14C>T) in IFITM5 has been identified in patients.(18, 19, 21, 22) OI type V presents with not only bone fragility and low bone mass, characteristic of other OI types, but also presents with the distinguishing features of hyperplastic callus formation and calcification of the interosseous membrane.(23) These type specific defects, where the callus contributes more to bone formation than would normally occur in the context of fractures, may constitute clues as to the underlying molecular mechanism(s) leading to altered fracture healing. Hence, we developed transgenic mice in order to assess the genetic contribution of the IFITM5 mutation and to serve as a mouse model of human OI type V.
The transgenic mice overexpressed Ifitm5 in osteoblasts, either with the c.−14C>T mutation or in the wild type context as controls for Ifitm5 overexpression in bone. The mutant Ifitm5 mice exhibited perinatal lethality, with severe defects of the long bones and ribs. We speculate that the severe rib deformities likely lead to respiratory failure in the neonatal mice. Skeletal preparations also showed that mineralization was delayed/abnormal and was associated with in utero fractures. This is consistent with previous literature of in vitro studies implying a possible function for Ifitm5 in bone mineralization.(27, 30) Furthermore, histological analysis clearly showed fractures, as well as abnormal long bone morphology. The cellular histology was altered, wherein bone cells appeared chondrocyte-like in shape, and Alcian blue staining was diffuse throughout the bone suggesting persistence of a cartilage-like matrix. This may be a result of cross-talk or induction of signaling pathways through the interaction of the extracellular domains of IFITM5 with other as of yet unknown proteins. Although we cannot determine if this causes the severe skeletal abnormalities, it could also be an important secondary component in disease pathology. In conjunction with the defects in mineralization, this provides a possible insight into the mechanism underlying Ifitm5 mutations in osteoblast differentiation and/or hyperplastic callus formation.
To further address mineralization in the context of the Ifitm5 mutation, we cultured osteoblasts derived from mice calvaria. Consistent with the in vivo phenotype, the osteoblast cultures showed a reduction in Alizarin red staining. Furthermore, quantitative real-time PCR performed on calvarial cDNA detected markedly reduced levels of Sp7, Col1a1, Bsp, and Ocn in the mutant samples. In contrast, both alkaline phosphatase staining and real-time PCR of Runx2 showed no differences between the samples. Collectively, these data suggest that Ifitm5 mutations do not affect very early differentiation, but rather play a role in late differentiation and/or function of osteoblasts.
IFITM5 topology studies suggest that the N-terminus of the protein is intracellular, while the C-terminus is extracellular.(29) This led us to believe that the two termini of the protein likely interact with other proteins and perform signaling functions, although the binding partners of IFITM5 are largely unknown. It has been shown that the mutant allele of Ifitm5 is expressed, and has an additional 5 amino acids on the N-terminus, with normal localization.(24, 29) Given that the localization is unaltered, it is reasonable to hypothesize that protein interactions are altered and this may affect signaling by IFITM5. However, in many other types of OI, the modifications of type I collagen are directly affected. Mass spectrometry analysis of mutant transgenic calvaria showed no changes in migration of the collagen, or changes at the specific residues that are altered in other forms of recessive OI. Mass spectrometry also showed normal collagen crosslinking patterns. This finding rules out an altered collagen structure and also rules out processing or trafficking as major consequences of the IFITM5 mutation.
In contrast, mice overexpressing wild type Ifitm5 in osteoblasts showed no defects in bone, and were comparable to non-transgenic littermates. Neonates showed no changes in skeletal preparations, radiographs, or bone histology. Furthermore, calvarial cultures were comparable to non-transgenic controls. Similarly μCT bone analysis of adult femurs or spines of these mice showed no difference with non-transgenic littermates. Taken together, these data suggest that the OI type V mutation is not a gain of normal function, as overexpression of wild type IFITM5 does not duplicate the findings in the mutant transgenic mice. Furthermore, it was previously published that mice deficient in Ifitm5 have only mild phenotype at the neonatal stage, including smaller size and slight bone dysplasia which normalized with growth. Adult mice deficient in Ifitm5 had no changes in μCT parameters or morphology (28), suggesting that OI type V mutations also do not act in a dominant negative or haploinsufficient nature.
Although we were unable to assess adult bone parameters in mutant Ifitm5 transgenic mice, and thus could not recapitulate the hyperplastic callus formation or interosseous membrane calcification, we believe this model provides important insight into the function of Ifitm5 in the context of the OI type V mutation. It is likely that protein interactions and downstream signaling cascades are altered by this mutant protein, which affects differentiation and mineralization in bone. Furthermore, we show that overexpression of wild type versus the OI V mutant Ifitm5 have distinctly different consequences. Thus, we conclude that the OI type V mutation (c.−14C>T) of Ifitm5 has a neomorphic effect in bone.
Supplementary Material
Acknowledgements
Funding: This work was supported by the NIH P01 HD070394 (BL, DE), F31DE022483 (CL), AR 037318 (DE), the BCM Intellectual and Developmental Disabilities Research Center (HD024064) from the Eunice Kennedy Shriver National Institute Of Child Health & Human Development, the BCM Advanced Technology Cores with funding from the NIH (AI036211, CA125123, and RR024574), the Rolanette and Berdon Lawrence Bone Disease Program of Texas, and the BCM Center for Skeletal Medicine and Biology.
We thank Megan Bagos for her analysis of the micro-CT in this manuscript and Frank Gannon for his analysis of the histological slides.
Footnotes
Disclosures
The authors have no conflicts of interest.
References
- 1.Wenstrup RJ, Willing MC, Starman BJ, Byers PH. Distinct biochemical phenotypes predict clinical severity in nonlethal variants of osteogenesis imperfecta. Am J Hum Genet. 1990 May;46(5):975–82. [PMC free article] [PubMed] [Google Scholar]
- 2.Rauch F, Glorieux FH. Osteogenesis imperfecta. Lancet. 2004 Apr 24;363(9418):1377–85. doi: 10.1016/S0140-6736(04)16051-0. [DOI] [PubMed] [Google Scholar]
- 3.Morello R, Bertin TK, Chen Y, Hicks J, Tonachini L, Monticone M, et al. CRTAP is required for prolyl 3- hydroxylation and mutations cause recessive osteogenesis imperfecta. Cell. 2006 Oct 20;127(2):291–304. doi: 10.1016/j.cell.2006.08.039. [DOI] [PubMed] [Google Scholar]
- 4.Marini JC, Cabral WA, Barnes AM. Null mutations in LEPRE1 and CRTAP cause severe recessive osteogenesis imperfecta. Cell Tissue Res. 2010 Jan;339(1):59–70. doi: 10.1007/s00441-009-0872-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baldridge D, Schwarze U, Morello R, Lennington J, Bertin TK, Pace JM, et al. CRTAP and LEPRE1 mutations in recessive osteogenesis imperfecta. Hum Mutat. 2008 Dec;29(12):1435–42. doi: 10.1002/humu.20799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pyott SM, Schwarze U, Christiansen HE, Pepin MG, Leistritz DF, Dineen R, et al. Mutations in PPIB (cyclophilin B) delay type I procollagen chain association and result in perinatal lethal to moderate osteogenesis imperfecta phenotypes. Hum Mol Genet. 2011 Apr 15;20(8):1595–609. doi: 10.1093/hmg/ddr037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kelley BP, Malfait F, Bonafe L, Baldridge D, Homan E, Symoens S, et al. Mutations in FKBP10 cause recessive osteogenesis imperfecta and Bruck syndrome. J Bone Miner Res. 2011 Mar;26(3):666–72. doi: 10.1002/jbmr.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alanay Y, Avaygan H, Camacho N, Utine GE, Boduroglu K, Aktas D, et al. Mutations in the gene encoding the RER protein FKBP65 cause autosomal-recessive osteogenesis imperfecta. Am J Hum Genet. 2010 Apr 9;86(4):551–9. doi: 10.1016/j.ajhg.2010.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christiansen HE, Schwarze U, Pyott SM, AlSwaid A, Al Balwi M, Alrasheed S, et al. Homozygosity for a missense mutation in SERPINH1, which encodes the collagen chaperone protein HSP47, results in severe recessive osteogenesis imperfecta. Am J Hum Genet. 2010 Mar 12;86(3):389–98. doi: 10.1016/j.ajhg.2010.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Homan EP, Rauch F, Grafe I, Lietman C, Doll JA, Dawson B, et al. Mutations in SERPINF1 cause osteogenesis imperfecta type VI. J Bone Miner Res. 2011 Dec;26(12):2798–803. doi: 10.1002/jbmr.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Becker J, Semler O, Gilissen C, Li Y, Bolz HJ, Giunta C, et al. Exome sequencing identifies truncating mutations in human SERPINF1 in autosomal-recessive osteogenesis imperfecta. Am J Hum Genet. 2011 Mar 11;88(3):362–71. doi: 10.1016/j.ajhg.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Asharani PV, Keupp K, Semler O, Wang W, Li Y, Thiele H, et al. Attenuated BMP1 function compromises osteogenesis, leading to bone fragility in humans and zebrafish. Am J Hum Genet. 2012 Apr 6;90(4):661–74. doi: 10.1016/j.ajhg.2012.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinez-Glez V, Valencia M, Caparros-Martin JA, Aglan M, Temtamy S, Tenorio J, et al. Identification of a mutation causing deficient BMP1/mTLD proteolytic activity in autosomal recessive osteogenesis imperfecta. Hum Mutat. 2012 Feb;33(2):343–50. doi: 10.1002/humu.21647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faqeih E, Shaheen R, Alkuraya FS. WNT1 mutation with recessive osteogenesis imperfecta and profound neurological phenotype. J Med Genet. 2013 Jul;50(7):491–2. doi: 10.1136/jmedgenet-2013-101750. [DOI] [PubMed] [Google Scholar]
- 15.Laine CM, Joeng KS, Campeau PM, Kiviranta R, Tarkkonen K, Grover M, et al. WNT1 mutations in early-onset osteoporosis and osteogenesis imperfecta. N Engl J Med. 2013 May 9;368(19):1809–16. doi: 10.1056/NEJMoa1215458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pyott SM, Tran TT, Leistritz DF, Pepin MG, Mendelsohn NJ, Temme RT, et al. WNT1 mutations in families affected by moderately severe and progressive recessive osteogenesis imperfecta. Am J Hum Genet. 2013 Apr 4;92(4):590–7. doi: 10.1016/j.ajhg.2013.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fahiminiya S, Majewski J, Mort J, Moffatt P, Glorieux FH, Rauch F. Mutations in WNT1 are a cause of osteogenesis imperfecta. J Med Genet. 2013 May;50(5):345–8. doi: 10.1136/jmedgenet-2013-101567. [DOI] [PubMed] [Google Scholar]
- 18.Cho TJ, Lee KE, Lee SK, Song SJ, Kim KJ, Jeon D, et al. A single recurrent mutation in the 5'-UTR of IFITM5 causes osteogenesis imperfecta type V. Am J Hum Genet. 2012 Aug 10;91(2):343–8. doi: 10.1016/j.ajhg.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rauch F, Moffatt P, Cheung M, Roughley P, Lalic L, Lund AM, et al. Osteogenesis imperfecta type V: marked phenotypic variability despite the presence of the IFITM5 c.−14C>T mutation in all patients. J Med Genet. 2013 Jan;50(1):21–4. doi: 10.1136/jmedgenet-2012-101307. [DOI] [PubMed] [Google Scholar]
- 20.Semler O, Garbes L, Keupp K, Swan D, Zimmermann K, Becker J, et al. A mutation in the 5'-UTR of IFITM5 creates an in-frame start codon and causes autosomal-dominant osteogenesis imperfecta type V with hyperplastic callus. Am J Hum Genet. 2012 Aug 10;91(2):349–57. doi: 10.1016/j.ajhg.2012.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shapiro JR, Lietman C, Grover M, Lu JT, Nagamani SC, Dawson BC, et al. Phenotypic variability of osteogenesis imperfecta type V caused by an IFITM5 mutation. J Bone Miner Res. 2013 Jul;28(7):1523–30. doi: 10.1002/jbmr.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takagi M, Sato S, Hara K, Tani C, Miyazaki O, Nishimura G, et al. A recurrent mutation in the 5'-UTR of IFITM5 causes osteogenesis imperfecta type V. Am J Med Genet A. 2013 Aug;161A(8):1980–2. doi: 10.1002/ajmg.a.36025. [DOI] [PubMed] [Google Scholar]
- 23.Glorieux FH, Rauch F, Plotkin H, Ward L, Travers R, Roughley P, et al. Type V osteogenesis imperfecta: a new form of brittle bone disease. J Bone Miner Res. 2000 Sep;15(9):1650–8. doi: 10.1359/jbmr.2000.15.9.1650. [DOI] [PubMed] [Google Scholar]
- 24.Lazarus S, McInerney-Leo AM, McKenzie FA, Baynam G, Broley S, Cavan BV, et al. The IFITM5 mutation c.−14C > T results in an elongated transcript expressed in human bone; and causes varying phenotypic severity of osteogenesis imperfecta type V. BMC Musculoskelet Disord. 2014;15:107. doi: 10.1186/1471-2474-15-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hickford D, Frankenberg S, Shaw G, Renfree MB. Evolution of vertebrate interferon inducible transmembrane proteins. BMC Genomics. 2012;13:155. doi: 10.1186/1471-2164-13-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Z, Liu J, Li M, Yang H, Zhang C. Evolutionary dynamics of the interferon-induced transmembrane gene family in vertebrates. PLoS One. 2012;7(11):e49265. doi: 10.1371/journal.pone.0049265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moffatt P, Gaumond MH, Salois P, Sellin K, Bessette MC, Godin E, et al. Bril: a novel bone-specific modulator of mineralization. J Bone Miner Res. 2008 Sep;23(9):1497–508. doi: 10.1359/jbmr.080412. [DOI] [PubMed] [Google Scholar]
- 28.Hanagata N, Li X, Morita H, Takemura T, Li J, Minowa T. Characterization of the osteoblast-specific transmembrane protein IFITM5 and analysis of IFITM5-deficient mice. J Bone Miner Metab. 2011 May;29(3):279–90. doi: 10.1007/s00774-010-0221-0. [DOI] [PubMed] [Google Scholar]
- 29.Patoine A, Gaumond MH, Jaiswal PK, Fassier F, Rauch F, Moffatt P. Topological Mapping of BRIL Reveals a Type II Orientation and Effects of Osteogenesis Imperfecta Mutations on Its Cellular Destination. J Bone Miner Res. 2014 Apr 9; doi: 10.1002/jbmr.2243. [DOI] [PubMed] [Google Scholar]
- 30.Hanagata N, Li X. Osteoblast-enriched membrane protein IFITM5 regulates the association of CD9 with an FKBP11-CD81-FPRP complex and stimulates expression of interferon-induced genes. Biochem Biophys Res Commun. 2011 Jun 10;409(3):378–84. doi: 10.1016/j.bbrc.2011.04.136. [DOI] [PubMed] [Google Scholar]
- 31.Zhou G, Zheng Q, Engin F, Munivez E, Chen Y, Sebald E, et al. Dominance of SOX9 function over RUNX2 during skeletogenesis. Proc Natl Acad Sci U S A. 2006 Dec 12;103(50):19004–9. doi: 10.1073/pnas.0605170103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–5. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 33.Hanna SL, Sherman NE, Kinter MT, Goldberg JB. Comparison of proteins expressed by Pseudomonas aeruginosa strains representing initial and chronic isolates from a cystic fibrosis patient: an analysis by 2-D gel electrophoresis and capillary column liquid chromatography-tandem mass spectrometry. Microbiology. 2000 Oct;146:2495–508. doi: 10.1099/00221287-146-10-2495. Pt 10. [DOI] [PubMed] [Google Scholar]
- 34.Hanson DA, Eyre DR. Molecular site specificity of pyridinoline and pyrrole cross-links in type I collagen of human bone. J Biol Chem. 1996 Oct 25;271(43):26508–16. doi: 10.1074/jbc.271.43.26508. [DOI] [PubMed] [Google Scholar]
- 35.Wu JJ, Woods PE, Eyre DR. Identification of cross-linking sites in bovine cartilage type IX collagen reveals an antiparallel type II-type IX molecular relationship and type IX to type IX bonding. J Biol Chem. 1992 Nov 15;267(32):23007–14. [PubMed] [Google Scholar]
- 36.Bouxsein ML, Boyd SK, Christiansen BA, Guldberg RE, Jepsen KJ, Muller R. Guidelines for assessment of bone microstructure in rodents using micro-computed tomography. J Bone Miner Res. 2010 Jul;25(7):1468–86. doi: 10.1002/jbmr.141. [DOI] [PubMed] [Google Scholar]
- 37.Baldridge D, Lennington J, Weis M, Homan EP, Jiang MM, Munivez E, et al. Generalized connective tissue disease in Crtap−/− mouse. PLoS One. 2010;5(5):e10560. doi: 10.1371/journal.pone.0010560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barnes AM, Cabral WA, Weis M, Makareeva E, Mertz EL, Leikin S, et al. Absence of FKBP10 in recessive type XI osteogenesis imperfecta leads to diminished collagen cross-linking and reduced collagen deposition in extracellular matrix. Hum Mutat. 2012 Nov;33(11):1589–98. doi: 10.1002/humu.22139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Choi JW, Sutor SL, Lindquist L, Evans GL, Madden BJ, Bergen HR, 3rd, et al. Severe osteogenesis imperfecta in cyclophilin B-deficient mice. PLoS Genet. 2009 Dec;5(12):e1000750. doi: 10.1371/journal.pgen.1000750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwarze U, Cundy T, Pyott SM, Christiansen HE, Hegde MR, Bank RA, et al. Mutations in FKBP10, which result in Bruck syndrome and recessive forms of osteogenesis imperfecta, inhibit the hydroxylation of telopeptide lysines in bone collagen. Hum Mol Genet. 2013 Jan 1;22(1):1–17. doi: 10.1093/hmg/dds371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lietman CD, Rajagopal A, Homan EP, Munivez E, Jiang MM, Bertin TK, et al. Connective tissue alterations in Fkbp10−/− mice. Hum Mol Genet. 2014 Apr 28; doi: 10.1093/hmg/ddu197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.