Abstract
In Aspergillus nidulans, the AcuK and AcuM transcription factors form a complex that regulates gluconeogenesis. In Aspergillus fumigatus, AcuM governs gluconeogenesis and iron acquisition in vitro and virulence in immunosuppressed mice. However, the function of AcuK was previously unknown. Through in vitro studies, we found that A. fumigatus ΔacuK single and ΔacuK ΔacuM double mutants had impaired gluconeogenesis and iron acquisition, similar to the ΔacuM mutant. Also, the ΔacuK, ΔacuM, and ΔacuK ΔacuM mutants had similar virulence defects in mice. However, the ΔacuK mutant had a milder defect in extracellular siderophore activity and induction of epithelial cell damage in vitro than did the ΔacuM mutant. Moreover, overexpression of acuM in the ΔacuK mutant altered expression of 3 genes and partially restored growth under iron-limited conditions, suggesting that AcuM can govern some genes independently of AcuK. Although the ΔacuK and ΔacuM mutants had very similar transcriptional profiles in vitro, their transcriptional profiles during murine pulmonary infection differed both from their in vitro profiles and from each other. While AcuK and AcuM governed the expression of only a few iron-responsive genes in vivo, they influenced the expression of other virulence-related genes, such as hexA and dvrA. Therefore, in A. fumigatus, while AcuK and AcuM likely function as part of the same complex, they can also function independently of each other. Furthermore, AcuK and AcuM have different target genes in vivo than in vitro, suggesting that in vivo infection stimulates unique transcriptional regulatory pathways in A. fumigatus.
INTRODUCTION
Aspergillus fumigatus causes a variety of diseases, including allergic bronchopulmonary aspergillosis, aspergilloma, invasive pulmonary aspergillosis, and hematogenously disseminated aspergillosis (1, 2). The incidence of invasive aspergillosis is rising due to the increasing number of immunocompromised patients, who are at risk for this disease (3, 4). Furthermore, because invasive aspergillosis remains difficult to diagnose and treat, this infection is still associated with significant mortality (5). One approach to developing new strategies to treat this frequently deadly infection is to target the factors that enable A. fumigatus to survive and proliferate within the host.
Iron is an essential trace element for most living organisms, including pathogenic microorganisms. Consequently, sequestration of free iron by the host is a key factor for inhibiting the virulence of microbial pathogens (6–8). Furthermore, most successful pathogens have evolved mechanisms to obtain iron while inside the host. For example, A. fumigatus acquires iron from the host by both reductive iron assimilation and secretion of siderophores (9–11). Siderophore secretion is more important for growth within the host, because A. fumigatus mutants with defects in siderophore synthesis have dramatically attenuated virulence, whereas those with defects in reductive iron assimilation do not (11–13).
Previously, we determined that the A. fumigatus AcuM Zn2Cys6 transcription factor governs both iron acquisition and gluconeogenesis (14). In vitro experiments indicated that AcuM controls the expression of genes in both the reductive iron assimilation and siderophore pathways. Also, an A. fumigatus ΔacuM deletion mutant had attenuated virulence in two different mouse models of invasive aspergillosis. The virulence defect of the ΔacuM mutant could be partially rescued by deletion of sreA, which encodes a transcription factor that normally represses expression of high-affinity iron acquisition genes. Deletion of sreA in the ΔacuM mutant restored siderophore production and iron uptake in vitro, suggesting that the virulence defect of the ΔacuM mutant may be due in part to reduced iron acquisition (14). AcuM is also present in Aspergillus nidulans, in which it governs gluconeogenesis but not iron acquisition (14, 15). In A. nidulans, AcuM forms a heterodimer with a related transcription factor, AcuK (16). This heterodimer binds DNA sequences in an interdependent manner, and deletion of either acuM or acuK results in similar transcriptional profiles and defects in gluconeogenesis (16).
The A. fumigatus genome contains an ortholog of A. nidulans acuK, Afu2g05830, but its biological function has not been investigated previously. In this study, we compared the role of A. fumigatus AcuK with that of AcuM in governing iron acquisition, pulmonary epithelial cell interactions, and virulence. In addition, we analyzed the downstream genes of these two transcription factors during in vitro growth and in the mouse model of invasive aspergillosis. The results of these investigations indicate that in A. fumigatus, while AcuK and AcuM likely function as part of the same complex, they also function independently of each other to regulate distinct target genes. In addition, both AcuK and AcuM have different target genes in vitro than they do in vivo. Furthermore, during A. fumigatus infection in mice, these transcription factors govern the expression of a limited number of genes involved in iron acquisition and a larger number of other virulence-related genes.
MATERIALS AND METHODS
Fungal strains and growth conditions.
The A. fumigatus strains used in the experiments are listed in Table 1. All fungal strains were maintained on Sabouraud dextrose agar (Difco) at 37°C for 5 to 7 days. Conidia were harvested by gently rinsing the colony surface with phosphate-buffered saline (PBS; Sigma-Aldrich) containing 0.1% Tween 80 (Sigma-Aldrich). The conidia in the resulting suspension were enumerated using a hemacytometer.
TABLE 1.
A. fumigatus strains used in the current study
| Strain | Genotype | Reference |
|---|---|---|
| Af293 | Wild type | 41 |
| Δacuk mutant | acuK::hph | This work |
| Δacuk+acuK mutant | acuK::hph + acuK | This work |
| ΔacuK gpdA-acuM-4 mutant | acuK::hph + pgpdA-acuM | This work |
| ΔacuK gpdA-acuM-5 mutant | acuK::hph + pgpdA-acuM | This work |
| ΔacuK gpdA-acuM-7 mutant | acuK::hph + pgpdA-acuM | This work |
| ΔacuM mutant | acuM::hph | 14 |
| ΔacuK ΔacuM mutant | acuK::ble acuM::hph | This work |
| Δuge3 mutant | uge3::hph | 30 |
Strain construction.
The ΔacuK mutant was generated by replacing the entire protein coding region of acuK (Afu2g05830) with the hph hygromycin resistance gene using the split-marker approach as described previously (14, 17). To generate the 5′ portion of the deletion cassette, 1,504 bp of the acuK upstream flanking sequence was amplified from A. fumigatus Af293 genomic DNA by high-fidelity PCR using primers AcuK-F2 and AcuK-F1. The sequences of all PCR primers used for strain construction are listed in Table S1 in the supplemental material. Next, the 3′ region of the hph resistance marker was amplified from plasmid pAN7-1 (18) using primers HYG-F and YG. The acuK upstream flanking sequence was combined with the 3′ portion of the resistance cassette by fusion PCR using primers AcuK-F1 and YG. To generate the 3′ portion of the deletion cassette, a DNA fragment containing 1,299 bp downstream of the acuK open reading frame was PCR amplified using primers AcuK-F4 and AcuK-F3. The resulting fragment was cloned into the XhoI-HindIII sites of plasmid pNLC106 (19), which contained the 5′ portion of the hph hygromycin resistance cassette, to yield plasmid pAcuK-HY. Next, a DNA fragment containing the downstream acuK flanking region and the 5′ portion of the hygromycin resistance marker was amplified by high-fidelity PCR from pAcuK-HY using primers AcuK-F4 and HY. Protoplasts of A. fumigatus Af293 were transformed with the 2 fragments containing the flanking sequences of acuK (20). The resulting hygromycin-resistant transformants were screened for deletion of acuK by whole-cell PCR.
A similar approach was used to delete acuK in the ΔacuM mutant, except that the ble phleomycin resistance cassette was used. To generate the 5′ portion of the deletion cassette, 1,554 bp of upstream flanking sequence was amplified using primers AcuK-P3 and AcuK-P4. Next, the 3′ portion of the phleomycin resistance cassette was amplified from plasmid p402 (21), which contained the ble phleomycin resistance gene, using primers LE and LE-R. Then the upstream flanking sequence was combined with the 3′ portion of the phleomycin resistance cassette by fusion PCR with primers AcuK-P4 and LE. The Gateway cloning system (Invitrogen) was used to construct the 3′ portion of the deletion cassette. Using high-fidelity PCR with primers AcuK-P1 and AcuK-P2, 1,299 bp of downstream flanking sequence was amplified. This fragment was cloned into the entry vector pENTR/D-TOPO and then transferred into the pBL vector according to the manufacturer's instructions (Invitrogen). A DNA fragment containing the downstream flanking sequence of acuK and the 5′ phleomycin cassette was amplified using primers AcuK-P1 and BL. Next, the two fragments containing the acuK flanking sequence and phleomycin resistance sequences were used to transform the ΔacuM mutant. The resulting phleomycin resistant colonies were screened by whole-cell PCR for the deletion of acuK.
To construct the ΔacuK+acuK complemented strain, a DNA fragment containing the acuK open reading frame along with 2,081 bp of upstream sequence and 537 bp of downstream sequences was amplified from Af293 genomic DNA by high-fidelity PCR using primers AcuK-ComF and AcuK-ComR. The resulting fragment was first cloned into pGEM-T Easy (Promega), then excised with XbaI/NotI, and ligated into plasmid p402. The resulting plasmid was used to transform the ΔacuK mutant. Next, whole-cell PCR with primers AcuK-ComF and AcuK-ComR was used to identify phleomycin-resistant clones that contained a wild-type allele of acuK.
ΔacuK strains that overexpressed acuM were constructed by transformation with a plasmid in which the expression of acuM was driven by the A. nidulans gpdA promoter. To construct this plasmid, 2,294 bp of the acuM open reading frame was PCR amplified from genomic DNA of A. fumigatus Af293 using primers AcuM-OE-F and AcuM-OE-R (see Table S1 in the supplemental material). The resulting fragment was cloned into the NcoI-SpeI sites of plasmid pGFP-Phleo (22), which was used to transform the ΔacuK mutant. Next, the acuM transcript levels in the phleomycin-resistant clones were determined by real-time PCR as described below.
RNA isolation and real time-PCR.
For RNA isolation, 50 ml of RPMI 1640 medium (Sigma-Aldrich) buffered to pH 7.0 with 4-morpholinepropanesulfonic acid was inoculated with conidia of the various A. fumigatus strains to achieve a final concentration of 5 × 105 conidia per ml. To obtain RNA for in vitro NanoString analysis under iron-limited conditions, the various strains were used to inoculate Aspergillus minimal medium (AMM) containing a 300 μM concentration of the iron chelator ferrozine (Sigma-Aldrich) (14). After incubation for 24 h in a shaking incubator at 37°C, the fungal cells were harvested by filtration. The total RNA was isolated using an RNeasy plant minikit (Qiagen), with minor modifications. The hyphae were added to an RNase-free 2-ml tube containing 500 μl of RLC from the kit and a mixture of 1/4-in. ceramic spheres and 0.1-mm silica spheres (MP Biomedicals). After being disrupted with a FastPrep FP120 beat beater (Thermo Electric Corp.), the samples were processed by following the manufacturer's instructions.
Fungal RNA was extracted from infected mouse lungs using the RNeasy minikit (Qiagen), with modifications. Approximately 2.4 ml of buffer RLT with 1% β-mercaptoethanol was added to each lung and the tissue was homogenized in an M tube (Miltenyi Biotec) using a gentleMACS dissociator (Miltenyi Biotec) on setting RNA_02.01. Next, the homogenate was mixed with an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1) and a half volume of zirconium beads (Ambion) and then vortexed with a Mini-Beadbeater (Biospec Products) for 3 min. After centrifugation, the aqueous phase was collected and mixed with an equal volume of 70% ethanol. The RNA was isolated from this mixture using an RNeasy spin column (Qiagen) by following the manufacturer's instructions.
For real-time PCR, RNA was reverse transcribed into cDNA using Moloney murine leukemia virus (MMLV) reverse transcriptase (Promega). Real-time PCR was performed by using the POWER SYBR green PCR master mix (Applied Biosystems) as previously described (14). The relative transcript level of each gene was normalized to tef1 or gpdA by the threshold cycle (2−ΔΔCT) method. The two control genes gave similar results. The primers used for real-time PCR are listed in Table S1 in the supplemental material.
Effects of different carbon sources on growth.
To investigate the growth of the different strains on different carbon sources, AMM agar was prepared without glucose and then supplemented with 0.5% ethanol, 50 mM acetate, 50 mM l-proline, or 1% glucose. Next, serial 10-fold dilutions of conidia in 5 μl of PBS were spotted onto the agar plates. After incubation at 37°C for 2 days, the plates were imaged for analysis of fungal growth. Each experiment was repeated three times.
Growth under iron-limited and iron-replete conditions.
To determine the capacity of the different strains to grow under iron limited conditions, conidia of the various A. fumigatus strains were added Sabouraud dextrose broth containing 30 μM phenanthroline (Sigma-Aldrich) at a final concentration of 2 × 106 conidia per ml. As a control, the organisms were also grown in the Sabouraud dextrose broth containing phenanthroline plus 1 mM FeSO4. After incubation in the dark at 37°C in a shaking incubator for 40 h, the mycelia were harvested, dried at 90 to 100°C for 5 days, and then weighed. Each A. fumigatus strain was tested in triplicate in 3 independent experiments.
Evaluation of extracellular siderophore production and iron uptake.
To measure the total extracellular siderophore activity of each strain, the chrome azurol S (CAS) colorimetric method was used as described previously (14). Briefly, 3 × 108 conidia of each fungal strain were inoculated in AMM with reduced phosphate (0.03% KH2PO4) and 2% glucose and incubated at 37°C for 40 h. After the resulting hyphae were harvested by filtration, their dry weight was determined. The siderophore activity in the culture filtrate was measured by adding 0.15 mM chrome azurol S, 0.015 mM FeCl3, 1.5 mM hexadecyl trimethyl ammonium bromide (HDTMA), and 1 M piperazine (pH 5.6) and determining the absorbance at 630 nm. Ferrichrome (Sigma-Aldrich) was used to generate a standard curve. Using this standard curve, the siderophore production of each fungal strain was calculated in terms of micromoles per gram (hyphal dry weight). To determine the capacity of each A. fumigatus strain to incorporate iron from the medium, a radiometric assay using 55FeCl3 was performed exactly as described previously (14, 23). Each A. fumigatus strain was tested in triplicate in 3 independent experiments.
Susceptibility to stressors.
To determine susceptibility to environmental stress, serial 10-fold dilutions of conidia from the different A. fumigatus strains ranging from 105 to 102 cells were spotted onto Sabauroud agar plates containing 300 μg/ml of calcofluor white, 200 μg/ml of Congo red, 7.5 mM Ca2+, 7.5 mM Mn2+, 7.5 mM Cu2+, 7.5 mM Zn2+, 200 mM Ca2+, 3 mM NaCl, 3 mM H2O2, or 0.01% SDS. After incubation of the plates at 37°C for 48 h, the growth of the various strains was analyzed.
Detection of surface-exposed galactosaminogalactan.
To detect the surface-exposed galactosaminogalactan, the strains were stained with fluorescein isothiocyanate (FITC)-labeled Wisteria floribunda lectin (WFL). A total of 106 conidia of each of the various strains in Dulbecco's modified Eagle's medium were added to 6-well plates and incubated at 37°C in 5% CO2 for 9 h. Next, the medium was replaced with 500 μl of PBS, after which the young hyphae were removed from the wells with a cell scraper. The hyphae were transferred to a 24-well plate containing BioCoat Cellware poly-d-lysine 12-mm round coverslips (BD Biosciences) and centrifuged at 216 × g for 10 min. After the samples were stained with FITC-WFL (Vector Laboratories) for 2 h at 4°C, they were washed twice with PBS and then fixed with 4% (wt/vol) paraformaldehyde at 4°C for 10 min. The coverslips were then mounted inverted onto glass slides and imaged by confocal microscopy.
Aniline blue staining to detect β-glucan.
For aniline blue staining, 2.5 × 105 conidia of each strain in Dulbecco's modified Eagle's medium were added to 24-well plates containing BioCoat Cellware poly-d-lysine 12-mm round coverslips (BD Biosciences). The plates were centrifuged at 216 × g for 10 min and incubated in 5% CO2 at 37°C for 9 h. Next, the wells were washed twice with 500 μl of PBS and then fixed with 4% (wt/vol) paraformaldehyde at 4°C for 10 min. The hyphae were washed twice with PBS and then stained with 0.05% aniline blue (Sigma-Aldrich) in PBS, pH 9.5, for 5 min at 4°C. The stain was aspirated, and coverslips were then mounted inverted onto glass slides and imaged by confocal microscopy.
Pulmonary epithelial cell damage assay.
The A549 type II pneumocyte line was purchased from the American Type Culture Collection (ATCC) and grown in F-12 K medium (ATCC) containing 10% fetal bovine serum (Gemini Bio-Products) and streptomycin and penicillin (Irvine Scientific) in 5% CO2 at 37°C. The extent of epithelial cell damage caused by the different strains was measured using our standard 51Cr release assay (24). A549 epithelial cell were grown to 95% confluence in a 24-well tissue culture plate and then labeled with 51Cr (ICN Biomedicals) overnight. The following day, the unincorporated 51Cr was removed and the cells were infected with 5 × 105 conidia in 1 ml of F-12 K medium per well. After incubation at 37°C in 5% CO2 for 20 h, the medium above the cells was collected. Next, the cells were lysed with 6 N NaOH and the wells were rinsed with Radiac wash (Biodex Medical Systems, Inc.). The lysate and rinses were combined, and the amount of 51Cr in the samples was determined by gamma counting. To measure the spontaneous release of 51Cr, uninfected A549 cells exposed to medium alone were processed in parallel. After adjusting for well-to-well differences in the incorporation of 51Cr, the percent specific release of 51Cr was calculated using the following formula: (experimental release − spontaneous release)/(total incorporation − spontaneous release). Each experiment was performed in triplicate three different times.
Mouse model of invasive pulmonary aspergillosis.
The virulence of the different A. fumigatus strains was determined in the corticosteroid-treated mouse model of invasive pulmonary aspergillosis (14, 24–26). Briefly, male BALB/c mice (Taconic Farms) were immunosuppressed with 10 mg of cortisone acetate administered subcutaneously every other day for 5 doses beginning at day −4 relative to infection. To infect the mice, 11 or 12 animals per strain were placed into an acrylic chamber, and 12 ml of a suspension containing 109 conidia per ml of PBS containing 0.1% Tween 80 was aerosolized into the chamber for 1 h. Control mice were immunosuppressed but not infected. Immediately after infection, 3 mice per group were sacrificed, after which the lungs were harvested, homogenized, and plated onto Sabouraud dextrose agar to determine the fungal inoculum, which was a mean of 3,131 ± 1,589 organisms per mouse and did not differ significantly among the various strains. The remaining mice (7 or 8 mice/group) were monitored for survival for 2 weeks.
To obtain infected lungs for NanoString analysis, 3 mice were inoculated as described above and then sacrificed after 4 days of infection. The lungs were harvested, immediately snap-frozen in liquid nitrogen, and stored at −80°C until RNA extraction. The animal studies were approved by the Institutional Animal Use and Care Committee at the Los Angeles Biomedical Research Institute and performed according to the National Institutes of Health guidelines for animal housing and care.
NanoString analysis.
For each NanoString assay, 10 μg of total tissue RNA isolated from a mouse lung or 100 ng of A. fumigatus RNA isolated from an in vitro culture was mixed with a NanoString codeset mix and incubated at 65°C overnight. The reaction mixes were loaded onto the NanoString nCounter Prep Station for binding and washing, and the resultant cartridge was transferred to the NanoString nCounter digital analyzer for scanning and data collection. A total of 600 fields were captured per sample. The raw data were first adjusted for binding efficiency and background subtraction following nCounter data analysis guidelines. To calculate gene expression ratios among different samples, we normalized adjusted raw counts using gpdA as the internal control gene. All expression ratios were calculated using mean values of three independent biological samples, and statistical significance was determined by the two-tailed Student t test (n = 3, P < 0.05, unless specified otherwise). A gene was considered to have a biologically significant difference in expression if the magnitude of the difference was at least 2-fold and the difference was statistically significant by the Student t test. The hierarchical clustering heat maps were generated using Multiexperimental Viewer 4.9.0 (27).
Statistical analysis.
Differences among the in vitro experimental groups were assessed using the Student t test. The survival data were analyzed by the log rank test. P values of ≤0.05 were considered significant.
RESULTS
AcuK function in growth.
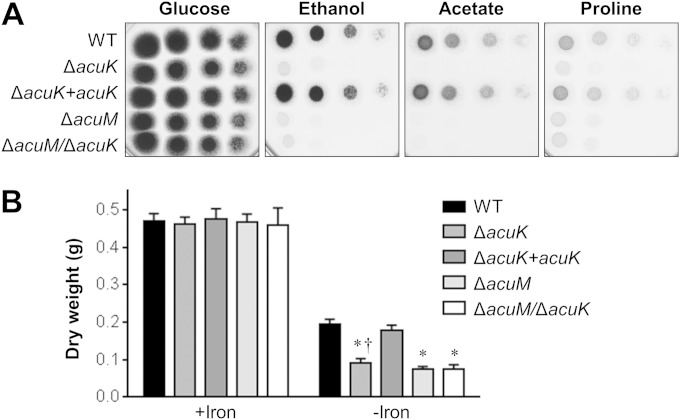
To analyze the function of acuK, we constructed an A. fumigatus ΔacuK single deletion mutant and a ΔacuK ΔacuM double mutant (Table 1). Using real-time PCR, we verified the absence of acuK transcript in the ΔacuK mutant and determined that deletion of acuK did not result in compensatory upregulation of acuM and vice versa (see Fig. S1 in the supplemental material). Previously, we determined that an A. fumigatus ΔacuM mutant was unable to grow on gluconeogenic carbon sources and had impaired growth under iron-limited conditions (14). Therefore, we compared the growth of ΔacuK and ΔacuM single mutants and an ΔacuK ΔacuM double mutant under these conditions. All three mutants grew very poorly on media in which ethanol, acetate, or proline was the carbon source (Fig. 1A). Notably, the growth defect of the ΔacuK mutant on these media was rescued by reintegration of an intact copy of acuK. Therefore, acuK, like acuM, is necessary for growth on gluconeogenic carbon sources.
FIG 1.
Effects of deletion of acuK and acuM on growth on gluconeogenic substrates and under iron-limited conditions. (A) Serial 10-fold dilutions of the indicated strains were plated on agar containing glucose, ethanol, acetate, or proline as the carbon source. The plates were imaged after incubation at 37°C for 48 h. (B) Dry weight of the indicated strains after 48 h of growth at 37°C in Sabouraud dextrose broth containing either 30 mM phenanthroline plus 1 mM FeCl2 (+Iron) or phenanthroline without FeCl2 (−Iron). Results are means ± SDs from 3 independent experiments, each performed in triplicate. *, P < 0.0001 compared to the wild-type strain (WT) grown under iron limitation; †, P < 0.0001 compared to the ΔacuK+acuK complemented strain grown under iron limitation.
Next, we determined the capacity of the various mutants to grow on medium in which the iron had been chelated with phenanthroline. Both the ΔacuK and ΔacuM single mutants had significantly reduced growth under iron-limited conditions compared to that of the wild-type strain (Fig. 1B). The growth defects of the ΔacuK and ΔacuM single mutants were similar, both to each other and to those of the ΔacuK ΔacuM double mutant. These results suggest that AcuK and AcuM function in the same pathway in governing growth under iron-limited conditions.
Dependence of siderophore synthesis and iron uptake on AcuK.
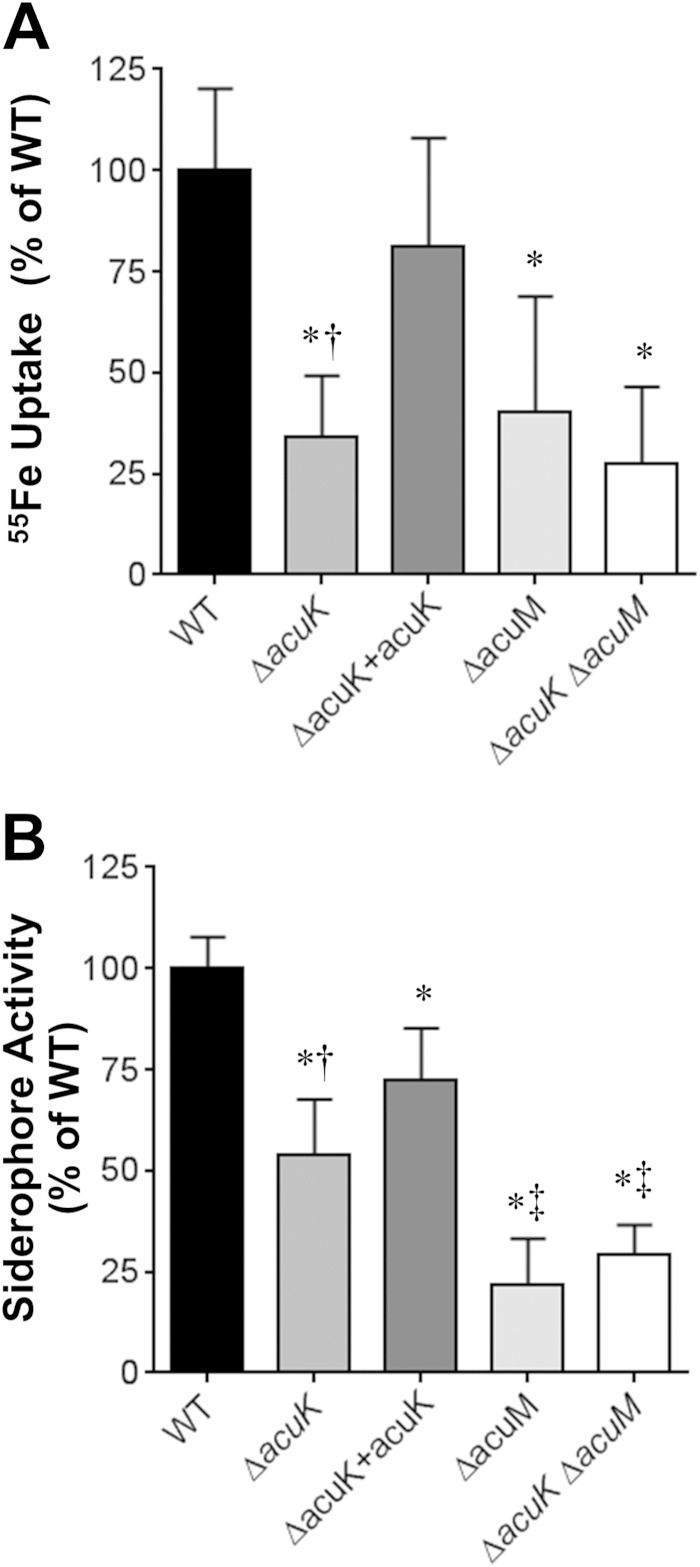
To investigate the cause of the growth defects of the different mutants under iron-limited conditions, we analyzed their iron uptake and production of extracellular siderophores. Compared to that of the wild-type strain, washed germlings of the ΔacuK mutant had significantly reduced capacity to incorporate 55FeCl3 from the medium (Fig. 2A). This defect in iron incorporation was similar to that of both the ΔacuM single mutant and the ΔacuK ΔacuM double mutant, and it was reversed when an intact copy of acuK was integrated into the ΔacuK mutant. Because the germlings were washed prior to being tested in the iron incorporation assay, the defective iron uptake of these mutants was likely the result of impaired reductive iron assimilation rather than reduced extracellular siderophore production.
FIG 2.
Roles of AcuK and AcuM in iron uptake and extracellular siderophore activity. (A) Relative incorporation of 55Fe after 1 h by washed germlings of the indicated strains. (B) Extracellular siderophore activity of the indicated A. fumigatus strains measured after 40 h of incubation. Results are means ± SDs from 3 biological replicates, each tested in triplicate. *, P < 0.01 compared to the wild-type strain; †, P < 0.04 compared to the ΔacuK+acuK complemented strain; ‡, P < 0.01 compared to the ΔacuK mutant.
Using the chrome azurol S assay (28), we determined that the ΔacuK mutant had a 46% reduction in extracellular siderophore activity (Fig. 2B). Interestingly, the extracellular siderophore activity of the ΔacuK mutant remained significantly greater than that of the ΔacuM mutant. Furthermore, the extracellular siderophore activity of the ΔacuK ΔacuM double mutant was similar to that of the ΔacuM mutant. While these results support the model that AcuK and AcuM function together as a heterodimer to govern iron acquisition and extracellular siderophore production, the greater defect in extracellular siderophore activity of the ΔacuM mutant suggests that AcuM may also function independently of AcuK.
Cell wall integrity and composition of the ΔacuK and ΔacuM mutants.
A. fumigatus AcuK and AcuM share 24% and 55% amino acid homology, respectively, with Candida albicans Cwt1, a Zn2Cys6 transcription factor that governs cell wall composition. The cell wall of a C. albicans cwt1Δ/Δ mutant has reduced β-glucan and increased mannan content, which cause this mutant to have increased susceptibility to the cell wall stressors Congo red, calcofluor white, and SDS (29). Therefore, we investigated whether AcuK or AcuM influences the cell wall composition of A. fumigatus. Using an agar dilution assay, we found that the ΔacuK, ΔacuM, and ΔacuK ΔacuM mutants displayed wild-type susceptibility to multiple stressors, including Congo red, SDS, caspofungin, divalent cations, and H2O2 (data not shown). In addition, we stained the surface of these mutants with FITC-labeled WFL to detect surface-exposed galactosaminogalactan, a glycan that plays a key role in mediating adherence to host constituents and masking fungal β-glucans (30). The staining of all three mutants was similar to that of the wild-type strain (see Fig. S2 in the supplemental material). The three mutants also had wild-type distribution of β-glucan in their cell walls, as determined by aniline blue staining (see Fig. S3 in the supplemental material). Collectively, these results indicate that AcuK and AcuM do not play a role in governing the composition of the A. fumigatus cell wall.
Impact of AcuK and AcuM on A. fumigatus damage to host cells.
Previously, we have found that the capacity of A. fumigatus mutants to damage host cells in vitro is predictive of their virulence in immunosuppressed mouse models of invasive pulmonary aspergillosis (24, 26, 31). Therefore, we assessed the capacity of the ΔacuK and ΔacuM single mutants and the ΔacuK ΔacuM double mutant to damage the A549 pulmonary epithelial cell line. All three mutants caused significantly less epithelial cell damage than the wild-type strain (Fig. 3). However, the ΔacuK mutant caused only 22% less damage than the wild-type strain, whereas both the ΔacuM and ΔacuK ΔacuM mutants caused approximately 34% less damage. Therefore, while AcuK and AcuM are both required for A. fumigatus to induce maximal epithelial cell damage, AcuM may play a greater role in stimulating this process. Again, these results suggest that AcuM has additional target genes that are not governed by the AcuK-AcuM heterodimer.
FIG 3.
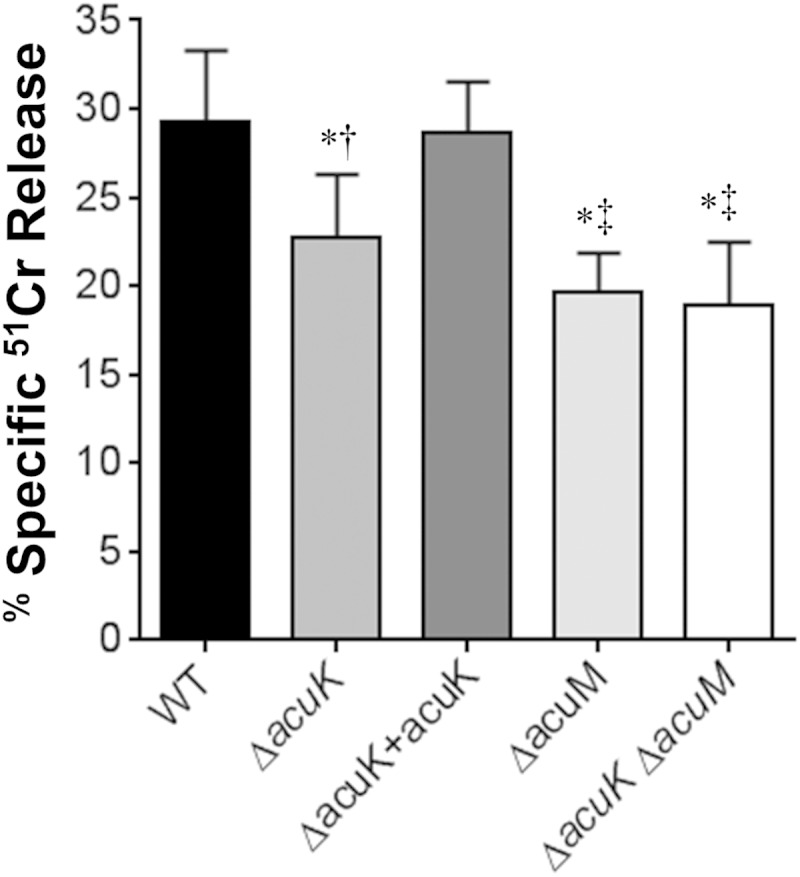
Effects of deletion of acuK and acuM on induction of pulmonary epithelial cell damage. The A549 pulmonary epithelial cell line was infected with the indicated strains of A. fumigatus for 20 h, after which the extent of epithelial cell damage was assessed by a 51Cr release assay. Results are means ± SDs from 3 independent experiments, each performed in triplicate. *, P < 0.01 compared to the wild-type strain; †, P < 0.01 compared to the ΔacuK+acuK complemented strain; ‡, P < 0.04 compared to the ΔacuK mutant.
AcuK function in virulence.
The impaired capacity of the ΔacuK mutant to grow under iron-limited conditions and induce epithelial cell damage suggested that this strain might have attenuated virulence. To test this hypothesis, we compared the virulence of the ΔacuK and ΔacuM mutants in a nonneutropenic mouse model of invasive aspergillosis. Mice infected with the ΔacuK mutant survived significantly longer than mice infected with either the wild-type strain or the ΔacuK+acuK complemented strain (Fig. 4). Consistent with our previous results (14), mice infected with the ΔacuM mutant also had prolonged survival, which was similar to that of mice infected with the ΔacuK mutant. In addition, while mice infected with the Δacuk ΔacuM double mutant survived longer than mice infected with the wild-type strain, their survival was similar to that of mice infected with the ΔacuK single mutant (Fig. 4B). These data indicate that both AcuK and AcuM are required for maximal virulence in the nonneutropenic mouse model of invasive pulmonary aspergillosis and that the simultaneous deletion of both AcuK and AcuM does not result in further attenuation in virulence.
FIG 4.
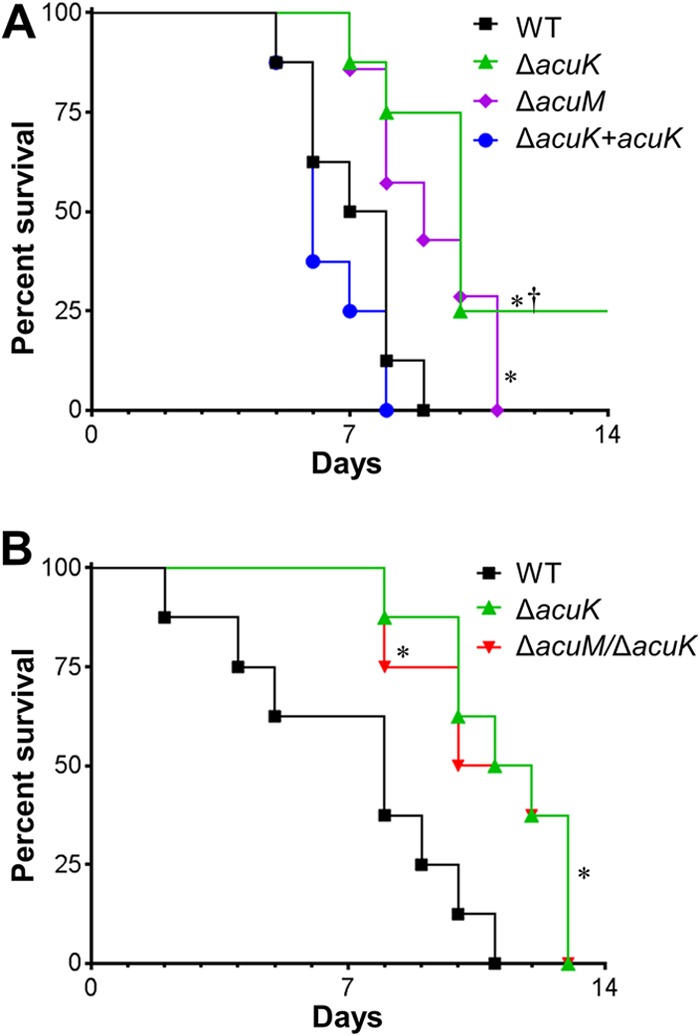
Deletion of acuK and/or acuM attenuates virulence in the mouse model of invasive pulmonary aspergillosis. Shown are survival times of mice that were immunosuppressed with cortisone acetate and then infected with the indicated A. fumigatus strains in an aerosol chamber. Results in both panels are from independent experiments using 7 or 8 mice per strain. *, P ≤ 0.02 compared to the wild-type strain; †, P < 0.001 compared to the ΔacuK+acuK complemented strain.
Analysis of AcuK- and AcuM-responsive gene expression.
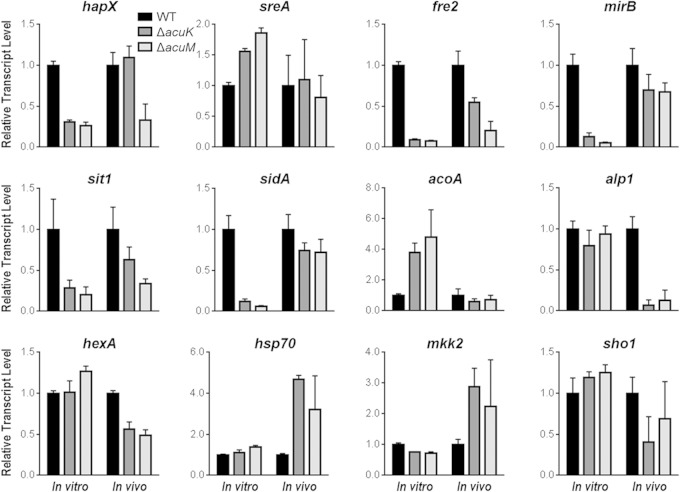
To gain insight into the downstream target genes regulated by AcuK and AcuM, we used real-time PCR to assay expression of a panel of 12 genes in the wild-type, ΔacuK, and ΔacuM strains. We selected genes related to carbon metabolism (acoA), iron acquisition (hapX, mirB, fre2, sidA, sit1, and sreA), cell wall and secretome function (alp1 and mkk2), and stress responses (hexA, hsp70, and sho1). Transcript levels, normalized to gpdA transcript levels to correct for differences in amounts of fungal mRNA, were analyzed under two growth conditions: (i) after 24 h of growth at 37°C in liquid Aspergillus minimal medium containing the iron chelator ferrozine (“in vitro” in Fig. 5) and (ii) after 4 days of pulmonary infection in mice (“in vivo” in Fig. 5). The data support three major conclusions (Fig. 5; see also Table S2 in the supplemental material). First, AcuK and AcuM govern expression of numerous carbon and iron metabolic genes, as we expected from our previous ΔacuM profiling study (14). Second, the gene expression defects are similar in the two mutants, as expected if AcuK and AcuM indeed function as a heterodimer like their A. nidulans orthologs (16). Third, the gene expression alterations manifested by each mutant under in vitro growth conditions are generally not recapitulated during infection. This result was unexpected, because the only study to date of A. fumigatus in which in vitro profiles were compared to invasive infection profiles found good agreement between mutant phenotypes in vitro and in vivo (32).
FIG 5.
Real-time PCR measurement of transcript levels in the ΔacuK and ΔacuM mutants during in vitro and in vivo growth. RNA levels of the indicated genes in the wild-type strain and ΔacuK and ΔacuM mutants were measured by real-time PCR using three independent cultures grown in vitro or from the lungs of 3 infected mice (in vivo). The RNA levels for each gene were normalized to internal control gpdA RNA levels and are presented as ratios to the mean RNA level for each gene in the wild-type strain.
To verify and extend these findings, we used a NanoString gene expression measurement platform to analyze the transcript levels of 97 genes in the same samples. As for the real time-PCR data, transcript levels in each sample were normalized to gpdA transcript levels. Transcript levels for 71 of the 97 genes were detectable in at least 11 of 12 samples and were thus included in the analysis.
There was overall good concordance between the real-time PCR and NanoString results (R2 = 0.68), with better agreement among in vitro samples than in vivo samples, as expected from the higher levels of fungal RNA in our in vitro samples (Fig. 5; see also Table S2 in the supplemental material).
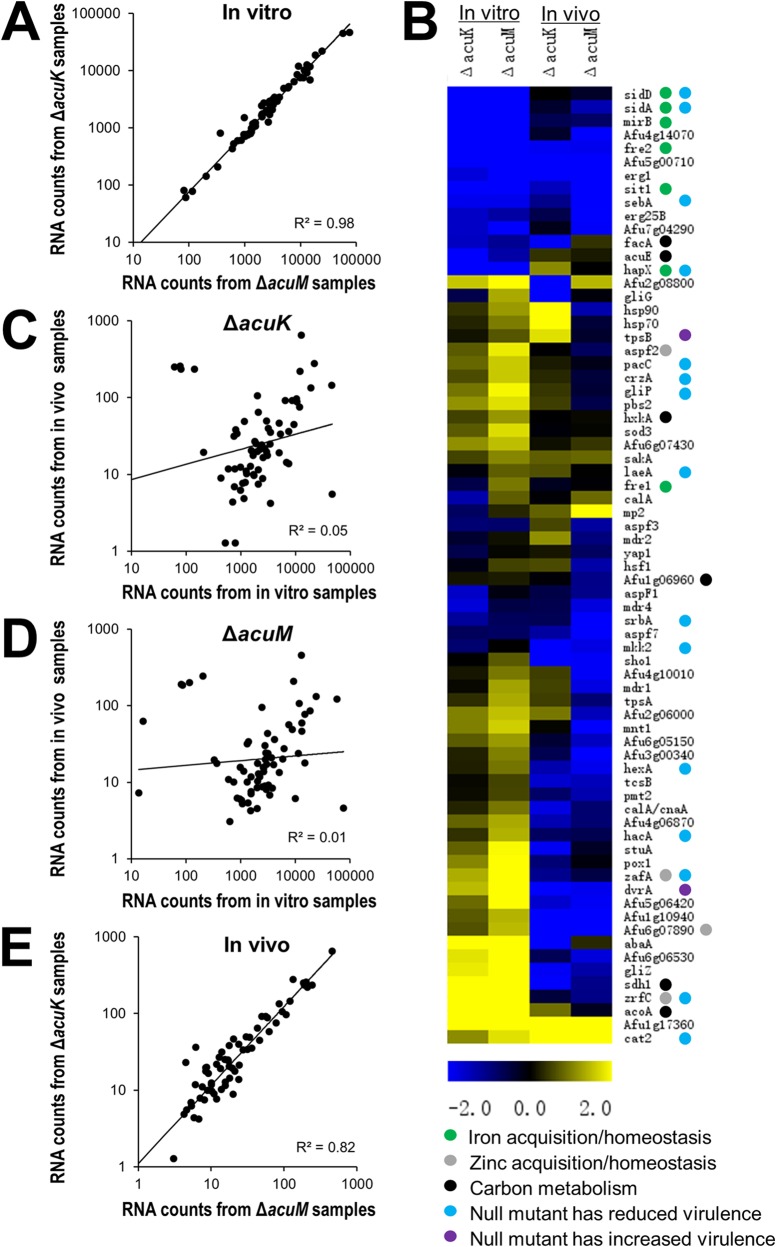
When grown in vitro, the ΔacuK and ΔacuM mutants had very similar transcriptional profiles (R2 = 0.98 [Fig. 6A]). Both mutants had strong downregulation of fre2, hapX, mirB, sidA, sidD, and sit1, which are iron responsive and/or involved in iron acquisition (Fig. 5 and 6B; see also Table S2 in the supplemental material). In addition, there was upregulation of acoA and sdh1, which specify carbon metabolism enzymes, and zafA and zrfC, which are involved in zinc acquisition. These results are similar to those obtained in our previous microarray analysis of the ΔacuM mutant (14). Thus, during in vitro growth, AcuK and AcuM govern the expression of many of the same genes, including those required for growth under iron-limited conditions.
FIG 6.
In vitro and in vivo transcriptional profiles of the ΔacuK and ΔacuM mutants. NanoString measurements were conducted on the indicated strains using three independent cultures grown in vitro or from the lungs of 3 infected mice (in vivo). The RNA levels for each gene were normalized to the internal control, gpdA. (A) Relative RNA levels in the ΔacuK and ΔacuM mutants during growth in vitro. Each symbol represents the mean mRNA level for a different gene, based on 3 biological replicates. (B) Heat map showing the relative expression ratios of the indicated genes in the A. fumigatus mutants compared to the wild-type strain grown under the same conditions. Upregulation is represented by yellow, and downregulation is represented by blue; the magnitude of regulation is reflected by color saturation. Colored circles next to each gene name indicate the known function of its product and the effects of deletion of that gene on virulence. Data are from the Aspergillus Genome Database (http://www.aspgd.org/). (C) Comparison of RNA levels of the ΔacuK mutants during growth in vitro and in vivo. (D) Comparison of RNA levels of the ΔacuM mutants during growth in vitro and in vivo. (E) Comparison of RNA levels of the ΔacuK and ΔacuM mutants during growth in vivo.
We also found that the ΔacuK and ΔacuM mutants had more divergent transcription profiles during growth in vivo (R2 = 0.82 [Fig. 6B to E]). For example, in the in vivo samples, expression of abaA (transcriptional regulator of conidiation) was dependent on AcuK but not AcuM, whereas expression of mnt1 (mannosyltransferase) was dependent on AcuM but not AcuK. Some of this divergence may have been due to variability as a result of the lower number of A. fumigatus transcripts measured in the mouse samples than in the in vitro samples. However, the phenotypic analysis also demonstrated some differences between the ΔacuK and ΔacuM mutants. Therefore, the aggregate data indicate that although AcuK and AcuM likely function as a heterodimer, each protein may also function independently of the other.
During pulmonary infection, the spectrum of AcuK- and AcuM-responsive genes was markedly different from that seen in vitro (Fig. 5 and 6). Although some core genes, such as Afu1g17360, Afu5g00710, erg1, and fre2, were AcuK and AcuM responsive both in vitro and in vivo, many genes were AcuK and AcuM responsive under only one of these conditions. For example, although the iron-related genes mirB, sidA, and sidD were downregulated in both mutants in vitro, they were expressed at near-wild-type levels in vivo. Similarly, acoA, sdh1, zafA, and zrfC were not upregulated in the ΔacuK or ΔacuM mutants in vivo. In contrast, Afu1g10940 (putative MAPKK), Afu6g07890 (zinc transporter), and alp1 (secreted alkaline serine protease) were only downregulated in vivo. Moreover, some genes that are known to impact virulence, including hexA (woronin body) (33) and dvrA (cell wall regulator) (24), had reduced expression only in the ΔacuK and ΔacuM mutants in vivo. These results provide strong evidence that AcuK and AcuM govern the expression of different genes when A. fumigatus grows in the lung than when grown in vitro. Furthermore, during the growth of A. fumigatus in vivo, AcuK and AcuM govern the expression of a much smaller subset of iron-responsive genes and a larger subset of genes that function in other virulence pathways. Thus, the virulence defects of these mutants may be due to a combination of impaired iron acquisition and dysregulation of additional virulence factors.
Evidence for AcuK-independent AcuM function.
Many of the results presented above indicate that the ΔacuM mutant has more severe defects than the ΔacuK mutant. We considered two models to explain those observations. The first model is that AcuM can function in the absence of AcuK to activate expression of many of the same genes that the AcuM-AcuK heterodimer activates. AcuM may do this poorly compared to AcuM-AcuK, and for that reason the ΔacuK mutant has prominent phenotypes. The second model is that AcuM can function in the absence of AcuK to activate different target genes from the AcuM-AcuK heterodimer, but these novel AcuM target genes have convergent functions with the AcuM-AcuK target genes. The novel AcuM target genes may not be completely functionally redundant with AcuM-AcuK target genes, and for that reason the ΔacuK mutant has prominent phenotypes.
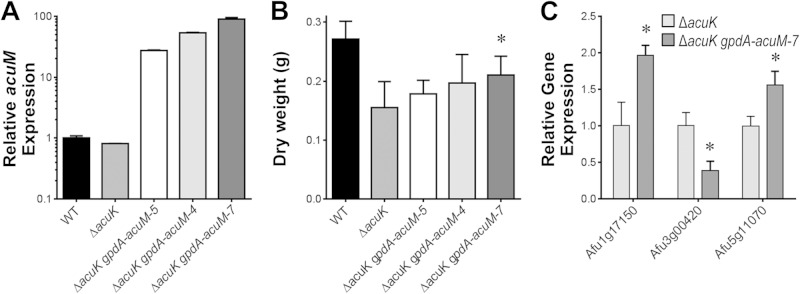
To test those models, we compared the phenotype of an ΔacuK mutant with those of ΔacuK mutants that overexpress acuM from the strong A. nidulans gpdA promoter (ΔacuK gpdA-acuM strains). We examined three ΔacuK gpdA-acuM strains in which acuM transcript levels ranged from 28-fold to 97-fold greater than in the wild-type strain (Fig. 7A). We tested the capacity of the acuM overexpression strains to grow on gluconeogenic carbon sources, damage A549 pulmonary epithelial cells, and grow under iron-limited conditions. None of the acuM overexpression strains were able to grow on gluconeogenic carbon sources, and they had the same epithelial cell damage defect as the ΔacuK parent strain (see Fig. S4 in the supplemental material). However, overexpression of acuM partially rescued the growth defect of the ΔacuK mutant under iron-limited conditions (Fig. 7B). In fact, the extent of growth in the various strains was proportional to the extent of acuM expression. Collectively, these results indicate that AcuM is able to function independently of AcuK to promote iron acquisition.
FIG 7.
Growth and gene expression profile of acuM overexpression strains under iron-limited conditions. (A) Relative acuM transcript levels in the indicated strains. Results are means ± SDs from 3 biological replicates, each performed in duplicate. (B) Dry weight of the indicated strains after growth for 48 h at 37°C in Sabouraud dextrose broth containing 30 mM phenanthroline. Results are means ± SDs from 3 independent experiments, each performed in duplicate. (C) Transcript levels of genes that were found to have significantly different expression in the ΔacuK gpdA-acum-7 overexpression strain compared to that in the ΔacuK mutant. Results are means ± SDs from NanoString analysis performed on 3 biological replicates. *, P < 0.03 compared to the ΔacuK strain.
In order to determine whether AcuM alone may activate the same genes as AcuM-AcuK, we examined RNA levels for the 95 genes whose expression was detected by our NanoString probes. We compared gene expression levels in the ΔacuK mutant and one ΔacuK gpdA-acuM strain grown in iron-limiting medium (see Table S3 in the supplemental material). These data confirmed that AcuM RNA levels were substantially elevated (>100-fold) in the ΔacuK gpdA-acuM strain. For 87 genes, there was no significant difference in expression between the two strains. These nonresponsive genes included 26 out of 28 genes whose expression had differed between the ΔacuK mutant and the wild-type strain. It thus seems clear that AcuM alone does not activate most of the same genes as AcuM-AcuK. In order to understand the possible mechanistic basis for the effects of acuM overexpression in the ΔacuK background, we examined specifically the genes whose expression differed between the ΔacuK and ΔacuK gpdA-acuM strains. We identified three genes with expression responses that seemed both biologically and statistically significant (≥1.5-fold change; P < 0.05): Afu1g17150, Afu3g00420, and Afu5g11070 (Fig. 7C). One of these genes, Afu5g11070, may have a functional connection to iron acquisition. Its A. nidulans ortholog, arcA, specifies an activator of arginine catabolic genes (34). The arginine breakdown product ornithine is essential for siderophore synthesis in aspergilli (13, 35). Therefore, acuM overexpression may improve growth of the ΔacuK mutant by increasing internal ornithine levels and, as a consequence, siderophore synthesis.
DISCUSSION
In A. nidulans, the AcuM and AcuK transcription factors form a heterodimer that is a key regulator of metabolic reprogramming in response to gluconeogenic carbon sources (15, 16). Previously, we found that in A. fumigatus, AcuM governs not only gluconeogenesis but also iron acquisition (14). In the current study, we determined that the function of A. fumigatus AcuK was largely similar to that of AcuM. Both the ΔacuK and ΔacuM mutants grew poorly on gluconeogenic substrates and under iron-limited conditions. Also, both mutants had decreased extracellular siderophore activity and reduced iron uptake in vitro and attenuated virulence in the mouse model of invasive aspergillosis. Importantly, the phenotype of the ΔacuK ΔacuM double mutant was virtually identical to that of the ΔacuM single mutant. Finally, the NanoString and real-time PCR data demonstrated that the ΔacuK and ΔacuM mutants had similar transcriptional profiles when A. fumigatus was grown in vitro. Collectively, these in vitro data are consistent with the model that AcuK and AcuM govern the expression of many genes by functioning as part of a complex in A. fumigatus, as they do in A. nidulans.
Although our data provide strong evidence that AcuK and AcuM govern the same biological functions and target genes, the phenotypic and transcriptional profiling data also suggest that AcuK and AcuM may function independently of one another. For example, the ΔacuM mutant had more severe phenotypic defects than the ΔacuK mutant in several assays. Also, the ΔacuM mutation had greater impact on expression of some target genes than the ΔacuK mutation (e.g., gliZ and Afu4g14070) and vice versa (e.g., calA and mdr4). These results are consistent with the possibility that AcuK and AcuM each have some activity in the absence of the other. We pursued this hypothesis to explore AcuM function in particular, because our phenotypic analysis implied that the independent function of AcuM may be biologically relevant. Indeed, we observed that AcuM overexpression could improve ΔacuK mutant growth under iron-limited conditions. The gene expression data led us to the idea that AcuM may promote ornithine accumulation and thus siderophore production, but this hypothesis is highly speculative. The most important conclusion from our gene expression analysis is that AcuM overexpression does not affect RNA levels of most AcuK-responsive genes. Therefore, the AcuK-independent and AcuK-dependent functions of AcuM may be largely distinct from one another.
A striking finding from our gene expression analysis was that that defects in either AcuK or AcuM cause different gene expression alterations during in vitro growth than they do during infection. For example, while both AcuK and AcuM governed the transcription of multiple genes involved in iron acquisition in vitro, they controlled the expression of only a subset of these genes in vivo. Also, other genes whose products are known to be required for maximal virulence, such as hexA (33) and srbA (36), had reduced transcript levels in the ΔacuK and/or ΔacuM mutants in vivo but not in vitro. These findings suggest that the virulence defects of the two mutants may reflect the composite of their effects on iron acquisition and on multiple virulence pathways. These results also demonstrate the importance of in vivo transcriptional profiling studies for identifying the downstream target genes of transcription factors that are relevant to virulence.
The transcriptional profile of wild-type A. fumigatus during the first 12 to 14 h of pulmonary infection in mice has been investigated previously by McDonagh et al. (37). Using microarray analysis, these authors found that in vivo infection induced the expression of more than 1,200 A. fumigatus genes compared to growth in liquid culture. While a minority of these in vivo-expressed genes was also upregulated by in vitro growth under iron-limited, nitrogen-limited, or alkaline conditions, over 1,000 genes were upregulated only in vivo. Thus, these results further support the conclusion that in vivo infection induces a unique transcriptional response in microbial pathogens. Although our NanoString experiments analyzed the expression level of a smaller number of genes, our results complement and extend the data of McDonagh et al. because the high sensitivity of the NanoString nCounter enabled us to assess A. fumigatus gene expression after infection was established rather than during the initiation of infection. Moreover, we were able to analyze the in vivo transcriptional response of two A. fumigatus mutants, even though the pulmonary fungal burden of mice infected with these mutants was lower than that of mice infected with the wild-type strain (14). In fact, 20 of the 71 genes that had detectable transcript counts in our in vivo NanoString study were below the limit of detection in the in vivo microarray analysis (37).
Will extensive differences between in vitro and in vivo gene expression impact of a transcription factor defect prove to be the exception or the rule for invasive pathogens? It is too soon to tell, of course, but we can assess the breadth of the issue from a survey of recent studies. For the C. albicans Bcr1 and Rim101 transcription factors, prior studies indicate that in vitro and in vivo profiles are fairly divergent (38, 39). For Cryptococcus neoformans Rim101 and A. fumigatus SrbA, it appears that the in vivo and in vitro profiles of null mutants are fairly similar (32, 40). We can now add A. fumigatus AcuM and AcuK to the group of transcription factors with divergent in vivo and in vitro mutant profiles. These groups are not absolute, because the assessment depends upon the number and nature of in vitro conditions tested, as well as the number of genes whose expression is assayed. However, it seems clear that the invasive infection environment causes modification of many regulatory interactions, likely because of the unique set of challenges it presents for pathogen survival.
Supplementary Material
ACKNOWLEDGMENTS
We thank David Villarreal for assistance with tissue culture.
This work was supported by grants R01AI073829 and R56AI111836 and contract N01-AI-30041 from the National Institutes of Health.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.02685-14.
REFERENCES
- 1.Hohl TM, Feldmesser M. 2007. Aspergillus fumigatus: principles of pathogenesis and host defense. Eukaryot Cell 6:1953–1963. doi: 10.1128/EC.00274-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Latgé JP. 1999. Aspergillus fumigatus and aspergillosis. Clin Microbiol Rev 12:310–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pappas PG, Alexander BD, Andes DR, Hadley S, Kauffman CA, Freifeld A, Anaissie EJ, Brumble LM, Herwaldt L, Ito J, Kontoyiannis DP, Lyon GM, Marr KA, Morrison VA, Park BJ, Patterson TF, Perl TM, Oster RA, Schuster MG, Walker R, Walsh TJ, Wannemuehler KA, Chiller TM. 2010. Invasive fungal infections among organ transplant recipients: results of the Transplant-Associated Infection Surveillance Network (TRANSNET). Clin Infect Dis 50:1101–1111. doi: 10.1086/651262. [DOI] [PubMed] [Google Scholar]
- 4.Neofytos D, Horn D, Anaissie E, Steinbach W, Olyaei A, Fishman J, Pfaller M, Chang C, Webster K, Marr K. 2009. Epidemiology and outcome of invasive fungal infection in adult hematopoietic stem cell transplant recipients: analysis of Multicenter Prospective Antifungal Therapy (PATH) Alliance registry. Clin Infect Dis 48:265–273. doi: 10.1086/595846. [DOI] [PubMed] [Google Scholar]
- 5.Baddley JW, Stephens JM, Ji X, Gao X, Schlamm HT, Tarallo M. 2013. Aspergillosis in intensive care unit (ICU) patients: epidemiology and economic outcomes. BMC Infect Dis 13:29. doi: 10.1186/1471-2334-13-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ganz T. 2009. Iron in innate immunity: starve the invaders. Curr Opin Immunol 21:63–67. doi: 10.1016/j.coi.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson L. 2008. Iron and siderophores in fungal-host interactions. Mycol Res 112:170–183. doi: 10.1016/j.mycres.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 8.Weinberg ED. 2009. Iron availability and infection. Biochim Biophys Acta 1 790:600–605. doi: 10.1016/j.bbagen.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 9.Haas H. 2012. Iron—a key nexus in the virulence of Aspergillus fumigatus. Front Microbiol 3:28. doi: 10.3389/fmicb.2012.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schrettl M, Haas H. 2011. Iron homeostasis—Achilles' heel of Aspergillus fumigatus? Curr Opin Microbiol 14:400–405. doi: 10.1016/j.mib.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schrettl M, Bignell E, Kragl C, Joechl C, Rogers T, Arst HN Jr, Haynes K, Haas H. 2004. Siderophore biosynthesis but not reductive iron assimilation is essential for Aspergillus fumigatus virulence. J Exp Med 200:1213–1219. doi: 10.1084/jem.20041242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schrettl M, Bignell E, Kragl C, Sabiha Y, Loss O, Eisendle M, Wallner A, Arst HN Jr, Haynes K, Haas H. 2007. Distinct roles for intra- and extracellular siderophores during Aspergillus fumigatus infection. PLoS Pathog 3(9):e128. doi: 10.1371/journal.ppat.0030128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hissen AH, Wan AN, Warwas ML, Pinto LJ, Moore MM. 2005. The Aspergillus fumigatus siderophore biosynthetic gene sidA, encoding L-ornithine N5-oxygenase, is required for virulence. Infect Immun 73:5493–5503. doi: 10.1128/IAI.73.9.5493-5503.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu H, Gravelat FN, Chiang LY, Chen D, Vanier G, Ejzykowicz DE, Ibrahim AS, Nierman WC, Sheppard DC, Filler SG. 2010. Aspergillus fumigatus AcuM regulates both iron acquisition and gluconeogenesis. Mol Microbiol 78:1038–1054. doi: 10.1111/j.1365-2958.2010.07389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hynes MJ, Szewczyk E, Murray SL, Suzuki Y, Davis MA, Sealy-Lewis HM. 2007. Transcriptional control of gluconeogenesis in Aspergillus nidulans. Genetics 176:139–150. doi: 10.1534/genetics.107.070904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suzuki Y, Murray SL, Wong KH, Davis MA, Hynes MJ. 2012. Reprogramming of carbon metabolism by the transcriptional activators AcuK and AcuM in Aspergillus nidulans. Mol Microbiol 84:942–964. doi: 10.1111/j.1365-2958.2012.08067.x. [DOI] [PubMed] [Google Scholar]
- 17.Sheppard DC, Doedt T, Chiang LY, Kim HS, Chen D, Nierman WC, Filler SG. 2005. The Aspergillus fumigatus StuA protein governs the up-regulation of a discrete transcriptional program during the acquisition of developmental competence. Mol Biol Cell 16:5866–5879. doi: 10.1091/mbc.E05-07-0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Punt PJ, Oliver RP, Dingemanse MA, Pouwels PH, van den Hondel CA. 1987. Transformation of Aspergillus based on the hygromycin B resistance marker from Escherichia coli. Gene 56:117–124. doi: 10.1016/0378-1119(87)90164-8. [DOI] [PubMed] [Google Scholar]
- 19.Catlett NL, Lee B-N, Yoder OC, Turgeon BG. 2003. Split-marker recombination for efficient targeted deletion of fungal genes. Fungal Genet Newsl 50:9–11. [Google Scholar]
- 20.Weidner G, d'Enfert C, Koch A, Mol PC, Brakhage AA. 1998. Development of a homologous transformation system for the human pathogenic fungus Aspergillus fumigatus based on the pyrG gene encoding orotidine 5′-monophosphate decarboxylase. Curr Genet 33:378–385. doi: 10.1007/s002940050350. [DOI] [PubMed] [Google Scholar]
- 21.Richie DL, Fuller KK, Fortwendel J, Miley MD, McCarthy JW, Feldmesser M, Rhodes JC, Askew DS. 2007. Unexpected link between metal ion deficiency and autophagy in Aspergillus fumigatus. Eukaryot Cell 6:2437–2447. doi: 10.1128/EC.00224-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Campoli P, Al Abdallah Q, Robitaille R, Solis NV, Fielhaber JA, Kristof AS, Laverdiere M, Filler SG, Sheppard DC. 2011. Concentration of antifungal agents within host cell membranes: a new paradigm governing the efficacy of prophylaxis. Antimicrob Agents Chemother 55:5732–5739. doi: 10.1128/AAC.00637-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fu Y, Lee H, Collins M, Tsai HF, Spellberg B, Edwards JE Jr, Kwon-Chung KJ, Ibrahim AS. 2004. Cloning and functional characterization of the Rhizopus oryzae high affinity iron permease (rFTR1) gene. FEMS Microbiol Lett 235:169–176. doi: 10.1016/j.femsle.2004.04.031. [DOI] [PubMed] [Google Scholar]
- 24.Ejzykowicz DE, Solis NV, Gravelat FN, Chabot J, Li X, Sheppard DC, Filler SG. 2010. Role of Aspergillus fumigatus DvrA in host cell interactions and virulence. Eukaryot Cell 9:1432–1440. doi: 10.1128/EC.00055-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sheppard DC, Rieg G, Chiang LY, Filler SG, Edwards JE Jr, Ibrahim AS. 2004. Novel inhalational murine model of invasive pulmonary aspergillosis. Antimicrob Agents Chemother 48:1908–1911. doi: 10.1128/AAC.48.5.1908-1911.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ejzykowicz DE, Cunha MM, Rozental S, Solis NV, Gravelat FN, Sheppard DC, Filler SG. 2009. The Aspergillus fumigatus transcription factor Ace2 governs pigment production, conidiation and virulence. Mol Microbiol 72:155–169. doi: 10.1111/j.1365-2958.2009.06631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J. 2003. TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34:374–378. [DOI] [PubMed] [Google Scholar]
- 28.Schwyn B, Neilands JB. 1987. Universal chemical assay for the detection and determination of siderophores. Anal Biochem 160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- 29.Moreno I, Pedreno Y, Maicas S, Sentandreu R, Herrero E, Valentin E. 2003. Characterization of a Candida albicans gene encoding a putative transcriptional factor required for cell wall integrity. FEMS Microbiol Lett 226:159–167. doi: 10.1016/S0378-1097(03)00588-3. [DOI] [PubMed] [Google Scholar]
- 30.Gravelat FN, Beauvais A, Liu H, Lee MJ, Snarr BD, Chen D, Xu W, Kravtsov I, Hoareau CM, Vanier G, Urb M, Campoli P, Al Abdallah Q, Lehoux M, Chabot JC, Ouimet MC, Baptista SD, Fritz JH, Nierman WC, Latge JP, Mitchell AP, Filler SG, Fontaine T, Sheppard DC. 2013. Aspergillus galactosaminogalactan mediates adherence to host constituents and conceals hyphal β-glucan from the immune system. PLoS Pathog 9(8):e1003575. doi: 10.1371/journal.ppat.1003575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gravelat FN, Ejzykowicz DE, Chiang LY, Chabot JC, Urb M, Macdonald KD, al-Bader N, Filler SG, Sheppard DC. 2010. Aspergillus fumigatus MedA governs adherence, host cell interactions and virulence. Cell Microbiol 12:473–488. doi: 10.1111/j.1462-5822.2009.01408.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chung D, Barker BM, Carey CC, Merriman B, Werner ER, Lechner BE, Dhingra S, Cheng C, Xu W, Blosser SJ, Morohashi K, Mazurie A, Mitchell TK, Haas H, Mitchell AP, Cramer RA. 2014. ChIP-seq and in vivo transcriptome analyses of the Aspergillus fumigatus SREBP SrbA reveals a new regulator of the fungal hypoxia response and virulence. PLoS Pathog 10(11):e1004487. doi: 10.1371/journal.ppat.1004487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beck J, Echtenacher B, Ebel F. 2013. Woronin bodies, their impact on stress resistance and virulence of the pathogenic mould Aspergillus fumigatus and their anchoring at the septal pore of filamentous Ascomycota. Mol Microbiol 89:857–871. doi: 10.1111/mmi.12316. [DOI] [PubMed] [Google Scholar]
- 34.Empel J, Sitkiewicz I, Andrukiewicz A, Lasocki K, Borsuk P, Weglenski P. 2001. arcA, the regulatory gene for the arginine catabolic pathway in Aspergillus nidulans. Mol Genet Genomics 266:591–597. doi: 10.1007/s004380100575. [DOI] [PubMed] [Google Scholar]
- 35.Eisendle M, Oberegger H, Zadra I, Haas H. 2003. The siderophore system is essential for viability of Aspergillus nidulans: functional analysis of two genes encoding l-ornithine N 5-monooxygenase (sidA) and a non-ribosomal peptide synthetase (sidC). Mol Microbiol 49:359–375. doi: 10.1046/j.1365-2958.2003.03586.x. [DOI] [PubMed] [Google Scholar]
- 36.Willger SD, Puttikamonkul S, Kim KH, Burritt JB, Grahl N, Metzler LJ, Barbuch R, Bard M, Lawrence CB, Cramer RA Jr. 2008. A sterol-regulatory element binding protein is required for cell polarity, hypoxia adaptation, azole drug resistance, and virulence in Aspergillus fumigatus. PLoS Pathog 4(11):e1000200. doi: 10.1371/journal.ppat.1000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McDonagh A, Fedorova ND, Crabtree J, Yu Y, Kim S, Chen D, Loss O, Cairns T, Goldman G, Armstrong-James D, Haynes K, Haas H, Schrettl M, May G, Nierman WC, Bignell E. 2008. Sub-telomere directed gene expression during initiation of invasive aspergillosis. PLoS Pathog 4(9):e1000154. doi: 10.1371/journal.ppat.1000154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fanning S, Xu W, Solis N, Woolford CA, Filler SG, Mitchell AP. 2012. Divergent targets of Candida albicans biofilm regulator Bcr1 in vitro and in vivo. Eukaryot Cell 11:896–904. doi: 10.1128/EC.00103-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheng S, Clancy CJ, Xu W, Schneider F, Hao B, Mitchell AP, Nguyen MH. 2013. Profiling of Candida albicans gene expression during intra-abdominal candidiasis identifies biologic processes involved in pathogenesis. J Infect Dis 208:1529–1537. doi: 10.1093/infdis/jit335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Meara TR, Xu W, Selvig KM, O'Meara MJ, Mitchell AP, Alspaugh JA. 2014. The Cryptococcus neoformans Rim101 transcription factor directly regulates genes required for adaptation to the host. Mol Cell Biol 34:673–684. doi: 10.1128/MCB.01359-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nierman WC, Pain A, Anderson MJ, Wortman JR, Kim HS, Arroyo J, Berriman M, Abe K, Archer DB, Bermejo C, Bennett J, Bowyer P, Chen D, Collins M, Coulsen R, Davies R, Dyer PS, Farman M, Fedorova N, Feldblyum TV, Fischer R, Fosker N, Fraser A, Garcia JL, Garcia MJ, Goble A, Goldman GH, Gomi K, Griffith-Jones S, Gwilliam R, Haas B, Haas H, Harris D, Horiuchi H, Huang J, Humphray S, Jimenez J, Keller N, Khouri H, Kitamoto K, Kobayashi T, Konzack S, Kulkarni R, Kumagai T, Lafon A, Latge JP, Li W, Lord A, Lu C, Majoros WH, May GS, Miller BL, Mohamoud Y, Molina M, Monod M, Mouyna I, Mulligan S, Murphy L, O'Neil S, Paulsen I, Penalva MA, Pertea M, Price C, Pritchard BL, Quail MA, Rabbinowitsch E, Rawlins N, Rajandream MA, Reichard U, Renauld H, Robson GD, Rodriguez de Cordoba S, Rodriguez-Pena JM, Ronning CM, Rutter S, Salzberg SL, Sanchez M, Sanchez-Ferrero JC, Saunders D, Seeger K, Squares R, Squares S, Takeuchi M, Tekaia F, Turner G, Vazquez de Aldana CR, Weidman J, White O, Woodward J, Yu JH, Fraser C, Galagan JE, Asai K, Machida M, Hall N, Barrell B, Denning DW. 2005. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 438:1151–1156. doi: 10.1038/nature04332. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.