Abstract
Cancer is an exceedingly complex disease that is orchestrated and driven by a combination of multiple aberrantly regulated processes. The nature and depth of involvement of individual events vary between cancer types, and in lung cancer, the deregulation of the epigenetic machinery, the tumor microenvironment and the immune system appear to be especially relevant. The contribution of microRNAs to carcinogenesis and cancer progression is well established with many reports and investigations describing the involvement of microRNAs in lung cancer, however most of these studies have concentrated on single microRNA-target relations and have not adequately addressed the complexity of their interactions. In this review, we focus, in part, on the role of microRNAs in the epigenetic regulation of lung cancer where they act as active molecules modulating enzymes that take part in methylation-mediated silencing and chromatin remodeling. Additionally, we highlight their contribution in controlling and modulating the tumor microenvironment and finally, we describe their role in the critical alteration of essential molecules that influence the immune system in lung cancer development and progression.
Keywords: microRNA, Lung cancer, Epigenetic regulation, Tumor microenvironment
Introduction
One of the most promising molecular entities to have impacted cancer research in recent times is the group of small non-coding RNAs known as microRNAs. Since the first miRNA gene called lin-4 was discovered in Caenorhabditis elegans [1], several others have been identified, and so far over 2500 miRs have been described in the human genome miRBase (www.mirbase.org) [2]. MicroRNAs have been described to be involved in several cellular processes including proliferation, development, metabolism, differentiation and apoptosis. Importantly, the deregulation of microRNA expression and function has been linked to many human pathological conditions, including cancer [3,4].
Classically, in cancer, microRNAs have been categorized as either oncogenic or tumor suppressor miRNAs. In the first instance, by post-transcriptionally lowering mRNA and subsequently protein levels of molecules with tumor suppressor functions, microRNAs assume an oncogenic function. Conversely, by targeting oncogenic molecules, they act as tumor suppressors and the altered expression levels of the microRNA(s) between tumor and normal states coupled with the function of the targeted molecule(s) dictate the resulting (end) phenotype. Beyond the classical regulation of oncogenes and tumor suppressor mRNAs, aberrantly expressed microRNAs could in further levels of complexity act: 1) to regulate the epigenetic machinery by a) directly modulating enzymes which take part in methylation-mediated silencing and chromatin remodeling [5] or b) be epigenetically regulated themselves [6]; 2) in a paracrine fashion via exosomes, microvesicles and protein complexes to influence the tumor microenvironment [7-10] and 3) to promote the release of mediators which activate pro- or anti-cancer immune activity [11,12]. These latter mechanisms of microRNA regulation are explained in detail.
MicroRNAs and epigenetic regulation in cancer
The epigenome is both dynamic and highly regulated in order to conserve normal expression patterns in cells. One of the most important components of expression regulation is the methylation of CpG islands within promoter regions both in normal as well as cancer cells [13,14]. The dynamic balance is maintained by a family of DNA methyltransferases: notably DNMT1 (maintenance DNMT) and DNMT3a and 3b (de novo DNMTs). Equally important in epigenetic regulation are covalent changes in histone tails that are orchestrated by histone methyltransferases (HMT), histone acetyltransferases (HAT), histone deacetylases (HDAC) [13,15] and polycomb proteins such as PRC 1 and PRC 2 [16]. MicroRNAs as a part of regulatory networks serve as significant targets for the epigenetic machinery and are also active players in modulating enzymes which take part in methylation mediated silencing and the chromatin remodeling machinery. MiR-127 was the first epigenetically regulated microRNA to be reported in a cancer entity [17]. Since then, many microRNAs were found to undergo epigenetic regulation in different cancer types including colorectal [18,19], urological [20], breast [21], ovarian [22,23], gastric [24] and pancreatic [25] as well as different types of haematological malignancies [26]. Most of the microRNAs are involved in crucial processes in the cell, thus, a great number of epigenetically regulated microRNAs (app. 45%) are implicated in several cancer types (eg., miR −124, −129, −34b at least 7 types, 34a −12 types), however, none of these have so far been found to be particularly involved in lung cancer [27].
MicroRNAs regulated by methylation
Hypermethylation is one of the most common epigenetic regulatory mechanisms and about half of the microRNA genes are subject to hypermethylation of CpG regions of their promoters [6], resulting in microRNA silencing or down-regulation. There is ample evidence supporting this mode of action, for instance, CpG island hypermethylation mediated silencing of miR-124a has been found in a wide range of lung cancer cell lines, including H358, CALU3, A549, A427, H2126 and H209. Interestingly, the epigenetic loss of microRNA-124a was functionally related to an activation of cyclin D kinase 6 (CDK6) and an inactivating phosphorylation of the retinoblastoma tumor suppressor protein (pRb) [28]. A similar type of regulation was also observed with miR-199a silencing in several cancer cell lines that occurred as a result of gene methylation [29]. This microRNA negatively regulates the proto-oncogene c-Met and its downstream effector ERK2, thus the hypermethylation of its promoter acts as an oncogenic switch. Moreover, transfection of miR-199a into cancer cells increases their pro-apoptotic ability, suggesting that miR-199a can be a potential therapeutic molecule. Ceppi and colleagues discovered that methylation mediated down-regulation of miR-200c correlated with a higher invasive potential of NSCLC cell lines [30] with additional experiments showing inhibition of in vitro invasion and in vivo metastasis. Accordingly, miR-200c mimics restored sensitivity to therapeutic compounds such as Cisplatin and Cetuximab. Invariably, an association between low miR-200c expression, poor grade differentiation, a higher degree of lymph node affectation and a lower endogenous E-cadherin expression was observed in a patient cohort. MiR-200c is an important EMT mediator where it suppresses EMT by down-regulating ZEB proteins. ZEB1 and ZEB2 proteins are also targets for miR-141 and miR-429, which have also been shown to be hypermethylated in lung cancer [31]. In a study by Heller et al. [32], miR-9-3 and miR-193a were found to be tumor-specifically methylated in NSCLC patients. Moreover, patients with miR-9-3 methylated NSCLC had a significantly shorter disease-free survival and shorter overall survival than the unmethylated control group. This data supports the idea, that miR-9-3 methylation could be an interesting prognostic parameter in patients with NSCLC. In two independent studies, Wang and colleagues [33] and Watanabe and colleagues [34] showed that promoter hypermethylation mediated silencing of the miR-34b/c is a common event in NSCLC. Additionally, Wang et al. showed a strong association between aberrant methylation of the miR-34b/c gene and a high probability of relapse, poor overall survival and disease-free survival in patients with lung cancer. Furthermore, Watanabe’s study revealed that miR-34b/c expression negatively correlated with the expression of c-Met. Similarly, they also showed that the tumor suppressor activity of miR-126 that inhibits invasion by directly targeting Crk is silenced in lung cancer cells by the DNA methylation of its host gene, EGFL7. Interestingly, it was also shown that the promoter methylation status of the miR-503 gene may influence sensitivity of NSCLC cells to cisplatin, suggesting that epigenetic interference might be a promising way to improve existing therapies [35].
Taken together, the hypermethylation of microRNA genes or promoter sequences is an important mechanism leading to the down-regulation of microRNA expression resulting in a reversal of oncogene suppression (c-Met, CDK6, Crk), repression of tumor suppressor genes (pRB), or activation of proteins involved in epithelial-to-mesenchymal transition (EMT) (ZEB1/2). However, the converse is also true, for instance, the high expression of miR-196a in lung cancer samples and cell lines was attributed to DNA demethylation [36]. Moreover, miR-196a up-regulation correlated with a more advanced clinical stage and lymph node metastasis in lung cancer. In vitro functional experiments demonstrated that higher miR-196a expression levels promote cell proliferation, migration and invasion by targeting and suppressing the homeobox protein -A5 (HOXA5) at the protein as well as at the mRNA level. An inverse relation was then demonstrated in NSCLC tissues.
MicroRNAs regulated by histone modifications
Aside methylation changes, histone modifications have also been implicated in the regulation of microRNA expression. Incoronato and colleagues [37] investigated the mechanism behind miR-212 silencing in lung cancer and by comparing Calu-1 (lung cancer cell line) and MRC5 (human fibroblasts), they found significant differences in the methylation status of H3K27 and H3K9 marks and in the acetylation status of H3K9, suggesting that modifications of histones on the miR-212 gene predicted transcriptional start sites contributing to its strong down-regulation in lung cancer. Additionally, an analysis of human tissue specimens of normal tissue and lung cancer revealed a correlation between miR-212 silencing and up-regulation of anti-apoptotic protein PED. Similarly, miR-373 silencing in lung cancer was found to be deacetylase-dependent with consequences on cell proliferation, migration, invasion and the expression of mesenchymal markers that appear to be modulated through IRAK2 and LAMP1 [38].
Interestingly, instances exist where certain microRNAs that have been established as key players in several cancer types including lung cancer [39], have been found to be regulated by epigenetic mechanisms in some of these tumor entities, but not in lung cancer. This is the case with miR-155, which in addition to being one of the most consistently regulated microRNAs in lung cancer, has equally significant prognostic significance [40,41]. Whereas miR-155 was found to be repressed by deacetylation of H2A and H3 through a mechanism mediated by BRCA1 in breast cancer [42], such reports are lacking in lung cancer, even though BRCA1 is not that unimportant in lung cancer [43].
MicroRNAs regulating the epigenetic machinery – Epi-microRNAs
MicroRNAs can also act as modifiers of the epigenetic network. These so-called epi-microRNAs contribute to altered epigenomic patterns in cancer cells, with the first evidence of such microRNA activity observed in lung cancer cell lines, where the miR-29 family (29a, 29b and 29c) was found to directly target the de novo DNA methyltransferases, DNMT-3a and DNMT-3b, inhibiting their activity. Consequently, this action resulted in a re-activation of silenced tumor suppressor genes (FHIT, WHOX), thus preventing tumor progression [5]. However, in contrast to the large number of studies conducted in different cancer types, very few reports exist on microRNAs that regulate the epigenome in lung cancer. Of the studies documented in lung cancer, was one where Xi et al. reported that the repression of miR-487b in cultured cells exposed to cigarette smoke, and in primary lung cancers correlated with an overexpression of the polycomb repressor proteins BMI1 and SUZ12 [44]. Interestingly, this state coincided with miR-487b gene methylation and de novo nucleosome occupancy in its area, indicating that the microRNA also epigenetically regulated itself. Besides its function of epigenetic regulation, miR-487b also directly targets WNT5A, KRAS and MYC, suggesting a multiple mode of action. Another exciting example of the regulatory role of microRNAs in lung cancer epigenetics involves miR-449 [45]. This study showed that the down-regulation of miR-449 might be responsible for an overexpression of HDAC1, subsequently inhibiting cell growth and promoting tumor suppression. Similar work by Jeon and colleagues also revealed a therapeutic potential for miR-449, showing that co-treatment with miR-449 and a HDAC1 inhibitor led to significantly higher growth reduction than with the HDAC1 inhibitor itself. Further examples demonstrating this regulatory principle include the work done by Cho and colleagues [46] where they showed that the down-regulation of miR-101 affected enhancer of zeste homolog 2 (EZH2), a member of the polycomb repressive complex causing its overexpression. EZH2 is known to be involved in the methylation of H3K27, bringing about gene silencing and in the process regulating several cancer-associated processes. They found that the general methylation status of H3K27 was significantly higher in metastatic lung cancer than in primary lung cancer tissue, with in vitro studies showing a significant impact on invasion. Furthermore, EZH2 was also shown to be a direct target of miR-138 [47] where experiments on several lung cancer cell lines, as well as tissue samples from patients with NSCLC revealed that the down-regulation of miR-138 resulted in an up-regulation of EZH2 inhibiting apoptosis, G0/G1 cell cycle arrest and promoting cell proliferation and tumor invasion. A summary of microRNAs involved in epigenetic regulation are presented in Table 1.
Table 1.
MicroRNAs involved in epigenetic regulatory mechanisms in lung cancer
| MicroRNA | Chromosome | Type of regulation | Target | Functional consequence | References |
|---|---|---|---|---|---|
| miR-124a-1 | 8 | Hypermethylation mediated silencing | CDK6 | Oncogene activation, repression of tumor suppression | Lujambio et al., 2007 [28] |
| miR-124a-2 | 8 | pRb | |||
| miR-124a-3 | 20 | ||||
| mir-199a-1 | 19 | Hypermethylation mediated silencing | c-Met | Anti-apoptotic function | Kim et al., 2008 [29] |
| mir-199a-2 | 1 | (ERK2) | Oncogene activation | ||
| miR-200c | 12 | Hypermethylation mediated down-regulation | ZEB1 | Promotion of EMT and metastasis | Ceppi et al., 2010 [30] |
| miR-141 | 12 | Hypermethylation | ZEB1/2 | Lopez Serra; Esteller, 2012 [31] | |
| miR-429 | 1 | Hypermethylation | ZEB1/2 | Lopez Serra; Esteller 2012 [31] | |
| miR-9-3 | 15 | Methylation mediated down-regulation | Heller et al., 2012 [32] | ||
| miR-193a | 17 | Methylation mediated down-regulation | Altered expression regulation of genes involved in Proliferation, apoptosis, differentiation and adhesion | Heller et al., 2012 [32] | |
| miR-34b/c | 11 | Promoter hypermethylation mediated silencing | (c-Met) | Oncogene activation, metastasis formation | Wang et al., 2011 [33] |
| Watanabe et al., 2011 [34] | |||||
| miR-503 | X | Promoter methylation mediated silencing | FANCA | Modulate sensitivity to cisplatin regulate apoptosis | Li et al., 2014[35] |
| miR-126 | 9 | Host gene promoter methylation mediated silencing | Crk | Oncogene activation | Watanabe et al., 2011 [34] |
| miR-196a-1 | 17 | Demethylation mediated up-regulation | HOXA5 | Promotion of cell proliferation, migration and invasion. | Liu Xiang-hua et al., 2012 [36] |
| miR-196a-2 | 12 | ||||
| miR-212 | 17 | Histone tail methylation mediated down-regulation | PED | Anti-apoptotic function | Incoronato et al., 2011 [37] |
| miR-373 | 19 | Histone acetylation mediated silencing | IRAK2 | Promotion of cell proliferation, migration, and invasion, Pro-EMT function | Seol et al., 2014 [38] |
| LAMP1 | |||||
| miR-29a | 7 | MicroRNA’s downregulation mediates DNMT | DNMT3a | Repression of tumor suppression | Fabbri et al., 2007 [5] |
| miR-29b-1 | 7 | Up-regulation | DNMT3b | ||
| miR-29b-2 | 1 | (FHIT | |||
| miR-29c | 1 | WHOX) | |||
| miR-487b | 14 | MicroRNA’s repression mediates target Genes overexpression | SUZ12, BMI1, WNT5A, MYC, K-ras | Promotion of tumor progression and metastasis | Xi et al.,2013 [44] |
| miR-449 | 5 | MicroRNA’s down-regulation mediates HDAC1 | HDAC1 | Repression of tumor suppression | Jeon et al., 2011 [45] |
| Up-regulation | Promotion of tumor metastasis | ||||
| miR-101-1 | 1 | MicroRNA’s down-regulation mediates EZH2 | EZH2 | Promotion of EMT and tumor metastasis | Cho et al., 2011 [46] |
| miR-101-2 | 9 | Up-regulation | (CDH1, MMP-2) | ||
| miR-138-1 | 3 | MicroRNA’s down-regulation mediates EZH2 | EZH2 | Promotion of proliferation, repression of apoptosis, Disruption of cell cycle arrest | Zhang et al., 2013 [47] |
| miR-138-2 | 16 | Up-regulation |
A compilation of the most significant microRNAs targeted by epigenetic regulatory mechanisms, including those that modulate epigenetic changes in lung cancer. The most important functional outcomes are described. Genes placed in brackets are not targeted directly.
MicroRNAs and the tumor microenvironment
The tumor microenvironment is an increasingly appreciated item of interest in cancer development and progression. During cancer progression, sequences of molecular changes arise in the tumor as well as in the neighboring tissue stroma. Since tumor development is driven by physiological responses to an aberrant stromal environment [48] the interaction between the tumor and stromal cells determines the outcome of progression [49]. The tumor microenvironment is a complex structure that in addition to the extracellular matrix (ECM) comprises many other cell types, including endothelial cells, inflammatory cells and most abundantly, fibroblasts [50] While normal fibroblasts have been found to prevent tumor progression [51] the so-called cancer associated fibroblasts (CAFs) generate an environment promoting tumor growth and invasiveness [52]. CAFs are characterized by specific changes in their secretory profile, e.g, chemokines that attract inflammatory cells. Together, they release growth factors that trigger cancer cell proliferation, inhibit apoptosis or indirectly stimulate angiogenesis [53]. Interestingly, one of the major mechanisms by which CAFs are activated is mediated by specific microRNAs, e.g. miR-21, miR-31, −214, and 155 [7,8]. There is ample evidence, that microRNAs are not only present in the cells, but also in extracellular space [54,55]. Importantly, extracellular microRNAs also play a very vital role in cell to cell communication [9] and are capable of contributing to microenvironment-associated processes such as angiogenesis, matrix degradation and stromal remodeling, all which support cancer progression and metastasis [10]. About 90% of microRNAs found in the ECM are bound to RNA- binding proteins (eg. AGO proteins, especially Ago2, NPM1 - nucleoplasmin 1 and GW 182 responsible for microRNA stabilization as well as secretion) and just 10% exist in extracellular vesicles such as exosomes, apoptotic bodies and, mainly microvesicles (MVs) [9,55,56]. Remarkably, most of the microRNAs that are differentially expressed in CAFs are down-regulated as compared with normal fibroblasts, suggesting that they act as suppressors of stromal activation [57]. Evidence for the contribution of microRNAs in lung cancer angiogenesis arising from a specific influence on the tumor microenvironment was hinted at in the study by Jusufovic and colleagues [58] who found significantly higher microvessel density (MVD) together with decreased expression levels of let-7b and miR-126 in tumor and surrounding tissue of 50 NSCLC patients as compared to corresponding normal tissues. Interestingly, there was no difference in expression between tumor and surrounding tissue for both microRNAs and MVD, indicating similar molecular changes in the tumor and surrounding cells in this context [58].
In addition to influencing the local microenvironment, tumor cells have also been shown to secrete exosomes at a significantly higher level compared to normal cells [59]. These so-called tumor-derived exosomes (TD exosomes) contain pro-tumorigenic components, in particular abundant microRNAs which influence and modify stromal cells and recruit them as cancer-associated stromal cells, which in turn influence cancer cells and the ECM contributing to cancer development and progression [10,59]. Additionally, a mechanism based on passive leakage has been proposed, in which microRNAs are released from the cells due to injury, chronic inflammation, necrosis, or from cells with very short half-lives, like platelets [60].
Notable instances of microRNA regulation of the tumor microenvironment in lung cancer include the repression of the angiogenic program by IL-8, ICAM and CXCL1 through the lung tumor cell mediated exosomal release of miR-192 with a consequent reduction in metastatic burden and tumor colonization [61]. In another study, TGF-β induced release of miR-183 by lung cancer cells was found to control DAP12, a critical stimulatory signal adaptor linked to numerous NKC receptors resulting in a suppression of NK cell function [62]. Still, Krysan and colleagues found the inflammatory molecule, prostaglandin E2 (PGE2) to mediate an up regulation of c-Myc in stromal cells, which in turn modulates the expression of the miR-17-92 cluster, that effectively targets the tumor suppressor PTEN. The end effect was an augmentation of apoptosis resistance in non-small cell lung cancer (NSCLC) cells [63]. In what appears to be an interesting example of the microenvironment of one organ affecting the dissemination of cells from a different organ, Subramani et al., found that miR-768-3p released by brain astrocytes targets K-ras, mitigating metastatic potential of lung cancer cells. The down-regulation of this microRNA seen in many brain tumors is thought to enhance K-ras expression, including its downstream effectors ERK1/2 and B-Raf, and thus promoting metastasis [64].
MicroRNAs and immune responses in lung cancer
In addition to the processes described above, it has become increasingly clear that microRNAs play multiple roles in the negative regulation of numerous immune responses triggered by rapidly proliferating neoplastic cells. The genetic instability of neoplastic cells is associated with alterations of antigenic patterns that may in turn lead to attenuated recognition of the tumor by immune cells [65]. Some of these alterations are largely influenced by actions mediated by a constellation of different microRNAs (Figure 1). An example of this is miR-9 that is over-expressed in many cancers including lung cancer [66]. MiR-9 is capable of down-regulating the transcription of the MHC class I gene, thereby preventing the recognition of tumor cells by the immune system [11,56]. On the other hand, a down-regulation of miR-29 expression in cancer leads to an up-regulation of B7-H3 gene transcription and the subsequent expression of this molecule on the surface of neoplastic cells [67]. Consequently, B7-H3 delivers an inhibitory signal to T cells and NK cells and therefore inhibits the anti-tumor activity mediated by these cells [68,69]. Similarly, miR-222 and −339 down-regulate the expression of intracellular cell adhesion molecule 1 (ICAM-1) on the surface of tumor cells [70]. The binding of ICAM - to lymphocyte function-associated antigen (LFA-1) has been shown to be essential for optimal activation of cytotoxic T cell (CTLs) [71], thus, miR-222 and −339 in cancer cells contribute to impaired CTL-mediated tumor cell lysis [70]. Moreover miRs-221 and −222 are overexpressed in NSCLC and act in part by targeting TIMP3 [72] which has known functions in controlling cytokine and growth factor homeostasis. Furthermore, several microRNAs, for instance miRs-21 and −126/126*, contribute to the modulation of the tumor microenvironment, which encompasses not only stroma cells but also soluble factors and signaling molecules [73-76] (Figure 1). Regulatory T lymphocytes (Tregs), are important components of the tumor stroma, and are known to predominantly exert immunosuppressive effects. A number of tumor-bound chemokines including CCL22 have been shown to recruit Tregs to modulate immune responses in cancer [77]. Notably, CCL22 production in cancer cells is regulated by miR-34a, which suppresses the expression of this chemokine. On the other hand, increased levels of TGF-β in the tumor milieu down-regulate the expression of miR-34a and thereby increase infiltration of immunosuppressive Tregs. The TGF-β - miR-34a - CCL22 axis is therefore important in modulating the tumor microenvironment [78]. Other chemokines such as CXCL12 (Sdf-1) and CCL2 (MCP-1) are secreted by various cancer types and function as autocrine growth factors as well as chemo-attractants for progenitor cells and monocytes/macrophages [12,79,80]. Interestingly, the transcription of the CXCL12 gene is controlled by miR126/miR126*. This microRNA pair directly inhibits the expression of CXCL12 and down-regulates the production of CCL2 in a CXCL12-dependent process in which stromal cells (mainly mesenchymal stem cells, MSCs) seem to play the crucial role. In cancer cells, decreased miR-126/126* expression results in an increased production and secretion of CXCL12 that in turn attracts MSCs to the tumor microenvironment and increases the secretion of CCL2 [75]. Subsequently, CCL2 recruits monocytes/macrophages to the site of neoplastic growth [81].
Figure 1.
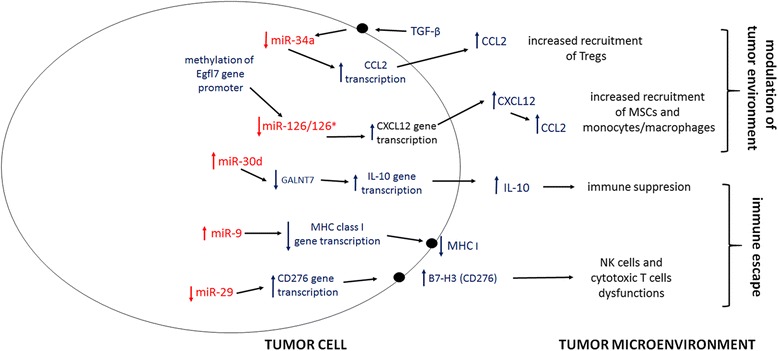
MicroRNA-mediated regulation of tumor microenvironment. The scheme demonstrates involvement of microRNAs in chemokine-mediated recruitment of regulatory T cells, monocytes, macrophages and MSCs and the role of microRNAs in tumor-related dysfunction of NK and cytotoxic T cells. All microRNAs are shown in red while gene targets and types of responses are shown in black. Arrows indicate directions of cause-and-effect relationships.
During migration, monocytes are exposed to many signals derived from cancer and stromal cells that induce their differentiation towards classically (M1) or alternatively activated (M2) macrophages. Tumor associated macrophages (TAMs) control the majority of immunological processes within tumors exerting both, regressive (M1) or progressive (M2) effects on the tumor development [82,83]. Unfortunately, the vast majority of TAMs exhibit an M2-like phenotype. Remarkably, a significant number of microRNAs like miRs-9, −21 and −146 are involved in the differentiation of the TAMs from peripheral blood monocytes (Figure 2). Both monocytes/macrophages and cancer cells are capable of releasing different pro- and anti-inflammatory cytokines like the immunosuppressive cytokine IL-10, which fosters tumor growth [84]. Substantial amounts of this cytokine are secreted by neoplastic cells including that of the lung [85,86]. Interestingly, the transcription of the IL-10 gene in tumor cells is indirectly regulated by miR-30d, which acts by directly repressing GALNT7. GALNT7 suppression leads to increased production and secretion of IL-10, which in turn promotes monocyte differentiation towards the M2 phenotype [87]. In addition, in human monocytes, IL-10 up-regulates miR-187 expression which down-regulates TNF-α production. Moreover, in macrophages, miR-187 indirectly decreases the expression of IL-6 and IL-12p40 via decreasing MAIL in an NFKBIZ-dependent manner [88]. Along similar lines, the differentiation of monocytes towards M2 macrophages can be induced by M-CSF. M-CSF signaling down-regulates miR-142-3p in monocytes which leads to increased Egr2 (Krox20) expression. This transcription factor can control differentiation of monocytes via activating macrophage-specific genes such as C-FMS, acting in cooperation with Spi-1/PU.1 [89].
Figure 2.
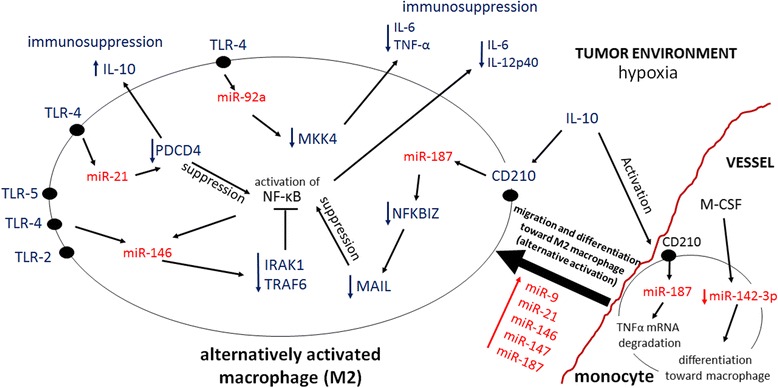
MicroRNA-mediated control of monocyte differentiation and tumor associated macrophages (TAMs) functions. All microRNAs are shown in red while gene targets and types of responses are shown in black. Arrows indicate directions of cause-and-effect relationships.
Many of the pro-tumorigenic outcomes of TAMs’ are controlled by microRNAs and depend on such factors like HMGB1 and IL-10 [88,90]. These mediators are secreted both by tumor cells and stroma and act via interactions with several cell surface receptors including toll-like receptors (TLR) and the IL-10 receptor. TLR2-, TLR-4- and TLR5-mediated signaling and NF-κβ activation in macrophages up-regulates the expression of miR-146 that in turn targets IRAK1 and TRAF6, proteins which are crucial components of interleukin-1- and TNF-related signaling pathways, respectively. More specifically, miR-146 acts via targeting IRAK1 and TRAF6, members of the TLR-NF-κβ pathway, and therefore contributes to controlling the expression of an array of inflammatory cytokine genes. MiR-146 therefore acts through a negative feedback loop and down-regulates cytokine signaling [91]. On another note, miR-92 is up-regulated following TLR-4 activation and in turn down-regulates the expression of TNF-α and IL-6 [92]. TLR4-mediated up-regulation of miR-21 suppresses programmed cell death protein-4 (PDCD4) expression. This mechanism may limit pro-inflammatory cytokine production in macrophages via suppressing NF-κβ activation, leading to an increased IL-10 production [90]. Notably, miR-21 and miR-29a are expressed at different levels in various cancers including lung cancer [93-95]. Both miR-21 and miR29a can act within cancer cells or can be delivered from cancer cells to macrophages by microvesicles and can subsequently bind intracellular TLR7 or TLR8. This microRNA-TLR interaction leads to the activation of the NF-κβ pathway and an increase in the production of the pro-inflammatory cytokines, IL-6 and TNF-α that in turn activate pro-metastatic responses in TAMs [96]. Some microRNAs also mediate a cross talk between cancer and stromal cells as in the case of TAM-derived miR-223, which down-regulates Mef2c expression in neoplastic cells, leading to an increase of cancer cell invasion, probably through the involvement of the miR-223 – Mef2c – β-catenin pathway [97].
Taken together, substantial evidence supports the dynamic role of microRNAs within the tumor microenvironment and with the immune system culminating in the promotion of tumor growth and immune evasion.
Conclusion
The role of microRNAs in the development and progression of lung cancer as well as other types of cancers is overwhelming; with several reports demonstrating the enormous impact microRNAs have on several cancer-associated processes.
In this review, we have elaborated on how microRNA-target interactions transcend beyond simple post-transcriptional regulation of oncogenes and/or tumor suppressor mRNAs to a more complex setting where they not only modulate networks and pathways, but also affect the tumor microenvironment, the epigenome and immune evasion. In lung cancer, their impact is very relevant, and to a large degree is deeply entrenched in several cascades, which impact lung cancer progression and metastasis. Inherently, microRNAs can target several molecules concurrently, and using this advantage together with the identification of the right molecule in the right context could have great potential, especially when used alongside other therapeutic modalities. Their involvement in several critical cancer associated processes makes them highly interesting molecules. Their effects transcend simple intracellular events, and are able to influence neighboring cells and even those at distant sites. Sufficient evidence indicates that they are significant modulators of our immune system which widens their scope as potential therapeutic agents.
Despite the tremendous amount of knowledge we have already on microRNAs, new publications keep coming out, describing yet other novel functions, making us marvel about the wide-reaching potential of these small molecules. In summary, microRNAs seem to be a very promising research target, however, the complexity of their actions makes keeping this promise very challenging.
Acknowledgements
The study was supported with the grant no. N N403 120440, financed by the National Science Centre (NCN) and the ‘Cancer/Mutagenesis’ research area of the Leading National Research Centre (KNOW). HA was supported by Deutsche Krebshilfe and Dr. Mildred-Scheel-Stiftung (together with MA), the Alfried Krupp von Bohlen und Halbach Foundation (Award for Young Full Professors) (Essen, Germany), Hella Bühler Foundation (Heidelberg, Germany), Dr. Ingrid zu Solms Foundation, (Frankfurt/Main, Germany), the FRONTIER Excellence Initiative of the University of Heidelberg, the Walter Schulz Foundation (Munich, Germany), the German-Israeli Project Cooperation DKFZ-MOST (Ministry of Science and Technology), the Wilhelm-Sander Foundation (Munich, Germany), and two grants of the HIPO/POP-Initiative, DKFZ Heidelberg.
Abbreviations
- CAF
Cancer associated fibroblasts
- CTL
Cytotoxic T-cell
- DNMT
DNA methyltransferase
- EMT
Epithelial to mesenchymal transition
- ECM
Extracellular matrix
- HAT
Histone acetyltransferase
- HDAC
Histone deacetylase
- HMT
Histone methyltransferase
- M-CSF
Macrophage colony stimulating factor
- MVD
Microvessel density
- MSC
Mesenchymal stem cells
- NKC
Natural killer cells
- NSCLC
Non-small cell lung cancer
- TAM
Tumor associated macrophages
- Tregs
Regulatory T-cells
Footnotes
Anna Maria Rusek, Mohammed Abba and Andrzej Eljaszewicz contributed equally to this work.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
AMR and HA drafted the greater part of the manuscript including Table 1. AE drafted “MicroRNAs and immune responses in lung cancer” section including Figures 1 and 2. MA drafted the part of “MicroRNAs and the tumor microenvironment” and also edited the manuscript. MM supervised and participated in writing and editing of the “MicroRNAs and immune responses in lung cancer” section. JN co-drafted parts of the review and participated in its design. HA, MA and JN coordinated and supervised the manuscript. All authors read and approved the final manuscript.
Contributor Information
Anna Maria Rusek, Email: a.rusek@dkfz-heidelberg.de.
Mohammed Abba, Email: m.abba@Dkfz-Heidelberg.de.
Andrzej Eljaszewicz, Email: andrzej.eljaszewicz@umb.edu.pl.
Marcin Moniuszko, Email: marcin.moniuszko@umb.edu.pl.
Jacek Niklinski, Email: niklinsj@umb.edu.pl.
Heike Allgayer, Email: heike.allgayer@medma.uni-heidelberg.de.
References
- 1.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 2.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annu Rev Med. 2009;60:167–79. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 4.Lee YS, Dutta A. MicroRNAs in cancer. Annu Rev Pathol. 2009;4:199–227. doi: 10.1146/annurev.pathol.4.110807.092222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci U S A. 2007;104:15805–10. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weber B, Stresemann C, Brueckner B, Lyko F. Methylation of human microRNA genes in normal and neoplastic cells. Cell Cycle. 2007;6:1001–5. doi: 10.4161/cc.6.9.4209. [DOI] [PubMed] [Google Scholar]
- 7.Mitra AK, Zillhardt M, Hua Y, Tiwari P, Murmann AE, Peter ME, et al. MicroRNAs reprogram normal fibroblasts into cancer-associated fibroblasts in ovarian cancer. Cancer Discov. 2012;2:1100–8. doi: 10.1158/2159-8290.CD-12-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yao Q, Cao S, Li C, Mengesha A, Kong B, Wei M. Micro-RNA-21 regulates TGF-beta-induced myofibroblast differentiation by targeting PDCD4 in tumor-stroma interaction. Int J Cancer. 2011;128:1783–92. doi: 10.1002/ijc.25506. [DOI] [PubMed] [Google Scholar]
- 9.Kosaka N, Yoshioka Y, Hagiwara K, Tominaga N, Katsuda T, Ochiya T. Trash or Treasure: extracellular microRNAs and cell-to-cell communication. Front Genet. 2013;4:173. doi: 10.3389/fgene.2013.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Su Y, Li X, Ji W, Sun B, Xu C, Li Z, et al. Small molecule with big role: MicroRNAs in cancer metastatic microenvironments. Cancer Lett. 2014;344:147–56. doi: 10.1016/j.canlet.2013.10.024. [DOI] [PubMed] [Google Scholar]
- 11.Hicklin DJ, Marincola FM, Ferrone S. HLA class I antigen downregulation in human cancers: T-cell immunotherapy revives an old story. Mol Med Today. 1999;5:178–86. doi: 10.1016/S1357-4310(99)01451-3. [DOI] [PubMed] [Google Scholar]
- 12.Balkwill F. Cancer and the chemokine network. Nat Rev Cancer. 2004;4:540–50. doi: 10.1038/nrc1388. [DOI] [PubMed] [Google Scholar]
- 13.Iorio MV, Piovan C, Croce CM. Interplay between microRNAs and the epigenetic machinery: an intricate network. Biochim Biophys Acta. 2010;1799:694–701. doi: 10.1016/j.bbagrm.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 14.Jakopovic M, Thomas A, Balasubramaniam S, Schrump D, Giaccone G, Bates SE. Targeting the epigenome in lung cancer: expanding approaches to epigenetic therapy. Front Oncol. 2013;3:261. doi: 10.3389/fonc.2013.00261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang IV, Schwartz DA. Epigenetic control of gene expression in the lung. Am J Respir Crit Care Med. 2011;183:1295–301. doi: 10.1164/rccm.201010-1579PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benetatos L, Vartholomatos G, Hatzimichael E. Polycomb group proteins and MYC: the cancer connection. Cell Mol Life Sci. 2014;71:257–69. doi: 10.1007/s00018-013-1426-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saito Y, Liang G, Egger G, Friedman JM, Chuang JC, Coetzee GA, et al. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–43. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 18.Toyota M, Suzuki H, Sasaki Y, Maruyama R, Imai K, Shinomura Y, et al. Epigenetic silencing of microRNA-34b/c and B-cell translocation gene 4 is associated with CpG island methylation in colorectal cancer. Cancer Res. 2008;68:4123–32. doi: 10.1158/0008-5472.CAN-08-0325. [DOI] [PubMed] [Google Scholar]
- 19.Tanaka T, Arai M, Wu S, Kanda T, Miyauchi H, Imazeki F, et al. Epigenetic silencing of microRNA-373 plays an important role in regulating cell proliferation in colon cancer. Oncol Rep. 2011;26:1329–35. doi: 10.3892/or.2011.1401. [DOI] [PubMed] [Google Scholar]
- 20.Liep J, Rabien A, Jung K. Feedback networks between microRNAs and epigenetic modifications in urological tumors. Epigenetics. 2012;7:315–25. doi: 10.4161/epi.19464. [DOI] [PubMed] [Google Scholar]
- 21.Vrba L, Munoz-Rodriguez JL, Stampfer MR, Futscher BW. miRNA gene promoters are frequent targets of aberrant DNA methylation in human breast cancer. PLoS One. 2013;8:e54398. doi: 10.1371/journal.pone.0054398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang C, Cai J, Wang Q, Tang H, Cao J, Wu L, et al. Epigenetic silencing of miR-130b in ovarian cancer promotes the development of multidrug resistance by targeting colony-stimulating factor 1. Gynecol Oncol. 2012;124:325–34. doi: 10.1016/j.ygyno.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 23.Iorio MV, Visone R, Di LG, Donati V, Petrocca F, Casalini P, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- 24.Tsai KW, Wu CW, Hu LY, Li SC, Liao YL, Lai CH, et al. Epigenetic regulation of miR-34b and miR-129 expression in gastric cancer. Int J Cancer. 2011;129:2600–10. doi: 10.1002/ijc.25919. [DOI] [PubMed] [Google Scholar]
- 25.Lee KH, Lotterman C, Karikari C, Omura N, Feldmann G, Habbe N, et al. Epigenetic silencing of MicroRNA miR-107 regulates cyclin-dependent kinase 6 expression in pancreatic cancer. Pancreatology. 2009;9:293–301. doi: 10.1159/000186051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pagano F, De ME, Grignani F, Nervi C. Epigenetic role of miRNAs in normal and leukemic hematopoiesis. Epigenomics. 2013;5:539–52. doi: 10.2217/epi.13.55. [DOI] [PubMed] [Google Scholar]
- 27.Kunej T, Godnic I, Ferdin J, Horvat S, Dovc P, Calin GA. Epigenetic regulation of microRNAs in cancer: an integrated review of literature. Mutat Res. 2011;717:77–84. doi: 10.1016/j.mrfmmm.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 28.Lujambio A, Ropero S, Ballestar E, Fraga MF, Cerrato C, Setien F, et al. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 2007;67:1424–9. doi: 10.1158/0008-5472.CAN-06-4218. [DOI] [PubMed] [Google Scholar]
- 29.Kim S, Lee UJ, Kim MN, Lee EJ, Kim JY, Lee MY, et al. MicroRNA miR-199a* regulates the MET proto-oncogene and the downstream extracellular signal-regulated kinase 2 (ERK2) J Biol Chem. 2008;283:18158–66. doi: 10.1074/jbc.M800186200. [DOI] [PubMed] [Google Scholar]
- 30.Ceppi P, Mudduluru G, Kumarswamy R, Rapa I, Scagliotti GV, Papotti M, et al. Loss of miR-200c expression induces an aggressive, invasive, and chemoresistant phenotype in non-small cell lung cancer. Mol Cancer Res. 2010;8:1207–16. doi: 10.1158/1541-7786.MCR-10-0052. [DOI] [PubMed] [Google Scholar]
- 31.Lopez-Serra P, Esteller M. DNA methylation-associated silencing of tumor-suppressor microRNAs in cancer. Oncogene. 2012;31:1609–22. doi: 10.1038/onc.2011.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heller G, Weinzierl M, Noll C, Babinsky V, Ziegler B, Altenberger C, et al. Genome-wide miRNA expression profiling identifies miR-9-3 and miR-193a as targets for DNA methylation in non-small cell lung cancers. Clin Cancer Res. 2012;18:1619–29. doi: 10.1158/1078-0432.CCR-11-2450. [DOI] [PubMed] [Google Scholar]
- 33.Wang Z, Chen Z, Gao Y, Li N, Li B, Tan F, et al. DNA hypermethylation of microRNA-34b/c has prognostic value for stage non-small cell lung cancer. Cancer Biol Ther. 2011;11:490–6. doi: 10.4161/cbt.11.5.14550. [DOI] [PubMed] [Google Scholar]
- 34.Watanabe K, Emoto N, Hamano E, Sunohara M, Kawakami M, Kage H, et al. Genome structure-based screening identified epigenetically silenced microRNA associated with invasiveness in non-small-cell lung cancer. Int J Cancer. 2012;130:2580–90. doi: 10.1002/ijc.26254. [DOI] [PubMed] [Google Scholar]
- 35.Li N, Zhang F, Li S, Zhou S. Epigenetic silencing of MicroRNA-503 regulates FANCA expression in non-small cell lung cancer cell. Biochem Biophys Res Commun. 2014;444:611–6. doi: 10.1016/j.bbrc.2014.01.103. [DOI] [PubMed] [Google Scholar]
- 36.Liu XH, Lu KH, Wang KM, Sun M, Zhang EB, Yang JS, et al. MicroRNA-196a promotes non-small cell lung cancer cell proliferation and invasion through targeting HOXA5. BMC Cancer. 2012;12:348. doi: 10.1186/1471-2407-12-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Incoronato M, Urso L, Portela A, Laukkanen MO, Soini Y, Quintavalle C, et al. Epigenetic regulation of miR-212 expression in lung cancer. PLoS One. 2011;6:e27722. doi: 10.1371/journal.pone.0027722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seol HS, Akiyama Y, Shimada S, Lee HJ, Kim TI, Chun SM, et al. Epigenetic silencing of microRNA-373 to epithelial-mesenchymal transition in non-small cell lung cancer through IRAK2 and LAMP1 axes. Cancer Lett. 2014;353:232–41. doi: 10.1016/j.canlet.2014.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen Z, Ma T, Huang C, Hu T, Li J. The pivotal role of microRNA-155 in the control of cancer. J Cell Physiol. 2014;229:545–50. doi: 10.1002/jcp.24492. [DOI] [PubMed] [Google Scholar]
- 40.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–98. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 41.Raponi M, Dossey L, Jatkoe T, Wu X, Chen G, Fan H, et al. MicroRNA classifiers for predicting prognosis of squamous cell lung cancer. Cancer Res. 2009;69:5776–83. doi: 10.1158/0008-5472.CAN-09-0587. [DOI] [PubMed] [Google Scholar]
- 42.Chang S, Wang RH, Akagi K, Kim KA, Martin BK, Cavallone L, et al. Tumor suppressor BRCA1 epigenetically controls oncogenic microRNA-155. Nat Med. 2011;17:1275–82. doi: 10.1038/nm.2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Akagi I, Okayama H, Schetter AJ, Robles AI, Kohno T, Bowman ED, et al. Combination of protein coding and noncoding gene expression as a robust prognostic classifier in stage I lung adenocarcinoma. Cancer Res. 2013;73:3821–32. doi: 10.1158/0008-5472.CAN-13-0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xi S, Xu H, Shan J, Tao Y, Hong JA, Inchauste S, et al. Cigarette smoke mediates epigenetic repression of miR-487b during pulmonary carcinogenesis. J Clin Invest. 2013;123:1241–61. doi: 10.1172/JCI61271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jeon HS, Lee SY, Lee EJ, Yun SC, Cha EJ, Choi E, et al. Combining microRNA-449a/b with a HDAC inhibitor has a synergistic effect on growth arrest in lung cancer. Lung Cancer. 2012;76:171–6. doi: 10.1016/j.lungcan.2011.10.012. [DOI] [PubMed] [Google Scholar]
- 46.Cho HM, Jeon HS, Lee SY, Jeong KJ, Park SY, Lee HY, et al. microRNA-101 inhibits lung cancer invasion through the regulation of enhancer of zeste homolog 2. Exp Ther Med. 2011;2:963–7. doi: 10.3892/etm.2011.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang H, Zhang H, Zhao M, Lv Z, Zhang X, Qin X, et al. MiR-138 inhibits tumor growth through repression of EZH2 in non-small cell lung cancer. Cell Physiol Biochem. 2013;31:56–65. doi: 10.1159/000343349. [DOI] [PubMed] [Google Scholar]
- 48.Barcellos-Hoff MH, Medina D. New highlights on stroma-epithelial interactions in breast cancer. Breast Cancer Res. 2005;7:33–6. doi: 10.1186/bcr972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kiaris H, Trimis G, Papavassiliou AG. Regulation of tumor-stromal fibroblast interactions: implications in anticancer therapy. Curr Med Chem. 2008;15:3062–7. doi: 10.2174/092986708786848596. [DOI] [PubMed] [Google Scholar]
- 50.Micke P, Ostman A. Exploring the tumour environment: cancer-associated fibroblasts as targets in cancer therapy. Expert Opin Ther Targets. 2005;9:1217–33. doi: 10.1517/14728222.9.6.1217. [DOI] [PubMed] [Google Scholar]
- 51.Paland N, Kamer I, Kogan-Sakin I, Madar S, Goldfinger N, Rotter V. Differential influence of normal and cancer-associated fibroblasts on the growth of human epithelial cells in an in vitro cocultivation model of prostate cancer. Mol Cancer Res. 2009;7:1212–23. doi: 10.1158/1541-7786.MCR-09-0073. [DOI] [PubMed] [Google Scholar]
- 52.Soon PS, Kim E, Pon CK, Gill AJ, Moore K, Spillane AJ, et al. Breast cancer-associated fibroblasts induce epithelial-to-mesenchymal transition in breast cancer cells. Endocr Relat Cancer. 2013;20:1–12. doi: 10.1530/ERC-12-0227. [DOI] [PubMed] [Google Scholar]
- 53.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–7. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu L, Yang BF, Ai J. MicroRNA transport: a new way in cell communication. J Cell Physiol. 2013;228:1713–9. doi: 10.1002/jcp.24344. [DOI] [PubMed] [Google Scholar]
- 55.Turchinovich A, Weiz L, Burwinkel B. Extracellular miRNAs: the mystery of their origin and function. Trends Biochem Sci. 2012;37:460–5. doi: 10.1016/j.tibs.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 56.Gao F, Zhao ZL, Zhao WT, Fan QR, Wang SC, Li J, et al. miR-9 modulates the expression of interferon-regulated genes and MHC class I molecules in human nasopharyngeal carcinoma cells. Biochem Biophys Res Commun. 2013;431:610–6. doi: 10.1016/j.bbrc.2012.12.097. [DOI] [PubMed] [Google Scholar]
- 57.Soon P, Kiaris H. MicroRNAs in the tumour microenvironment: big role for small players. Endocr Relat Cancer. 2013;20:R257–67. doi: 10.1530/ERC-13-0119. [DOI] [PubMed] [Google Scholar]
- 58.Jusufovic E, Rijavec M, Keser D, Korosec P, Sodja E, Iljazovic E, et al. let-7b and miR-126 are down-regulated in tumor tissue and correlate with microvessel density and survival outcomes in non–small–cell lung cancer. PLoS One. 2012;7:e45577. doi: 10.1371/journal.pone.0045577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hannafon BN, Ding WQ. Intercellular Communication by Exosome-Derived microRNAs in Cancer. Int J Mol Sci. 2013;14:14240–69. doi: 10.3390/ijms140714240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Redis RS, Calin S, Yang Y, You MJ, Calin GA. Cell-to-cell miRNA transfer: from body homeostasis to therapy. Pharmacol Ther. 2012;136:169–74. doi: 10.1016/j.pharmthera.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Valencia K, Luis-Ravelo D, Bovy N, Anton I, Martinez-Canarias S, Zandueta C, et al. miRNA cargo within exosome-like vesicle transfer influences metastatic bone colonization. Mol Oncol. 2014;8:689–703. doi: 10.1016/j.molonc.2014.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Donatelli SS, Zhou JM, Gilvary DL, Eksioglu EA, Chen X, Cress WD, et al. TGF-beta-inducible microRNA-183 silences tumor-associated natural killer cells. Proc Natl Acad Sci U S A. 2014;111:4203–8. doi: 10.1073/pnas.1319269111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Krysan K, Kusko R, Grogan T, O’Hearn J, Reckamp KL, Walser TC, et al. PGE2-driven expression of c-Myc and oncomiR-17-92 contributes to apoptosis resistance in NSCLC. Mol Cancer Res. 2014;12:765–74. doi: 10.1158/1541-7786.MCR-13-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Subramani A, Alsidawi S, Jagannathan S, Sumita K, Sasaki AT, Aronow B, et al. The brain microenvironment negatively regulates miRNA-768-3p to promote K-ras expression and lung cancer metastasis. Sci Rep. 2013;3:2392. doi: 10.1038/srep02392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–67. doi: 10.1016/0092-8674(90)90186-I. [DOI] [PubMed] [Google Scholar]
- 66.Xu T, Liu X, Han L, Shen H, Liu L, Shu Y. Up-regulation of miR-9 expression as a poor prognostic biomarker in patients with non-small cell lung cancer. Clin Transl Oncol. 2014;16:469–75. doi: 10.1007/s12094-013-1106-1. [DOI] [PubMed] [Google Scholar]
- 67.Xu H, Cheung IY, Guo HF, Cheung NK. MicroRNA miR-29 modulates expression of immunoinhibitory molecule B7-H3: potential implications for immune based therapy of human solid tumors. Cancer Res. 2009;69:6275–81. doi: 10.1158/0008-5472.CAN-08-4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen C, Shen Y, Qu QX, Chen XQ, Zhang XG, Huang JA. Induced expression of B7-H3 on the lung cancer cells and macrophages suppresses T-cell mediating anti-tumor immune response. Exp Cell Res. 2013;319:96–102. doi: 10.1016/j.yexcr.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 69.Lemke D, Pfenning PN, Sahm F, Klein AC, Kempf T, Warnken U, et al. Costimulatory protein 4IgB7H3 drives the malignant phenotype of glioblastoma by mediating immune escape and invasiveness. Clin Cancer Res. 2012;18:105–17. doi: 10.1158/1078-0432.CCR-11-0880. [DOI] [PubMed] [Google Scholar]
- 70.Ueda R, Kohanbash G, Sasaki K, Fujita M, Zhu X, Kastenhuber ER, et al. Dicer-regulated microRNAs 222 and 339 promote resistance of cancer cells to cytotoxic T-lymphocytes by down-regulation of ICAM-1. Proc Natl Acad Sci U S A. 2009;106:10746–51. doi: 10.1073/pnas.0811817106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Christensen JP, Marker O, Thomsen AR. T-cell-mediated immunity to lymphocytic choriomeningitis virus in beta2-integrin (CD18)- and ICAM-1 (CD54)-deficient mice. J Virol. 1996;70:8997–9002. doi: 10.1128/jvi.70.12.8997-9002.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Garofalo M, Di LG, Romano G, Nuovo G, Suh SS, Ngankeu A, et al. miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell. 2009;16:498–509. doi: 10.1016/j.ccr.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 73.Bullock MD, Pickard KM, Nielsen BS, Sayan AE, Jenei V, Mellone M, et al. Pleiotropic actions of miR-21 highlight the critical role of deregulated stromal microRNAs during colorectal cancer progression. Cell Death Dis. 2013;4:e684. doi: 10.1038/cddis.2013.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nouraee N, Van RK, Vasei M, Semnani S, Samaei NM, Naghshvar F, et al. Expression, tissue distribution and function of miR-21 in esophageal squamous cell carcinoma. PLoS One. 2013;8:e73009. doi: 10.1371/journal.pone.0073009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang Y, Yang P, Sun T, Li D, Xu X, Rui Y, et al. miR-126 and miR-126* repress recruitment of mesenchymal stem cells and inflammatory monocytes to inhibit breast cancer metastasis. Nat Cell Biol. 2013;15:284–94. doi: 10.1038/ncb2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sun X, Wang ZM, Song Y, Tai XH, Ji WY, Gu H. MicroRNA-126 modulates the tumor microenvironment by targeting calmodulin-regulated spectrin-associated protein 1 (Camsap1) Int J Oncol. 2014;44:1678–84. doi: 10.3892/ijo.2014.2321. [DOI] [PubMed] [Google Scholar]
- 77.Nishikawa H, Sakaguchi S. Regulatory T cells in tumor immunity. Int J Cancer. 2010;127:759–67. doi: 10.1002/ijc.25429. [DOI] [PubMed] [Google Scholar]
- 78.Yang P, Li QJ, Feng Y, Zhang Y, Markowitz GJ, Ning S, et al. TGF-beta-miR-34a-CCL22 signaling-induced Treg cell recruitment promotes venous metastases of HBV-positive hepatocellular carcinoma. Cancer Cell. 2012;22:291–303. doi: 10.1016/j.ccr.2012.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fridlender ZG, Kapoor V, Buchlis G, Cheng G, Sun J, Wang LC, et al. Monocyte chemoattractant protein-1 blockade inhibits lung cancer tumor growth by altering macrophage phenotype and activating CD8+ cells. Am J Respir Cell Mol Biol. 2011;44:230–7. doi: 10.1165/rcmb.2010-0080OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li M, Knight DA, Snyder A, Smyth MJ, Stewart TJ. A role for CCL2 in both tumor progression and immunosurveillance. Oncoimmunology. 2013;2:e25474. doi: 10.4161/onci.25474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Qian BZ, Li J, Zhang H, Kitamura T, Zhang J, Campion LR, et al. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature. 2011;475:222–5. doi: 10.1038/nature10138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mantovani A, Sozzani S, Locati M, Allavena P, Sica A. Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 2002;23:549–55. doi: 10.1016/S1471-4906(02)02302-5. [DOI] [PubMed] [Google Scholar]
- 83.Eljaszewicz A, Wiese M, Helmin-Basa A, Jankowski M, Gackowska L, Kubiszewska I, et al. Collaborating with the enemy: function of macrophages in the development of neoplastic disease. Mediators Inflamm. 2013;2013:831387. doi: 10.1155/2013/831387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tanikawa T, Wilke CM, Kryczek I, Chen GY, Kao J, Nunez G, et al. Interleukin-10 ablation promotes tumor development, growth, and metastasis. Cancer Res. 2012;72:420–9. doi: 10.1158/0008-5472.CAN-10-4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mocellin S, Wang E, Marincola FM. Cytokines and immune response in the tumor microenvironment. J Immunother. 2001;24:392–407. doi: 10.1097/00002371-200109000-00002. [DOI] [PubMed] [Google Scholar]
- 86.Hatanaka H, Abe Y, Kamiya T, Morino F, Nagata J, Tokunaga T, et al. Clinical implications of interleukin (IL)-10 induced by non-small-cell lung cancer. Ann Oncol. 2000;11:815–9. doi: 10.1023/A:1008375208574. [DOI] [PubMed] [Google Scholar]
- 87.Gaziel-Sovran A, Segura MF, Di MR, Collins MK, Hanniford D, de ME V-S, et al. miR-30b/30d regulation of GalNAc transferases enhances invasion and immunosuppression during metastasis. Cancer Cell. 2011;20:104–18. doi: 10.1016/j.ccr.2011.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rossato M, Curtale G, Tamassia N, Castellucci M, Mori L, Gasperini S, et al. IL-10-induced microRNA-187 negatively regulates TNF-alpha, IL-6, and IL-12p40 production in TLR4-stimulated monocytes. Proc Natl Acad Sci U S A. 2012;109:E3101–10. doi: 10.1073/pnas.1209100109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lagrange B, Martin RZ, Droin N, Aucagne R, Paggetti J, Largeot A, et al. A role for miR-142-3p in colony-stimulating factor 1-induced monocyte differentiation into macrophages. Biochim Biophys Acta. 1833;2013:1936–46. doi: 10.1016/j.bbamcr.2013.04.007. [DOI] [PubMed] [Google Scholar]
- 90.Sheedy FJ, Palsson-McDermott E, Hennessy EJ, Martin C, O’Leary JJ, Ruan Q, et al. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat Immunol. 2010;11:141–7. doi: 10.1038/ni.1828. [DOI] [PubMed] [Google Scholar]
- 91.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103:12481–6. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lai L, Song Y, Liu Y, Chen Q, Han Q, Chen W, et al. MicroRNA-92a negatively regulates Toll-like receptor (TLR)-triggered inflammatory response in macrophages by targeting MKK4 kinase. J Biol Chem. 2013;288:7956–67. doi: 10.1074/jbc.M112.445429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Barkley LR, Santocanale C. MicroRNA-29a regulates the benzo[a]pyrene dihydrodiol epoxide-induced DNA damage response through Cdc7 kinase in lung cancer cells. Oncogenesis. 2013;2:e57. doi: 10.1038/oncsis.2013.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Capodanno A, Boldrini L, Proietti A, Ali G, Pelliccioni S, Niccoli C, et al. Let-7 g and miR-21 expression in non-small cell lung cancer: correlation with clinicopathological and molecular features. Int J Oncol. 2013;43:765–74. doi: 10.3892/ijo.2013.2003. [DOI] [PubMed] [Google Scholar]
- 95.Liu ZL, Wang H, Liu J, Wang ZX. MicroRNA-21 (miR-21) expression promotes growth, metastasis, and chemo- or radioresistance in non-small cell lung cancer cells by targeting PTEN. Mol Cell Biochem. 2013;372:35–45. doi: 10.1007/s11010-012-1443-3. [DOI] [PubMed] [Google Scholar]
- 96.Fabbri M, Paone A, Calore F, Galli R, Gaudio E, Santhanam R, et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci U S A. 2012;109:E2110–6. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yang M, Chen J, Su F, Yu B, Su F, Lin L, et al. Microvesicles secreted by macrophages shuttle invasion-potentiating microRNAs into breast cancer cells. Mol Cancer. 2011;10:117. doi: 10.1186/1476-4598-10-117. [DOI] [PMC free article] [PubMed] [Google Scholar]


