Abstract
Mesenchymal stem cells (MSCs) show great promise in blood vessel restoration and vascularization enhancement in many therapeutic situations. Typically, the co-implantation of MSCs with vascular endothelial cells (ECs) is effective for the induction of functional vascularization in vivo, indicating its potential applications in regenerative medicine. The effects of MSCs-ECs-induced vascularization can be modeled in vitro, providing simplified models for understanding their underlying communication. In this article, a contact coculture model in vitro and an RNA-seq approach were employed to reveal the active crosstalk between MSCs and ECs within a short time period at both morphological and transcriptional levels. The RNA-seq results suggested that angiogenic genes were significantly induced upon coculture, and this prevascularization commitment might require the NF-κB signaling. NF-κB blocking and interleukin (IL) neutralization experiments demonstrated that MSCs potentially secreted IL factors including IL1β and IL6 to modulate NF-κB signaling and downstream chemokines during coculture. Conversely, RNA-seq results indicated that the MSCs were regulated by the coculture environment to a smooth muscle commitment within this short period, which largely induced myocardin, the myogenic co-transcriptional factor. These findings demonstrate the mutual molecular mechanism of MSCs-ECs-induced prevascularization commitment in a quick response.
Introduction
Stem cell transplantation has shown increasing therapeutic potential for treatment of pathological situations tissue restoration [1–3]. Mesenchymal stem cells (MSCs), the stromal progenitor cells found in the bone marrow [3], were recently proved effective in vascular enhancement and protection [4,5]. Since tissue engineering is currently limited by the inability to adequately vascularize tissues surrounding the engineered constructs [6,7], the co-implantation of MSCs with human endothelial cells (ECs) recently succeed in creation of fully functional blood vessels under different circumstances in vivo [8–10], which could serve as a good solution for controlled vascularization. However, the cellular and molecular bases of MSCs-ECs interaction and their joint influence on endothelial vascularization were not fully understood.
Previous research has revealed the active interaction between MSCs and ECs in different situations. Culture of MSCs in human umbilical vein endothelial cell (HUVEC)-conditioned media (CM) promoted the activity of the enhanced alkaline phosphatase (ALP), indicating that EC-secreted growth factors might prompt the osteogenic differentiation of MSCs in vitro [11,12]. However, MSCs were also profoundly affected by ECs via direct cell–cell contact. The prolonged MSC-EC crosstalk in contact coculture in vitro stimulated the proliferation of MSCs [13]. Under other circumstances, direct contact with vascular ECs resulted in an increased proportion of myogenic phenotypes in MSCs [14–17]. Tissue engineering-based research in both two-dimensional (2D) and three-dimensional cocultures in vitro revealed that the active interaction between ECs and MSCs (or osteoprogenitor and fibroblast) induced the formation of tube-like cell aggregation structures. This result indicates that angiogenesis and prevascularization are involved in the cell–cell communication and remodeling of the functional blood vessel development in vivo [5,18–26]. Moreover, MSCs could repress ECs by regulating cytokine-induced leukocyte recruitment [27], while the activated ECs could regulate the MSCs-to-ECs transmigration in a leukocyte-like mechanism [28,29]. However, the innate mechanism of MSCs-ECs interaction-dependent vascularization remains elusive.
In this article, therefore, we focus on revealing the internal molecular mechanisms in cell coupling and reciprocal ECs–MSCs interactions and thus on better understanding the functional vascularization effect induced by the MSCs-ECs coculture environment. As long-term vascularization was responsible for the combinatory effects of complex and nutrient micro- environment in vivo, we simplified a 2D MSCs-ECs coculture model in vitro to reason whether a simple direct MSCs–ECs interaction is sufficient to stimulate prevascularization at early period. Conditions for MSCs-ECs direct coculture were preliminarily optimized, including respective seeding densities and culture medium formulation, which guarantee the suitable culture conditions.
As we focused on the early interaction within MSC-EC coculture, we comprehensively analyzed the transcriptome by RNA-seq for both cell types individually, followed by flow cytometer separation after 6, 12, and 24 h of coculture. We aim to monitor the potential ECs-MSCs communications at molecular level. The results revealed that even during a relatively short period, the coculture promoted the angiogenesis-related gene activation in HUVECs. Further analysis demonstrated that the angiogenesis-related NF-κB signaling was modulated mainly in HUVECs, which was at least partially stimulated by two interleukins (ILs) IL1β and IL6. Both IL1β and IL6 were mainly secreted by MSCs in a contact-dependent manner. Conversely, this short-period interaction also promoted the early smooth muscle commitment of MSCs partially by modulation of TGF-β. This was correlated with the physiological situations that recruited mural cell types support EC functionalization in the development and repair of blood vessels [8].
Materials and Methods
Cell culture conditions and separation by fluorescence activated cell sortor
Human MSCs (HMSCs) with stable green fluorescent protein (GFP) expression from passage 5 (OriCell™ HMSC-GFP; Cyagen) were tested for CD29, CD44, and CD105 positive, and CD34, CD45 negative, and expanded in HMSC basal media with 10% fetal bovine serum, 1% v/v penicillin/streptomycin (HUXMX-90011; Cyagen); HUVECs (Lonza) obtained at passage 2–3, maintained and proliferated in EGM-2 basal medium (Lonza).). The innate angiogenic potential of HUVEC was tested via Matrigel™ tube formation assay (Supplementary Fig. S1A; Supplementary Data are available online at www.liebertpub.com/scd). HMSCs within passage 8 and HUVECs within passage 5 were used during all experiments.
For coculture, complete media for both cell types were mixed, and the most adequate medium ratio was preselected to finally set the ratio of HMSC basal medium to EGM-2 medium at 1:1. Both coculture groups and monoculture groups were maintained in this coculture medium, and these media were changed every other day. Seeding density between HMSCs and HUVECs was optimized and applied at about 4×104 and 1.3×104 cells/cm2 in all experiments based on a quick and obvious observation of previous reported cell aggregation structure formation. Controlled indirect coculture of HMSCs on the plate bottom and HUVECs on the upside of the filter was conducted with a 24 mm Transwell (0.4 μm pore) Polyester Membrane Insert (Corning Corporation).
During fluorescence activated cell sortor (FACS)-based cell sorting, cells from each group were trypsinized and washed thrice with phosphate buffer solution (PBS). GFP-positive HMSC populations were immediately isolated with a BD FACS cell sorter (Aril III; BD Biosciences). During CD31+ cell sorting, the GFP-negative populations were incubated with Cy3-conjugated anti-CD31 antibody (anti-CD31-Cy3; Sino Biologicals) and sorted with BD FACS. The sorted co-HMSCs and co-HUVECs were immediately collected and used in downstream RNA extraction or protein extraction.
Observation and quantification of cell aggregation structures
To observe cell aggregation structures, both static images and time lapse movies were captured by an Eclipse Ti microscope system (Nikon Cooperation) with phase-contrast field and GFP florescence channels. We quantified angiogenic cell aggregation structures with a cell aggregate index calculated by ImageJ™. Coculture-induced aggregation trend was quantified as follows:
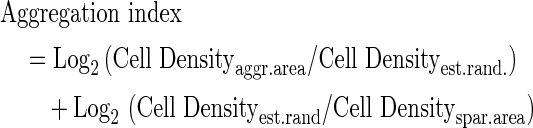 |
where Cell Density=cell counts/spreading areas. Aggr.area means the aggregating area in which the cocultured cells aggregate and tend to form cell aggregation structures, and the density is higher than the randomly distributed density; Spar.area mean the sparsely distributed area, in which cells are sparsely located in the cocultured environment, and the density is lower than the randomly distributed density; Est.rand means estimated absolutely random distribution, or the estimated cell density for cells randomly distributed in the culture dishes for a defined seeding cell count.
Analysis of gene expression by high-throughput RNA sequencing and quantitative polymerase chain reaction
For both high throughput transcriptome analysis and quantitative polymerase chain reaction (qPCR), RNA was isolated from at least three groups per experimental setting, including monocultures and FACS-sorted counterparts. RNA was extracted by a Total mRNA Isolation Kit (Tiangen). The quality and quantity of isolated RNAs were determined with a NanoDrop 2000 analyzer (NanoDrop Technologies) and a Bioanalyzer 2100 (Agilent).
During high throughput transcriptome analysis, the RNA integrity number in all cases ranged from 8.9 to 10, indicating minimal degradation. The Ploy(A)-based mRNA was enriched with NEBNext™ Poly(A) mRNA Magnetic Isolation Module (New England Biolabs) following the manufacture's instruction. CDNA libraries were constructed with a NEBNext Ultra™ Directional RNA Library Prep Kit for Illumina™ (New England Biolabs). The quantity of the constructed libraries was further confirmed by Bioanalyzer 2100. Pair-end RNA-seq procedures were conducted with Illumina 2000 (Illumina) following the manufacturer's instructions. RNA-seq data were analyzed as reported [30]. Heatmap constructions were synthesized by R 3.0.2 (R Foundation for Statistical Computing) with a package “gplots” as reported [30]. Gene ontology (GO) was analyzed by DAVID Bioinformatics Resources 6.7 as reported [31].
For qPCR analysis, cDNA was synthesized using a Fastquant RT Kit (Tiangen) and real-time PCR, and mRNA was analyzed by a SuperReal PreMix (SYBR Green) kit (Tiangen) and a Real-time 7500 Fast machine (Applied Biosystems). All primers used for real-time PCR are listed in Supplementary Table S1. All data were analyzed by Comparative ΔΔCt.
Analysis of NF-κB activation by nuclear-cytoplasmic extraction and western blot
The trypsinized cells and FACS-sorted cells were immediately washed with PBS, and the nuclear- cytoplasm was extracted by NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Scientific) following the manufacture's instruction. The extracted cytoplasmic and nucleus proteins were preserved immediately at −80°C until use.
Western blot for detection of NF-κB activation was performed based on the p65 protein cytoplasm-to-nuclei translocation following the standard western blot protocol. Both cytoplasmic and nucleus p65 were detected by anti-p65 monoclonal antibody (Sino Biologicals) and were normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH; Sigma-Aldrich). The secondary antibody conjugated with horseradish peroxidase (HRP) (Sigma-Aldrich) was used for blotting. The blots were visualized by electrogenerated chemiluminescence (ECL) reagents (Pierce Biotechnology, Thermo Scientific).
Coculture environment interfered with chemical inhibitor, siRNA and neutralization antibodies
For PTDC-specific blockage of NF-κB signaling, a given concentration of PTDC (Sigma-Aldrich) was added into the culture medium 1 h after cell attachment. Control groups were treated with an equivalent volume of dimethyl sulfoxide (DMSO), which was used for PTDC dilution. For RNAi treatment, siRNA against target genes of interest and sramble siRNA were all purchased from GenePharma Company. SiRNA was transfected with Lipofectamine LTX and PLUS™ Reagent with 35 nM siRNA following the manufacture's instruction for both cell types separately 1 day before coculture. For IL1β, IL6, and TGF-β neutralization experiments, 50–200 μg/mL neutralizing antibodies against human IL1β, IL6, and TGF-β (Sino Biologicals) were added during the coculture; control groups were treated with an equivalent amount of IgG (Sigma-Aldrich).
Evaluation of protein concentration in culture media by enzyme-linked immunosorbent assay
Cytokines IL1β and IL6 in the supernatants of media after different treatments were collected and preserved immediately at −80°C until use. Experiments were conducted using commercial enzyme-linked immunosorbent assay (ELISA) kits (Boster Bioscience) following the manufacture's instruction. Briefly, 100 μM of the supernatant of each sample was added to the anti-IL1β/IL6 antibody in the precoated 96-well plates, followed by incubation at 37°C for 90 min. Solutions were then discarded, and capture antibodies were added for another 60-min incubation. The plates were then washed by PBS and incubated with an avidin-biotin complex working solution for 30 min. Chromogenic reaction was then performed with TMB, and the wavelength of 450 nm was read. Raw reads were converted to the protein concentrations according to the standard sample curves.
Statistical analysis
Data were shown as mean±standard deviation. Experiments between two groups were analyzed using Student's t-test. Significance was evaluated using one-way analysis of variance. *P<0.01 or **P<0.05 were considered significant.
Results
Establishment of HMSCs-HUVECs coculture system in vitro
To understand the potential of MSC-EC communication during vascularization, we simplified a 2D coculture model. The primary HMSCs stably transfected with GFP and the primary GFP-negative HUVECs were used in the coculture experiments. The seeding ratio of HUVECs- to-HMSCs was optimized at about 3:1, at the density around 4×104 and 1.3×104 cells/cm2 respectively, and the culture media formulations were optimized to support both monocultures of two cell types and cocultured conditions.
Coculture promoted formation of cell aggregation structures
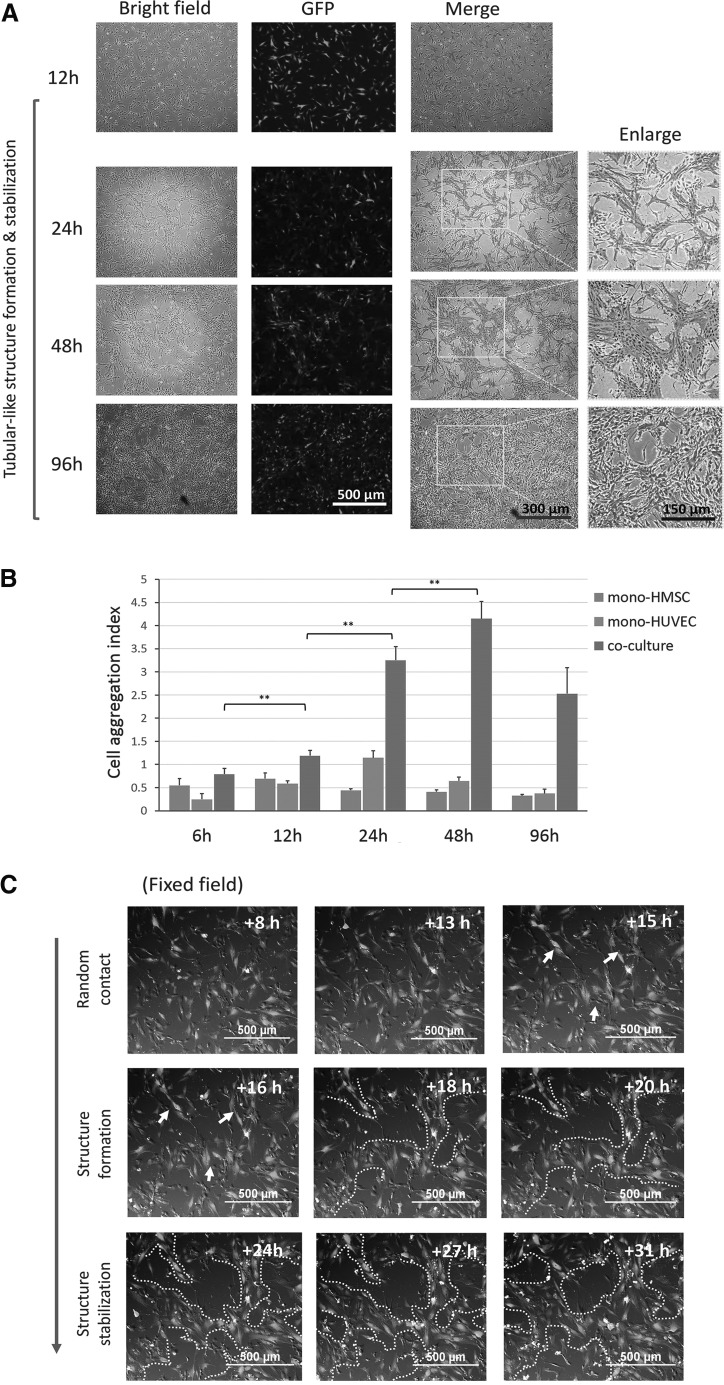
In the experimental conditions, we clearly observed and quantified cell aggregation structures caused by the HUVECs-HMSCs mutual recruitment (Fig. 1A, B). In this high density coculture mode, cell aggregation structures were gradually formed within 24 h and stabilized for more than 4 days (Fig. 1A), and then their clear morphology was gradually overwhelmed by the coculture owing to the outgrowth of both types of cells. Real-time monitoring further confirmed that this structure was formed mainly between 12 and 24 h (Fig. 1C). During this process, HUVECs behaved more actively than HMSCs, and these cell aggregation structures were mainly attributed to the active migration and rearrangement between HUVECs and MSCs (Supplementary Video S1). This morphological arrangement strictly depended on the contact coculture mode, since the indirect coculture using Transwell™ or CM did not induce the formation of cell aggregation structures (Supplementary Fig. S1B).
FIG. 1.
Coculture promoted cell aggregation structure formation. (A) Representative images of HMSC-GFP and HUVEC coculture within 96 h. Cell aggregation structure was clearly observed at 24 h and further stabilized. Images at 24, 48, and 96 h were enlarged. Cell aggregation structures were indicated as dot-dash lines. Scale bars were indicated. (B) Cell aggregation structure formation trends were quantified by the cell aggregation in coculture and monoculture of HMSCs and HUVECs at indicated time points. >3 independent fields were randomly selected for quantification, and data were expressed as mean+SD (C) Representative time lapse snapshots showed gradual formation of the cell aggregation structure from the indicated time intervals in fixed filed. Early cell aggregation trends were indicated as arrow; Cell aggregation structures formation process was indicated as white dot dash lines. Scale bar were indicated. ** Indicates P value <0.01. GFP, green fluorescent protein; HMSC, human mesenchymal stem cell; HUVEC, human umbilical vein endothelial cell; SD, standard deviation.
RNA-seq revealed differential gene expression profiles and angiogenic genes induction
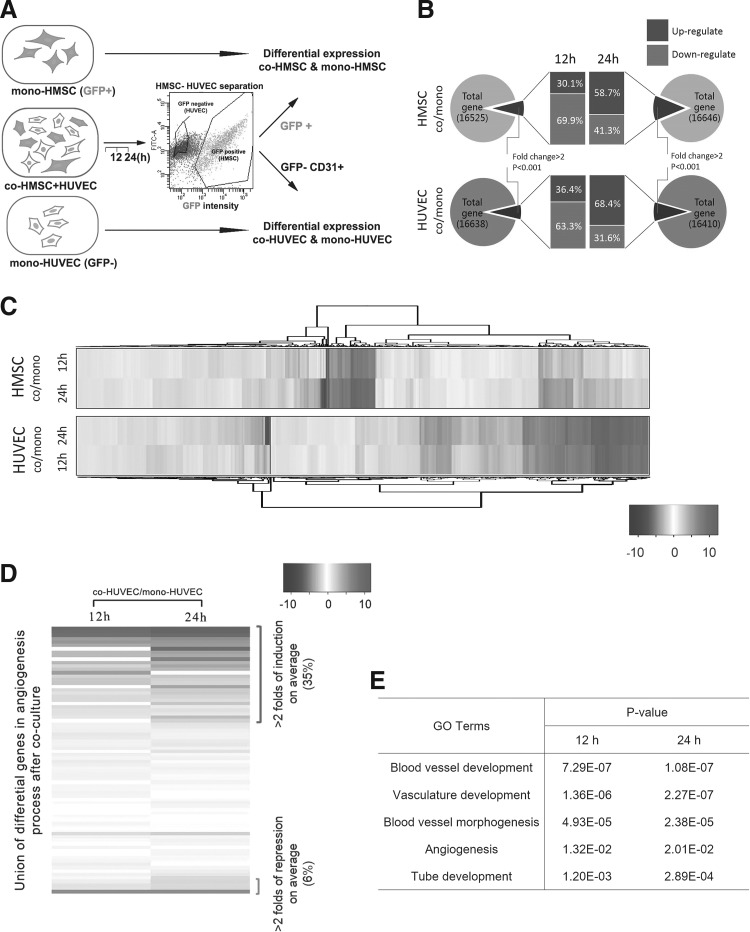
As angiogenic cell aggregation structure was spontaneously formed within an interval from 12 to 24 h, we next mainly performed the coculture system within 24 h, and immediately separated the GFP-positive cells as an HMSC population and the GFP-negative cells as an HUVEC population via FACS. The purity of the HUVEC population, was further validated by an EC surface marker CD31 (Fig. 2A). As angiogenic cell aggregation structures were spontaneously formed within from 12 to 24 h, we used FACS to sort HMSCs and HUVECs (co-MSCs & co-HUVECs hereafter) after 12 and 24 h of coculture, and their monoculture controls (mono-MSCs & mono-HUVECs) for detailed transcriptome analysis through RNA-seq (Fig. 2A and Supplementary Fig. S2). A total of >36 million raw reads were obtained for each sample with ∼95% integrity after filtering. Collectively, ∼80% of the reads were mapped to >16,000 genes in each sample, and >60% of reads covered at least 80% of genes (Supplementary Tables S2–S6). For co-MSCs, 8.27% and 13.88% of total genes were significantly modified compared with the monoculture controls for 12 and 24 h respectively (fold change >2, P<0.001). For co-HUVECs, 7.63% and 9.36% of total genes were modified significantly compared with monoculture control for 12 and 24 h respectively (Fig. 2B and Supplementary Tables S7–S10). Hierarchical clustering of RNA-seq data demonstrated that the morphological changes were accompanied by dynamic changes in cell transcriptome (Fig. 2C). Notably, among the significantly induced expression profiles, a proportion of genes including CDH5, PECAM1, TGFB family, and PDGF family (Supplementary Tables S6–S9) were well correlated with a previous report in a similar situation [32].
FIG. 2.
Coculture dynamically regulated the transcriptome alteration in both MSCs and HUVECs and raised angiogenic gene expression in HUVECs. (A) Schematic of FACS-based cell sorting after coculture and RNA-seq analysis. Co-HMSC was sorted based on GFP+ and co-HUVEC was sorted based on GFP− and CD31+. Transcriptome alteration was compared between cocultured MSC/HUVEC and their monoculture counterparts. (B) Statistical analysis of gene differential expression after coculture for 12 and 24 h separately in HMSCs and HUVECs. (C) Hierarchical clustering of RNA-seq data within genes both significantly altered in 12 and 24 h coculture for HMSCs and HUVECs. (D) Significant proportion of angiogenic related genes were elevated upon coculture in HUVECs. Heatmap was ordered via the fold of induction in Log2 scale. Significantly induced genes and repressed genes were indicated. (E) Gene ontology (GO) analysis on vascularization and angiogenesis-related process. Correlation of listed relevant process in 12 and 24 h coculture in HUVECs was evaluated by P value. FACS, fluorescence activated cell sortor; MSC, mesenchymal stem cell.
Consistent with our cell aggregation structure formation morphologically, the unbiased RNA-seq data analysis revealed that a large proportion of angiogenesis-related factors were expressed differentially and significantly (26/78 genes >2-fold induction on average; 5/78 >2-fold suppression, Supplementary Table S11) within 12 and 24 h in HUVECs (Fig. 2D), including PLAU [21] and fibroblast growth factor (FGF) [33], which were involved in the formation of cell aggregation structures in a similar context. However, unexpectedly classical angiogenic stimulators ANGPT and vascular endothelial growth factor (VEGF) family [28,30,41,42] were not obviously induced in both cell types within this short period. The GO analysis in differential gene expression revealed significant correlation between angiogenesis and vascularization (Fig. 2E and Supplementary Tables S12 and S13), indicating that direct coculture of MSCs and ECs significantly induced the pro-angiogenic effect, even within a short period. As a large proportion of the angiogenesis-related genes encoded the secretion molecules, we then collected the culture media from 12- and 24-h coculture groups. However, incubation of monocultured HUVECs with these CMs did not induce the formation of cell aggregation structures (data not shown), indicating that this prevascularization morphology was dependent on direct contact and might be associated with chemotaxis gradient.
NF-κB participated in coculture-induced formation of cell aggregation structures
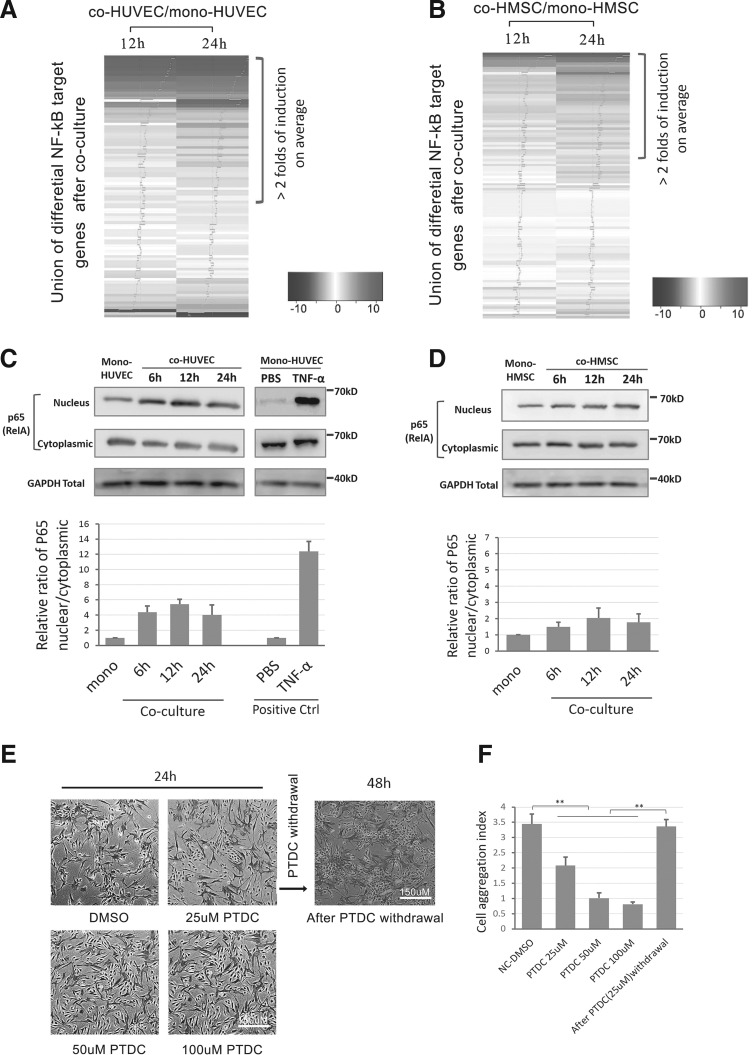
Since HMSCs-HUVECs coculture could induce the early pro-angiogenic effect in a quick response, we further interrogated the potential evidence of signaling pathway activation from gene expression profiles. We first referred to two classic endothelial angiogenesis-related pathways both in vitro and in vivo: VEGF signaling [6,7,22] and angiopoietin/TIE2 signaling [34,35]. However, the RNA-seq results did not show significant elevation of upstream ligands or receptors for these pathways in co-HUVECs within 24 h. Alternatively, unbiased analysis demonstrated that a large proportion of experimentally validated NF-κB target genes (mainly based on www.bu.edu/NF-κB/gene-resources/target genes) were upregulated upon coculture for 12 and 24 h in both co-HUVECs and co-HMSCs (Fig. 3A, B). NF-κB signaling was related to EC activation-induced cell–cell aggregation and angiogenesis [31,32]. Notably, activation of NF-κB target genes was more significant in co-HUVECs (Supplementary Table S14) compared with co-HMSCs (Supplementary Table S15). Therefore, we hypothesized that the NF-κB signaling pathway might be stimulated upon our coculture conditions and participate in angiogenic induction.
FIG. 3.
NF-κB participated in coculture-induced cell aggregation structure formation. (A, B) NF-kB target genes differential expression in co-HMSCs and co-HUVECs within 12 and 24 h. Heatmap was ordered via the fold of induction in Log2 scale. Fold of expression was indicated as Blue tracer in Heatmap. Genes with >2-folds of induction were indicated. (C) Top: Western blot of p65 (RelA) nucleus translocation in co-HUVECs and mono-HUVECs. GAPDH was used as a loading control for each group and TNF-α (10 ng/mL) treated HUVEC was used as a positive control; bottom: quantification of relative nucleus/cytoplasmic ratio of p65 via signal intensity (the ratio in coculture groups were normalized to the relative ratio in monoculture control groups, and hereafter). (D) Top: western blot of p65 (RelA) nucleus translocation in co-HMSCs and mono-HMSCs after coculture within 24 h; bottom: quantification of nucleus/cytoplasmic ratio of p65 via signal intensity. (E) Representative images of PTDC treatment (25–100 and 25 μM withdrawal) on the influence of cell aggregation structure formation in coculture for 24 h. (F) Quantification of cell aggregation structure formation after PTDC treatment and withdrawal. ** Indicates P value <0.01. GAPDH, glyceraldehyde 3-phosphate dehydrogenase; TNF-α, tumor necrosis factor-α.
To validate the involvement of NF-κB signaling in the coculture, we evaluated the NF-κB signaling activation based on the nucleus translocation of NF-κB large subunit P65 (RelA) within 24 h. Accordingly, the translocation of P65 nucleus was improved upon coculture in HUVECs compared with the monoculture counterparts within 6, 12, and 24 h separately (Fig. 2C), although these translocation trends are not as significant as transient activation by tumor necrosis factor (TNF)-α (a classic positive control for NF-κB activation). Similarly, co-HMSCs shared similar but even moderate trends compared to co-HUVECs in the coculture environment (Fig. 2D).
We next interrogated whether the inhibition of NF-κB could block the coculture-induced formation of angiogenic cell aggregation structures. An NF-κB signaling specific inhibitor pyrrolidine dithiocarbamate (PTDC) could effectively block the P65 nucleus translocation raised by coculture (Supplementary Fig. S3). Accordingly, attenuation of NF-κB signaling through a specific inhibitor pyrrolidine dithiocarbamate (PTDC) effectively suppressed the formation of cell aggregation structures in a dose-dependent manner in the coculture environment, but this suppression was reversible following the withdrawal of PTDC (Fig. 3E, F). Collectively, the NF-κB signaling was activated and required for the coculture-induced angiogenic process.
IL1β and IL6 were mainly elevated by HMSCs in a direct contact manner
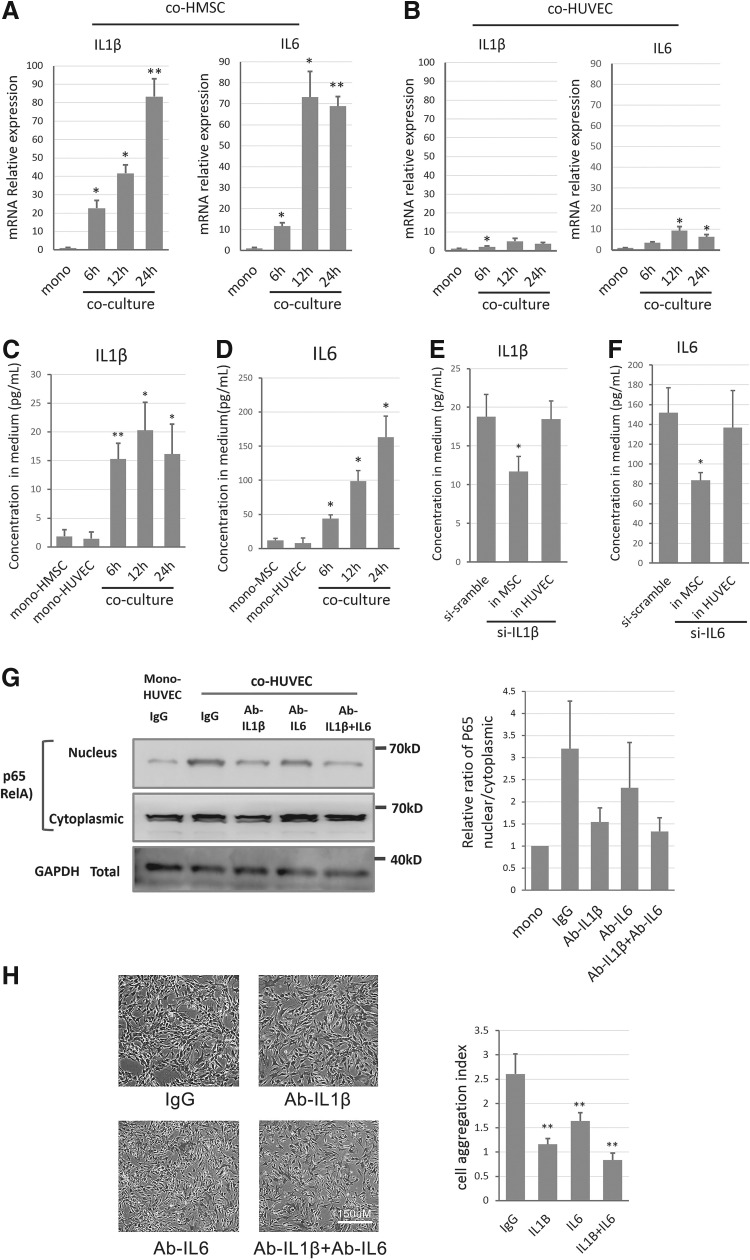
As NF-κB signaling pathways can be activated by various external signals and maintained by feedback loops, we screened the candidates that may be responsible for NF-κB activation in RNA-seq data from both cell types. Among the known factors regulating NF-κB signaling, IL1β (Log2 ratio fold change:10.8/12.4) and IL6 (Log2 ratio fold change:5.3/4.6) were significantly inductive in HMSCs in both 12-h and 24-h coculture groups (Supplementary Table S6 and S7). qPCR validated that both IL1β and IL6 were significantly induced in co-HMSCs in a time-dependent manner (Fig. 4A, B). In contrast, IL1β and IL6 were only moderately induced in co-HUVECs (Fig. 4A, B), and their basal levels were significantly lower than in HMSCs. We then excluded the possibility that the elevation of the two ILs specifically in co-HMSCs was due to the innate signals of ECs or the indirect communication with ECs, since neither Transwell-based indirect coculture nor mono-HUVEC CM could achieve such significant induction (Supplementary Fig. S4A, B), demonstrating that the transcriptional induction of IL1β or IL6 was caused by direct contact with HUVECs in coculture.
FIG. 4.
HMSCs secreted IL1β and IL6 to regulate cell aggregation structure formation potentially through regulating NF-κB in HUVECs. (A, B) Relative mRNA expressions of IL1β and IL6 in HMSCs and HUVECs after coculture within 24 h. (C, D) Secretion levels of IL1β and IL6 in culture medium after coculture within 24 h. (E, F) Effect of siRNA targeting IL1β and IL6 on secretion of IL1β and IL6 in culture medium. MSCs and ECs were pretreated with siRNA targeting IL1β, IL6, or negative scramble siRNA as indicated. (G) Effect of neutralizing IL1β, IL6, and both on coculture-induced NF-kB activation in co-HUVECs. Left: representative western blot analysis of p65 (RelA) nucleus translocation in co-HUVEC at 24 h after treatment of scramble IgG, Anti-IL1β, Anti-IL6, or both; right: quantification of nucleus/cytoplasmic ratio of p65 via signal intensity. (H) Left: representative images of coculture-induced cell aggregation structure formation at 24 h after neutralizing IL1β, IL6, or both in medium. Right: cell aggregation structure trends were quantified for each indicated groups. * Indicates P-value <0.05, ** indicates P-value <0.01. IL, interleukin.
We further tested the secretion of both factors via standard sandwich ELISA, and they were induced in the supernatant of cocultured medium (Fig. 4C, D), which was correlated with their mRNA levels. To further elucidate whether the increased secretion of IL1β and IL6 was caused by co-HMSCs, we applied siRNAs against IL1β and IL6 separately to reduce their mRNA expressions. As a result, the high-level IL1β in the supernatant of coculture groups was significantly reduced in the HMSCs, but not significantly in the HUVECs (Fig. 4E). Similar results were found in IL6 RNAi tests (Fig. 4F). These evidences suggested that IL1β and IL6 were mainly elevated by co-HMSCs.
IL1β and IL6 served as the NF-κB signaling potential regulators in coculture environment
We then interrogated whether the co-HMSC-secreted IL1β and IL6 regulated the activation of NF-κB signaling for co-HUVECs in the coculture environment. We used neutralizing antibodies against IL1B and IL6 to separately or simultaneously block their activities in the supernatant. As expected, blocking IL1β, IL6, or both effectively suppressed the NF-κB nucleus translocation induced by the coculture (Fig. 4G), and restricted the coculture-induced formation of angiogenic cell aggregation structures (Fig. 4H), which mimicked the effects of NF-κB attenuation by PTDC treatment.
Since some ILs including IL1β and IL6 are transcriptionally regulated by NF-κB, which may create a positive feedback loop to chronically activate the signaling pathway [36,37], we further tested whether IL1β and IL6 could be self-regulated via the NF-κB signaling. Interestingly, the attenuation of NF-κB suppressed the expressions of IL1β and IL6 at both mRNA and protein levels in the supernatant (Supplementary Fig. S5A–D); meanwhile, blocking their activity in the supernatant by neutralizing antibodies also decreased their mRNA transcription (Supplementary Fig. S5A, B). Collectively, the co-HMSCs-elevated IL1β and IL6 served as the positive regulators for coculture-induced early vascularization by modulating NF-κB signaling and they were regulated in a positive feedback manner.
NF-κB-dependent formation of cell aggregation structures was associated with angiogenic chemokines
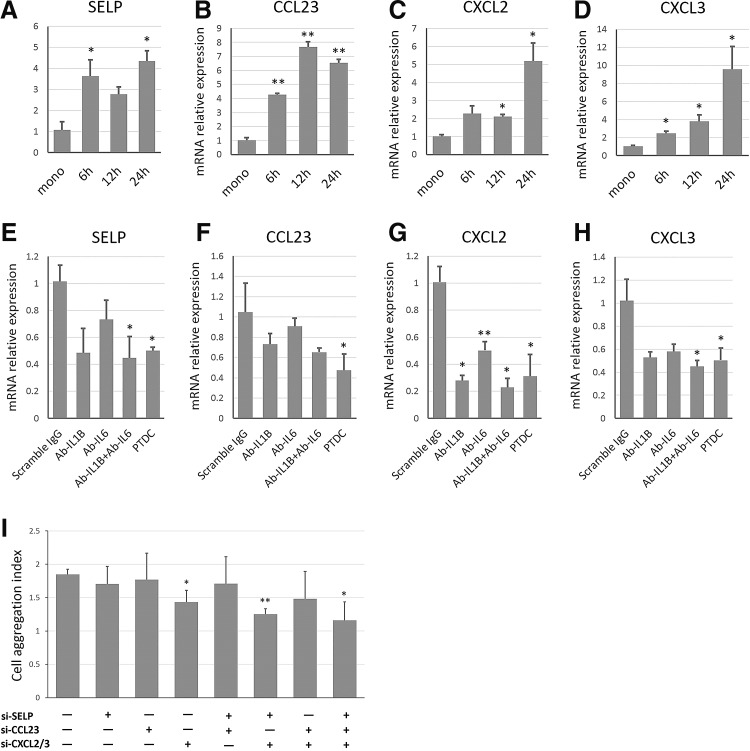
The NF-κB signaling in the downstream targets at a series of genes, including a proportion of chemokines, surface receptors, and secreted molecules that may promote angiogenesis both in vitro and in vivo [38]. Angiogenesis is commonly raised by HUVEC activation with complex chemokine regulation [39]. We thus screened the differentially expressed genes that were overlapped with coculture-stimulated NF-κB targets, and reported the angiogenic genes and the chemotaxics-related genes. To clarify the effects of these regulated genes during the coculture-induced early vascularization, we selected four significantly induced candidates, P-selectin (SELP), CC chemokine ligand23 (CCL23), and CXC chemokine ligands 2/3 (CXCL2 and CXCL3) for further study.
qPCR confirmed that the levels of SELP, CCL23, CXCL2, and CXCL3 significantly increased in a time-dependent manner (Fig. 5A–D). Then, we interrogated whether these genes were modulated by NF-κB signaling. Inhibition of NF-κB by 25 μM PTDC abolished the induction of these genes upon coculture in co-HUVECs (Fig. 5E–H); neutralizing antibodies against IL1B and IL6 in the supernatant also separately or simultaneously suppressed the induction of these genes (Fig. 5E–H). Collectively, these chemotaxics-related genes were potentially regulated by the IL-mediated NF-κB signaling.
FIG. 5.
NF-κB-dependent cell aggregation structure formation was associated with angiogenic chemotaxis-related genes. (A–D) Relative mRNA expressions of SELP, CCL23, CXCL2, and CXCL3 respectively in HUVECs after coculture induction within 24 h. (E–H) Effects of PTDC (25 uM) treatment and neutralizing IL1β, IL6, or both on mRNA expression of SELP, CCL23, CXCL2, and CXCL3 in coculture respectively. (I) Effects of selective and cooperative RNAi of SELP, CCL23, CXCL2, and CXCL3 on the formation of cell aggregation structure. “+” stands for RNAi of indicated genes; “−” for nonspecific scramble siRNA control. ≥3 independent experimental fields were randomly selected for quantification, and data were represented as mean+SD. *P<0.05 and **P<0.01 versus control group.
To further demonstrate whether these NF-κB-modulated angiogenic chemotaxics-related genes might actively participate in early vascularization, we selectively or cooperatively knocked down the overlapped candidates and evaluated their potential contributions to the formation of cell aggregation structures. As expected, the selective or cooperative inhibition partially suppressed the formation of cell aggregation structures (Fig. 5I), which further indicated that these chemotaxics-related genes might function as a cocktail in the downstream for regulation of early prevascularization.
HMSCs were endowed with early smooth muscle differentiation upon coculture
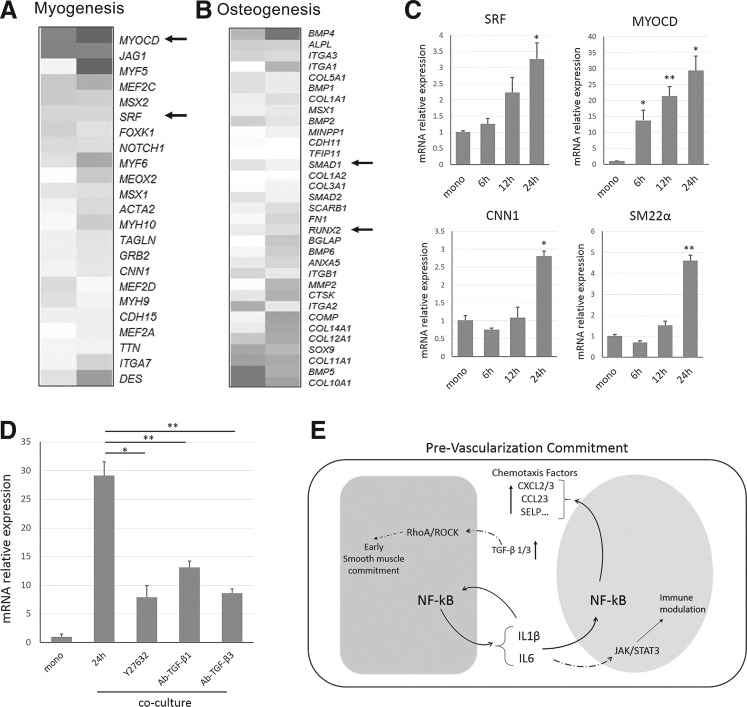
Besides the effects of HMSCs on the angiogenic activation of HUVECs in the coculture environment, we also analyzed the potential differentiation trends of HMSCs induced by coculture with terminally differentiated HUVECs. As MSCs were multipotent and showed both osteoblastic and myogenic trends under the influence of ECs in different situations, we comprehensively evaluated the multiple expression patterns on osteoblastic and myogenic differentiation markers under our experimental conditions. The unbiased analysis revealed that a proportion of the smooth muscle markers were induced within 24 h, including master transcriptional factors and cofactors for myogenesis (Fig. 6A and Supplementary Table S16). However, the osteoblast differentiation master transcriptional factors were not significantly induced within this short period, although alkaline phosphatase tissue-nonspecific isoform (ALPL) was overexpressed (Fig. 6B and Supplementary Table S17), which was consistent with other literatures [12,13, 26].
FIG. 6.
HMSC was endowed with early smooth muscle differentiation trends upon coculture. (A, B) Myogenic and osteoblastic differentiation markers were represented as in the heatmaps; heatmaps were ordered via the fold of induction in Log2 scale. Master regulatory factors in myogenesis and osteogeneis process were indicated by arrow. (C) Relative mRNA expressions of SRF, MYOCD, CNN1, and SM22a in HMSC after coculture within 24 h. (D) TGF-β1 and TGF-β3 might regulate myogenic differentiation via mediating MYOCD through RhoA/ROCK pathway. Relative mRNA expressions of MYOCD with different treatments were indicated. (E) Schematic of reciprocal mechanisms in HMSC-HUVEC contact communication mediated prevascularization. * Indicates P-value <0.05, ** indicates P-value <0.01.
We selectively validated the most-induced myogenic markers, including serum response factor (SRF), myocardin (MYOCD), calponin (CNN1), and smooth muscle 22α (SM22α) by qPCR (Fig. 6C), and the results were consistent with the expression trend in RNA-seq data. The induction of these genes was also mainly contact-dependent in the coculture (Supplementary Fig. S6A–D). CNN1 and SM22a were reported as smooth muscle-specific markers, but we did not detect any significant induction of skeleton or cardiac muscle markers (MyoD, MyoG, NKX2.5, a-MHC, b-MHC, and cTnT) either from RNA-seq data or qPCR (data not shown). Thus, we supposed this coculture tended to induce smooth muscle fate commitment within 24 h.
Noticeably, MYOCD, which encoded an SRF co-transcriptional factor, was the most significantly induced among these markers (Fig. 6C). As myogenic specific genes are transcriptionally activated by the SRF-MYOCD complex predominately and this mechanism was governed by the RhoA/ROCK signaling in the upstream [40–42], we treated the coculture environment with a specific RhoA/ROCK inhibitor, Y27632. The treatment with Y27632 did not obviously disturb the formation of cell aggregation structures, but suppressed the MYOCD elevation during coculture (Fig. 6D), indicating that RhoA/ROCK might also participate in the coculture-induced early myogenic differentiation of HMSCs. RhoA/ROCK signaling can be activated by various external signals, including TGF-β1 and TGF-β3 [43,44], which were significantly upregulated in both co-MSCs and co-HUVECs (Supplementary Fig. S7). Neutralization of TGF-β1 or TGF-β3 during the coculture partially repressed the induction of MYOCD (Fig. 6D), demonstrating that TGF-β1 and TGF-β3 might be two external signals induced during coculture, and act on HMSCs to trigger the early myogenic differentiation commitment within short period.
Discussion
Contact coculture of ECs with MSCs can induce vascularization both in vitro and in vivo [20,45–47], and thus is a promising solution to controlled vascularization enhancement in therapeutic needs. Here, we uncovered the underlying communication mechanism in MSCs-ECs coculture at molecular level by scrutinizing their transcriptional profiles separately. For the real therapeutic application, certain specific cell types such as endothelial colony-forming cell, which can be readily isolated from human cord blood and have a higher proliferative potential than mature EC, may become more ideal EC sources [48]. Therefore, our data here may be also illustrative for MSC coculture with these specific ECs.
To obtain precise gene expression profiles without mixing with the other type of cells after intensive contact coculture, our preliminary studies focused on a strict separation of two cell types. In previous works studying the direct contact interaction between heterogonous cell types, cell types after coculture were sorted by either labeling cell types with cell tracker dyes [49,50] or using specific surface antigens [13]. Our preliminary test indicates that these separation methods might yield a small false positive or false negative rate in the separated cell populations, and in the downstream, might affect the mRNA profiling such as secreting factors and differentiation markers, in which the expression levels might be highly varied. To maximize the precision in this process, we applied GFP stably expressed primary HMSCs in the coculture experiment for sorting HMSCs with FACS, and further chose the anti-CD31-positive (an EC-specific surface marker) subpopulation in the GFP-negative population as ECs. This separation procedure maximally guaranteed the sorting purity after intensive cell–cell interaction. During RNA sequencing, we also strictly excluded those differentially expressed genes that were potentially caused by slight mixture of the other type of cells.
Based on the RNA-seq data, the unbiased analysis revealed that a proportion of angiogenesis- related genes including cytokines, chemokines, and adhesion molecules were significantly upregulated, which was consistent well with the formation of angiogenic cell aggregation structures observed in our coculture model in vitro and other similar experimental settings [13,22,32]. Although at least several weeks will be taken by general transplantation to form possible functional blood vessels in vivo [8–10,17], evidence from this work suggested that direct contact communication between HMSCs and HUVECs even in a short period (within 24 h) was potent enough for commitment of early prevascularization. To determine the key regulators mediating this process, we first inspected the gene expression levels of the most common angiogenic factors such as VEGF [51], FGF [33], and ANGPT family [34] that were widely reported as critical for angiogenesis induction. As a result, none of these factors was significantly induced in the coculture condition within this short time period. Alternatively, a proportion of most significantly induced genes were among the NF-κB targets, which remind us the activation of NF-κB signaling. Unbiased NF-κB target analysis based on a public database indicated that a significant number of NF-κB-regulated genes were upregulated from our RNA-seq data, which strengthened our hypothesis. We proved its activation by determining an increased p65 translocation into the nucleus upon coculture, since phosphorylated IκB can liberate the p65 in the cytoplasm so that p65 will shutter into the nuclear compartment upon the activation of NF-κB signaling [52]. NF-κB plays a key role in regulating a wide range of physiological processes, including inflammation, cancer progression, synaptic plasticity, and angiogenic response [52–54]. NF-κB may regulate the angiogenic potential and directional cell migration via stimulating adhesion molecules and cytokines, especially in tumor models and immune response [27,29,54,55]. In this work, we found NF-κB signaling was also involved in the prevascularization induced by ECs-MSCs coculture.
We then found that HMSCs secreted two potent ILs, IL1β and IL6, which were responsible for the activation of NF-κB in co-HUVECs (also moderately in co-HMSCs). These two factors were among the reported NF-κB activators only in both types of cells that were significantly induced both at mRNA and secreted protein levels, and their induction was strictly regulated by a contact coculture model. Considering that only direct contact coculture (rather than Transwell or conditioned medium) induce the ILs expression and following process, it is likely that interplay with heterotypic cell junction communication attribute to the initial ILs expression in MSC. Literatures on coculture experiments provided evidence for this hypothesis, for example gap junction channel which consisted of Cx43 was observed in contact HUVEC and osteoprogenitor cell coculture situation, and a gap junction blocker could effectively block the contact coculture induced cell differentiation [56]. Besides, as IL1β and IL6 belong to the important immune regulators [52], it is also possible that initial raising of these IL factors may be caused by a quick immune response between heterogeneous cell interactions.
Neutralizing these two ILs significantly abolished the NF-κB activation and cell aggregation formation in the coculture environment. Both IL1β and IL6 may be expressed in MSCs [27,57,58]. IL1β is regarded as a typical NF-κB activator for long time, and its elevation potently supports the NF-κB activation. In our context, IL1β was more potent in activation of NF-κB than IL6. IL6 may function in a more intricate manner, and was classically regarded as an activator of JAK/STAT3 signaling, which can also regulate angiogenesis parallel to NF-κB. In this work, we also demonstrated that IL6 could concomitantly function with IL1β to induce and maintain the NF-κB signaling as reported in intestinal epithelia [59]. One possible mechanism of the IL-6-participated NF-κB activation is attributed to the crosslink between STAT3 and NF-κB [59–61]. Interestingly, IL1β and IL6 are both NF-κB targets and activators, and there was a NF-κB positive feedback loop in this context, suggesting this loop might be essential for sustainable NF-κB activation during the coculture-induced prevascularization. Our evidences collectively demonstrated that HMSCs could activate NF-κB signaling in neighbor HUVECs via a paracrine mechanism.
Notably, MSCs were increasingly recognized as capable of dampening allogeneic immune response during transplantation by weakening the recruitment of innate immune cells [2,27]. Although NF-κB is widely regarded as the proinflammatory signaling in many other contexts [53], our high-throughput data revealed that this coculture-based vascularization model with NF-κB participation did not actually evoke most relevant proinflammatory cytokines, including vascular cell adhesion molecule, intercellular adhesion molecule, and selectin E [62] in ECs. On one hand, we inferred that NF-κB induced in this context was relatively mild compared with TNF-α-mediated transient activation and thus might not be deleterious to both cell types or enhance inflammation. On the other hand, IL6 was also immunosuppressive via modulating JAK/STAT3 signaling. In the circumstance of MSC-controlled immunosuppressive machinery, MSCs secreted IL6 as anti-immflamatory effect through activating SOCS3 via JAK/STAT3 in ECs and thereby alleviated the recruitment of neutrophils or lymphocytes [27]. Therefore, we supposed that IL6 was likely involved in both JAK/STAT3 and NF-κB signaling as dual functions: IL6 participates in angiogenesis via NF-κB and safeguards immunosuppression in the coculture.
Since angiogenic cell aggregation structures were formed by cell–cell recruitment, we focused on those significantly induced NF-κB targets that may be responsible for chemotaxis. We selected four chemotaxis-related molecules for further study, as all these genes were tightly regulated by IL1β- and IL6-potentiated NF-κB signaling. The results showed that these molecules might cooperatively contribute to the dynamic cell–cell recruitments. However, incubation of monocultured HUVECs in the coculture conditioned medium failed to induce the formation of cell aggregation structures, indicating the complexity of contact-dependent communication and the importance of chemotaxis machinery, even though this conditioned medium might contain higher concentrations of angiogenic molecules and IL1β/IL6.
Many coculture models indicated that MSC differentiation can be affected by various heterogenous cells, while MSC differentiation induction by ECs is intricate [11–14,18,49]. In this work, the comprehensive gene expression profiling after strict separation demonstrated that intensive MSCs-ECs interaction within 24 h might be sufficient to determine MSC commitment toward early smooth muscle lineage, but there was no obvious osteogenic trend in this context. We validated that the typical smooth muscle markers including SRF, MYOCD, CNN1, and SM22α were elevated only in direct contact manner. MYOCD, which encodes a SRF co-transcriptional factor, was identified as the most significantly induced one among these markers, consistent with a previous observation in HMSCs and a mouse EC coculture model [17]. SRF-MYOCD was the master transcriptional factor activating the myogenic genes [63–65], which in the upstream were regulated by RhoA/ROCK signaling [41,42,44]. We hypothesized that the coculture stimulated the secretion of signaling factors and the remodeling of extracellular matrix, which collectively activated the RhoA/ROCK-dependent myogenic differentiation. TGF-β1 and TGF-β3 among external RhoA/ROCK signaling activators [43,44] were upregulated in both co-HMSCs and co-HUVECs, and the subsequent experimental evidence suggested that TGF-β1/3 might participate in the coculture-raised smooth muscle differentiation through modulating MYOCD expression. Overall, this early identity commitment of MSCs stimulated by coculture may be essential for further development into the mural cell types so as to surround and stabilize the inner vascular wall composed mainly by ECs in long-term transplantation in vivo [8,15,16].
Supplementary Material
Acknowledgments
This research was supported by Natural Sciences Foundation of China (grant no. 31170940), National High Technology Research and Development (863 Program) (grant no. 2012AA020503, 2013AA020301, and 2012AA02A700), 973 Basic Research Fund (grant no. 2012CB725204) and Tsinghua University Initiative Scientific Research Program (no. 20131089199). We also acknowledge the China National Center for Protein Sciences Beijing for providing the facility support.
Author Disclosure Statement
All authors declared no conflicts of interest.
References
- 1.Abedin M, Tintut Y. and Demer LL. (2004). Mesenchymal stem cells and the artery wall. Circ Res 95:671–676 [DOI] [PubMed] [Google Scholar]
- 2.Williams AR. and Hare JM. (2011). Mesenchymal stem cells: biology, pathophysiology, translational findings, and therapeutic implications for cardiac disease. Circ Res 109:923–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pontikoglou C, Deschaseaux F, Sensebe L. and Papadaki HA. (2011). Bone marrow mesenchymal stem cells: biological properties and their role in hematopoiesis and hematopoietic stem cell transplantation. Stem Cell Rev 7:569–589 [DOI] [PubMed] [Google Scholar]
- 4.Pati S, Khakoo AY, Zhao J, Jimenez F, Gerber MH, Harting M, Redell JB, Grill R, Matsuo Y, et al. (2011). Human mesenchymal stem cells inhibit vascular permeability by modulating vascular endothelial cadherin/beta-catenin signaling. Stem Cells Dev 20:89–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duffy GP, Ahsan T, O'Brien T, Barry F. and Nerem RM. (2009). Bone marrow-derived mesenchymal stem cells promote angiogenic processes in a time- and dose-dependent manner in vitro. Tissue Eng Part A 15:2459–2470 [DOI] [PubMed] [Google Scholar]
- 6.Auger FA, Gibot L. and Lacroix D. (2013). The pivotal role of vascularization in tissue engineering. Annu Rev Biomed Eng 15:177–200 [DOI] [PubMed] [Google Scholar]
- 7.Lovett M, Lee K, Edwards A. and Kaplan DL. (2009). Vascularization strategies for tissue engineering. Tissue Eng Part B Rev 15:353–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koike N, Fukumura D, Gralla O, Au P, Schechner JS. and Jain RK. (2004). Tissue engineering: creation of long-lasting blood vessels. Nature 428:138–139 [DOI] [PubMed] [Google Scholar]
- 9.Wang ZZ, Au P, Chen T, Shao Y, Daheron LM, Bai H, Arzigian M, Fukumura D, Jain RK. and Scadden DT. (2007). Endothelial cells derived from human embryonic stem cells form durable blood vessels in vivo. Nat Biotechnol 25:317–318 [DOI] [PubMed] [Google Scholar]
- 10.Samuel R, Daheron L, Liao S, Vardam T, Kamoun WS, Batista A, Buecker C, Schafer R, Han X, et al. (2013). Generation of functionally competent and durable engineered blood vessels from human induced pluripotent stem cells. Proc Natl Acad Sci U S A 110:12774–12779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saleh FA, Whyte M, Ashton P. and Genever PG. (2011). Regulation of mesenchymal stem cell activity by endothelial cells. Stem Cells Dev 20:391–403 [DOI] [PubMed] [Google Scholar]
- 12.Kaigler D, Krebsbach PH, West ER, Horger K, Huang YC. and Mooney DJ. (2005). Endothelial cell modulation of bone marrow stromal cell osteogenic potential. FASEB J 19:665–667 [DOI] [PubMed] [Google Scholar]
- 13.Bidarra SJ, Barrias CC, Barbosa MA, Soares R, Amedee J. and Granja PL. (2011). Phenotypic and proliferative modulation of human mesenchymal stem cells via crosstalk with endothelial cells. Stem Cell Res 7:186–197 [DOI] [PubMed] [Google Scholar]
- 14.Ball SG, Shuttleworth AC. and Kielty CM. (2004). Direct cell contact influences bone marrow mesenchymal stem cell fate. Int J Biochem Cell Biol 36:714–727 [DOI] [PubMed] [Google Scholar]
- 15.Toma C, Pittenger MF, Cahill KS, Byrne BJ. and Kessler PD. (2002). Human mesenchymal stem cells differentiate to a cardiomyocyte phenotype in the adult murine heart. Circulation 105:93–98 [DOI] [PubMed] [Google Scholar]
- 16.Tsuji H, Miyoshi S, Ikegami Y, Hida N, Asada H, Togashi I, Suzuki J, Satake M, Nakamizo H, et al. (2010). Xenografted human amniotic membrane-derived mesenchymal stem cells are immunologically tolerated and transdifferentiated into cardiomyocytes. Circ Res 106:1613–1623 [DOI] [PubMed] [Google Scholar]
- 17.Au P, Tam J, Fukumura D. and Jain RK. (2008). Bone marrow-derived mesenchymal stem cells facilitate engineering of long-lasting functional vasculature. Blood 111:4551–4558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grellier M, Bordenave L. and Amedee J. (2009). Cell-to-cell communication between osteogenic and endothelial lineages: implications for tissue engineering. Trends Biotechnol 27:562–571 [DOI] [PubMed] [Google Scholar]
- 19.Chen DY, Wei HJ, Lin KJ, Huang CC, Wang CC, Wu CT, Chao KT, Chen KJ, Chang Y. and Sung HW. (2013). Three-dimensional cell aggregates composed of HUVECs and cbMSCs for therapeutic neovascularization in a mouse model of hindlimb ischemia. Biomaterials 34:1995–2004 [DOI] [PubMed] [Google Scholar]
- 20.Lee WY, Tsai HW, Chiang JH, Hwang SM, Chen DY, Hsu LW, Hung YW, Chang Y. and Sung HW. (2011). Core-shell cell bodies composed of human cbMSCs and HUVECs for functional vasculogenesis. Biomaterials 32:8446–8455 [DOI] [PubMed] [Google Scholar]
- 21.Li H, Daculsi R, Grellier M, Bareille R, Bourget C, Remy M. and Amedee J. (2011). The role of vascular actors in two dimensional dialogue of human bone marrow stromal cell and endothelial cell for inducing self-assembled network. PLoS One 6:e16767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grellier M, Ferreira-Tojais N, Bourget C, Bareille R, Guillemot F. and Amedee J. (2009). Role of vascular endothelial growth factor in the communication between human osteoprogenitors and endothelial cells. J Cell Biochem 106:390–398 [DOI] [PubMed] [Google Scholar]
- 23.Carrion B, Kong YP, Kaigler D. and Putnam AJ. (2013). Bone marrow-derived mesenchymal stem cells enhance angiogenesis via their alpha6beta1 integrin receptor. Exp Cell Res 319:2964–2976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Melero-Martin JM, De Obaldia ME, Kang SY, Khan ZA, Yuan L, Oettgen P. and Bischoff J. (2008). Engineering robust and functional vascular networks in vivo with human adult and cord blood-derived progenitor cells. Circ Res 103:194–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kunz-Schughart LA, Schroeder JA, Wondrak M, van Rey F, Lehle K, Hofstaedter F. and Wheatley DN. (2006). Potential of fibroblasts to regulate the formation of three-dimensional vessel-like structures from endothelial cells in vitro. Am J Physiol Cell Physiol 290:C1385–C1398 [DOI] [PubMed] [Google Scholar]
- 26.Li H, Xue K, Kong N, Liu K. and Chang J. (2014). Silicate bioceramics enhanced vascularization and osteogenesis through stimulating interactions between endothelia cells and bone marrow stromal cells. Biomaterials 35:3803–3818 [DOI] [PubMed] [Google Scholar]
- 27.Luu NT, McGettrick HM, Buckley CD, Newsome P, Ed RG, Frampton J. and Nash GB. (2013). Crosstalk between mesenchymal stem cells and endothelial cells leads to down-regulation of cytokine-induced leukocyte recruitment. Stem Cells 31:2690–2702 [DOI] [PubMed] [Google Scholar]
- 28.Matsushita T, Kibayashi T, Katayama T, Yamashita Y, Suzuki S, Kawamata J, Honmou O, Minami M. and Shimohama S. (2011). Mesenchymal stem cells transmigrate across brain microvascular endothelial cell monolayers through transiently formed inter-endothelial gaps. Neurosci Lett 502:41–45 [DOI] [PubMed] [Google Scholar]
- 29.Teo GS, Ankrum JA, Martinelli R, Boetto SE, Simms K, Sciuto TE, Dvorak AM, Karp JM. and Carman CV. (2012). Mesenchymal stem cells transmigrate between and directly through tumor necrosis factor-alpha-activated endothelial cells via both leukocyte-like and novel mechanisms. Stem Cells 30:2472–2486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL. and Pachter L. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7:562–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang DW, Sherman BT. and Lempicki RA. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57 [DOI] [PubMed] [Google Scholar]
- 32.Aguirre A, Planell JA. and Engel E. (2010). Dynamics of bone marrow-derived endothelial progenitor cell/mesenchymal stem cell interaction in co-culture and its implications in angiogenesis. Biochem Biophys Res Commun 400:284–291 [DOI] [PubMed] [Google Scholar]
- 33.Tsuda S, Ohtsuru A, Yamashita S, Kanetake H. and Kanda S. (2002). Role of c-Fyn in FGF-2-mediated tube-like structure formation by murine brain capillary endothelial cells. Biochem Biophys Res Commun 290:1354–1360 [DOI] [PubMed] [Google Scholar]
- 34.Mochizuki Y, Nakamura T, Kanetake H. and Kanda S. (2002). Angiopoietin 2 stimulates migration and tube-like structure formation of murine brain capillary endothelial cells through c-Fes and c-Fyn. J Cell Sci 115:175–183 [DOI] [PubMed] [Google Scholar]
- 35.Zacharek A, Chen J, Cui X, Li A, Li Y, Roberts C, Feng Y, Gao Q. and Chopp M. (2007). Angiopoietin1/Tie2 and VEGF/Flk1 induced by MSC treatment amplifies angiogenesis and vascular stabilization after stroke. J Cereb Blood Flow Metab 27:1684–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ogura H, Murakami M, Okuyama Y, Tsuruoka M, Kitabayashi C, Kanamoto M, Nishihara M, Iwakura Y. and Hirano T. (2008). Interleukin-17 promotes autoimmunity by triggering a positive-feedback loop via interleukin-6 induction. Immunity 29:628–636 [DOI] [PubMed] [Google Scholar]
- 37.Pugazhenthi S, Zhang Y, Bouchard R. and Mahaffey G. (2013). Induction of an inflammatory loop by interleukin-1beta and tumor necrosis factor-alpha involves NF-kB and STAT-1 in differentiated human neuroprogenitor cells. PLoS One 8:e69585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pahl HL. (1999). Activators and target genes of Rel/NF-kappa B transcription factors. Oncogene 18:6853–6866 [DOI] [PubMed] [Google Scholar]
- 39.Dimberg A. (2010). Chemokines in angiogenesis. In: The Chemokine System in Experimental and Clinical Hematology. Springer Berlin Heidelberg, pp 59–80 [Google Scholar]
- 40.Pagiatakis C, Gordon JW, Ehyai S. and McDermott JC. (2012). A novel RhoA/ROCK-CPI-17-MEF2C signaling pathway regulates vascular smooth muscle cell gene expression. J Biol Chem 287:8361–8370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wei L, Zhou W, Croissant JD, Johansen FE, Prywes R, Balasubramanyam A. and Schwartz RJ. (1998). RhoA signaling via serum response factor plays an obligatory role in myogenic differentiation. J Biol Chem 273:30287–30294 [DOI] [PubMed] [Google Scholar]
- 42.Jeon ES, Park WS, Lee MJ, Kim YM, Han J. and Kim JH. (2008). A Rho kinase/myocardin-related transcription factor-A-dependent mechanism underlies the sphingosylphosphorylcholine-induced differentiation of mesenchymal stem cells into contractile smooth muscle cells. Circ Res 103:635–642 [DOI] [PubMed] [Google Scholar]
- 43.Chen S. and Lechleider RJ. (2004). Transforming growth factor-beta-induced differentiation of smooth muscle from a neural crest stem cell line. Circ Res 94:1195–1202 [DOI] [PubMed] [Google Scholar]
- 44.Chen S, Crawford M, Day RM, Briones VR, Leader JE, Jose PA. and Lechleider RJ. (2006). RhoA modulates Smad signaling during transforming growth factor-beta-induced smooth muscle differentiation. J Biol Chem 281:1765–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fuchs S, Hofmann A. and Kirkpatrick C. (2007). Microvessel-like structures from outgrowth endothelial cells from human peripheral blood in 2-dimensional and 3-dimensional co-cultures with osteoblastic lineage cells. Tissue Eng 13:2577–2588 [DOI] [PubMed] [Google Scholar]
- 46.Baiguera S. and Ribatti D. (2013). Endothelialization approaches for viable engineered tissues. Angiogenesis 16:1–14 [DOI] [PubMed] [Google Scholar]
- 47.Hofmann A, Ritz U, Verrier S, Eglin D, Alini M, Fuchs S, Kirkpatrick CJ. and Rommens PM. (2008). The effect of human osteoblasts on proliferation and neo-vessel formation of human umbilical vein endothelial cells in a long-term 3D co-culture on polyurethane scaffolds. Biomaterials 29:4217–4226 [DOI] [PubMed] [Google Scholar]
- 48.Egorova AD, DeRuiter MC, de Boer HC, van de Pas S, Gittenberger-de Groot AC, van Zonneveld AJ, Poelmann RE. and Hierck BP. (2012). Endothelial colony-forming cells show a mature transcriptional response to shear stress. In Vitro Cell Dev Biol Anim 48:21–29 [DOI] [PubMed] [Google Scholar]
- 49.Xu J, Liu X, Chen J, Zacharek A, Cui X, Savant-Bhonsale S, Chopp M. and Liu Z. (2010). Cell-cell interaction promotes rat marrow stromal cell differentiation into endothelial cell via activation of TACE/TNF-alpha signaling. Cell Transplant 19:43–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Plotnikov EY, Khryapenkova TG, Vasileva AK, Marey MV, Galkina SI, Isaev NK, Sheval EV, Polyakov VY, Sukhikh GT. and Zorov DB. (2008). Cell-to-cell cross-talk between mesenchymal stem cells and cardiomyocytes in co-culture. J Cell Mol Med 12:1622–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cross MJ. and Claesson-Welsh L. (2001). FGF and VEGF function in angiogenesis: signalling pathways, biological responses and therapeutic inhibition. Trends Pharmacol Sci 22:201–207 [DOI] [PubMed] [Google Scholar]
- 52.Oeckinghaus A, Hayden MS. and Ghosh S. (2011). Crosstalk in NF-kappaB signaling pathways. Nat Immunol 12:695–708 [DOI] [PubMed] [Google Scholar]
- 53.Baker RG, Hayden MS. and Ghosh S. (2011). NF-kappaB, inflammation, and metabolic disease. Cell Metab 13:11–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wani AA, Jafarnejad SM, Zhou J. and Li G. (2011). Integrin-linked kinase regulates melanoma angiogenesis by activating NF-kappaB/interleukin-6 signaling pathway. Oncogene 30:2778–2788 [DOI] [PubMed] [Google Scholar]
- 55.Uchibori R, Tsukahara T, Mizuguchi H, Saga Y, Urabe M, Mizukami H, Kume A. and Ozawa K. (2013). NF-kappaB activity regulates mesenchymal stem cell accumulation at tumor sites. Cancer Res 73:364–372 [DOI] [PubMed] [Google Scholar]
- 56.Villars F, Guillotin B, Amedee T, Dutoya S, Bordenave L, Bareille R. and Amedee J. (2002). Effect of HUVEC on human osteoprogenitor cell differentiation needs heterotypic gap junction communication. Am J Physiol Cell Physiol 282:C775–C785 [DOI] [PubMed] [Google Scholar]
- 57.Kim J, Breunig MJ, Escalante LE, Bhatia N, Denu RA, Dollar BA, Stein AP, Hanson SE, Naderi N, et al. (2012). Biologic and immunomodulatory properties of mesenchymal stromal cells derived from human pancreatic islets. Cytotherapy 14:925–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bartosh TJ, Ylostalo JH, Bazhanov N, Kuhlman J. and Prockop DJ. (2013). Dynamic compaction of human mesenchymal stem/precursor cells into spheres self-activates caspase-dependent IL1 signaling to enhance secretion of modulators of inflammation and immunity (PGE2, TSG6, and STC1). Stem Cells 31:2443–2456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang L, Walia B, Evans J, Gewirtz AT, Merlin D. and Sitaraman SV. (2003). IL-6 induces NF-kappa B activation in the intestinal epithelia. J Immunol 171:3194–3201 [DOI] [PubMed] [Google Scholar]
- 60.Han SS, Yun H, Son DJ, Tompkins VS, Peng L, Chung ST, Kim JS, Park ES. and Janz S. (2010). NF-kappaB/STAT3/PI3K signaling crosstalk in iMyc E mu B lymphoma. Mol Cancer 9:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang J, Liao X, Agarwal MK, Barnes L, Auron PE. and Stark GR. (2007). Unphosphorylated STAT3 accumulates in response to IL-6 and activates transcription by binding to NFkappaB. Genes Dev 21:1396–1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Koch AE, Halloran MM, Haskell CJ, Shah MR. and Polverini PJ. (1995). Angiogenesis mediated by soluble forms of E-selectin and vascular cell adhesion molecule-1. Nature 376:517–519 [DOI] [PubMed] [Google Scholar]
- 63.Yoshida T, Sinha S, Dandre F, Wamhoff BR, Hoofnagle MH, Kremer BE, Wang DZ, Olson EN. and Owens GK. (2003). Myocardin is a key regulator of CArG-dependent transcription of multiple smooth muscle marker genes. Circ Res 92:856–864 [DOI] [PubMed] [Google Scholar]
- 64.Xie WB, Li Z, Miano JM, Long X. and Chen SY. (2011). Smad3-mediated myocardin silencing: a novel mechanism governing the initiation of smooth muscle differentiation. J Biol Chem 286:15050–15057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen J, Kitchen CM, Streb JW. and Miano JM. (2002). Myocardin, a component of a molecular switch for smooth muscle differentiation. J Mol Cell Cardiol 34:1345–1356 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.