Abstract
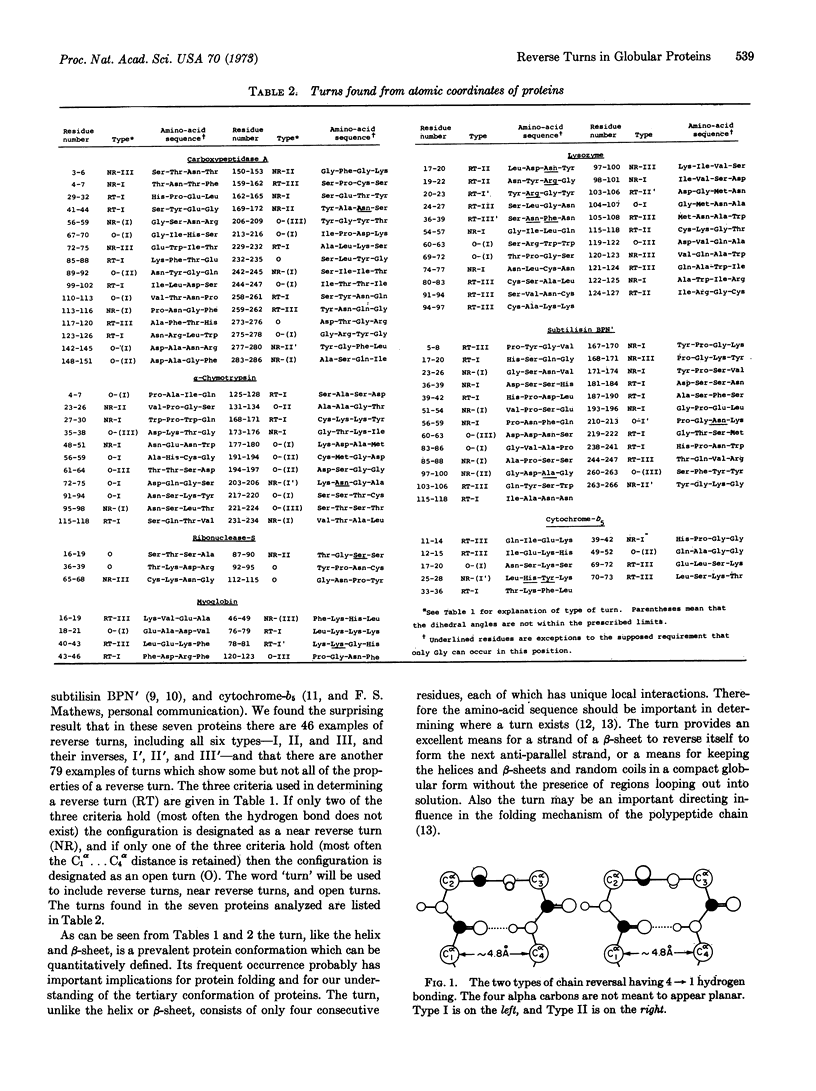
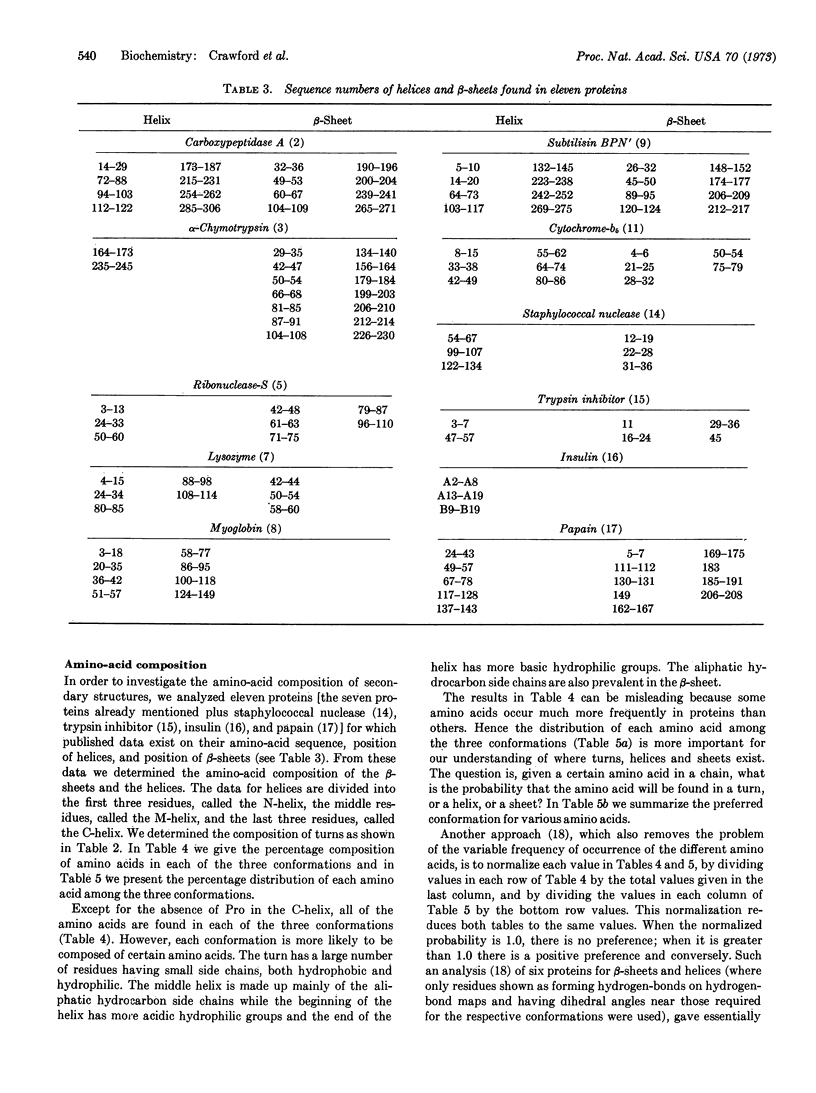
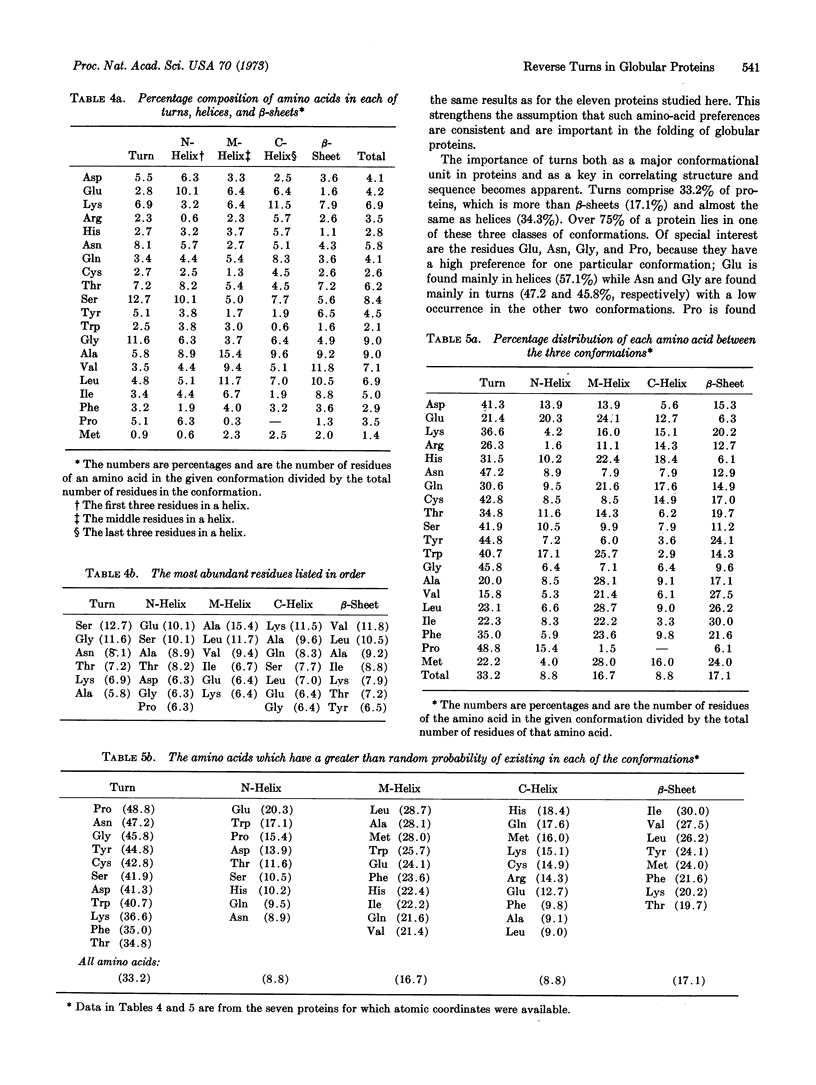
The reverse turn, involving four consecutive amino acids, as a tertiary conformation in globular proteins is defined in terms of dihedral angles, the C1α...C4α distance and the O1...H-N4 hydrogen bond distance. In seven proteins we find 125 examples of turns, comprising 33% of the amino acids in these proteins, as compared with 34% of the residues forming helices and only 17% forming β-sheets. The amino-acid compositions of turns, helices, and β-sheets are analyzed in some detail. We find Asn and Gly mainly in turns, Pro in turns (and at the beginning of helices), and Glu in helices. In these turns a statistical survey indicates that 19% of Asp residues are in the first position, 33% of Pro residues are in the second position, 24% of Asn residues are in the third position, and 26% of Trp residues are in the fourth position.
Keywords: amino-acid composition, helices, β-sheets
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alden R. A., Birktoft J. J., Kraut J., Robertus J. D., Wright C. S. Atomic coordinates for subtilisin BPN' (or Novo). Biochem Biophys Res Commun. 1971 Oct 15;45(2):337–344. doi: 10.1016/0006-291x(71)90823-0. [DOI] [PubMed] [Google Scholar]
- Birktoft J. J., Matthews B. W., Blow D. M. Atomic co-ordinates for tosyl-alpha-chymotrypsin. Biochem Biophys Res Commun. 1969 Jul 7;36(1):131–137. doi: 10.1016/0006-291x(69)90659-7. [DOI] [PubMed] [Google Scholar]
- Blake C. C., Mair G. A., North A. C., Phillips D. C., Sarma V. R. On the conformation of the hen egg-white lysozyme molecule. Proc R Soc Lond B Biol Sci. 1967 Apr 18;167(1009):365–377. doi: 10.1098/rspb.1967.0034. [DOI] [PubMed] [Google Scholar]
- Blundell T. L., Dodson G. G., Dodson E., Hodgkin D. C., Vijayan M. X-ray analysis and the structure of insulin. Recent Prog Horm Res. 1971;27:1–40. doi: 10.1016/b978-0-12-571127-2.50025-0. [DOI] [PubMed] [Google Scholar]
- Kuntz I. D. Protein folding. J Am Chem Soc. 1972 May 31;94(11):4009–4012. doi: 10.1021/ja00766a060. [DOI] [PubMed] [Google Scholar]
- Lewis P. N., Momany F. A., Scheraga H. A. Folding of polypeptide chains in proteins: a proposed mechanism for folding. Proc Natl Acad Sci U S A. 1971 Sep;68(9):2293–2297. doi: 10.1073/pnas.68.9.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews F. S., Argos P., Levine M. The structure of cytochrome b 5 at 2.0 Angstrom resolution. Cold Spring Harb Symp Quant Biol. 1972;36:387–395. doi: 10.1101/sqb.1972.036.01.050. [DOI] [PubMed] [Google Scholar]
- Quiocho F. A., Lipscomb W. N. Carboxypeptidase A: a protein and an enzyme. Adv Protein Chem. 1971;25:1–78. doi: 10.1016/s0065-3233(08)60278-8. [DOI] [PubMed] [Google Scholar]
- Venkatachalam C. M. Stereochemical criteria for polypeptides and proteins. V. Conformation of a system of three linked peptide units. Biopolymers. 1968 Oct;6(10):1425–1436. doi: 10.1002/bip.1968.360061006. [DOI] [PubMed] [Google Scholar]
- Wyckoff H. W., Tsernoglou D., Hanson A. W., Knox J. R., Lee B., Richards F. M. The three-dimensional structure of ribonuclease-S. Interpretation of an electron density map at a nominal resolution of 2 A. J Biol Chem. 1970 Jan 25;245(2):305–328. [PubMed] [Google Scholar]


