Abstract
The nuclear envelope consists of inner and outer nuclear membranes. While the outer membrane is an extension of the endoplasmic reticulum, the inner nuclear membrane represents a unique membranous environment containing specific proteins. The mechanisms of integral inner nuclear membrane protein degradation are unknown. Here we investigated the turnover of Asi2, an integral INM protein in Saccharomyces cerevisiae. We report that Asi2 is degraded by the proteasome and independent of the vacuole exhibiting a half-life of ≈ 45 min. Asi2 exhibits enhanced stability in mutants lacking the E2 ubiquitin conjugating enzymes Ubc6 or Ubc7, or the E3 ubiquitin ligase Doa10. Consistently, Asi2 is post-translationally modified by poly-ubiquitylation in a Ubc7- and Doa10-dependent manner. Importantly Asi2 degradation is significantly reduced in a sts1-2 mutant that fails to accumulate proteasomes in the nucleus, indicating that Asi2 is degraded in the nucleus. Our results reveal a molecular pathway that affects the stability of integral proteins of the inner nuclear membrane and indicate that Asi2 is subject to protein quality control in the nucleus.
Keywords: ERAD, nuclear membrane, nuclear proteasome, protein degradation, ubiquitylation
Introduction
The confinement of the genome within the nucleus by the nuclear envelope is a hallmark of eukaryotic cells. The nuclear envelope is composed of two membrane layers, the inner and the outer nuclear membrane, which are connected at the sites of nuclear pore complexes (NPC) (Hetzer, 2010). The outer nuclear membrane (ONM) is a direct extension of the endoplasmic reticulum (ER) membrane and its protein composition closely resembles that of the ER membrane. In contrast, the inner nuclear membrane (INM) contains many specific proteins, which are involved in nuclear processes, such as chromatin organization, control of gene expression and DNA damage repair (Heessen and Fornerod, 2007; Mekhail et al., 2008; Oza et al., 2009; Schober et al., 2009). Although the ONM and INM form a continuous membrane system, the NPCs that assemble at the junction of the two membranes establish a physical barrier limiting diffusion and free exchange of integral membrane proteins between the ER/ONM and INM. Several models have been proposed to mechanistically explain the transport of membrane proteins from the site of their synthesis and membrane insertion at the ER to their final destination in the INM. These include the diffusion – nuclear retention model and the nuclear localization signal (NLS)-mediated transport through peripheral channels of the NPC (reviewed in (Zuleger et al., 2012)) and, more recently, the NLS mediated transport through the central NPC channel (Meinema et al., 2011). In contrast to their nuclear targeting, the fate of the integral membrane proteins once they have reached the INM is less clear. Whether integral INM proteins are subject to turnover, and if so, what degradative pathway is involved, remains an intriguing and unsolved biological question.
Protein degradation is a key regulatory mechanism controlling cellular processes like gene transcription and cell cycle progression, and is also crucial in protein quality control pathways protecting cells from accumulation of aberrant proteins (Goldberg, 2003). The majority of cellular proteins exhibit characteristic rates of turnover with varying half-lives ranging from a few minutes to a few days (Belle et al., 2006; Ciechanover, 2007; Yen et al., 2008). However, studies in Caenorhabditis elegans and rat brain revealed that some components of the nuclear membrane – the scaffold nucleoporins embedded in the nuclear pore membrane - are extremely long lived proteins that appear stable for the entire life-span of a metazoan organism without any significant turnover (D’Angelo et al., 2009; Savas et al., 2012). It remains unclear whether the stability of these proteins is due to the inability of the degradation machinery to gain access to them, or because the components involved in NE-associated protein degradation are missing in the nucleus.
Cells possess two major sites of protein degradation; the lysosome (or its yeast analogue the vacuole) and the proteasome, a large multi-catalytic protease complex that degrades poly-ubiquitylated proteins (Ciechanover, 2007). Proteasomal degradation substrates include misfolded or damaged proteins, as well as properly folded proteins whose levels need to be downregulated, such as cell cycle regulators or transcription factors. Protein ubiquitylation is required as a signal for proteasomal targeting and is mediated by three classes of enzymes, ubiquitin (Ub)-activating enzyme (E1), Ub-conjugating enzyme (Ubc or E2) and a substrate specific Ub-protein ligase (E3). While a large portion of proteasomes localizes to the nucleus (Enenkel et al., 1998; Peters et al., 1994; Russell et al., 1999) and protein degradation clearly can occur in the nucleus, the best characterized protein quality control (PQC) degradation pathways are localized in the cytoplasm (Goldberg, 2003). There are only a limited number of studies providing evidence for degradation-mediated PQC in the nucleus (Furth et al., 2011; Gardner et al., 2005; Iwata et al., 2009). In particular, the molecular pathways that may govern the turnover of integral membrane proteins of the INM are not known.
Doa10 is an integral membrane E3 ubiquitin ligase that partially localizes to the INM, where it participates in the degradation of a soluble non-membrane bound nuclear transcription factor Matα2 (Deng and Hochstrasser, 2006). Doa10 also mediates degradation of a mutant kinetochore protein Ndc10-2 (Furth et al., 2011; Ravid et al., 2006). Furthermore, Doa10 has a well-established role as one of the two major ubiquitin ligases facilitating ER-associated degradation (ERAD). ERAD primarily targets damaged or misfolded proteins in the ER membrane and lumen to the proteasomes, but it also regulates physiological levels of some correctly folded wild-type proteins (Hegde and Ploegh, 2010). Doa10 functions with E2 enzymes Ubc6 and Ubc7 (Swanson et al., 2001), which have also been found at the INM (Deng and Hochstrasser, 2006). Ubc6 is an integral membrane protein, whereas Ubc7 lacks transmembrane domains and is tethered to the membrane via interaction with an integral membrane protein, Cue1. Interestingly, Ubc6 is itself a substrate for Doa10-dependent degradation (Swanson et al., 2001; Walter et al., 2001). Membrane extraction of ubiquitylated ER membrane protein substrates is mediated by the ATPase Cdc48 protein complex or in some cases by the proteasome itself (Vembar and Brodsky, 2008). Cdc48 is known to enter the nucleus in a cell cycle dependent manner (Madeo et al., 1998). Mammalian cells possess TEB4 (Kreft et al., 2006), an orthologue of Doa10, and there are orthologues of Ubc6 and Ubc7 in mammalian cells as well (Kostova et al., 2007; Vembar and Brodsky, 2008).
In this study we analyze degradation of Asi2, an integral INM protein in Saccharomyces cerevisiae. Asi2 is a 33 kDa protein with two membrane-spanning segments and a large 26 kDa N-terminal domain oriented towards the nucleoplasm (Zargari et al., 2007). Asi2 is a negative regulatory component of the amino acid induced Ssy1-Ptr3-Ssy5 (SPS) sensing pathway (Forsberg et al., 2001; Ljungdahl and Daignan-Fornier, 2012). In the absence of inducing amino acids, Asi2 functions together with two other INM proteins, Asi1 and Asi3, to prevent promoter binding of transcription factors Stp1 and Stp2 (Boban et al., 2006; Zargari et al., 2007). We report that Asi2 is ubiquitylated in an Ubc6, Ubc7, and Doa10-dependent manner and is subsequently targeted for degradation by proteasomes in the nucleus. This study represents the first example of a mechanism for the turnover of an integral component of the INM.
Results
Asi2 exhibits turnover independent of SPS-sensor signaling
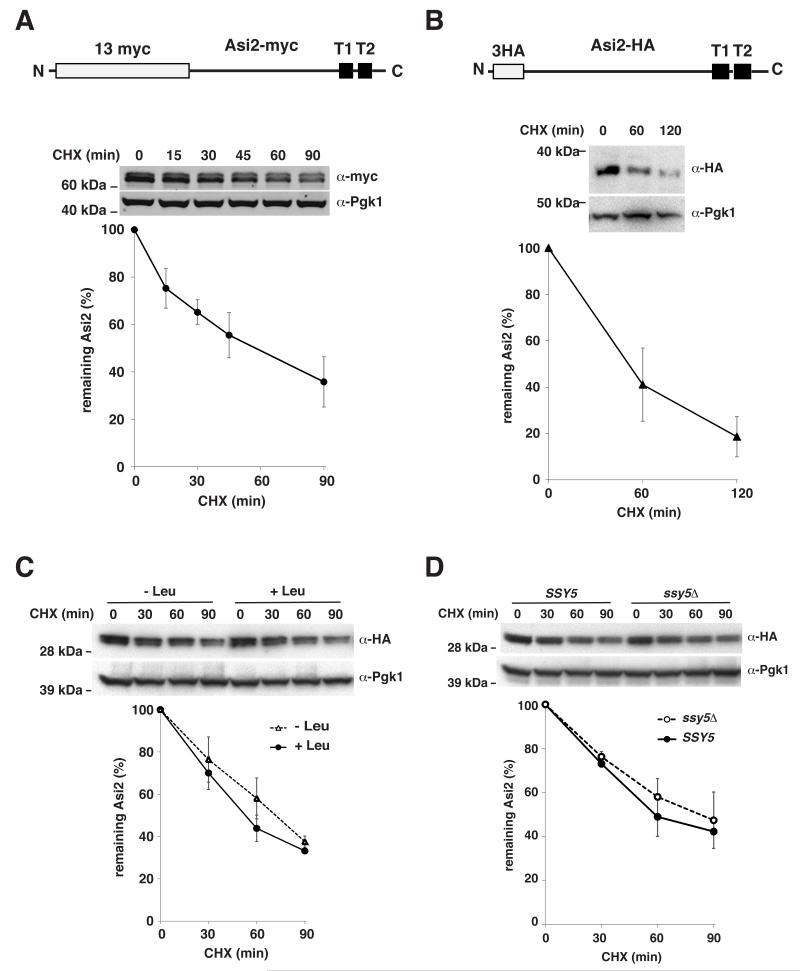
In order to characterize the degradation of the INM protein Asi2 we first investigated the turnover rate of functional Asi2 proteins carrying N-terminal myc or HA epitope tags (Zargari et al., 2007). During cycloheximide (0.36 mM) treatment for up to 120 min to inhibit new protein synthesis, we followed the decrease in protein levels and determined the half-life of myc-tagged and HA-tagged Asi2 (Fig. 1). Asi2 protein levels were analyzed by immunoblotting with anti-myc or anti-HA antibodies. Asi2-myc exhibited a half-life of 43 min (Fig. 1A). A slightly lower degradation rate (half-life, 51 min) was measured for HA-epitope tagged Asi2 (Fig. 1B). Asi2 resolves as two specific protein bands, suggesting that Asi2 is subject to post-translational modification (Zargari et al., 2007); the nature of the modification is currently unknown. Although Asi2 has two potential N-glycosylation sites, treatment with endoglycosidase-H (endo-H) does not affect the electrophoretic mobility of either of the Asi2 bands (Zargari et al., 2007).
Fig. 1. Asi2 protein is constitutively turned over.
Asi2 protein stability was assessed by cycloheximide (CHX) chase. Cells were harvested at indicated time points after CHX addition and assayed by immunoblotting with anti-myc, anti-HA and anti-Pgk1 antibodies. Pgk1 is a stable protein used as a control. Graphs represent percentage of remaining Asi2 after CHX addition. Average values and standard deviation of indicated number of independent samples are shown. P-values are listed in Table IV, supplementary material. 13myc and 3HA epitope-tagged Asi2 constructs are represented schematically in A and B. Epitope tags and transmembrane segments (T1, T2) are indicated. (A) CHX chase of Asi2-myc (pMB55) in asi2Δ strain (MBY163). Asi2-myc protein half-life: 43 min (n=3). (B) CHX chase of Asi2-HA (pMS1) expressed in asi2Δ strain (MBY163). Asi2-HA protein half-life: 51 min. (n=3). (C) Cells (PLY1340) expressing Asi2-HA (pMS01) were grown in the SD medium and where indicated, 1.3 mM L-leucine was added for 1 hour to induce the SPS sensor. CHX chase was performed. Asi2 protein half-life: 50 min (SD+Leu) and 63 min (SD-Leu), n=3. (D) Asi2-HA (pMS01) stability was examined by CHX chase in SSY5 (PLY1340) and ssy5Δ (PLY1632) strain lacking functional SPS sensor. Asi2 protein half-life: 56 min (SSY5, n=2) and 63 min (ssy5Δ, n=3).
Asi2 functions as a negative regulator of the amino acid-induced SPS-sensor dependent pathway regulating gene expression (Zargari et al., 2007). Although the precise mechanism has yet to be determined, Asi2 appears to function by restricting access of the transcription factors Stp1 and Stp2 to SPS-sensor regulated promoters. In cells lacking Asi2, Stp1 and Stp2 inappropriately bind promoters and ectopically induce gene expression (Boban et al., 2006; Zargari et al., 2007). We examined the possibility that the stability of Asi2 is coupled to amino acid-induced signaling. If so, we posited that under amino acid-inducing conditions (+Leu), Asi2 would exhibit decreased stability. This would remove Asi2 as an inhibitory component and would be expected to contribute to SPS-sensor signaling. Using two approaches we compared Asi2-HA protein stability in non-induced and amino acid-induced cells. First, we tested Asi2-HA protein stability in the absence and presence of extracellular amino acids (Fig. 1C). We observed only a minor difference in Asi2 degradation rates. In the second approach, we examined Asi2-HA protein stability in a ssy5Δ mutant, which lacks the Stp1/Stp2 processing endoprotease component of the SPS-sensor (Andréasson et al., 2006; Andréasson and Ljungdahl, 2002; Pfirrmann et al., 2010) and is defective in SPS-sensor-dependent signaling (Fig. 1D). Again Asi2 protein stability was only nominally increased in the ssy5Δ mutant as compared to the wild-type (Fig. 1D). Together these results indicate that Asi2 protein degradation is independent of the SPS-sensor signaling.
Asi2 is degraded by the ubiquitin-proteasome system and independent of the vacuole
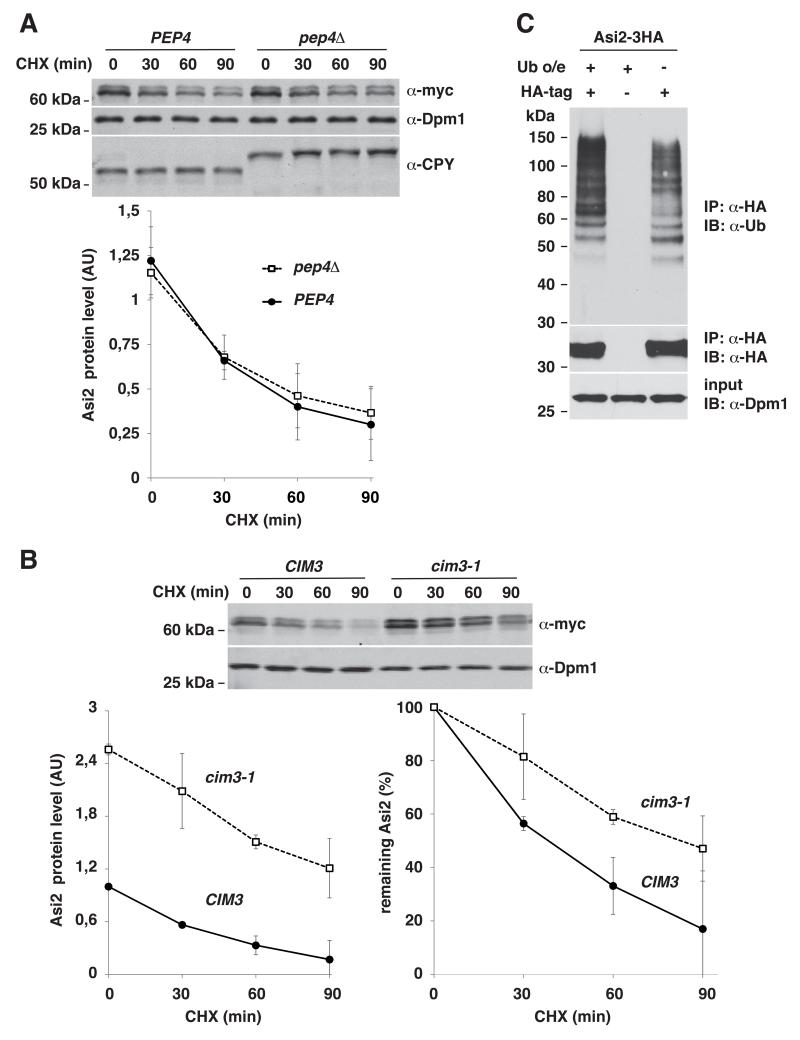
Different pathways could account for Asi2 protein degradation. First, we tested whether vacuolar function is necessary for Asi2 degradation. We analyzed Asi2 protein stability in a pep4Δ mutant yeast strain with impaired vacuolar function. Pep4 is a vacuolar aspartic proteinase required for proteolytic activation of many vacuolar zymogens, such as carboxypeptidase Y (CPY) (Ammerer et al., 1986; Woolford et al., 1986). Zymogens and proteins normally degraded in the vacuole are stabilized in the pep4Δ deletion mutant. We found that Asi2 stability was not affected in the pep4Δ mutant, indicating that the vacuole is not involved in Asi2 protein degradation (Fig. 2A).
Fig. 2. Asi2 protein is degraded by the ubiquitin-proteasome system.
Cycloheximide (CHX) chase (A, B) was performed as described in Fig. 1. Graphs represent Asi2 protein levels in each strain (arbitrary units, AU) or percentage of remaining Asi2. Average values and standard deviation of three independent samples are shown. P-values are listed in Table IV, supplementary material. (A) Asi2 protein degradation is independent of PEP4. CHX chase of Asi2-myc (pMB55) in a PEP4 (MBY163) and pep4Δ strain (MBY167) was assayed by immunoblotting with anti-myc and anti-Dpm1 antibodies. Cleavage of vacuolar carboxypeptidase Y (CPY), a Pep4 substrate, was examined using anti-CPY antibody. Dpm1 is a stable protein used as a control. (B) Asi2 protein is stabilized in a cim3-1 mutant. CHX chase of Asi2-myc (pMB55) expressed in CIM3 (MBY178) and cim3-1 thermosensitive mutant (MBY179). Growing cultures (27°C) were concentrated to OD600 1.5, incubated for 15 min at 27°C and following a 30 min incubation at 37°C, CHX was added. Immunoblotting was done as in (A). Asi2 protein half-life: 35 min (CIM3) and 81 min (cim3-1). (C) Asi2 protein is poly-ubiquitylated. Immunoprecipitation of HA-tagged Asi2 (pMB3) or untagged Asi2 (pMB128) expressed in cim3-1 strain (PLY1348) carrying ubiquitin-overexpression (Ub o/e) plasmid (myc-Ub/LEU2 2μ) or empty vector (pRS315) was performed using anti-HA antibody (IP: α-HA). Immunoblot (IB) analysis was done using anti-HA, anti-ubiquitin and anti-Dpm1 antibodies.
Next, we tested whether Asi2 is degraded by the proteasome. We examined Asi2 protein stability in a yeast strain carrying a temperature-sensitive cim3-1 mutation that impairs the activity of the Rpt6 (Cim3) ATPase in the proteasomal regulatory particle (Ghislain et al., 1993; Schork et al., 1995). At the restrictive temperature of 37 °C, Asi2 protein levels were significantly increased and Asi2 turnover was greatly slowed in the cim3-1 mutant (half-life, 81 min), as compared to the wild-type (half-life, 35 min). These findings are consistent with Asi2 being a substrate for proteasomal degradation (Fig. 2B).
As proteins are usually targeted to the proteasome by post-translational poly-ubiquitin modification, we examined whether Asi2 is ubiquitylated. To enrich ubiquitylated forms of Asi2 in the cell, Asi2-HA was expressed in a cim3-1 mutant (see above) with or without overexpression of ubiquitin. Anti-ubiquitin immunoblot analysis of the immunoprecipitated Asi2 clearly showed that Asi2 is modified by poly-ubiquitylation (Fig. 2C). Notably, ubiquitylated species of Asi2 were detected both in cells overexpressing ubiquitin from a plasmid and in cells expressing endogenous levels of ubiquitin (Fig. 2C, lanes 1 and 3, respectively). Together these data indicate that Asi2 is targeted for degradation by the ubiquitin-proteasome pathway.
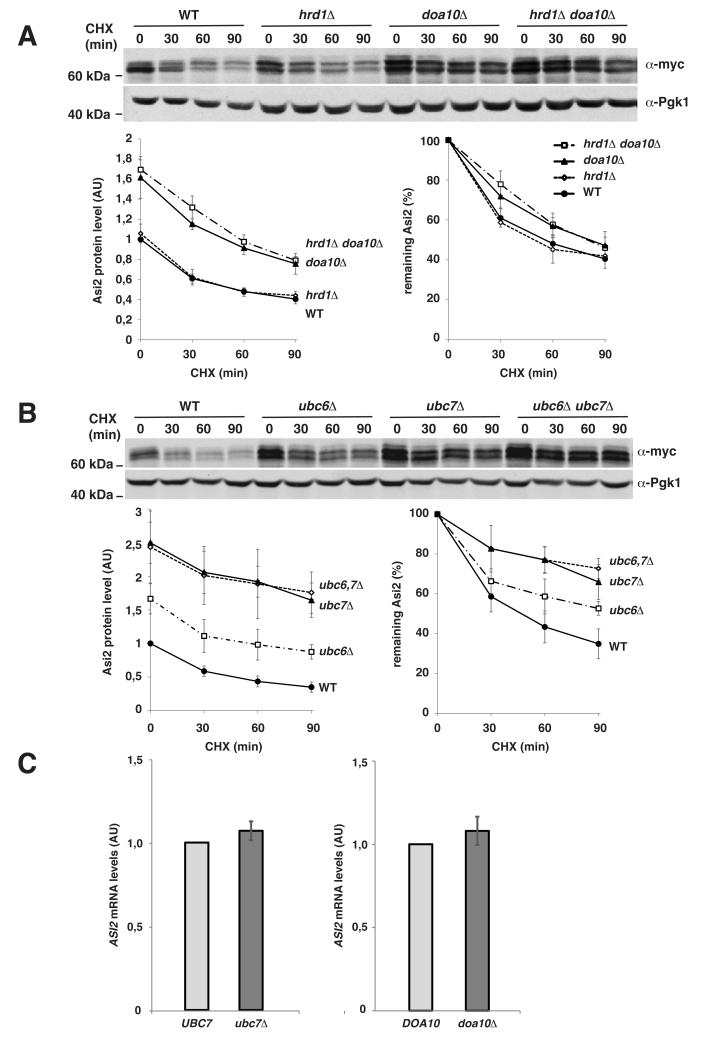
Asi2 degradation is impaired in mutants lacking ubiquitylation components Doa10, Ubc6 and Ubc7
Since Asi2 is an integral component of the INM we tested the requirement of the ER-associated E3 ubiquitin ligase Doa10 that is known to partially localize also to the INM (Deng and Hochstrasser, 2006). We examined whether Asi2 degradation required Doa10 and its associated E2 enzymes Ubc6 and Ubc7 (Swanson et al., 2001). We also examined Asi2 stability in a mutant lacking E3 ubiquitin ligase Hrd1, an integral membrane protein of the ER involved in ubiquitylation of several ERAD substrates (Vembar and Brodsky, 2008), that functions primarily with E2 enzyme Ubc7 (Bays et al., 2001; Deak and Wolf, 2001). In contrast to Doa10, Hrd1 has not been found in the INM (Deng and Hochstrasser, 2006). While the deletion of HRD1 did not significantly alter Asi2 stability, Asi2 was stabilized in the deletion mutant lacking DOA10 (Fig. 3A). Furthermore, the stability of Asi2 was similar in the doa10 Δ mutant and in the doa10Δ hrd1Δ double mutant cells. These data indicate that a Doa10-dependent pathway contributes to targeting Asi2 for degradation. Next, we examined stability of Asi2 in ubc6Δ, ubc7Δ single and ubc6Δ ubc7Δ double mutants. Consistent with our finding that Asi2 is stabilized in a doa10Δ mutant, Asi2 was also stabilized in mutants lacking either Ubc6 or Ubc7 (Fig. 3B). These latter observations are consistent with yeast two hybrid data indicating that Ubc6 and Ubc7 may interact in a protein complex (Chen et al., 1993). However, we noted that Asi2 is more stable in the ubc7Δ than in ubc6Δ mutant. The underlying basis for the greater stability in the ubc7Δ mutant is not clear.
Fig. 3. Asi2 is targeted for degradation by Doa10, Ubc6 and Ubc7-dependent ubiquitylation machinery.
(A, B) Immunoblot analysis of cycloheximide (CHX) chase was done as described in Fig. 1. Graphs show Asi2 protein levels in each strain (arbitrary units, AU) and percentage of remaining Asi2. Data represent average values (n=3). Standard deviation is indicated. P-values are listed in Table IV, supplementary material. (A) Asi2 stability in doa10Δ and hrd1Δ mutants. Asi2-myc (pMB108) was expressed in WT (MBY163), hrd1Δ (MBY164), doa10Δ (MBY165) and doa10Δ hrd1Δ (MBY166) strains. Asi2 protein half-life: 36 min (WT), 39 min (hrd1Δ), 53 min (doa10Δ) and 67 min (doa10Δ hrd1Δ). (B) Asi2 stability in ubc6Δ and ubc7 Δ mutants. Asi2-myc (pMB55) was expressed in WT (MBY159), ubc6 Δ (MBY160), ubc7Δ (MBY161) and ubc6Δ ubc7Δ (MBY162) strains. Asi2 protein half-life: 36 min (WT), 45 min (ubc6Δ), 86 min (ubc7Δ) and 86 min (ubc6Δ ubc7 Δ). (C) ASI2 mRNA levels were unaffected in ubc7Δ and doa10Δ mutants. Relative ASI2 mRNA levels were determined using RT-PCR and compared in UBC7 (MBY159) versus ubc7Δ (MBY161), and DOA10 (MBY163) versus doa10Δ (MBY165) strains expressing Asi2-HA from pMS01 plasmid. Data represents relative ASI2 mRNA concentration (arbitrary units, AU). Average value and standard deviation of two independent samples is shown.
To test the possibility that the elevated levels of Asi2 in ERAD mutants were due to secondary and unanticipated consequences of increased ASI2 transcription, we assessed ASI2 mRNA levels in doa10Δ and ubc7Δ mutants using real time quantitative PCR (Fig. 3-C). ASI2 mRNA levels were similar in wild-type cells and doa10Δ and ubc7Δ mutants. The results indicate that the elevated levels of Asi2 in the cells lacking Doa10 and Ubc7 are due to enhanced Asi2 protein stability rather than elevated transcription.
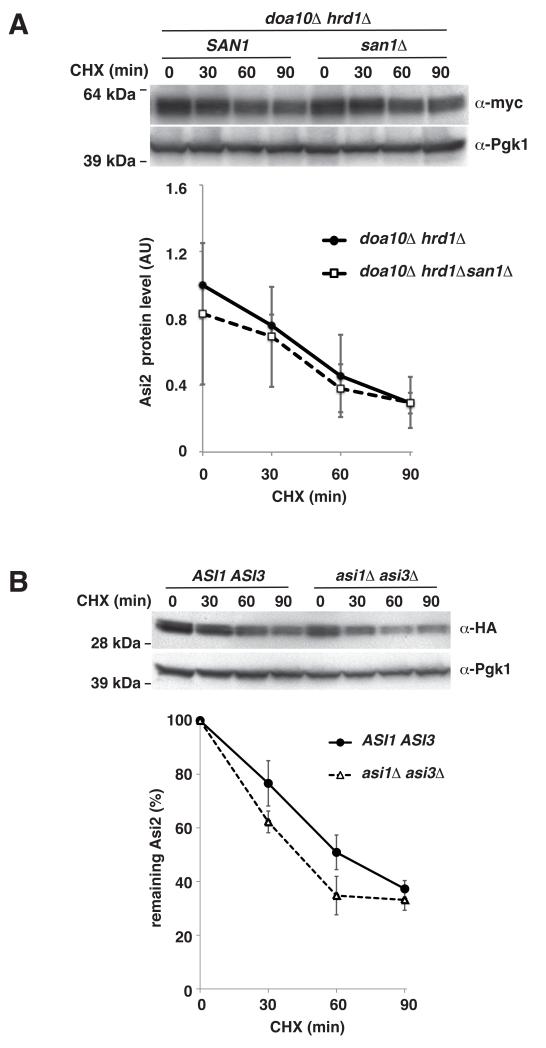
Asi2 was only partially stabilized in a doa10Δ mutant (see Fig. 3A), thus additional E3 ligases are likely to be involved in targeting Asi2 for degradation. San1 was recently discovered as an E3 ubiquitin ligase involved in degradation of aberrant nuclear proteins (Gardner et al., 2005; Rosenbaum et al., 2011). Therefore, we tested Asi2 stability in a doa10Δ hrd1Δ mutant lacking SAN1. The deletion of SAN1 did not enhance the stability of Asi2 (Fig. 4A). Therefore it is unlikely that San1 participates in Asi2 degradation. Consequently, an additional and yet to be identified E3 ubiquitin ligase appears to function in parallel with Doa10, or may engage in the absence of Doa10, to target Asi2 for degradation.
Fig. 4. Asi2 degradation is not mediated by San1, Asi1 or Asi3.
Cycloheximide (CHX) chase was carried out as described in Fig. 1. Immunoblotting was performed with anti-myc, anti-HA and anti-Pgk1 antibodies. Data represent average values of three independent samples. Standard deviation is indicated. P-values are listed in Table IV, supplementary material. (A) Asi2 protein levels are unaffected in mutant lacking ubiquitin ligase San1. Asi2-myc (pMB108) stability in doa10Δ hrd1Δ (MBY166) and doa10Δ hrd1Δ san1Δ (MPY143) mutants was analyzed. (B) Asi2 protein degradation rate is increased in a mutant lacking Asi1 and Asi3. Asi2-HA (pMS01) stability in ASI1 ASI3 (PLY1340) and asi1Δ asi3Δ (PLY1346) was analyzed. Asi2 protein half-life: 64 min (ASI1 ASI3) and 40 min (asi1Δ asi3Δ).
We have previously found that Asi2 functions in concert with two other INM proteins, Asi1 and Asi3, presumably in a protein complex at the INM (Zargari et al., 2007). Asi1 and Asi3 possess RING domains similar to those found in E3 RING-finger ligases. Therefore we tested the possibility that Asi1 and Asi3 are involved in ubiquitylation of Asi2 by examining Asi2 protein stability in a double asi1Δ asi3Δ mutant. Consistent with the Asi proteins functioning together within a complex, Asi2 degradation was enhanced in the asi1Δ asi3Δ mutant as compared to wild-type (Fig. 4B). This result clearly indicates that neither Asi1 nor Asi3 function as E3 ligases that ubiquitylate Asi2.
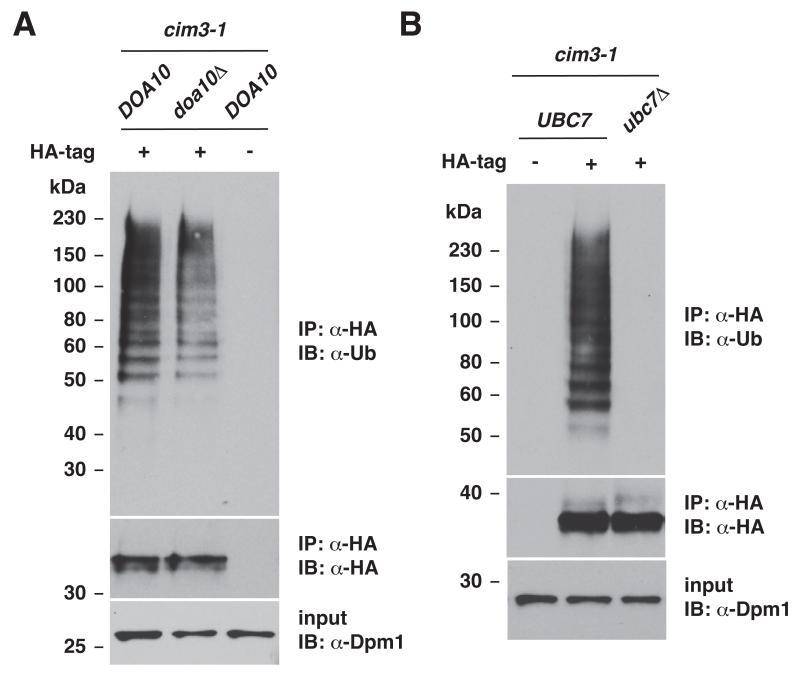
Asi2 is poly-ubiquitylated in a Ubc7- and Doa10-dependent manner
To test whether Asi2 protein stabilization in doa10Δ and ubc7Δ deletion mutants (see Fig. 3) is a direct consequence of impaired Asi2 ubiquitylation in the mutant strains lacking specific E3 or E2 enzyme, we examined Asi2 ubiquitylation status in the mutants lacking E3 ligase Doa10 and E2 enzyme Ubc7 (Fig. 5). To enrich ubiquitylated species and thus facilitate detection of ubiquitylated proteins, the experiment was performed with yeast strains carrying cim3-1 mutation, which impairs proteasomal degradation downstream of protein ubiquitylation. By analyzing the ubiquitylation status of immunoprecipitated Asi2 in immunoblots using anti-ubiquitin antibody we found that Asi2 was less ubiquitylated in the cim3-1 doa10Δ mutant as compared to the cim3-1 mutant expressing functional Doa10 (Fig. 5A). Moreover, Asi2 ubiquitylation was almost completely abolished in the mutant lacking Ubc7 (Fig. 5B). The diminished level of Asi2 ubiquitylation correlates well with the enhanced stability of Asi2 in doa10Δ and ubc7Δ mutants (Figs. 3A and B, respectively). Together the data demonstrate that Asi2 is a novel substrate for Doa10-Ubc7-Ubc6 dependent ubiquitylation and degradation.
Fig. 5. Asi2 ubiquitylation is decreased in mutants lacking Doa10 and Ubc7.
Immunoblot analysis of immunoprecipitation experiments. (A) Strains cim3-1 DOA10 (PLY1348) and cim3-1 doa10 Δ (MBY226) carrying ubiquitin-overexpression plasmid (myc-Ub/LEU2 2μ) and either a plasmid expressing epitope-tagged Asi2-HA (pMB3) or a plasmid expressing untagged Asi2 (pMB128) were analyzed. Plasmid pMB128 expressing untagged Asi2 serves as a control for unspecific immunoprecipitation. Immunoprecipitation was performed using anti-HA antibody (IP: α-HA) and immunoblotting (IB) was done using anti-HA or anti-ubiquitin antibodies, as indicated. To verify that a similar amount of protein was used as a starting material for the immunoprecipitation, input samples immunoblotted with anti-Dpm1 antibody are shown. (B) Experiment was done as in (A) using cim3-1 DOA10 (PLY1348) and cim3-1 ubc7Δ (MBY225) strains.
Asi2 is degraded by proteasomes localized in the nucleus
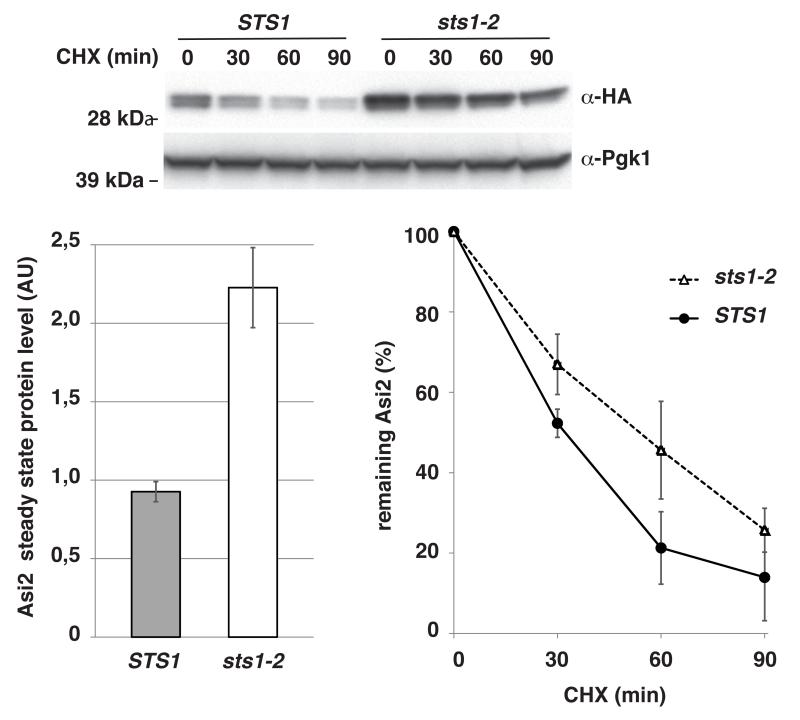
In yeast, proteasomes are also localized in the nucleus (Enenkel et al., 1998; Peters et al., 1994; Russell et al., 1999). The degradation of nuclear substrates requires efficient targeting of proteasomal subunits to the nucleus (Chen et al., 2011). Nuclear targeting of the proteasomes depends on a functional Sts1, which interacts with proteasomal components and the nuclear import factor Srp1 (Chen et al., 2011; Romero-Perez et al., 2007). As shown by (Chen et al., 2011), proteasomes fail to accumulate in the nucleus of cells carrying the temperature sensitive sts1-2 mutation when grown at the restrictive temperature of 37 °C (Fig. S1A in supplementary material). Asi2 is substantially stabilized in the sts1-2 mutant grown at the restrictive temperature (half-life, 45 min) as compared to wild-type cells (half-life, 31 min) (Fig. 6), consistent with Asi2 being targeted for proteasomal degradation in the nucleus. As a positive control we tested ΔssPrA, a misfolded protein that has been shown to be degraded in the nucleus (Prasad et al., 2010); ΔssPrA was stabilized in sts1-2 mutant at the restrictive temperature (Fig. S1B in supplementary material). In contrast and as previously shown by (Chen et al., 2011), a cytoplasmic proteasomal substrate Ura3-HA-SL17 (Gilon et al., 1998; Ravid et al., 2006) was not stabilized in the sts1-2 mutant (Ravid et al., 2006) (Fig. S1C in supplementary material).
Fig. 6. Asi2 protein is stabilized in the sts1-2 mutant exhibiting impaired proteasome accumulation in the nucleus.
Cycloheximide (CHX) chase of Asi2-HA (pMS01) in a STS1 (NA10) and sts1-2 (NA25) strain was assessed by immunoblotting with anti-HA and anti-Pgk1 antibodies. Cells were grown at 26°C to OD600 0.8, transferred to medium pre-warmed to 37°C (OD600 0.7) and incubated at 37°C for 4 hours before CHX was added. Steady state protein levels at t=0 (p = 0.009, two-tailed T-test, type 3) and the percentage of remaining Asi2 after CHX addition are shown. Data represent average values (n=3). Standard deviation is indicated. P-values of data in time course are listed in Table IV, supplementary material. Asi2 protein half-life: 31 min (STS1) and 45 min (sts1-2).
Discussion
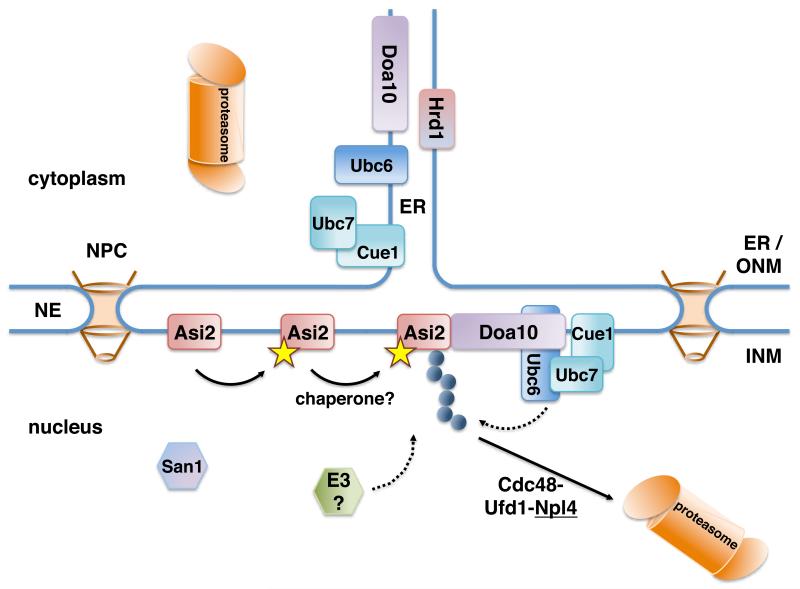
In this study we addressed the molecular basis of INM protein turnover by examining the stability of Asi2, a well-characterized integral INM protein in yeast (Zargari et al., 2007). Our results clearly demonstrate that Asi2 is subject to turnover and is targeted to the proteasomes in the nucleus by a Ubc6-Ubc7-Doa10-dependent ubiquitylation pathway. While the Doa10-associated integral membrane E2 enzyme Ubc6 is targeted by Doa10-dependent degradation (Swanson et al., 2001; Walter et al., 2001), presumably also in the INM, our study shows for the first time that a bona-fide integral INM protein, which is not itself a component of the ubiquitylation machinery is delivered to the nuclear proteasomes. Based on our data, we propose a model of Asi2 protein degradation in the nucleus (Fig. 7).
Fig. 7. A molecular model for INM protein Asi2 degradation.
A putative Asi2 degradation signal (yellow star) may be exposed upon damage or upon changes of its molecular environment at the INM. INM-localized Asi2 is poly-ubiquitylated (blue circles) by the components of ERAD pathway localized in the nucleus (E3 ligase Doa10 and E2 enzymes Ubc6 and Ubc7). The potential involvement of other E3 ligases and chaperones, which may facilitate Asi2 ubiquitylation, is indicated. ER-localized Hrd1 E3 ligase and nuclear San1 are not involved in Asi2 degradation. Poly-ubiquitylated Asi2 is targeted to the nuclear proteasomes, presumably in a manner dependent on the activity of the Cdc48-Ufd1-Npl4 complex (Hitchcock et al., 2003).
This model is supported by our finding that Asi2 is stabilized in the sts1-2 mutant, which exhibits impaired nuclear accumulation of proteasomes. This result indicates that a substantial pool of Asi2 is degraded by the proteasomes localized in the nucleus. Consistent with Asi2 being recognized by the ubiquitylation machinery in the nucleus, Doa10 and associated E2 enzymes Ubc6 and Ubc7 have been found to localize to both ER membrane and the INM (Deng and Hochstrasser, 2006). Nuclear localization of Doa10 is required for degradation of a transcription factor MATα2 (Deng and Hochstrasser, 2006). In contrast to Doa10, Hrd1 seems to localize exclusively to the ER (Deng and Hochstrasser, 2006). The fact that Hrd1 is physically separated from the INM is consistent with Asi2 not being a substrate for Hrd1-dependent ubiquitylation and with the degradation of Asi2 in the nucleus.
Asi2 protein stabilization correlated well with the decrease in Asi2 ubiquitylation in doa10Δ and ubc7Δ mutant strains. However, the fact that Asi2 is still degraded in doa10Δ mutants suggests that a Doa10-independent pathway exists that is capable of targeting Asi2 for degradation. This additional pathway could function in parallel with Doa10, or become engaged in Asi2 turnover only after Doa10 function is impaired. We found that the E3 ligase San1, previously reported in soluble nuclear protein quality control, is not involved in this parallel pathway. Delivery of integral membrane proteins to the proteasome includes a membrane extraction step, which is in many cases mediated by the Cdc48-Ufd1-Npl4 complex (Wolf and Stolz, 2012). In support of our results, Asi2 was found enriched in an npl4 mutant strain in a proteome-wide screen for ubiquitylated membrane proteins (Hitchcock et al., 2003). Newly synthesized proteins are subject to protein quality control (Duttler et al., 2013). A minor fraction of these, comprising mostly cytoplasmic proteins, is ubiquitylated co-translationally (Duttler et al., 2013). A recent study indicates that Sts1 links proteasomes to the Srp1-bound nascent polypeptide chains and therefore might be involved in co-translational protein degradation (Ha et al., 2014), however, further investigation is necessary to clarify the potential role of Sts1in this process. Although we cannot fully exclude that a small pool of Asi2 is degraded in the ER immediately after its synthesis, our finding that Asi2 protein continues to be degraded at later time points after new protein synthesis has been inhibited by cycloheximide suggests that Asi2 degradation is not limited to the quality control of newly synthesized Asi2 at the ER. A substantial fraction of the cellular pool of Asi2 is therefore degraded after having been correctly targeted to the INM.
Previous studies have identified Mps2-1, a mutant form of spindle pole body protein Mps2, as a substrate of Doa10-dependent degradation pathway (McBratney and Winey, 2002; Ravid et al., 2006). While wild-type Mps2 is an integral membrane protein that localizes to the site of the spindle pole body insertion into the nuclear envelope (Jaspersen and Winey, 2004), the thermolabile Mps2-1 mutant protein is degraded at the non-permissive temperature in a Doa10-dependent manner (McBratney and Winey, 2002; Ravid et al., 2006). In contrast to our results for Asi2, the published data indicate that mutant Mps2-1 degradation occurs directly after protein synthesis as a protein quality control mechanism in the ER membrane.
Many integral membrane proteins are known to be degraded in the vacuole (lysosome) (Davies et al., 2009; Hegde and Ploegh, 2010). Taking into account that the INM is structurally and functionally distinct from the ER membrane (English and Voeltz, 2013), vacuolar degradation of an INM protein in yeast could be done by a process called piecemeal microautophagy of the nucleus (PMN). In yeast, PMN has been shown to occur when a segment of the nuclear envelope is directly sequestered by local invagination of the vacuolar membrane, followed by the release of a nuclear envelope-derived vesicle into the vacuolar lumen (Roberts et al., 2003). Our data show that vacuolar function is not required for Asi2 turnover under normal growth conditions, which is in agreement with previous reports that PMN is induced to higher levels during starvation, but is very low under normal growth conditions (Roberts et al., 2003). The result also indicates that Asi2 is not delivered to the vacuole via membrane trafficking from the ONM/ER membrane.
Unlike in metazoans, where the NE is disassembled at the onset of mitosis and re-assembled around separated sister chromatids in telophase, yeast, filamentous fungi and some protists undergo closed mitosis in which the NE remains intact throughout the cell cycle (Anderson and Hetzer, 2008; Cohen et al., 2001; Ribeiro et al., 2002). Thus, the only way for yeast INM proteins to become accessible for the cytoplasmic degradation machinery is by retrograde transport from the INM back to the ONM/ER membrane. Although passive diffusion out of the nucleus has been observed for artificial INM reporter proteins upon blocking karyopherin mediated transport across the NPC, a native INM protein remained localized in the nucleus and only very slowly redistributed to the ER, even under conditions when active nuclear import was inhibited (Meinema et al., 2013). Moreover, an active nuclear export of membrane proteins was never observed (Meinema et al., 2013). In human cells lacking lamin A, INM protein emerin is mislocalized to the ER membrane and targeted for proteasomal degradation (Muchir et al., 2006).
Doa10-mediated Asi2 protein degradation may represent a nuclear protein quality control pathway. Signals that may mediate Asi2 recognition and targeting for degradation are currently unknown. Amphipathic helices have been predicted in some Doa10 substrates, such as in the Deg1 region of MATα2 (Johnson et al., 1998) and in Ndc10 (Furth et al., 2011). Asi2 region encompassing amino acid residues 49 to 66 is predicted to form an amphipathic helix, hence it is tempting to speculate that this region is involved in targeting Asi2 for degradation. As previously suggested for aberrant nuclear protein substrates of San1 (Gardner et al., 2005), degradation signals in Asi2 might become exposed when damaged or upon changes of its molecular environment at the INM. For instance, a loss of an interaction partner in the nucleus may uncover Asi2 degradation signals. Asi2 functions together with two other integral membrane proteins at the INM, Asi1 and Asi3, (Zargari et al., 2007), which may affect Asi2 protein stability. Indeed, the rate of Asi2 protein degradation is faster in cells lacking Asi1 and Asi3, suggesting that interaction of Asi2 with Asi1 and Asi3 might protect Asi2 from degradation. Moreover, since Asi2 is a component of the SPS sensor pathway (Zargari et al., 2007), we considered a possibility that Asi2 protein degradation has a regulatory role in SPS sensor signaling. However, the Asi2 protein levels were similar under both inducing and non-inducing conditions for the SPS-sensor pathway, indicating that the suppressive role of Asi2 in SPS signaling is not modulated via Asi2 protein stability.
In conclusion, this study addresses for the first time the turnover of a native, functional INM protein. It identifies Asi2 as the first bona-fide integral INM protein targeted to the proteasomal degradation in the nucleus and reveals the molecular pathway that mediates Asi2 ubiquitylation. As accumulation of aberrant proteins in the nucleus is a likely cause of several diseases (Brais, 2003; Jana and Nukina, 2003; Orr and Zoghbi, 2001; Peters et al., 1999), elucidating mechanisms of NE-associated protein degradation may contribute to better understanding of disease mechanisms.
Materials and Methods
Yeast growth media
Standard yeast culture media such as Yeast extract-Peptone-Dextrose (YPD) medium, ammonia-based synthetic minimal dextrose (SD) medium and ammonia-based synthetic complex dextrose (SC) medium were prepared as described (Andréasson and Ljungdahl, 2002). Antibiotic selections were made on solid YPD containing 200 mg/l G418, 300 mg/l hygromycin B or 100 mg/l clonNAT. When needed, 1 g/l 5-fluoroorotic acid (5-FOA) was added to SCD medium. Cells were grown at 30°C unless indicated otherwise.
Yeast strains
Yeast Saccharomyces cerevisiae strains used are listed in Table I in supplementary material. All strains except cim3-1 and sts1-2 mutant strains (MBY178, MBY179, NA10 and NA25) are isogenic descendants of the S288c-derived strain AA255/PLY115 (Antebi and Fink, 1992). Recombinant DNA work was done by standard methods. Genomic manipulation of yeast strains was done by using homologous recombination of DNA fragments transformed into yeast strains. FGY256 was constructed by transforming strain PLY127 with SphI-SalI DNA fragment containing hrd1Δ::URA3 (Bays et al., 2001). Gene DOA10 was deleted in PLY127 and FGY256 using PCR-amplified doa10Δ::natMX cassette creating JKY28 and JKY38, respectively. MBY159-167 were constructed by deleting ASI2 in strains PLY123, FGY205, FGY206, JKY36, PLY127, FGY256, JKY28, JKY38 and FGY217, respectively, using asi2Δ::kanMX cassette that was PCR-amplified from plasmid pUG6 by primers prMB310 and prMB311. To construct MBY178 and MBY179, ASI2 was deleted in CIM3 (CAY220) and cim3-1 (PLY1348 - CMY763) strain respectively, using asi2Δ::kanMX cassette as described above. CAY220 was constructed by transforming PLY1348 with plasmid pRS316 and a PCR product encompassing a complete RPT6 (CIM3) ORF obtained from genomic YMH233 DNA, selecting transformants at 37°C, followed by curing from plasmid on medium containing 5-FOA. Strain MPY143 was constructed by transforming MBY166 with san1Δ::hphMX cassette that was PCR-amplified using primers prMB515 and prMB516 and plasmid pAG32 as a template. Crosses and subsequent tetrad analysis were performed to verify 2:2 segregation of deletion markers. Primer sequences are listed in Table III in supplementary material.
Plasmids
Plasmids used are listed in Table II in supplementary material. Plasmid pMS1 (pAZ042-2) was constructed by ligating XmnI/SpeI-cut plasmid pMB03 with XhoI/StuI-cut plasmid pRS316. Plasmid pMB108 was constructed using homologous recombination, by transforming yeast with PvuI-cut pMB55 and XbaI/HindIII-cut pRS317 and selecting Lys+ colonies. Plasmid pMB122 was created by ligating a large fragment from BsrGI-cut pMS1 with the small fragment of similarly cut pPL741. Plasmid pMB128 was created by ligating a large fragment from XhoI/NotI-cut plasmid pMB3 with the small fragment of similarly cut pMB122.
Cycloheximide chase and immunoblot analysis
Cells were grown in SC at 30°C unless indicated otherwise to OD600 0.6 - 0.9, pelleted by centrifugation and resuspended in fresh SC to OD600 1.5. After 20 min incubation at 30 °C, cycloheximide (CHX) was added to the culture to a final concentration of 100 μg/ml. Samples were harvested at indicated time points after addition of cycloheximide by placing 1.5 ml culture aliquots to the ice bath. Total protein was extracted as described (Silve et al., 1991). In short, cells were harvested by centrifugation, resuspended in 250 μl ice-cold 1.85 M NaOH containing 7% β-mercaptoethanol and incubated for 10 min on ice. Protein was precipitated by adding 250 μl cold 50 % TCA, followed by 10 min incubation on ice and centrifugation. Protein pellet was washed with 1M Tris and resuspended in sample buffer (100 mM Tris-Cl pH 6.8, 4 mM EDTA, 4% SDS, 2% β-mercaptoethanol, 10% glycerol, 0.02% bromophenol blue). Proteins were denatured by incubation at 37 °C for 10 min, resolved by SDS-PAGE and analyzed by immunoblotting. The following primary antibodies were used for immunoblotting: anti-myc (mouse monoclonal 9E10, Roche, dilution 1/2000), anti-HA (rat monoclonal 3F10, HRP-conjugated, Roche, 1/3000, unless otherwise indicated), anti-Pgk1 (mouse monoclonal 22C5, Invitrogen, dilution 1/20000), anti-Dpm1 (mouse monoclonal 5C5, Molecular Probes, dilution 1/500) and anti-CPY (mouse monoclonal 10A5, Molecular Probes, dilution 1/4000). Secondary antibody was conjugated to IRDye® fluorescent dyes (LI-COR). Immunoreactive bands were visualized and the signal was quantified by Oddyssey® Infrared Imaging System (LI-COR Biosciences). In Fig. 1 (B, C, D), Fig. 4B, Fig. 6 and Fig. S1A enhanced chemiluminescence detection was used and quantified using BIO-RAD or LAS1000 (Fuji) system. The sum of the signal intensities of both Asi2-immunoreactive bands was normalized to the signal of stable protein control Dpm1 or Pgk1. In each experiment average values and standard deviation of three independent samples, unless otherwise indicated, were calculated. Percentage of protein present before CHX addition (time point 0 min) and, where appropriate, quantification of protein levels in each strain (arbitrary units) are shown. P-values for data sets in graphs are shown in Table IV in supplementary material. Protein half-life was calculated using the data of the 30 minutes time point after the addition of cycloheximide.
Testing protein ubiquitylation
Ubiquitylation assay was performed based on protocol by (Furth et al., 2011) with some modifications. Yeast strains impaired in proteasomal degradation (cim3-1 mutant) expressing HA-tagged Asi2 from a 2μ plasmid (pMB3) or untagged Asi2 (pMB128) and, where indicated, carrying ubiquitin overexpression plasmid (Yep181-CUP1-myc-Ub/ LEU2 2μ) were grown in selective SC. Over-night cultures (28°C) were diluted to OD600 0.27 - 0.30 in selective SC medium containing 100 μM CuSO4 and cells were incubated at 28°C for two hours, followed by incubation at restrictive temperature of 37°C for two to three hours. Around 5×108 cells (OD600, 20) were harvested by centrifugation at 4°C, cell pellet was washed in 1 ml ice cold water. Cells were resuspended in 800 μl ice cold water, 250 μl solution of ice cold 1.85 M NaOH and 7% β-mercaptoethanol was added and incubated on ice for 15 min, followed by addition of 250 μl ice cold 50% TCA, incubation on ice for 15 min and centrifugation (10 min, 13000 rpm, 4°C). Protein pellet was washed with 500 μl Tris. At this point samples can be frozen at −80°C. Samples were thawed on ice and protein pellet was resuspended in 200 μl denaturation solution (2% SDS, 2 mM EDTA, 25 mM Tris pH7.5, Protease Inhibitor Cocktail (Roche), 0.5 mg/ml Pefabloc (Roche), 100 μM MG132 (Enzo) and 5 mM N-ethyl-maleimide (NEM)). Proteins were denatured at 65 °C for 10 min and insoluble cell debris was removed by two rounds of centrifugation (5 min, 13000 rpm, room temperature). Cleared protein lysate was diluted 1/5 with IP-dilution buffer (50 mM Tris pH 7.5, 2 mM EDTA, 100 mM NaCl, 1.2 % Triton, Protease Inhibitor Cocktail, Pefabloc, MG132 and NEM in concentrations as above). Lysate was incubated at 4°C for 1 hour with gentle rotation, followed by removal of insoluble material by 10 min centrifugation. 20 μl of sample (input) were mixed with 20 μl of sample buffer (8% SDS, 200 mM Tris pH 6.8, 8 mM EDTA, 20% glycerol, bromophenol blue, 4% β-mercaptoethanol) and frozen until analysis by immunoblot. The rest of the sample is mixed with 25 μl of anti-HA affinity matrix (clone 3F10, Roche) that was previously prepared by mixing with 10-20 μl IP-dilution buffer. Samples are immunoprecipitated for 1.5 hours at 4°C with gentle rotation, matrix is pelleted by centrifugation. Washings are done as following: two times with washing solution A (50 mM Tris pH 7.5, 2 mM EDTA, 100 mM NaCl, 1 % Triton, 0.4% SDS), once washing solution B (50 mM Tris pH 7.5, 2 mM EDTA, 350 mM NaCl, 1 % Triton) including 10 min incubation period at 4°C with gentle rotation, once washing solution A. Immunoprecipitated protein is eluted by adding 50 μl sample buffer (100 mM Tris pH 6.8, 4 mM EDTA, 5% SDS, 8M urea, 2% β-mercaptoethanol) to the pelleted matrix and 10 min incubation at 65°C. Around 10-20 μl of samples is separated on SDS-PAGE gels (8-16 % gradient). For analysis of immunoprecipitated Asi2-HA, eluate is diluted 1/20 with sample buffer. Immunoblotting analysis was done using anti-HA 12CA5 antibody (1/1000, a gift from Ogris lab, MFPL Vienna) and anti-ubiquitin P4D1 antibody (1/1000, Santa Cruz). Protein levels in input samples were assessed using anti-Dpm1 antibody (1/500, Molecular Probes, clone 5C5). Immunoreactive bands were visualized by enhanced chemiluminescence detection.
RNA isolation and real time PCR
Around 107 yeast cells exponentially growing in SC medium were harvested by centrifugation. RNA was isolated using RiboPure™ Yeast Kit and treated with Turbo-DNase (Ambion) according to manufacturer’s instructions. 3 μg of isolated RNA was examined by electrophoresis on a 1% Guanidine Thiocyanate Agarose Gel (10mM). 1 μg of RNA was used for cDNA synthesis with oligo (dT)12-19 (Invitrogen) using SuperScript® III Reverse Transcriptase (Life Technologies) according to manufacturer’s instructions. The absence of DNA contamination was confirmed using minus reverse transcriptase samples as a negative control. The qPCR reaction was prepared using Kapa Sybr®Fast qPCR Master Mix (KapaBiosystems). cDNA mixtures were diluted 1/20 and 5 μl was used in a total qPCR reaction volume of 20 μl. Following primer pairs were used: KR65asi2fwd and KR66asi2rev, TAF10fwd and TAF10rev (Table III in supplementary material). Assays were conducted in triplicates on a Corbett Research RotorGene machine.
Supplementary Material
Acknowledgements
We are grateful to Kicki Ryman (Ljungdahl laboratory, Stockholm University) for performing RT-PCR experiments, Egon Ogris (MFPL, Vienna) for the gift of 12CA5 antibody, Helle Ulrich (University of Mainz) for the ubiquitin-plasmid, Claes Andréasson (Stockholm University) for the ΔssPrA-HA plasmid and to Mark Hochstrasser and Kiran Madura for plasmids and yeast strains. We acknowledge grant support from the Swedish Research Council (to POL), the Austrian Science Fund, grant number FWF P23805-B20 (to RF) and EMBO Long-Term Fellowship (to MB).
Footnotes
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Ammerer G, Hunter CP, Rothman JH, Saari GC, Valls LA, Stevens TH. PEP4 gene of Saccharomyces cerevisiae encodes proteinase A, a vacuolar enzyme required for processing of vacuolar precursors. Mol. Cell. Bio.J. 1986;6:2490–2499. doi: 10.1128/mcb.6.7.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DJ, Hetzer MW. The life cycle of the metazoan nuclear envelope. Curr. Opin. Cell Biol. 2008;20:386–392. doi: 10.1016/j.ceb.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andréasson C, Heessen S, Ljungdahl PO. Regulation of transcription factor latency by receptor-activated proteolysis. Genes Dev. 2006;20:1563–1568. doi: 10.1101/gad.374206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andréasson C, Ljungdahl PO. Receptor-mediated endoproteolytic activation of two transcription factors in yeast. Genes Dev. 2002;16:3158–3172. doi: 10.1101/gad.239202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antebi A, Fink GR. The yeast Ca(2+)-ATPase homologue, PMR1, is required for normal Golgi function and localizes in a novel Golgi-like distribution. Mol. Biol. Cell. 1992;3:633–654. doi: 10.1091/mbc.3.6.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bays NW, Gardner RG, Seelig LP, Joazeiro CA, Hampton RY. Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nat. Cell Biol. 2001;3:24–29. doi: 10.1038/35050524. [DOI] [PubMed] [Google Scholar]
- Belle A, Tanay A, Bitincka L, Shamir R, O’Shea EK. Quantification of protein half-lives in the budding yeast proteome. Proc. Natl. Acad. Sci. USA. 2006;103:13004–13009. doi: 10.1073/pnas.0605420103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boban M, Zargari A, Andréasson C, Heessen S, Thyberg J, Ljungdahl PO. Asi1 is an inner nuclear membrane protein that restricts promoter access of two latent transcription factors. J. Cell Bio.J. 2006;173:695–707. doi: 10.1083/jcb.200601011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brais B. Oculopharyngeal muscular dystrophy: a late-onset polyalanine disease. Cytogene.t Genome Res. 2003;100:252–260. doi: 10.1159/000072861. [DOI] [PubMed] [Google Scholar]
- Chen L, Romero L, Chuang SM, Tournier V, Joshi KK, Lee JA, Kovvali G, Madura K. Sts1 plays a key role in targeting proteasomes to the nucleus. J. Biol. Chem. 2011;286:3104–3118. doi: 10.1074/jbc.M110.135863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Johnson P, Sommer T, Jentsch S, Hochstrasser M. Multiple ubiquitin-conjugating enzymes participate in the in vivo degradation of the yeast MAT alpha 2 repressor. Cell. 1993;74:357–369. doi: 10.1016/0092-8674(93)90426-q. [DOI] [PubMed] [Google Scholar]
- Ciechanover A. Intracellular protein degradation from a vague idea through the lysosome and the ubiquitin-proteasome system and on to human diseases and drug targeting: Nobel Lecture, December 8, 2004. Ann. N. Y. Acad. Sc.i. 2007;1116:1–28. doi: 10.1196/annals.1402.078. [DOI] [PubMed] [Google Scholar]
- Cohen M, Lee KK, Wilson KL, Gruenbaum Y. Transcriptional repression, apoptosis, human disease and the functional evolution of the nuclear lamina. Trends Biochem. Sci. 2001;26:41–47. doi: 10.1016/s0968-0004(00)01727-8. [DOI] [PubMed] [Google Scholar]
- D’Angelo MA, Raices M, Panowski SH, Hetzer MW. Age-dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell. 2009;136:284–295. doi: 10.1016/j.cell.2008.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies BA, Lee JR, Oestreich AJ, Katzmann DJ. Membrane protein targeting to the MVB/lysosome. Chem. Rev. 2009;109:1575–1586. doi: 10.1021/cr800473s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deak PM, Wolf DH. Membrane topology and function of Der3/Hrd1p as a ubiquitin-protein ligase (E3) involved in endoplasmic reticulum degradation. J. Biol. Chem. 2001;276:10663–10669. doi: 10.1074/jbc.M008608200. [DOI] [PubMed] [Google Scholar]
- Deng M, Hochstrasser M. Spatially regulated ubiquitin ligation by an ER/nuclear membrane ligase. Nature. 2006;443:827–831. doi: 10.1038/nature05170. [DOI] [PubMed] [Google Scholar]
- Duttler S, Pechmann S, Frydman J. Principles of cotranslational ubiquitination and quality control at the ribosome. Mol. Cell. 2013;50:379–393. doi: 10.1016/j.molcel.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enenkel C, Lehmann A, Kloetzel PM. Subcellular distribution of proteasomes implicates a major location of protein degradation in the nuclear envelope-ER network in yeast. EMBO J. 1998;17:6144–6154. doi: 10.1093/emboj/17.21.6144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English AR, Voeltz GK. Endoplasmic reticulum structure and interconnections with other organelles. Cold Spring Harb. Perspect. Biol. 2013;5:a013227. doi: 10.1101/cshperspect.a013227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg H, Hammar M, Andréasson C, Moliner A, Ljungdahl PO. Suppressors of ssy1 and ptr3 null mutations define novel amino acid sensor-independent genes in Saccharomyces cerevisiae. Genetics. 2001;158:973–988. doi: 10.1093/genetics/158.3.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furth N, Gertman O, Shiber A, Alfassy OS, Cohen I, Rosenberg MM, Doron NK, Friedler A, Ravid T. Exposure of bipartite hydrophobic signal triggers nuclear quality control of Ndc10 at the endoplasmic reticulum/nuclear envelope. Mol. Biol. Cell. 2011;22:4726–4739. doi: 10.1091/mbc.E11-05-0463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner RG, Nelson ZW, Gottschling DE. Degradation-mediated protein quality control in the nucleus. Cell. 2005;120:803–815. doi: 10.1016/j.cell.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Ghislain M, Udvardy A, Mann C. S. cerevisiae 26S protease mutants arrest cell division in G2/metaphase. Nature. 1993;366:358–362. doi: 10.1038/366358a0. [DOI] [PubMed] [Google Scholar]
- Gilon T, Chomsky O, Kulka RG. Degradation signals for ubiquitin system proteolysis in Saccharomyces cerevisiae. EMBO J. 1998;17:2759–2766. doi: 10.1093/emboj/17.10.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AL. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426:895–899. doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- Ha SW, Ju D, Xie Y. Nuclear import factor Srp1 and its associated protein Sts1 couple ribosome-bound nascent polypeptides to proteasomes for cotranslational degradation. J. Biol. Chem. 2014;289:2701–2710. doi: 10.1074/jbc.M113.524926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heessen S, Fornerod M. The inner nuclear envelope as a transcription factor resting place. EMBO Rep. 2007;8:914–919. doi: 10.1038/sj.embor.7401075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde RS, Ploegh HL. Quality and quantity control at the endoplasmic reticulum. Curr. Opin. Cell Bio.l. 2010;22:1–10. doi: 10.1016/j.ceb.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzer MW. The nuclear envelope. Cold Spring Harb Perspect Biol. 2010;2:a000539. doi: 10.1101/cshperspect.a000539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitchcock AL, Auld K, Gygi SP, Silver PA. A subset of membrane-associated proteins is ubiquitinated in response to mutations in the endoplasmic reticulum degradation machinery. Proc. Natl. Acad. Sci. USA. 2003;100:12735–12740. doi: 10.1073/pnas.2135500100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata A, Nagashima Y, Matsumoto L, Suzuki T, Yamanaka T, Date H, Deoka K, Nukina N, Tsuji S. Intranuclear degradation of polyglutamine aggregates by the ubiquitin-proteasome system. J. Biol. Chem. 2009;284:9796–9803. doi: 10.1074/jbc.M809739200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jana NR, Nukina N. Recent advances in understanding the pathogenesis of polyglutamine diseases: involvement of molecular chaperones and ubiquitin-proteasome pathway. J. Chem. Neuroanat. 2003;26:95–101. doi: 10.1016/s0891-0618(03)00029-2. [DOI] [PubMed] [Google Scholar]
- Jaspersen SL, Winey M. The budding yeast spindle pole body: structure, duplication, and function. Annu. Rev. Cell Dev. Biol. 2004;20:1–28. doi: 10.1146/annurev.cellbio.20.022003.114106. [DOI] [PubMed] [Google Scholar]
- Johnson PR, Swanson R, Rakhilina L, Hochstrasser M. Degradation signal masking by heterodimerization of MATalpha2 and MATa1 blocks their mutual destruction by the ubiquitin-proteasome pathway. Cell. 1998;94:217–227. doi: 10.1016/s0092-8674(00)81421-x. [DOI] [PubMed] [Google Scholar]
- Kostova Z, Tsai YC, Weissman AM. Ubiquitin ligases, critical mediators of endoplasmic reticulum-associated degradation. Semin. Cell. Dev. Biol. 2007;18:770–779. doi: 10.1016/j.semcdb.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreft SG, Wang L, Hochstrasser M. Membrane topology of the yeast endoplasmic reticulum-localized ubiquitin ligase Doa10 and comparison with its human ortholog TEB4 (MARCH-VI) J. Biol. Chem. 2006;281:4646–4653. doi: 10.1074/jbc.M512215200. [DOI] [PubMed] [Google Scholar]
- Ljungdahl PO, Daignan-Fornier B. Regulation of amino acid, nucleotide, and phosphate metabolism in Saccharomyces cerevisiae. Genetics. 2012;190:885–929. doi: 10.1534/genetics.111.133306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeo F, Schlauer J, Zischka H, Mecke D, Frohlich KU. Tyrosine phosphorylation regulates cell cycle-dependent nuclear localization of Cdc48p. Mol. Biol. Cell. 1998;9:131–141. doi: 10.1091/mbc.9.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBratney S, Winey M. Mutant membrane protein of the budding yeast spindle pole body is targeted to the endoplasmic reticulum degradation pathway. Genetics. 2002;162:567–578. doi: 10.1093/genetics/162.2.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinema AC, Laba JK, Hapsari RA, Otten R, Mulder FA, Kralt A, van den Bogaart G, Lusk CP, Poolman B, Veenhoff LM. Long unfolded linkers facilitate membrane protein import through the nuclear pore complex. Science. 2011;333:90–93. doi: 10.1126/science.1205741. [DOI] [PubMed] [Google Scholar]
- Meinema AC, Poolman B, Veenhoff LM. Quantitative analysis of membrane protein transport across the nuclear pore complex. Traffic. 2013;14:487–501. doi: 10.1111/tra.12048. [DOI] [PubMed] [Google Scholar]
- Mekhail K, Seebacher J, Gygi SP, Moazed D. Role for perinuclear chromosome tethering in maintenance of genome stability. Nature. 2008;456:667–670. doi: 10.1038/nature07460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchir A, Massart C, van Engelen BG, Lammens M, Bonne G, Worman HJ. Proteasome-mediated degradation of integral inner nuclear membrane protein emerin in fibroblasts lacking A-type lamins. Biochem. Biophy.s Res. Commun. 2006;351:1011–1017. doi: 10.1016/j.bbrc.2006.10.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HT, Zoghbi HY. SCA1 molecular genetics: a history of a 13 year collaboration against glutamines. Hum. Mol. Genet. 2001;10:2307–2311. doi: 10.1093/hmg/10.20.2307. [DOI] [PubMed] [Google Scholar]
- Oza P, Jaspersen SL, Miele A, Dekker J, Peterson CL. Mechanisms that regulate localization of a DNA double-strand break to the nuclear periphery. Genes Dev. 2009;23:912–27. doi: 10.1101/gad.1782209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM, Franke WW, Kleinschmidt JA. Distinct 19 S and 20 S subcomplexes of the 26 S proteasome and their distribution in the nucleus and the cytoplasm. J. Biol. Chem. 1994;269:7709–7718. [PubMed] [Google Scholar]
- Peters MF, Nucifora FC, Jr., Kushi J, Seaman HC, Cooper JK, Herring WJ, Dawson VL, Dawson TM, Ross CA. Nuclear targeting of mutant Huntingtin increases toxicity. Mol. Cell Neurosci. 1999;14:121–128. doi: 10.1006/mcne.1999.0773. [DOI] [PubMed] [Google Scholar]
- Pfirrmann T, Heessen S, Omnus DJ, Andreasson C, Ljungdahl PO. The prodomain of Ssy5 protease controls receptor-activated proteolysis of transcription factor Stp1. Mol. Cell Biol. 2010;30:3299–3309. doi: 10.1128/MCB.00323-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad R, Kawaguchi S, Ng DT. A nucleus-based quality control mechanism for cytosolic proteins. Mol. Biol. Cell. 2010;21:2117–2127. doi: 10.1091/mbc.E10-02-0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravid T, Kreft SG, Hochstrasser M. Membrane and soluble substrates of the Doa10 ubiquitin ligase are degraded by distinct pathways. EMBO J. 2006;25:533–543. doi: 10.1038/sj.emboj.7600946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro KC, Mariante RM, Coutinho LL, Benchimol M. Nucleus behavior during the closed mitosis of Tritrichomonas foetus. Biol. Cell. 2002;94:289–301. doi: 10.1016/s0248-4900(02)01206-6. [DOI] [PubMed] [Google Scholar]
- Roberts P, Moshitch-Moshkovitz S, Kvam E, O’Toole E, Winey M, Goldfarb DS. Piecemeal microautophagy of nucleus in Saccharomyces cerevisiae. Mol. Biol. Cell. 2003;14:129–141. doi: 10.1091/mbc.E02-08-0483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Perez L, Chen L, Lambertson D, Madura K. Sts1 can overcome the loss of Rad23 and Rpn10 and represents a novel regulator of the ubiquitin/proteasome pathway. J. Biol. Chem. 2007;282:35574–35582. doi: 10.1074/jbc.M704857200. [DOI] [PubMed] [Google Scholar]
- Rosenbaum JC, Fredrickson EK, Oeser ML, Garrett-Engele CM, Locke MN, Richardson LA, Nelson ZW, Hetrick ED, Milac TI, Gottschling DE, et al. Disorder targets misorder in nuclear quality control degradation: a disordered ubiquitin ligase directly recognizes its misfolded substrates. Mol. Cell. 2011;41:93–106. doi: 10.1016/j.molcel.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell SJ, Steger KA, Johnston SA. Subcellular localization, stoichiometry, and protein levels of 26 S proteasome subunits in yeast. J. Biol. Chem. 1999;274:21943–21952. doi: 10.1074/jbc.274.31.21943. [DOI] [PubMed] [Google Scholar]
- Savas JN, Toyama BH, Xu T, Yates JR, 3rd, Hetzer MW. Extremely long-lived nuclear pore proteins in the rat brain. Science. 2012;335:942. doi: 10.1126/science.1217421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schober H, Ferreira H, Kalck V, Gehlen LR, Gasser SM. Yeast telomerase and the SUN domain protein Mps3 anchor telomeres and repress subtelomeric recombination. Genes Dev. 2009;23:928–938. doi: 10.1101/gad.1787509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schork SM, Thumm M, Wolf DH. Catabolite inactivation of fructose-1,6-bisphosphatase of Saccharomyces cerevisiae. Degradation occurs via the ubiquitin pathway. J. Biol. Chem. 1995;270:26446–26450. doi: 10.1074/jbc.270.44.26446. [DOI] [PubMed] [Google Scholar]
- Silve S, Volland C, Garnier C, Jund R, Chevallier MR, Haguenauer-Tsapis R. Membrane insertion of uracil permease, a polytopic yeast plasma membrane protein. Mol. Cell. Biol. 1991;11:1114–1124. doi: 10.1128/mcb.11.2.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson R, Locher M, Hochstrasser M. A conserved ubiquitin ligase of the nuclear envelope/endoplasmic reticulum that functions in both ER-associated and Matalpha2 repressor degradation. Genes Dev. 2001;15:2660–2674. doi: 10.1101/gad.933301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vembar SS, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Nat. Rev. Mol. Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J, Urban J, Volkwein C, Sommer T. Sec61p-independent degradation of the tail-anchored ER membrane protein Ubc6p. EMBO J. 2001;20:3124–3131. doi: 10.1093/emboj/20.12.3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf DH, Stolz A. The Cdc48 machine in endoplasmic reticulum associated protein degradation. Biochim. Biophys. Acta. 2012;1823:117–124. doi: 10.1016/j.bbamcr.2011.09.002. [DOI] [PubMed] [Google Scholar]
- Woolford CA, Daniels LB, Park FJ, Jones EW, Van Arsdell JN, Innis MA. The PEP4 gene encodes an aspartyl protease implicated in the posttranslational regulation of Saccharomyces cerevisiae vacuolar hydrolases. Mol. Cell Biol. 1986;6:2500–2510. doi: 10.1128/mcb.6.7.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen HC, Xu Q, Chou DM, Zhao Z, Elledge SJ. Global protein stability profiling in mammalian cells. Science. 2008;322:918–923. doi: 10.1126/science.1160489. [DOI] [PubMed] [Google Scholar]
- Zargari A, Boban M, Heessen S, Andréasson C, Thyberg J, Ljungdahl PO. Inner nuclear membrane proteins Asi1, Asi2, and Asi3 function in concert to maintain the latent properties of transcription factors Stp1 and Stp2. J. Biol. Chem. 2007;282:594–605. doi: 10.1074/jbc.M609201200. [DOI] [PubMed] [Google Scholar]
- Zuleger N, Kerr AR, Schirmer EC. Many mechanisms, one entrance: membrane protein translocation into the nucleus. Cell. Mol. Life Sci. 2012;69:2205–2216. doi: 10.1007/s00018-012-0929-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.