Abstract
The mechanisms whereby the crucial pluripotency transcription factor Oct4 regulates target gene expression are incompletely understood. Using an assay system based on partially differentiated embryonic stem cells, we show that Oct4 opposes the accumulation of local H3K9me2 and subsequent Dnmt3a-mediated DNA methylation. Upon binding DNA, Oct4 recruits the histone lysine demethylase Jmjd1c. Chromatin immunoprecipitation (ChIP) time course experiments identify a stepwise Oct4 mechanism involving Jmjd1c recruitment and H3K9me2 demethylation, transient FACT (facilitates chromatin transactions) complex recruitment, and nucleosome depletion. Genome-wide and targeted ChIP confirms binding of newly synthesized Oct4, together with Jmjd1c and FACT, to the Pou5f1 enhancer and a small number of other Oct4 targets, including the Nanog promoter. Histone demethylation is required for both FACT recruitment and H3 depletion. Jmjd1c is required to induce endogenous Oct4 expression and fully reprogram fibroblasts to pluripotency, indicating that the assay system identifies functional Oct4 cofactors. These findings indicate that Oct4 sequentially recruits activities that catalyze histone demethylation and depletion.
INTRODUCTION
Oct4/Pou5f1 is an indispensable component of the regulatory circuitry controlling the establishment and maintenance of pluripotent, undifferentiated inner cell mass and epiblast cells, as well as primordial germ cells, germ line progenitor cells, and cultured embryonic stem cells (ESCs) (1–3). Its loss accompanies ESC differentiation and loss of pluripotency (4). Oct4 is also one of a small group of pluripotency regulators, including Nanog and Sox2, which reinforce each other's expression (5), while simultaneously regulating other parameters of ESC function such as developmental gene poising. Oct4 is widely used to generate induced pluripotent stem cells (iPSCs) from somatic cells (6, 7). The critical role of Oct4 in establishing and maintaining pluripotency places priority on determining how it functions. Studies have identified functional interactions between Oct4 and Paf1/PD2, a component of the PAF transcription elongation complex (8); Wdr5, a core subunit of the MLL and SET1 histone methyltransferase complexes (9); and XPC, a DNA repair protein (10). Other work has demonstrated an association between Oct4 binding and nucleosome depletion at some targets (11). In inhibitory contexts, Oct4 has been associated with ESET/Setdb1 and NuRD (12–14).
Oct1, an Oct4 paralog, regulates transcription by associating with two chromatin modifying activities, NuRD and Jmjd1a, in a mutually exclusive fashion (15). NuRD is a repressive complex that also associates with Oct4 (16, 17). Jmjd1a (also known as KDM3A and Jhdm2a) is a histone lysine demethylase that catalyzes the removal of repressive histone H3 lysine 9 dimethyl (H3K9me2) marks to potentiate gene activity.
The homology between Oct1 and Oct4 (18), as well as the known role of H3K9me2 in pluripotency (19, 20), suggested that Oct4 may use a similar mechanism. Here, we establish a system in which Oct4 protein is re-expressed in differentiating ESCs that have lost endogenous Oct4 expression. We found that Oct4 catalyzes the loss of H3K9me2 and blocks the deposition of DNA methylation marks by Dnmt3a at the Pou5f1 distal enhancer. Oct4 recruits Jmjd1c, a histone lysine demethylase and paralog of Jmjd1a, to Pou5f1. Using chromatin immunoprecipitation (ChIP) time course assays, we show that Jmjd1c recruitment and H3K9me2 loss precede depletion of H3 itself and that transient recruitment of the FACT histone chaperone complex coincides with H3 loss. We show that nucleosome demethylation is required for FACT recruitment and H3 depletion and further show that this mechanism acts at multiple Oct4 targets, including targets directly involved in pluripotency. Lastly, we show that efficient generation of reprogrammed iPSCs that express endogenous Oct4 requires Jmjd1c. These findings thus identify an essential mechanism in which Oct4 sequentially recruits different activities to alter local chromatin.
MATERIALS AND METHODS
Cell culture.
tetON-Oct4 doxycycline-inducible ESCs (21) were obtained from K. Hochedlinger. Cells were cultured in Dulbecco modified Eagle medium, 15% tetracycline-free fetal bovine calf serum (HyClone), 100 U of penicillin/ml, 100 μg of streptomycin/ml, 2 mM l-glutamine (Life Technologies), 50 μM β-mercaptoethanol (Sigma), 1,000 U of leukemia inhibitor factor (LIF; Chemicon)/ml, 0.1 mM minimal essential medium nonessential amino acids, and 1 mM sodium pyruvate. Cells were cultured at 37°C and 5% CO2 in a humidified atmosphere. Retinoic acid (RA; Sigma) was used at 1.5 μg/ml, and doxycycline (Sigma) was used at 2 μg/ml. ESCs were separated from feeder fibroblasts by allowing feeders to adhere to tissue culture plastic prior to RA-mediated differentiation. Dimethyloxalylglycine (DMOG; EMD Biosciences) was used at 1 mM.
ChIP.
Oct1, Jmjd1a, H3, and H3K9me2 ChIP experiments were performed as described previously (15). Enrichment was quantified relative to an isotype control antibody, nonspecific control region, and input DNA titration as described previously (22). The isotype control for rabbit antibodies (Oct4, Jmjd1c, Dnmt3a, histone H3, and H3K9me2) was normal rabbit IgG (sc-2027; Santa Cruz), and the isotype control for mouse monoclonal antibodies (SSRP1, Spt16) was normal mouse IgG2b (sc-3879; Santa Cruz). The control primer pairs came from the mouse Actb locus: b-actin-F (TGTTACCAACTGGGACGACA) and b-actin-R (CTATGGGAGAACGGCAGAAG). Oct4 and Jmjd1c ChIP assays were performed with antibodies from Abcam (ab19857 and ab31215), and SSRP1 and Spt16ChIP assays were performed with antibodies from BioLegend (catalog numbers 609702 and 607002). The oligonucleotide sequences were as follows: Pou5f1 enhancer (forward, 5′-CCCCAGGGAGGTTGAGAGTT; reverse, 5′-AAGGGCTAGGACGAGAGGGA), Nanog promoter (forward, 5′-CTCAGATCCCCCACTTGACCTG; reverse, 5′-GCAAGACACCAACCAAATCAGC); Vamp1 promoter (forward, 5′-TGTGGGTTTTATGGTCTCAG; reverse, 5′-GACTGCTGTCATTCTTACCG); and Slain2 enhancer (forward, 5′-CAAAGAACAAAGCAGCAAAC; reverse, 5′-CGCAGACCTTACTACGACAG).
Quantitative reverse transcription-PCR (qRT-PCR).
RNA was isolated using TRIzol (Life Technologies), followed by RNeasy purification (Qiagen) using the RNA cleanup procedure. cDNA was synthesized using SuperScript III and random hexamers (Life Technologies). The primers for amplification of endogenous mouse Pou5f1 were as follows: forward, 5′-CTAGAGAAGGATGTGGTTCGAGTATGGTTC; and reverse, 5′-TATCTACTGTGTGTCCCAGTCTTTATTT. The sequences for quantification of mouse Gata4 were taken from Pachernik et al. (23).
Bisulfite sequencing.
Bisulfite DNA modification and analysis was performed as published elsewhere (15). The primers used were mouse Pou5f1 enhancer forward (5′-ATAGATAGGATTGTTGGGTTGT) and reverse (5′-CACAAAACTTCCTCAATAACAA) and mouse Pou5f1 promoter forward (5′-ATGGGTTGAAATATTGGGTTTA) and reverse (5′-AATCTAAAACCAAATATCCAACCA).
Aza-IP.
TetON-Oct4 ESCs were differentiated with RA (1.5 μg/ml) for 12 days. Aza-IP (a variant of ChIP without formaldehyde cross-linking used to identify methyltransferases linked to nucleic acids) was performed at the indicated time points similarly to conventional ChIP, except that 10 μM 5-AzadC (Sigma) was added to the culture medium for the final 8 h of differentiation at the indicated time points. At each time point, the cells were collected without formaldehyde fixation and processed for ChIP. Doxycycline (2 μg/ml) was added to the culture medium for 12 h to induce expression of ectopic Oct4. Dnmt3a antibodies were purchased from Abcam (catalog no. ab2850).
Coimmunoprecipitation.
tetON-Oct4 ESCs in a 10-cm dish were treated with trypsin and replated for 4 h at 37°C to allow feeder fibroblasts to adhere. ESCs were collected, rinsed with ice-cold phosphate-buffered saline (PBS), and lysed in ice-cold lysis buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM NaVO4) supplemented with protease inhibitor cocktail (Roche). Differentiating cells were directly collected and lysed similarly. The debris was cleared by centrifugation (16,000 × g, 5 min, 4°C) in a microcentrifuge. Protein G-magnetic beads (Active Motif) were washed twice with binding buffer (PBS, 0.5% bovine serum albumin, 0.5% Tween 20) containing protease inhibitors, followed by incubation with 5 μg of specific or isotype control antibody for 2 h with rotation at 4°C. Beads were washed twice with binding buffer to remove excess antibody, resuspended in a 250-μl binding buffer, and added to the cell lysates. The lysate-bead mixture was incubated with rotation overnight at 4°C and collected using a magnetic particle concentrator. Immunoprecipitates were washed three times with lysis buffer. Bound proteins denatured by adding 30 μl of sample buffer and boiling the mixture for 5 min. Protein was resolved by SDS–10% PAGE and analyzed by immunoblotting. Oct4 (sc-8628; Santa Cruz) and Jmjd1c (ab31215; Abcam) antibodies were used for immunoprecipitation (IP) and Western blot analyses.
ChIP-Seq.
ChIP-Seq was performed as described previously (24). Samples were sequenced on an Illumina HiSeq 2000 instrument. Jmjd1c ChIP-Seq used an antibody from Millipore (catalog no. 17-10262). All other antibodies were the same as those used in conventional ChIP. Subsequent analysis was performed similarly as described previously (25). Briefly, for low-level sequence read processing, single-end 25-bp Illumina fastq sequence files were aligned to the mouse mm9/NCBI Build 37 genome using Bowtie (http://bowtie-bio.sourceforge.net). Reads aligning to multiple locations were reduced to one randomly selected alignment with Picard's MarkDuplicates utility (http://broadinstitute.github.io/picard/). Relative read coverage tracks were generated using the USeq Sam2USeq application (http://useq.sourceforge.net/cmdLnMenus.html#Sam2USeq). Bam alignments were converted to binary PointData for subsequent enrichment analysis using the USeq SamParser utility (http://useq.sourceforge.net/cmdLnMenus.html#SamParser). Candidate peaks were identified using a sliding window to score 200-bp overlapping windows for ChIP versus input alignment count enrichment using a binomial P value test (USeq ScanSeqs [http://useq.sourceforge.net/cmdLnMenus.html#ScanSeqs] and EnrichedRegionMaker [http://useq.sourceforge.net/cmdLnMenus.html#EnrichedRegionMaker]) (26). To control for the bimodal peak shift due to fragment end sequencing, each alignment was shifted 100 bp. P values were converted to false discovery rates (FDRs) using Storey's QValue package in R (27). Overlapping windows that exceeded both an FDR of ≤5% and a log2 ratio of ≥1.0 were merged using the EnrichedRegionMaker. Those intersecting satellite repeats as defined by RepeatMasker were eliminated. For some comparisons, more stringent thresholds were applied, i.e., an FDR of ≤1% and a log2 of ≥1.585.
iPSC generation and RNAi.
hSTEMCCA and mSTEMCCA lentiviral constructs expressing human or mouse Oct4, Sox2, Klf4, and c-Myc were obtained from R. Mostoslavsky. Lentiviral constructs expressing either scrambled shRNA or shRNA against Jmjd1c were obtained from Applied Biological Materials. The green fluorescent protein (GFP) cassette was removed from Jmjd1c shRNA constructs by digestion with BamHI and religation. Lentiviruses were produced by cotransfecting 293T cells with 1.7 μg of packaging plasmid (pMDLg/pRRE), 1.7 μg of envelope plasmid (pVSVG), and 1.7 μg of RNA export plasmid (pRSV-Rev), along with 5 μg of either hSTEMCCA, mSTEMCCA, scrambled shRNA, or Jmjd1c shRNA plasmid. A total of 105 Oct4-GFP mouse embryonic fibroblasts (MEFs) (28) generated from Pou5f1tm2Jae/J mice (Jackson Laboratories) were plated on gelatin-coated six-well plates, allowed to attach for 12 to 16 h, and then cotransduced with hSTEMCCA and either scrambled or Jmjd1c shRNA-expressing lentiviruses. Lentiviral transductions were performed on two consecutive days in the presence of 4 μg of Polybrene (Sigma)/ml. Transduced MEFs were selected the following day with 4 μM puromycin in ESC medium for 4 days. On day 7, Oct4-GFP MEFs were treated with trypsin and transferred to gelatin-coated 10-cm dishes with irradiated feeder fibroblast cells. The medium was changed daily. For human STEMCCA-infected cells, the iPSC colony number and the green fluorescence were scored on day 14. All experiments were performed in quadruplicate. GFP epifluorescence and phase-contrast images were captured using an Olympus IX51 inverted microscope (×10 objective) and a Lumenera Infinity 2 camera. Mouse STEMCCA lentiviruses were similarly used, with some modifications. Oct4-GFP MEFs were transduced and selected in six-well plates, without transfer to 10-cm dishes. The MEFs were allowed to grow in six-well dishes for 24 days rather than 14 days. The iPSC colonies were scored on day 24. The Jmjd1c knockdown shRNA sequences were as follows: scrambled, 5′-GGGTGAACTCACGTCAGAA; Jmjd1c-A, 5′-CCAGAGTAACTACTTCACTACTTTATCTA; Jmjd1c-B, 5′-TCTGACTGTTTCTTCTACTAATGCACTCC; Jmjd1c-C, 5′-GTGCCGAGAGTGCAGACTTATCCGGAGTA; and Jmjd1c-D, 5′-GCCCTGGAAGTGCAAGTACAGACAGCAGG.
Accession number.
ChIPseq raw and processed data files have been deposited in the GEO Sequence Read Archive database under accession number GSE65192.
RESULTS
Oct4 blocks deposition of DNA methyl marks at Pou5f1 in differentiating ESCs.
To study regulation by Oct4 in the absence of Oct1, we studied the Pou5f1 distal enhancer. Oct4 positively regulates its own synthesis via binding to a site in this region (29). Despite the fact that they are coexpressed in ESCs, Oct4 but not Oct1 interacts with the distal enhancer (30). Two CpGs flank the Oct4 binding site (Fig. 1A). ESC differentiation is known to result in DNA methylation of the Pou5f1 promoter and enhancer regions (31, 32). We hypothesized that loss of Oct4 during differentiation enables DNA methylation near the Oct4 binding site at the Pou5f1 enhancer region and that therefore ectopic Oct4 expression would maintain a local demethylated state.
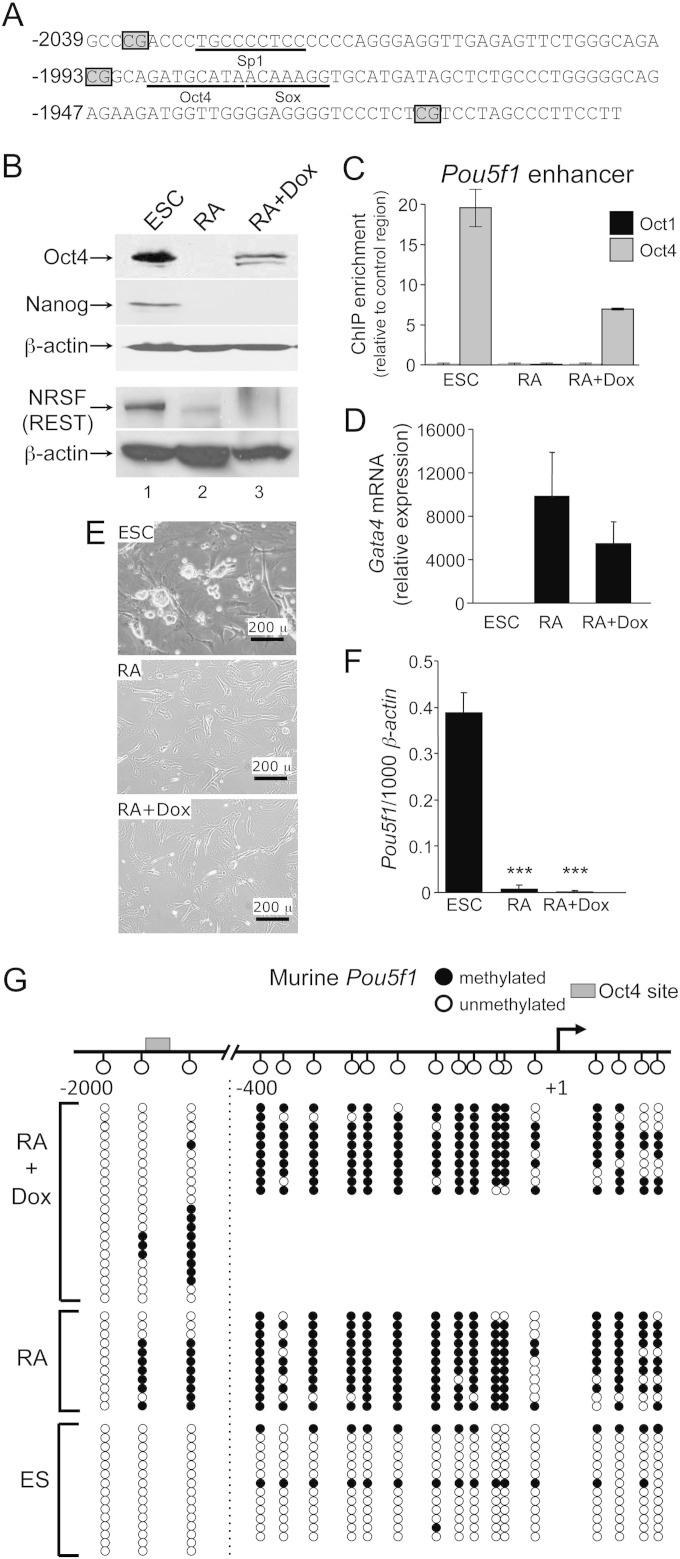
FIG 1.
ESCs can be differentiated normally with low-level Oct4. (A) Sequence of the murine Pou5f1 enhancer region. CpG positions are boxed. Known transcription factor binding sites are underlined. (B) Oct4 Western blots of lysates from tetON-Oct4 ESCs (21) and differentiated ESCs cultured for 16 days in medium lacking LIF and containing RA, with or without doxycycline. Experiments were performed in the absence and presence of doxycycline (Dox). Oct4, Nanog, and REST protein expression levels were evaluated. β-Actin was used as a loading control. (C) Oct1 and Oct4 ChIP enrichment at the Pou5f1 distal enhancer. Differentiation was for 12 days. Values represent averages of triplicate experiments. Error bars depict the standard deviations of the mean. (D) Gata4 mRNA expression was determined by qRT-PCR under similar conditions to (C). mRNA levels were normalized to β-actin and changes in expression are represented as the fold change (n = 3). Error bars depict the standard deviations. (E) Phase microscopy images of cultured tetON-Oct4 ESCs. The upper left panel shows cells propagated in ESC culture medium. The upper right panel shows cells differentiated for 16 days in medium lacking LIF and containing RA. The bottom panel shows cells also cultured in doxycycline. Images were taken using an Olympus IX51 inverted microscope at ×40 magnification with a Lumenera Infinity 2 camera. (F) Pou5f1 mRNA expression levels as determined by qRT-PCR using primers specific for the endogenous allele (n = 3). Error bars depict the standard deviations. (G) Bisulfite sequencing analysis of the murine Pou5f1 enhancer and promoter regions. A schematic is shown at the top. The Oct4 binding site is boxed. Filled circles indicate DNA methylation, and open circles indicate unmethylated DNA for a particular sequenced clone. Sequence reads on either side of dashed line were separately amplified, miniprepped, and sequenced.
To test this hypothesis, we used ESCs carrying a doxycycline-inducible Oct4 cassette (21). Mice generated from these “tetON-Oct4” ESCs are viable and phenotypically normal, but upon doxycycline treatment manifest hyperplasia in gut stem cell compartments, resulting in a disruption of normal tissue function (21). In tetON-Oct4 ESCs differentiated by LIF withdrawal and RA addition, ectopic Oct4 was expressed at a relatively low level compared to endogenous Oct4, as evidenced by Western blotting (Fig. 1B, lane 3). Tetracycline-free serum was used with tetON-Oct4 ESCs, and RA-differentiated cells show no evidence of “leaky” Oct4 expression (lane 2). When doxycycline was present, the Oct4 that is produced interacted with the Pou5f1 distal enhancer (Fig. 1C). In contrast endogenous Oct1, which is coexpressed with Oct4 in ESCs and continues to be expressed in differentiated cells, does not interact with this site. This result is consistent with prior findings (30).
RA treatment resulted in ESC differentiation regardless of doxycycline administration and exogenous Oct4 expression, as measured by the downregulation of Nanog and NRSF/REST (Fig. 1B) and the induction of Gata4 mRNA (Fig. 1D). REST inhibition indicates that the cells still undergo neuronal differentiation in the presence of RA and doxycycline. In addition, the differentiated cells were morphologically equivalent in phase microscopy (Fig. 1E). As a further control, we also determined the status of endogenous Pou5f1 mRNA expression. The doxycycline-inducible Oct4 allele contains a truncated 3′ untranslated region, allowing specific interrogation of endogenous expression using qRT-PCR (see Materials and Methods). Endogenous Pou5f1 was silenced regardless of the presence of doxycycline (Fig. 1F). These data indicate that, in this system, RA-mediated ESC differentiation occurs in the presence of doxycycline treatment and ectopic Oct4 expression.
Consistent with Pou5f1 silencing, after 16 days of differentiation, DNA methylation at the promoter-proximal Pou5f1 CpGs was unaffected by ectopic Oct4 (Fig. 1G, right side). In contrast to the promoter region, ectopic Oct4 expression reduced DNA methylation at the CpGs adjacent to the Oct4 site within the Pou5f1 enhancer (Fig. 1G, left side). The effect was strongest at the CpG closest to the Oct4 site (64% versus 14% methylation with low-level Oct4, the Fisher exact test P value = 0.059), CpG 2, and less pronounced at a more distant CpG, CpG 1 (73% versus 45%, P value = 0.266). Because Oct4 directly interacts with the Pou5f1 enhancer but not with the Pou5f1 proximal promoter, these results suggest that, like Oct1 (15), Oct4 can act locally to maintain a state of reduced DNA methylation. Pou5f1 is transcriptionally silent regardless of ectopic Oct4 expression (Fig. 1F), indicating that Oct4's effects on DNA methylation are independent of ongoing transcription.
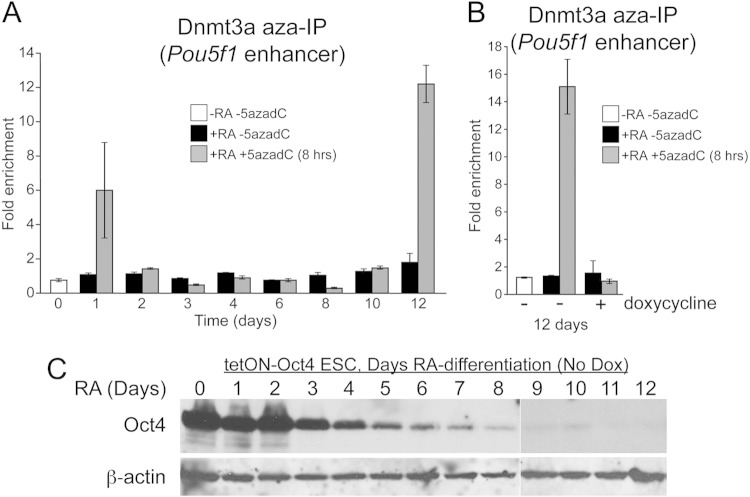
Dnmt3a methylates DNA at the Pou5f1 locus during ESC differentiation (33–35). Methylation of the Pou5f1 locus during differentiation is thought to initiate at the promoter and proximal enhancer regions and spread into the distal enhancer containing the Oct4 binding site (33, 36). DNA methylation is promoted by H3K9 methylation and G9a, a histone H3K9 methyltransferase (34). The decreased DNA methylation associated with local Oct4 binding was consistent with two models in which either (i) Oct4 recruits activities that catalyze DNA demethylation or (ii) Oct4 maintains a chromatin state refractory to Dnmt3a-mediated deposition of DNA methyl marks. In the first model, Dnmt3a would be predicted to deposit DNA methyl marks on the DNA, which would then be removed by activities such as Tet proteins associated with Oct4. In the second model, DNA methyl groups would not be deposited. To discriminate between these models, we used a method in which we could directly capture Dnmt3a associated with the Pou5f1 enhancer and tested whether ectopic Oct4 expression blocks association and capture. 5-Aza-2′-deoxycytidine (5-AzadC; decitabine) is a deoxycytosine analog that incorporates into replicating DNA. Because of the two-step nature of DNA methylation, with an intermediate in which the methyltransferase is covalently linked to the cytosine moiety, DNA methyltransferases become trapped in covalent linkages with 5-AzadC moieties in the DNA (37–39). This covalent link can be used in a variant of ChIP (without formaldehyde cross-linking) to identify methyltransferases linked to nucleic acids (Aza-IP) (40). We differentiated tetON-Oct4 ESCs (without doxycycline) and observed two waves of Dnmt3a activity near the Oct4 binding site: one at very short time points after RA treatment and a larger one at 12 days (Fig. 2A). The signals were dependent on an 8-h pulse of 5-AzadC. 12 days is the same point at which de novo methyltransferases associate with the Pou5f1 enhancer in conventional ChIP (11). The signal at 12 days was eliminated by doxycycline treatment and ectopic Oct4 expression (Fig. 2B), indicating that Oct4 expression blocks Dnmt3a engagement with enhancer DNA. Endogenous Oct4 protein expression was mostly quenched by day 5; however, prolonged exposure indicated that residual protein was detectable through 10 days (Fig. 2C, lane 11). These results are consistent with a model in which Oct4 decreases DNA methylation by preventing association of Dnmt3a rather than by catalyzing DNA demethylation. Dnmt3a enrichment monitored by conventional ChIP-qPCR showed the same trends (see Fig. S1 in the supplemental material). Because prior work indicated that Oct1 regulates DNA methylation indirectly through Jmjd1a recruitment and control of H3K9me2 (15), we pursued a model in which Oct4 also regulates DNA methylation indirectly through control of H3K9 methylation.
FIG 2.
Oct4 prevents DNA methyl mark deposition at Pou5f1. (A) Dnmt3a reactivity with the Pou5f1 distal enhancer region was assessed by Aza-IP using differentiating ESCs. Enrichment in normal ESCs (day 0) lacking a 5-AzadC pulse is arbitrarily set to 1.0, and the relative enrichment for 1 μM RA-treated cells with or without a 5-AzadC pulse is shown. (B) tetON-Oct4 ESCs differentiated for 12 days with RA with or without the addition of doxycycline were used in Aza-IP assays. Enrichment in normal ESCs (day 0) lacking a 5-AzadC pulse is arbitrarily set to 1.0, and the relative enrichment for RA-treated cells with or without a 5-AzadC pulse is shown. (C) tetON-Oct4 ESCs were differentiated by removing LIF and adding RA. A 12-day time course is shown. Oct4 expression was probed by Western blotting. β-Actin was used as a loading control. No doxycycline was present.
Sequential Oct4-mediated Jmjd1c recruitment, histone demethylation, and nucleosome depletion.
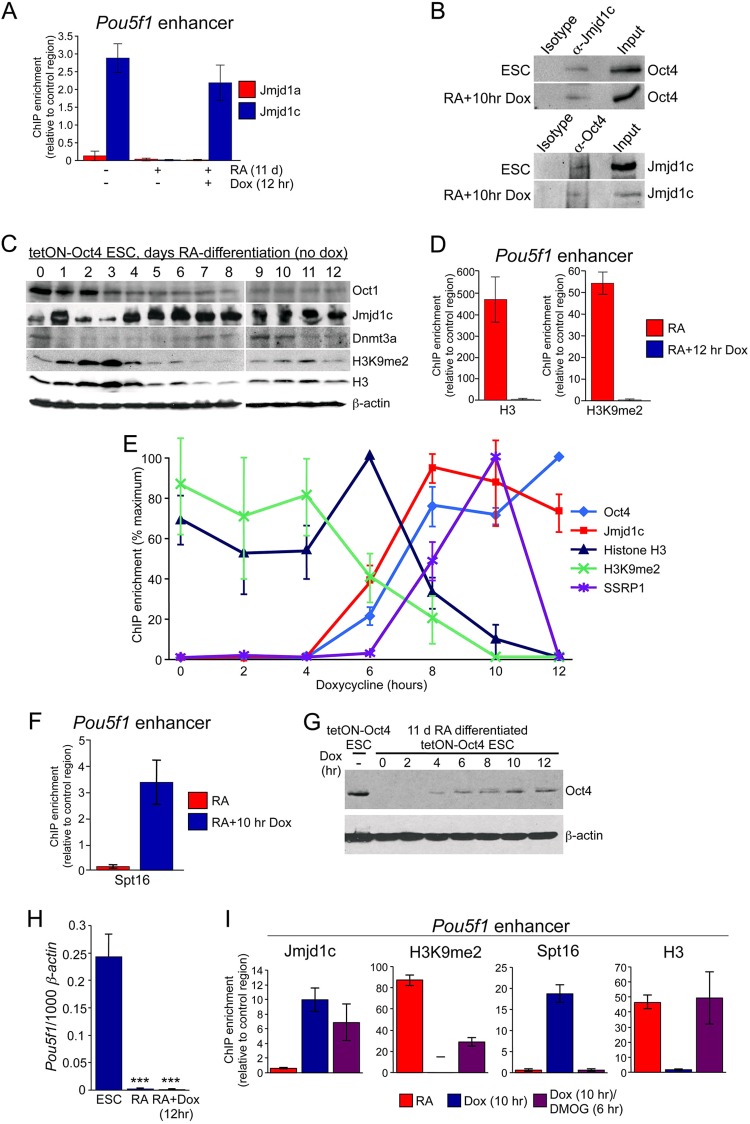
We first tested whether Jmjd1a associates with Oct4 at the Pou5f1 enhancer. Relative to isotype control antibodies and an intergenic control region, no enrichment was detected at Pou5f1 in ESCs or in RA differentiated cells at 11 days given a 12-h pulse of doxycycline (Fig. 3A). We therefore tested whether Oct4 recruits paralogs of Jmjd1a. Jmjd1c is a lysine demethylase necessary for normal mouse testes development (41). In contrast to Jmjd1a, Jmjd1c robustly associated with the Oct4 site in the Pou5f1 distal enhancer in undifferentiated ESCs but not in differentiating cells that have lost endogenous Oct4 (Fig. 3A). A 12-h pulse of doxycycline and ectopic Oct4 expression was sufficient to induce Jmjd1c recruitment, indicating that Oct4 recruits Jmjd1c to the Pou5f1 enhancer. Unlike Jmjd1a, Oct4 has been reported to interact with Jmjd1c in a large screen (42). We confirmed this finding by immunoprecipitating Oct4 or Jmjd1c from ESC extracts, as well as extracts from 11-day RA differentiated cells given a 12-h pulse of doxycycline, and probing for the other coprecipitated protein (Fig. 3B). Although levels fluctuate with neuronal differentiation, significant levels of Oct1, Jmjd1c, Dnmt3a, and H3K9me2 were present at the 11-day time point (Fig. 3C). Jmjd1c is known to be expressed in multiple adult neuronal cell types (43).
FIG 3.
Oct4 associates with Jmjd1c and FACT at Pou5f1. (A) Jmjd1a and Jmjd1c ChIP enrichment was measured at the Pou5f1 enhancer in tetON-Oct4 ESCs, ESCs undergoing RA-mediated differentiation for 11 days, and similarly differentiated ESCs treated with doxycycline for 12 h to induce Oct4. (B) Coimmunoprecipitation assay using undifferentiated tetON-Oct4 ESCs (without doxycycline in the medium). Lysates were immunoprecipitated with anti-Jmjd1c antibodies or with an isotype control, followed by Western blotting with Oct4 antibodies. (C) Differentiation time course of Oct1, Jmjd1c, Dmnt3a, and H3K9me2 protein expression (without doxycycline). Western blot data are shown. Histone H3 and β-actin were used as loading controls. (D) Histone H3 and H3K9me2 ChIP enrichment in 11-day-differentiated ESCs (with or without 12 h of doxycycline treatment to induce ectopic Oct4 expression). (E) A ChIP time course was performed using ESCs differentiated for 11 days with RA and subsequently induced using doxycycline for the indicated times. ChIP was performed at the Pou5f1 enhancer with the indicated antibodies. For each antibody, the ChIP signal is shown as a percentage of the maximum (100%). Each experiment was performed in biological triplicates. Error bars depict the standard deviations. In cases where the same time point showed maximum signal for all biological and technical replicates (Oct4, histone H3, and SSRP1), the signal was 100%, and the standard deviation for that time point was 0. In other cases (Jmjd1c and H3K9me2) the maximum signal was at different times for different replicates, and an error was derived at all time points. (F) A similar ChIP experiment was performed at the 0- and 10-h time points with antibodies against the FACT subunit Spt16. (G) A Western blot was performed with antibodies against Oct4 to monitor production of protein over the same time course. ESCs and endogenous Oct4 are shown as a positive control. β-Actin was used as a loading control. (H) Endogenous Pou5f1 expression was monitored in partially differentiated ESCs and the same cells treated with doxycycline for 12 h using qRT-PCR. Undifferentiated ESCs were used as a control. (I) ChIP was performed at the Pou5f1 enhancer in 11-day RA-differentiated ESCs and after 10 h of induction with doxycycline as in panel D; however, DMOG was also added after 4 h of doxycycline treatment. Cells were therefore exposed to doxycycline for 10 h and to DMOG for 6 h. Jmjd1c, H3K9me2, Spt16, and H3 antibodies were used.
We tested whether Oct4 and Jmjd1c association coincide with loss of H3K9me2. ESCs partially differentiated for 11 days possessed significant levels of H3 and H3K9me2 at the Pou5f1 distal enhancer (Fig. 3D). In contrast, 12 h of doxycycline treatment and ectopic Oct4 expression eliminated enrichment for not only H3K9me2 but also H3. H3 depletion is consistent with findings that Oct4 binding can correlate with nucleosome-free regions (11).
Provided chromatin changes occur synchronously in the population, the culture system described above can be used to probe dynamics by performing ChIP time courses. We differentiated ESCs for 11 days and then provided doxycycline to induce Oct4, calculating the ChIP enrichment for each antibody as a percentage relative to the maximum enrichment. At day 11 (0 h of doxycycline administration), Oct4 and Jmjd1c were absent and H3K9me2 was present (Fig. 3E). Both Oct4 and Jmjd1c became detectable at 6 h (light blue and red lines). In contrast, at 6 h there was no association with another chromatin-modifying activity, the FACT complex (purple line). Concomitant with Jmjd1c association, H3K9me2 enrichment began to decline at 6 h (green line), reaching zero by 10 h. In the case of H3, no diminution was apparent until 8 h (dark blue line). The transient increase in signal at 6 h was likely due to improved epitope availability as K9me2 was removed. H3 reached zero enrichment at 12 h. Loss of H3 therefore lagged H3K9me2 loss by 2 h, suggesting that K9me2 demethylation is requisite for nucleosome eviction.
The FACT complex (Spt16+SSRP1) is a regulator of nucleosome function associated with transcription initiation and elongation (44, 45). FACT is abundant, but local enrichment has been associated with nucleosome depletion as part of transcription initiation processes in other contexts (46–48). Studies confirm interactions between Oct4 and FACT (17, 42), although no functional role has been ascribed. We performed ChIP with antibodies against SSRP1 to determine whether FACT is recruited to the Pou5f1 distal enhancer. No SSRP1 was detected at Pou5f1 at 0 h, or at 6 h when Oct4 and Jmjd1c became detectable (Fig. 3E). SSRP1 association rose at 8 h, coinciding with the loss of H3. Association peaked at 10 h. Unexpectedly, SSRP1 was completely lost at 12 h, indicating not only that FACT may mediate the loss of H3 but also that it may require H3 to stably associate. Non-normalized ChIP enrichment is shown in Fig. S2 in the supplemental material. We also identified significant ChIP signal using antibodies to the other FACT complex component, Spt16, at the peak 10-h time point but not at t = 0 (Fig. 3F). Oct4 protein was induced with kinetics that slightly preceded the ChIP signal (Fig. 3G). Despite the chromatin changes mediated by Oct4, endogenous Pou5f1 was not induced (Fig. 3H). Nanog expression was also unaltered (see Fig. S3 in the supplemental material). These results suggest that newly bound Oct4 acts locally at Pou5f1 by first recruiting Jmjd1c and promoting histone demethylation and then promoting FACT recruitment and the depletion of local nucleosomes. Subsequently, FACT is released. These changes are not indirect consequences of changes in Pou5f1 expression but rather result from the immediate binding of Oct4 and associated factors.
FACT recruitment and H3 depletion at Pou5f1 both require dioxygenase activity.
The findings described above provide a portrait of changes to chromatin and associated modifying activities as a function of time. To provide evidence for a causal relationship, we used DMOG, an inhibitor of dioxygenase activity, which is required to demethylate H3K9. We performed a similar RA-mediated differentiation/re-expression assay in the presence of added DMOG or vehicle control. Because DMOG has multiple effects (such as mimicking hypoxia), we incubated cells for the minimum possible time. DMOG was added at 4 h, when Oct4 protein is detectable (Fig. 3G) but not bound to Pou5f1 (Fig. 3E). DMOG was present throughout the remaining 6 h of the assay. DMOG treatment had no effect on Jmjd1c recruitment by Oct4 (Fig. 3I). In contrast and as expected, DMOG significantly blocked H3K9me2 demethylation, resulting in substantial H3K9me2 retention at 10 h compared to the control. DMOG also blocked Spt16 binding, strongly suggesting that complete H3K9me2 demethylation is a prerequisite for FACT association. In addition, H3 depletion was blocked, suggesting that depletion requires demethylation and/or FACT recruitment.
The Oct4-Jmjd1c-FACT pathway operates at multiple Oct4 target genes.
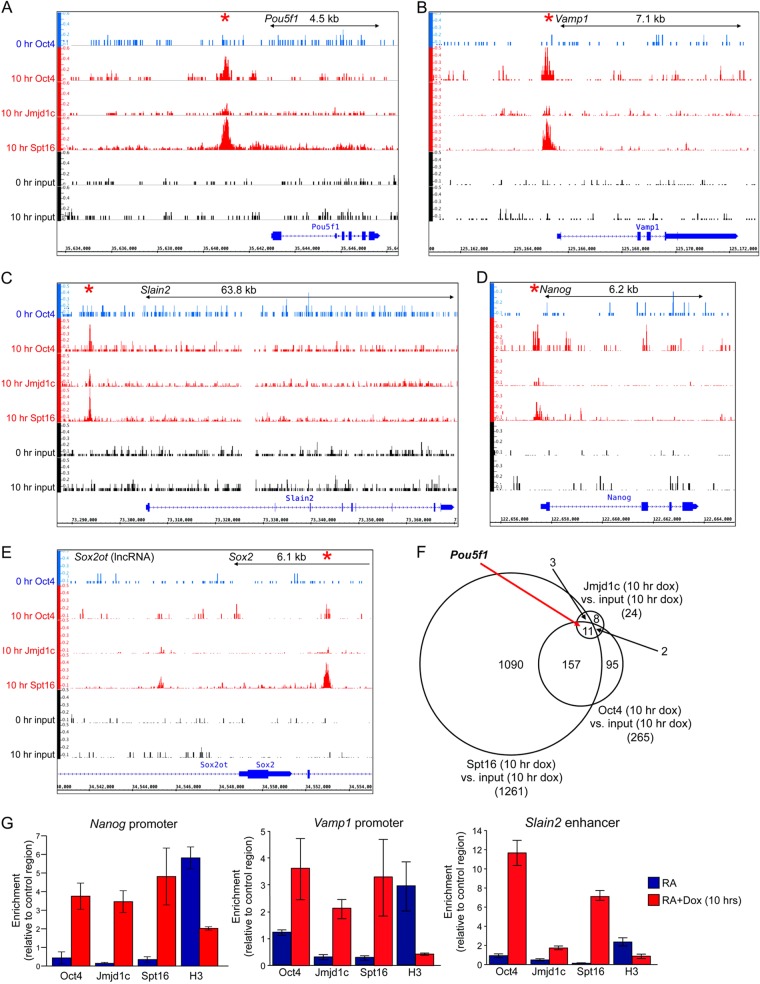
To understand whether this assay system can be used to broadly measure Oct4 binding and chromatin-modifying activity, we performed ChIP-Seq. We differentiated ESCs for 11 days using RA and then induced ectopic Oct4 with doxycycline for 10 h to assess Oct4, Jmjd1c, and Spt16 (FACT complex) binding. Between 31 and 50 million sequence reads were generated (average 49.5 million), ca. 65% of which uniquely aligned to the mouse reference genome.
We identified sites specifically bound by newly synthesized Oct4 at t = 10 h compared to the input control (see Table S1 in the supplemental material). Using a 2-fold ratio and a Q-value FDR = 0.05 cutoffs, we identified 265 specific Oct4-bound regions, 72% of which were genic based on a transcription start site distance cutoff of 10 kb. A high frequency (no less than 73%) of these binding events correlate with known Oct4 target genes based on one murine ESC data set (49). Examples include Pou5f1, Vamp1, Slain2, and the pluripotency gene Nanog (Fig. 4A to D). We also analyzed the core pluripotency Oct4 target Sox2, visually identifying Oct4 enrichment at the Sox2 promoter (Fig. 4E). This result suggests that additional biological targets may be missed in this analysis. We also compared the set of Oct4 targets with Jmjd1c and Spt16 (FACT) binding events. FACT is abundant and known to associate with many genomic regions in yeast, such as sites of high gene activity (50). Approximately 1,100 FACT binding events were identified (see Table S2 in the supplemental material), including 65% of all identified Oct4 peaks, including Pou5f1, Vamp1, Slain2, and Nanog (Fig. 4F). Jmjd1c produced less enrichment, yielding only 24 specific binding events with the same statistical algorithms and cutoffs (see Table S3 in the supplemental material). Nevertheless, Pou5f1 was objectively identified as a common Oct4, Jmjd1c, and Spt16 target (Fig. 4F). Using qPCR, we confirmed specific Oct4 binding and identified a similar pattern of cofactor recruitment and H3 depletion at the Nanog and Vamp1 promoters and Slain2 enhancer (Fig. 4G).
FIG 4.
Oct4, Jmjd1c, and FACT coassociate with multiple genomic targets. (A) Relative alignment depth coverage tracks of ChIP-Seq data corresponding to the murine Pou5f1 locus and surrounding region. Oct4 enrichment is shown in blue in 11-day RA-differentiated ESCs in the absence of doxycycline (0 h Oct4) or in red after a 10-h doxycycline treatment (10 h Oct4). Jmjd1c and Spt16 enrichment is shown below in red. Input controls for each time point are shown below in black. Relative depth tracks are scaled the same and represented as per-base FPKMs (fragments per kilobase of exon per million fragments). (B to E) Similar data for Vamp1 (B), Slain2 (C), Nanog (D), and Sox2 (E). A region of upstream noncoding RNA expression (Sox2ot) is also shown. (F) Venn diagram depicting target identification for Oct4, Spt16, and Jmjd1c ChIPseq, as well as overlap. The peak calling was based on a 2-fold cutoff with a Q-value FDR of 0.05. (G) ChIP enrichment for Oct4, Jmjd1c, Spt16, and histone H3 was determined by qPCR similarly to Fig. 3; however, a ChIP primer pair spanning the Oct4 site in the murine Nanog, Vamp1, and Slain2 promoters was used in place of Pou5f1.
Jmjd1c is required for iPSC reprogramming.
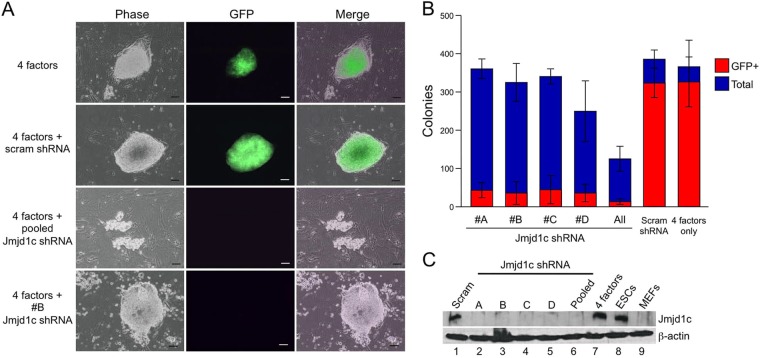
If Jmjd1c helps execute Oct4's transcription functions, then it should be necessary for processes known to require Oct4. Most prominent of these is the ability to reprogram somatic cells to pluripotency (6, 51). We used lentiviral shRNA knockdown to ablate Jmjd1c in Oct4-GFP MEFs and tested iPSC formation using the 4-in-1 human STEMCCA vector system (52). Oct4-GFP fibroblasts fluoresce green upon reprogramming and activation of endogenous Oct4.
Prior work suggested that Jmjd1c knockdown only minimally reduces (∼20%) iPSC generation efficiency (19). However, knockdown efficacy was not described in the present study. Furthermore, iPSC morphology was used to assess reprogramming. Using human STEMCCA and Oct4-GFP MEFs, together with four different Jmjd1c shRNAs, we also observed iPSC-like colonies. However, the colonies lacked green fluorescence (Fig. 5A), indicating that they were incompletely reprogrammed and still reliant on virally expressed rather than endogenous Oct4. A scrambled shRNA lentivirus had no effect on GFP+ iPSC colony emergence. The strongest effects were observed when the five shRNAs were combined (Fig. 5A and B). Collected data from quadruplicate experiments are quantified in Fig. 5B. Additional images are shown in Fig. S4A in the supplemental material. We also reprogrammed cells using the mouse reprogramming factors, generating similar results. Images of the colonies generated are shown in Fig. S4B. This reprogramming method generated fewer colonies with iPSC-like morphology upon Jmjd1c knockdown. Collected data from quadruplicate experiments are shown in Fig. S4C in the supplemental material. Western blotting indicated that the Jmjd1c protein was expressed robustly in mouse ESCs but poorly in the target Oct4-GFP MEFs (Fig. 5C). iPSC reprogramming resulted in Jmjd1c induction. Specific shRNA knockdown but not scrambled shRNA blocked induction of Jmjd1c (Fig. 5C). These results indicate that Jmjd1c enables formation of bona fide iPSCs and may act in a late step in the reprogramming process.
FIG 5.
Jmjd1c is required to completely reprogram MEFs to pluripotency. (A) Uninfected primary Oct4-GFP MEFs (passage 5) or MEFs cotransduced with empty vector or Jmjd1c shRNA-encoding lentiviruses were infected with hSTEMCCA encoding human Oct4, Sox2, Klf4, and c-Myc. Cells were selected with 4 μM puromycin in ESC medium and plated on irradiated feeder fibroblasts. Images were obtained at 14 days posttransduction. Scale bars, 100 μm. (B) Quantification of total colonies and GFP+ colonies observed in a 10-cm dish. Averages were taken from four biological replicates. Error bars indicate ± the standard deviations. (C) Oct4-GFP MEFs or mouse ESCs were probed for Jmjd1c by Western blotting (lanes 8 and 9). In lanes 1 to 7, MEFs infected with hSTEMCCA and reprogrammed for 14 days and then collected and probed for Jmjd1c. Lanes 1 to 6 were additionally infected with Jmjd1c knockdown lentiviruses or scrambled controls. β-Actin was used as a loading control.
DISCUSSION
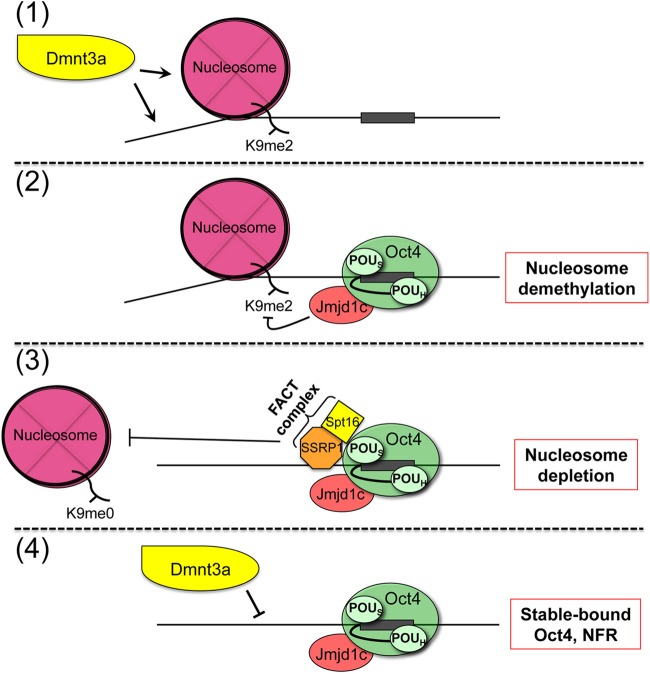
Although prior work has provided insights into how Oct4 regulates transcription (9–13, 53), critical components of Oct4's downstream functions remain unknown. We describe an experimental platform that allows collection of synchronous Oct4 binding and local chromatin remodeling data. It offers advantages over using established ESCs, because early changes mediated by initial Oct4 binding and transiently recruited activities can be visualized. It also offers advantages over using fully differentiated cells, in which induction of pluripotency targets by exogenous factors is typically inefficient, variable and slow. Partially reprogrammed iPSCs (or “pre-iPSCs”), which have been used with success in other settings, fail to demonstrate Oct4 binding to key late target genes, including Pou5f1, despite the fact that Oct4 is expressed from the reprogramming cocktail (19). Using this system, we identified a sequential mechanism in which first Oct4 recruits the histone lysine demethylase Jmjd1c to chromatin occupied by a nucleosome that contains H3K9me2, then Jmjd1c demethylates H3K9me2, and finally the FACT complex is transiently recruited and local H3 is depleted. The resulting chromatin is refractory to DNA methylation by Dnmt3a (Fig. 6). This mechanism operates at three critical Oct4 pluripotency targets, Pou5f1, Sox2, and Nanog. These findings are consistent with the known pioneer activity of Oct4 (54).
FIG 6.
(Step 1) Model mechanism for sequential chromatin changes mediated by Jmjd1c and FACT recruited to Oct4 at pluripotency targets. (Step 2) Prior to Oct4 induction, target chromatin is characterized by H3K9me2 and the ability of Dnmt3a to methylate CpG DNA). (Step 3) After Oct4 expression, Oct4 and Jmjd1c associate and mediate H3K9 demethylation. (Step 4) FACT is subsequently recruited and mediates nucleosome depletion. FACT dissociates, and Oct4-occupied DNA refractory to methylation by Dnmt3a remains.
Oct1, an Oct4 paralog, recruits the KDM3 family member Jmjd1a to remove H3K9me2 from local chromatin (15). Pou5f1 (the gene encoding Oct4) is occupied by Oct4 but not by Oct1 (30), allowing for the analysis of Oct4 function in the absence of Oct1. It is not clear why Jmjd1c is selectively recruited by Oct4; however, Jmjd1c is larger than Jmjd1a (∼280 kDa versus ∼148 kDa) and contains a distinct N terminus (55). Recent work suggests that Jmjd1c associates with the transcription factor Nanog (56), but evidence for a functional role as a Nanog cofactor is lacking. Jmjd1c is important for maintenance of spermatogonial germ cells (57), and rare germ line Jmjd1c variants are associated with certain germ cell tumors (58). In addition, Jmjd1c is a downstream Oct4 target (59), suggesting a possible regulatory loop. We show that Jmjd1c RNAi limits iPSC reprogramming, indicating that the system described in this report identifies physiologically important Oct4 coregulators. The mark Jmjd1c acts on, H3K9me2, presents a barrier to iPSC generation that must be overcome by one or more reprogramming factors (19). Interestingly, Jmjd1c knockdown impacts differentiation but not maintenance of established ESCs (60). Therefore, Jmjd1c appears to be a critical regulator of both ESC establishment and differentiation but not maintenance.
Subsequent to K9 demethylation, FACT is recruited to Oct4/Jmjd1c/DNA complexes. FACT is known to associate with Oct4 (17, 42), providing a basis for this recruitment. FACT recruitment is concomitant with H3 depletion, suggesting a mechanism in which locally enriched FACT mediates local nucleosome depletion. Consistently, suppression of H3 demethylation blocks FACT recruitment and also depletion of H3. FACT enrichment is lost following depletion of H3. In yeast, transient FACT has been identified as a key component of a sequential transcription initiation cascade that culminates in nucleosome displacement (44, 45, 48). In metazoans, both Drosophila GAGA factor and mammalian HSF1 have been shown to associate with FACT as part of their transcription function (46, 47). Recently, the mammalian transcription factor myogenin was also shown to recruit FACT and displace nucleosomes as part of a transcription initiation (not elongation) process (61). Knockdown of either FACT subunit impairs ESC maintenance and impairs mouse survival at the early blastocyst stage (62, 63), a finding consistent with a role in pluripotency.
We performed genome-wide studies of newly synthesized Oct4 binding events, as well as binding of Jmjd1c and Spt16 (FACT). A prior study in yeast identified Spt16 binding events, demonstrating preferential localization to transcription units (50). Our analysis of mouse cells also identified thousands of Spt16 binding events; however, we also identified focal Spt16 enrichment at a specific subset of Oct4 target genes, including Pou5f1, Nanog, Vamp1, and Slain2. Ectopic Oct4 expression and Jmjd1c recruitment is also associated with loss of H3. Loss of H3 is consistent with findings that Oct4 occupancy can correlate with nucleosome-free regions (11). Other work has shown that many Oct4 sites are enriched rather than depleted for nucleosomes (54), which is consistent with the finding that this mechanism only acts at the majority of Oct4 targets. H3 loss provides a plausible mechanism for the observed block to DNA methylation observed with ectopic Oct4 expression, because Dnmt3-containing complexes are thought to associate with nucleosomes to initiate de novo DNA methylation (64, 65).
In conclusion, we have developed a system in which Oct4 expression is induced in partially differentiated ESCs. Induction results in uniform binding and temporally synchronous chromatin remodeling, enabling the discovery of activities that functionally collaborate with Oct4. Using this system, we identify Jmjd1c and the FACT complex as activities associated with Oct4 binding and local chromatin remodeling. This method should provide a valuable tool for studying Oct4's transcriptional functions.
Supplementary Material
ACKNOWLEDGMENTS
We thank T. Formosa and D. Stillman for critical reading of the manuscript. We thank B. Cairns for assistance with the Aza-IP. We thank K. Hochedlinger for the doxycycline-inducible Oct4 ESCs. We thank W. Voth and D. Stillman for assistance with the ChIP. G. Mostoslavsky provided the human and mouse STEMCCA vectors.
This study was supported by grant R01AI100873 (to D.T.) and an institutional seed grant.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01105-14.
REFERENCES
- 1.Jerabek S, Merino F, Scholer HR, Cojocaru V. 2014. OCT4: dynamic DNA binding pioneers stem cell pluripotency. Biochim Biophys Acta 1839:138–154. doi: 10.1016/j.bbagrm.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 2.Tantin D. 2013. Oct transcription factors in development and stem cells: insights and mechanisms. Development 140:2857–2866. doi: 10.1242/dev.095927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Radzisheuskaya A, Silva JC. 2014. Do all roads lead to Oct4? The emerging concepts of induced pluripotency. Trends Cell Biol 24:275–284. doi: 10.1016/j.tcb.2013.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Scholer H, Smith A. 1998. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 95:379–391. doi: 10.1016/S0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- 5.Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, Gifford DK, Melton DA, Jaenisch R, Young RA. 2005. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Okita K, Ichisaka T, Yamanaka S. 2007. Generation of germline-competent induced pluripotent stem cells. Nature 448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 7.Li Y, Zhang Q, Yin X, Yang W, Du Y, Hou P, Ge J, Liu C, Zhang W, Zhang X, Wu Y, Li H, Liu K, Wu C, Song Z, Zhao Y, Shi Y, Deng H. 2011. Generation of iPSCs from mouse fibroblasts with a single gene, Oct4, and small molecules. Cell Res 21:196–204. doi: 10.1038/cr.2010.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ponnusamy MP, Deb S, Dey P, Chakraborty S, Rachagani S, Senapati S, Batra SK. 2009. RNA polymerase II associated factor 1/PD2 maintains self-renewal by its interaction with Oct3/4 in mouse embryonic stem cells. Stem Cells 27:3001–3011. doi: 10.1002/stem.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ang YS, Tsai SY, Lee DF, Monk J, Su J, Ratnakumar K, Ding J, Ge Y, Darr H, Chang B, Wang J, Rendl M, Bernstein E, Schaniel C, Lemischka IR. 2011. Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell 145:183–197. doi: 10.1016/j.cell.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fong YW, Inouye C, Yamaguchi T, Cattoglio C, Grubisic I, Tjian R. 2011. A DNA repair complex functions as an oct4/sox2 coactivator in embryonic stem cells. Cell 147:120–131. doi: 10.1016/j.cell.2011.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.You JS, Kelly TK, De Carvalho DD, Taberlay PC, Liang G, Jones PA. 2011. OCT4 establishes and maintains nucleosome-depleted regions that provide additional layers of epigenetic regulation of its target genes. Proc Natl Acad Sci U S A 108:14497–14502. doi: 10.1073/pnas.1111309108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yeap LS, Hayashi K, Surani MA. 2009. ERG-associated protein with SET domain (ESET)-Oct4 interaction regulates pluripotency and represses the trophectoderm lineage. Epigenet Chromatin 2:12. doi: 10.1186/1756-8935-2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuan P, Han J, Guo G, Orlov YL, Huss M, Loh YH, Yaw LP, Robson P, Lim B, Ng HH. 2009. Eset partners with Oct4 to restrict extraembryonic trophoblast lineage potential in embryonic stem cells. Genes Dev 23:2507–2520. doi: 10.1101/gad.1831909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liang J, Wan M, Zhang Y, Gu P, Xin H, Jung SY, Qin J, Wong J, Cooney AJ, Liu D, Songyang Z. 2008. Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat Cell Biol 10:731–739. doi: 10.1038/ncb1736. [DOI] [PubMed] [Google Scholar]
- 15.Shakya A, Kang J, Chumley J, Williams MA, Tantin D. 2011. Oct1 is a switchable, bipotential stabilizer of repressed and inducible transcriptional states. J Biol Chem 286:450–459. doi: 10.1074/jbc.M110.174045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van den Berg DL, Snoek T, Mullin NP, Yates A, Bezstarosti K, Demmers J, Chambers I, Poot RA. 2010. An Oct4-centered protein interaction network in embryonic stem cells. Cell Stem Cell 6:369–381. doi: 10.1016/j.stem.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pardo M, Lang B, Yu L, Prosser H, Bradley A, Babu MM, Choudhary J. 2010. An expanded Oct4 interaction network: implications for stem cell biology, development, and disease. Cell Stem Cell 6:382–395. doi: 10.1016/j.stem.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang J, Shakya A, Tantin D. 2009. Stem cells, stress, metabolism and cancer: a drama in two Octs. Trends Biochem Sci 34:491–499. doi: 10.1016/j.tibs.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 19.Chen J, Liu H, Liu J, Qi J, Wei B, Yang J, Liang H, Chen Y, Wu Y, Guo L, Zhu J, Zhao X, Peng T, Zhang Y, Chen S, Li X, Li D, Wang T, Pei D. 2013. H3K9 methylation is a barrier during somatic cell reprogramming into iPSCs. Nat Genet 45:34–42. doi: 10.1038/ng.2491. [DOI] [PubMed] [Google Scholar]
- 20.Sridharan R, Gonzales-Cope M, Chronis C, Bonora G, McKee R, Huang C, Patel S, Lopez D, Mishra N, Pellegrini M, Carey M, Garcia BA, Plath K. 2013. Proteomic and genomic approaches reveal critical functions of H3K9 methylation and heterochromatin protein-1gamma in reprogramming to pluripotency. Nat Cell Biol 15:872–882. doi: 10.1038/ncb2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hochedlinger K, Yamada Y, Beard C, Jaenisch R. 2005. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell 121:465–477. doi: 10.1016/j.cell.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 22.Tantin D, Voth WP, Shakya A. 2013. Efficient chromatin immunoprecipitation using limiting amounts of biomass. J Vis Exp 75:e50064. doi: 10.3791/50064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pachernik J, Bryja V, Esner M, Kubala L, Dvorak P, Hampl A. 2005. Neural differentiation of pluripotent mouse embryonal carcinoma cells by retinoic acid: inhibitory effect of serum. Physiol Res 54:115–122. [DOI] [PubMed] [Google Scholar]
- 24.Ram O, Goren A, Amit I, Shoresh N, Yosef N, Ernst J, Kellis M, Gymrek M, Issner R, Coyne M, Durham T, Zhang X, Donaghey J, Epstein CB, Regev A, Bernstein BE. 2011. Combinatorial patterning of chromatin regulators uncovered by genome-wide location analysis in human cells. Cell 147:1628–1639. doi: 10.1016/j.cell.2011.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kang J, Gemberling M, Nakamura M, Whitby FG, Handa H, Fairbrother WG, Tantin D. 2009. A general mechanism for transcription regulation by Oct1 and Oct4 in response to genotoxic and oxidative stress. Genes Dev 23:208–222. doi: 10.1101/gad.1750709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nix DA, Courdy SJ, Boucher KM. 2008. Empirical methods for controlling false positives and estimating confidence in ChIP-Seq peaks. BMC Bioinformatics 9:523. doi: 10.1186/1471-2105-9-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Storey JD. 2002. A direct approach to false discovery rates. J R Stat Soc Ser B 64:479–498. doi: 10.1111/1467-9868.00346. [DOI] [Google Scholar]
- 28.Lengner CJ, Camargo FD, Hochedlinger K, Welstead GG, Zaidi S, Gokhale S, Scholer HR, Tomilin A, Jaenisch R. 2007. Oct4 expression is not required for mouse somatic stem cell self-renewal. Cell Stem Cell 1:403–415. doi: 10.1016/j.stem.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chew JL, Loh YH, Zhang W, Chen X, Tam WL, Yeap LS, Li P, Ang YS, Lim B, Robson P, Ng HH. 2005. Reciprocal transcriptional regulation of Pou5f1 and Sox2 via the Oct4/Sox2 complex in embryonic stem cells. Mol Cell Biol 25:6031–6046. doi: 10.1128/MCB.25.14.6031-6046.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ferraris L, Stewart AP, Kang J, DeSimone AM, Gemberling M, Tantin D, Fairbrother WG. 2011. Combinatorial binding of transcription factors in the pluripotency regions of the genome. Genome Res 21:1055–1064. doi: 10.1101/gr.115824.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hattori N, Nishino K, Ko YG, Ohgane J, Tanaka S, Shiota K. 2004. Epigenetic control of mouse Oct-4 gene expression in embryonic stem cells and trophoblast stem cells. J Biol Chem 279:17063–17069. doi: 10.1074/jbc.M309002200. [DOI] [PubMed] [Google Scholar]
- 32.Gidekel S, Bergman Y. 2002. A unique developmental pattern of Oct-3/4 DNA methylation is controlled by a cis-demodification element. J Biol Chem 277:34521–34530. doi: 10.1074/jbc.M203338200. [DOI] [PubMed] [Google Scholar]
- 33.Athanasiadou R, de Sousa D, Myant K, Merusi C, Stancheva I, Bird A. 2010. Targeting of de novo DNA methylation throughout the Oct-4 gene regulatory region in differentiating embryonic stem cells. PLoS One 5:e9937. doi: 10.1371/journal.pone.0009937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feldman N, Gerson A, Fang J, Li E, Zhang Y, Shinkai Y, Cedar H, Bergman Y. 2006. G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat Cell Biol 8:188–194. doi: 10.1038/ncb1353. [DOI] [PubMed] [Google Scholar]
- 35.Li JY, Pu MT, Hirasawa R, Li BZ, Huang YN, Zeng R, Jing NH, Chen T, Li E, Sasaki H, Xu GL. 2007. Synergistic function of DNA methyltransferases Dnmt3a and Dnmt3b in the methylation of Oct4 and Nanog. Mol Cell Biol 27:8748–8759. doi: 10.1128/MCB.01380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sato N, Kondo M, Arai K. 2006. The orphan nuclear receptor GCNF recruits DNA methyltransferase for Oct-3/4 silencing. Biochem Biophys Res Commun 344:845–851. doi: 10.1016/j.bbrc.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 37.Juttermann R, Li E, Jaenisch R. 1994. Toxicity of 5-aza-2′-deoxycytidine to mammalian cells is mediated primarily by covalent trapping of DNA methyltransferase rather than DNA demethylation. Proc Natl Acad Sci U S A 91:11797–11801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu K, Wang YF, Cantemir C, Muller MT. 2003. Endogenous assays of DNA methyltransferases: evidence for differential activities of DNMT1, DNMT2, and DNMT3 in mammalian cells in vivo. Mol Cell Biol 23:2709–2719. doi: 10.1128/MCB.23.8.2709-2719.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schermelleh L, Spada F, Easwaran HP, Zolghadr K, Margot JB, Cardoso MC, Leonhardt H. 2005. Trapped in action: direct visualization of DNA methyltransferase activity in living cells. Nat Methods 2:751–756. doi: 10.1038/nmeth794. [DOI] [PubMed] [Google Scholar]
- 40.Khoddami V, Cairns BR. 2013. Identification of direct targets and modified bases of RNA cytosine methyltransferases. Nat Biotechnol 31:458–464. doi: 10.1038/nbt.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim SM, Kim JY, Choe NW, Cho IH, Kim JR, Kim DW, Seol JE, Lee SE, Kook H, Nam KI, Bhak YY, Seo SB. 2010. Regulation of mouse steroidogenesis by WHISTLE and JMJD1C through histone methylation balance. Nucleic Acids Res 38:6389–6403. doi: 10.1093/nar/gkq491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ding J, Xu H, Faiola F, Ma'ayan A, Wang J. 2011. Oct4 links multiple epigenetic pathways to the pluripotency network. Cell Res 22:155–167. doi: 10.1038/cr.2011.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith SM, Kimyon RS, Watters JJ. 2014. Cell-type-specific Jumonji histone demethylase gene expression in the healthy rat CNS: detection by a novel flow cytometry method. ASN Neuro 6:193–207. doi: 10.1042/AN20130050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Formosa T. 2012. The role of FACT in making and breaking nucleosomes. Biochim Biophys Acta 1819:247–255. doi: 10.1016/j.bbagrm.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Winkler DD, Luger K. 2011. The histone chaperone FACT: structural insights and mechanisms for nucleosome reorganization. J Biol Chem 286:18369–18374. doi: 10.1074/jbc.R110.180778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fujimoto M, Takaki E, Takii R, Tan K, Prakasam R, Hayashida N, Iemura S, Natsume T, Nakai A. 2012. RPA assists HSF1 access to nucleosomal DNA by recruiting histone chaperone FACT. Mol Cell 48:182–194. doi: 10.1016/j.molcel.2012.07.026. [DOI] [PubMed] [Google Scholar]
- 47.Shimojima T, Okada M, Nakayama T, Ueda H, Okawa K, Iwamatsu A, Handa H, Hirose S. 2003. Drosophila FACT contributes to Hox gene expression through physical and functional interactions with GAGA factor. Genes Dev 17:1605–1616. doi: 10.1101/gad.1086803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takahata S, Yu Y, Stillman DJ. 2009. FACT and Asf1 regulate nucleosome dynamics and coactivator binding at the HO promoter. Mol Cell 34:405–415. doi: 10.1016/j.molcel.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, Loh YH, Yeo C, Narang V, Ramamoorthy G, Leong B, Shahab A, Ruan Y, Bourque G, Sung WK, Clarke ND, Wei CL, Ng HH. 2008. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 50.Mayer A, Lidschreiber M, Siebert M, Leike K, Soding J, Cramer P. 2010. Uniform transitions of the general RNA polymerase II transcription complex. Nat Struct Mol Biol 17:1272–1278. doi: 10.1038/nsmb.1903. [DOI] [PubMed] [Google Scholar]
- 51.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. 2007. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 52.Somers A, Jean JC, Sommer CA, Omari A, Ford CC, Mills JA, Ying L, Sommer AG, Jean JM, Smith BW, Lafyatis R, Demierre MF, Weiss DJ, French DL, Gadue P, Murphy GJ, Mostoslavsky G, Kotton DN. 2010. Generation of transgene-free lung disease-specific human induced pluripotent stem cells using a single excisable lentiviral stem cell cassette. Stem Cells 28:1728–1740. doi: 10.1002/stem.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.You JS, De Carvalho DD, Dai C, Liu M, Pandiyan K, Zhou XJ, Liang G, Jones PA. 2013. SNF5 is an essential executor of epigenetic regulation during differentiation. PLoS Genet 9:e1003459. doi: 10.1371/journal.pgen.1003459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Soufi A, Donahue G, Zaret KS. 2012. Facilitators and impediments of the pluripotency reprogramming factors' initial engagement with the genome. Cell 151:994–1004. doi: 10.1016/j.cell.2012.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cloos PA, Christensen J, Agger K, Helin K. 2008. Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes Dev 22:1115–1140. doi: 10.1101/gad.1652908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Costa Y, Ding J, Theunissen TW, Faiola F, Hore TA, Shliaha PV, Fidalgo M, Saunders A, Lawrence M, Dietmann S, Das S, Levasseur DN, Li Z, Xu M, Reik W, Silva JC, Wang J. 2013. NANOG-dependent function of TET1 and TET2 in establishment of pluripotency. Nature 495:370–374. doi: 10.1038/nature11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kuroki S, Akiyoshi M, Tokura M, Miyachi H, Nakai Y, Kimura H, Shinkai Y, Tachibana M. 2013. JMJD1C, a JmjC domain-containing protein, is required for long-term maintenance of male germ cells in mice. Biol Reprod 89:93. doi: 10.1095/biolreprod.113.108597. [DOI] [PubMed] [Google Scholar]
- 58.Wang L, Yamaguchi S, Burstein MD, Terashima K, Chang K, Ng HK, Nakamura H, He Z, Doddapaneni H, Lewis L, Wang M, Suzuki T, Nishikawa R, Natsume A, Terasaka S, Dauser R, Whitehead W, Adekunle A, Sun J, Qiao Y, Marth G, Muzny DM, Gibbs RA, Leal SM, Wheeler DA, Lau CC. 2014. Novel somatic and germline mutations in intracranial germ cell tumours. Nature 511:241–245. doi: 10.1038/nature13296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Katoh M, Katoh M. 2007. Comparative integromics on JMJD1C gene encoding histone demethylase: conserved POU5F1 binding site elucidating mechanism of JMJD1C expression in undifferentiated ES cells and diffuse-type gastric cancer. Int J Oncol 31:219–223. doi: 10.3892/ijo.31.1.219. [DOI] [PubMed] [Google Scholar]
- 60.Wang J, Park JW, Drissi H, Wang X, Xu RH. 2014. Epigenetic regulation of miR-302 by JMJD1C inhibits neural differentiation of human embryonic stem cells. J Biol Chem 289:2384–2395. doi: 10.1074/jbc.M113.535799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lolis AA, Londhe P, Beggs BC, Byrum SD, Tackett AJ, Davie JK. 2013. Myogenin recruits the histone chaperone facilitates chromatin transcription (FACT) to promote nucleosome disassembly at muscle-specific genes. J Biol Chem 288:7676–7687. doi: 10.1074/jbc.M112.426718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cao S, Bendall H, Hicks GG, Nashabi A, Sakano H, Shinkai Y, Gariglio M, Oltz EM, Ruley HE. 2003. The high-mobility-group box protein SSRP1/T160 is essential for cell viability in day 3.5 mouse embryos. Mol Cell Biol 23:5301–5307. doi: 10.1128/MCB.23.15.5301-5307.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gaspar-Maia A, Alajem A, Polesso F, Sridharan R, Mason MJ, Heidersbach A, Ramalho-Santos J, McManus MT, Plath K, Meshorer E, Ramalho-Santos M. 2009. Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature 460:863–868. doi: 10.1038/nature08212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, Cheng X, Bestor TH. 2007. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jeong S, Liang G, Sharma S, Lin JC, Choi SH, Han H, Yoo CB, Egger G, Yang AS, Jones PA. 2009. Selective anchoring of DNA methyltransferases 3A and 3B to nucleosomes containing methylated DNA. Mol Cell Biol 29:5366–5376. doi: 10.1128/MCB.00484-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.