Abstract
Fluorescence microscopy is a method commonly used to examine individual differences between bacterial cells, yet many studies still lack a quantitative analysis of fluorescence microscopy data. Here we introduce some simple tools that microbiologists can use to analyze and compare their microscopy images. We show how image data can be converted to distribution data. These data can be subjected to a cluster analysis that makes it possible to objectively compare microscopy images. The distribution data can further be analyzed using distribution fitting. We illustrate our methods by scrutinizing two independently acquired data sets, each containing microscopy images of a doubly labeled Bacillus subtilis strain. For the first data set, we examined the expression of srfA and tapA, two genes which are expressed in surfactin-producing and matrix-producing cells, respectively. For the second data set, we examined the expression of eps and tapA; these genes are expressed in matrix-producing cells. We show that srfA is expressed by all cells in the population, a finding which contrasts with a previously reported bimodal distribution of srfA expression. In addition, we show that eps and tapA do not always have the same expression profiles, despite being expressed in the same cell type: both operons are expressed in cell chains, while single cells mainly express eps. These findings exemplify that the quantification and comparison of microscopy data can yield insights that otherwise would go unnoticed.
INTRODUCTION
Bacterial cells can develop into distinct and discrete phenotypic states, which are typically referred to as cell types (1–4). Even when cells experience nearly identical environmental conditions, differentiation is possible (i.e., probabilistic cell differentiation) due to regulatory feedback loops that amplify inherent cellular noise (5–8). In many cases, however, cell differentiation is triggered by environmental changes (2, 9–12). Besides responding to environmental conditions, cells can also modify their environment. For example, they can produce extracellular polysaccharides, communication signals, and antimicrobials (13–15). The feedback between cells and their environment drives colony development (14, 16).
In Bacillus subtilis, cell behavior is often studied in the context of colony development (17). B. subtilis cells can differentiate into a number of cell types, and each of them is associated with a unique set of phenotypes (1, 2, 18). The regulatory mechanisms underlying cell differentiation are often studied using time-lapse fluorescence microscopy, in which gene expression is monitored using fluorescent reporters (8, 18–21). Microscopy images can be analyzed using advanced image-analysis software, which allows the detailed quantification of gene expression along time (22, 23). In this way, Veening and colleagues (24) showed that the timing of sporulation in B. subtilis depends on epigenetic inheritance. In a similar way, Levine and colleagues (25) showed that positive-feedback loops affect the timing of sporulation as well.
Time-lapse microscopy is ideal for studying microcolonies, consisting of colonies with up to a few hundred cells, but not for studying macroscopic colonies, where cell numbers are much higher (20, 26). For those experiments, alternative methods, such as flow cytometry or colony thin sectioning, are used (27, 28). In some cases, macroscopic colonies can still be subjected to microscopy, but only when images are taken at the colony edge, where there is a monolayer of cells, or when the colonies are dissected before microscopy (28–33). Studies that perform microscopy on macroscopic colonies typically show qualitative results, such as representative images with fluorescent overlays (e.g., see the work of Fall et al. [29] and López et al. [32]). Even though these qualitative results are valuable, they are difficult to compare to quantitative data, such as flow cytometry data. In addition, it is also difficult to compare microscopy images between different studies without any form of quantification. In this study, we introduce some tools that will help microbiologists to quantify and compare their microscopy data.
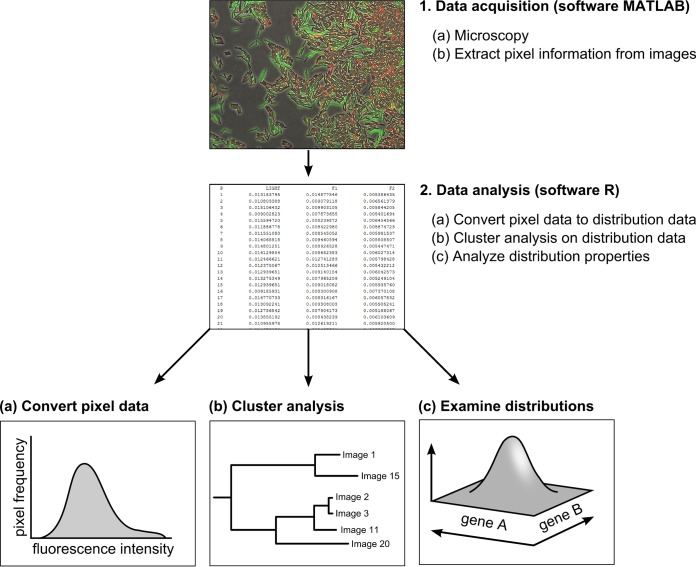
We illustrate our methods by scrutinizing two independent data sets, which are introduced in the next section. For each data set, we examined a doubly labeled B. subtilis strain by fluorescence microscopy. The microscopy images are analyzed in two steps: (i) data acquisition (step 1) and (ii) data analysis (step 2) (Fig. 1). During data acquisition, microscopy images are segmented into cells and background. Pixel data are extracted from the cells and used for further data analysis. By focusing our analysis on data on the fluorescence intensity of pixels as opposed to the segmentation of each cell separately and evaluation of the total intensity per cell, this analysis can be performed at a much higher rate than other types of image analyses that require cell segmentation. Data analysis consists of multiple phases (Fig. 1). Pixel data are first converted to distribution data. Distribution data are subsequently used for a cluster analysis that allows one to compare microscopy images. In addition, distribution data are analyzed to examine how gene expression is distributed. We give an extensive description of all the image-analysis steps and provide in the supplemental material all information that is necessary to use our methods (e.g., a user manual and programs). We hope to stimulate microbiologists to use our methods to analyze their own microscopy data.
FIG 1.
Overview of work flow. Schematic diagram of work flow consisting of two steps: data acquisition in MatLab (step 1) and data analysis in R (step 2). In the first step, microscopy images are converted to tables containing the fluorescent values of pixels associated with cells. In the second step, data are analyzed by converting pixel information (fluorescence intensity per pixel as a proxy for gene expression) (a), performing cluster analysis (b), and examining the gene expression distributions (c).
MATERIALS AND METHODS
The challenge: gene expression analysis in Bacillus subtilis colonies.
We illustrate our methods by examining two doubly labeled Bacillus subtilis strains with PsrfA-yfp-PtapA-cfp and Peps-yfp-PtapA-cfp labels. We were interested in the gene expression patterns in two cell types: surfactin-producing and matrix-producing cells. Surfactin-producing cells secrete surfactin, a lipopeptide that functions as a surfactant and facilitates colony expansion under a number of growth conditions (34–37). In addition, surfactin acts as a communication signal (32, 38) and antimicrobial (39). Matrix-producing cells secrete two important matrix components: the structural protein TasA and an extracellular polysaccharide (EPS) (35, 40, 41). TasA forms amyloid-like fibers that are anchored to the cell wall through TapA, while EPS functions as glue that facilitates cell-to-cell adherence (41–45). Matrix production is essential for biofilm formation and responsible for the wrinkly morphology that is observed in B. subtilis biofilms (35, 46). We examined the coexpression of genes that are essential for either one of these cell types. For the first data set, we examined the coexpression of srfA and tapA (using PsrfA-yfp and PtapA-cfp), genes expressed in surfactin-producing and matrix-producing cells, respectively. For the second data set, we studied the coexpression of the tapA and epsA-epsO (epsA-O) operons (using Peps-yfp and PtapA-cfp), essential for TasA and EPS production, respectively (41). Both operons are expressed in matrix-producing cells and are in part controlled by the same regulatory proteins (17).
To obtain the first and second data sets, strains were cultured for 3 and 22 h, respectively, on a solid growth medium that induces sliding motility (29). After those times, microscopy was performed (see the Materials and Methods in the supplemental material for a detailed description). The different durations of colony growth were necessary because surfactin is mainly produced at the onset of colony growth, while matrix production is upregulated later. For both data sets, only the colony edge, where there was a monolayer of cells, was subjected to microscopy. The colony edge was extracted by cutting a piece of the agar, which was subsequently flipped onto a glass-bottomed well, sandwiching the cells between the coverslip and an agar pad. This glass-bottomed well was placed on an inverted microscope for subsequent examination.
Each data set was analyzed independently using the same methods, summarized above, with two goals in mind. First, we wanted to compare the microscopy images within each data set. Images were taken along the colony edge, where the environmental conditions that cells encounter might vary. We therefore wondered if cells would behave the same along the colony edge or, instead, show differences in their gene expression. Second, we wanted to examine how gene expression is distributed. Is there a unimodal or a bimodal distribution, and how does the expression of one gene depend on that of the other gene? To address these questions, we used a pixel-based proxy of gene expression (see below for a description of our proxy). In the first data set, one expects that gene expression is mutually exclusive, because srfA and tapA are expressed in different cell types (32). In the second data set, gene expression is expected to correlate, because eps and tapA are expressed in the same cell type. Although the data sets were primarily analyzed to illustrate our methods, the analyses resulted in some interesting empirical insights that are discussed below.
RESULTS
Data acquisition: image segmentation.
Microscopy images were processed in two steps: (i) data acquisition (step 1) and (ii) data analysis (step 2) (Fig. 1). In the first step, pixel data were extracted from regions of the microscopy images that correspond to cells. For this, the images were segmented, without delineating each individual cell, into two regions: cells and background. There are various programs available to perform image segmentation (e.g., MicrobeTracker and Schnitzcell [22, 23, 47, 48]). These programs vary in their functionality. For image segmentation, we developed our own Matlab program, which was specifically adjusted to our needs (see the supplemental material). The program is quick and can analyze tens to hundreds of microscopy images within a short amount of time. It displays the fluorescent and phase-contrast images in one window, and it saves all data to a format that can be further analyzed with other software (for details, see the information in the supplemental material). After segmentation, pixel information was saved to text files. In our case, two fluorescence intensity values corresponding to the genes that were monitored (for data set 1, cyan fluorescent protein [CFP] for tapA expression and yellow fluorescent protein [YFP] for srfA expression; for data set 2, CFP for tapA expression and YFP for epsA-O expression) were saved per pixel. This pixel information formed the raw data that were used for data analysis.
One major difference between our program and other image-analysis software is that we use the fluorescence intensity of pixels instead of fluorescence data at the cell level (i.e., the sum of fluorescence intensities per cell) as a proxy for gene expression. A major advantage of this pixel-based approach is that image analysis is much quicker; however, this is achieved at the expense of accuracy. Our proxy for gene expression—the fluorescence intensity of pixels—corresponds to the concentration of the transcript product (assuming that there is no bottleneck in translation). The concentration of the transcript product is determined by the gene expression (i.e., transcription rate), cell size, and degradation of the transcript product. Assuming that degradation of the transcript product is quick (mRNA is unstable), the transcription rate and cell size influence the fluorescence intensity per pixel. Our proxy of gene expression ignores cell-specific information (e.g., cell size) and therefore relies on two assumptions: (i) the transcript product is homogeneously distributed throughout the cell, and (ii) the variation in gene expression is independent of the variation in cell size. In contrast to our pixel-based proxy, a cell-based proxy does account for the cell size and therefore does not depend on the assumptions mentioned above.
The best method of image analysis (pixel based versus cell based) depends on the research question that is addressed and the type of data. When one examines small changes in gene expression during cell cycles, the cell-based approach is favored over the pixel-based approach. However, when one addresses questions on the occurrence of different cell types in a large number of images—as in this study—the less accurate, but quicker, pixel-based approach can be favored (i.e., we are interested in differences in the baseline levels of gene expression and not in small gene expression changes over a cell cycle). Independently of the approach that is used, one should always be aware of its underlying assumptions. When assumptions are violated, one could draw incorrect conclusions. Here, we performed data analysis on our pixel-based image data, yet the same analysis tools can be applied using cell-based image data, as illustrated in Fig. S5 to S7 in the supplemental material. For our microscopy data, the pixel-based and cell-based image analyses yielded similar results (see Fig. S5 to S7 in the supplemental material).
Data analysis: comparing microscopy images.
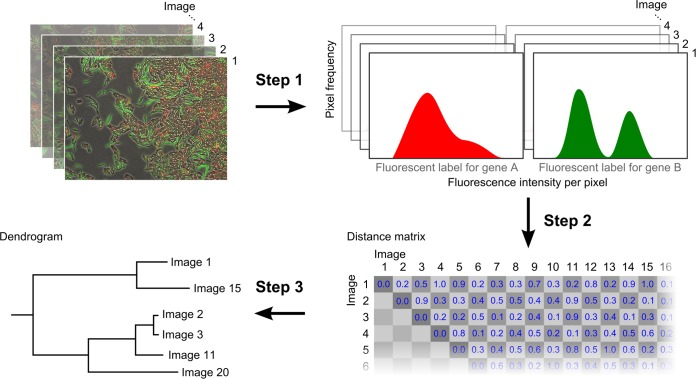
After data acquisition, we began data analysis by comparing the pixel information from images within each data set. Images were compared using a cluster analysis that resulted in the construction of a dendrogram, a tree-like diagram that shows how images are related (Fig. 1b). Cluster analysis was performed in three steps (Fig. 2). First, we converted the pixel information that was stored after segmentation to a workable data format (step 1 in Fig. 2). That is, we converted pixel information to distribution data. This conversion was necessary not only for comparing microscopy images but also for characterizing how genes were expressed within each image. The distribution of fluorescence intensity values was interpreted as the distribution of gene expression levels (see the supplemental material for a description of cell-based analysis). Since each pixel corresponds to two fluorescence intensities, one for each gene, an image could be converted to a two-dimensional distribution of fluorescence intensity levels. This distribution showed how strongly the genes were expressed and, in addition, how their expression covaried. The data were further simplified by ignoring the covariance, thereby converting each image to two distributions of gene expression (step 1 in Fig. 2), one associated with each gene (in this way, one implicitly assumes that both genes are expressed independently and therefore can be treated as such).
FIG 2.
Cluster analysis to compare microscopy images. The construction of a dendrogram is divided into three steps. First, microscopy images are converted to distributions of fluorescence intensity (step 1 arrow transition). We used a pixel-based measure of fluorescence intensity, but a cell-based measure can be used as well. Then, the distributions associated with the images are compared and a distance matrix is calculated (step 2 arrow transition). The distance matrix summarizes how different the microscopy images are from each other. Finally, a dendrogram is constructed from the distance matrix using a clustering algorithm (step 3 arrow transition).
After converting the pixel data to distribution data, we compared the images by comparing their associated distributions (step 2 in Fig. 2). The dissimilarity between two images was determined by use of a distance metric, which measures the distance between their associated fluorescence intensity distributions. By calculating the distance between each pairwise combination of images using the same distance metric, one can construct a distance matrix that summarizes the difference between all microscopy images. The distance metric should be chosen such that the distance matrix is symmetric, meaning that the difference between image A and image B is equal to the difference between image B and image A. Furthermore, the diagonal of the matrix should contain only zeros, meaning that identical images do not differ (i.e., image A is the same as image A, image B is the same as image B, etc.). There are a number of distance metrics available to assess the distance between two distributions. Any of these metrics reduces the difference between two continuous or discrete data distributions to a single value. This process inevitably results in the loss of information. Some distance metrics account more for the shape of the distribution, whereas others account more for the location of the distribution. In the supplemental material, we discuss five distance metrics: Euclidian distance, Jeffreys divergence, Jensen-Shannon divergence, Earth mover's distance, and the Cramér-von Mises statistics. For our analysis, we used the Cramér-von Mises statistics, which is based on the cumulative distribution function and therefore accounts for both the location and the shape of the distribution. This metric can be applied only to one-dimensional data distributions. We therefore calculated the distance between two images by comparing the fluorescence intensity distributions of each gene reporter separately; the sum of both distance values is used for the distance matrix.
The distance matrix can subsequently be converted to a dendrogram using a clustering algorithm (step 3 in Fig. 2). There are different clustering algorithms available (see the supplemental material). We applied a commonly used clustering algorithm that hierarchically clusters the images on the basis of their similarity (i.e., the hierarchical clustering algorithm). Figure 2 gives an overview of the computational steps that are involved in the cluster analysis. We performed the cluster analysis using R, a programming language and working environment that is specifically designed for statistical computing and plotting (http://www.r-project.org). The R script that we used for cluster analysis is included in the supplemental material with a user manual. Finally, it is important to note that the outcome of the clustering analysis strongly depends on both the distance metric and the clustering algorithm (steps 2 and 3 in Fig. 2). We illustrate this in the supplemental material by analyzing our data sets using five different distance metrics and two clustering algorithms (see Fig. S2 to S4 in the supplemental material).
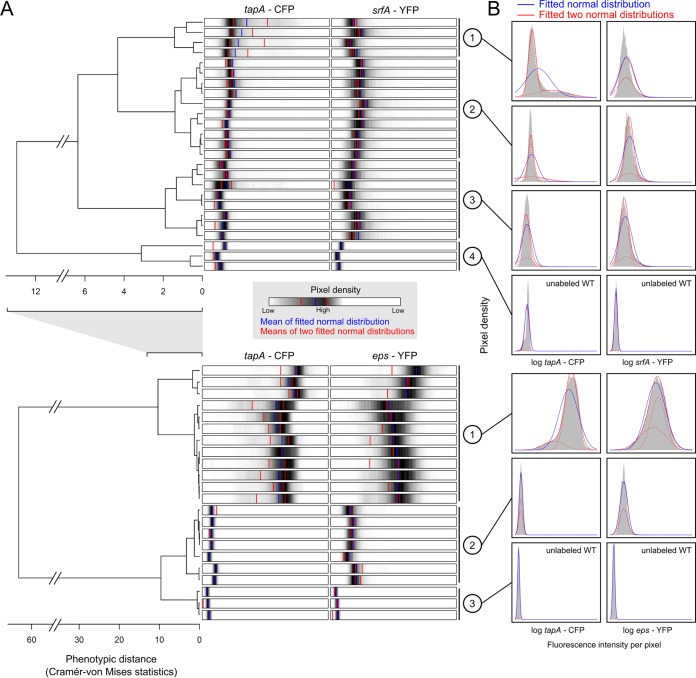
Figure 3A shows the outcome of the cluster analysis for both data sets (top, data set 1 [PtapA-cfp PsrfA-yfp]; bottom, data set 2 [PtapA-cfp Peps-yfp]). For the first data set, images were coarsely divided into three phenotypic clusters (i.e., three sets of microscopy images that showed more or less the same fluorescence intensity distributions). A fourth cluster corresponded to the unlabeled wild-type strain, which was included as a control. For the second data set, images were divided into two clusters. These clusters were more distinct from each other than those observed in the first data set (see the differences in the x axis). The third cluster in the second data set corresponded to the unlabeled wild-type strain. Each tip in the dendrogram corresponds to a single image. To characterize the images, we showed the corresponding fluorescence intensity distribution for each image. The distributions are shown as density bars, in which the highest pixel densities are colored black. We also characterized the clusters by showing, for each cluster separately, the fluorescence intensity distributions for the collective image data corresponding to a cluster (Fig. 3B).
FIG 3.
Hierarchical clustering of microscopy images and the corresponding fluorescence intensity distributions. (A) The upper and lower dendrograms show the results of the data analyses for the first (PtapA-cfp PsrfA-yfp) and second (PtapA-cfp Peps-yfp) data sets, respectively. For each dendrogram, the distance between microscopy images is represented by the length of the branches; each tip corresponds to one image. Every image is characterized by two horizontal bars shown next to the corresponding dendrogram tip. Each bar shows the fluorescence intensity distribution for one of the labeled genes, i.e., the pixel density distribution over the range of log-transformed fluorescence intensities (the range is the same within each data set but not between data sets). High pixel densities are shown in black, and low pixel densities are shown in white. The blue vertical line and the two red vertical lines correspond to the mean fluorescence intensity level based on fitting of either a normal distribution or two normal distributions (with each red line corresponding to the mean of one of the two normal distributions) to the image data, respectively. (B) The distribution of fluorescence intensity (gray) is shown for each phenotypic cluster. A phenotypic cluster is a collection of images that are shown to be closely related according to the dendrogram. For each phenotypic cluster, a normal distribution (blue) and two normal distributions (red lines; the dotted lines show the separate normal distributions) are fitted on the collective image data using a minimal log-likelihood procedure. Clusters 4 and 3 of the first and second data sets, respectively, represent microscopy images from the unlabeled control wild-type (WT) strain.
Data analysis: characterizing the fluorescence intensity distributions.
The fluorescence intensity distributions were further examined by fitting probability distributions (e.g., the normal distribution) to the observed data (Fig. 1c). Since we use pixel fluorescence intensity as a proxy for gene expression, we refer to these distributions as gene expression distributions, although they are actually an approximation thereof. By fitting the probability distributions, the gene expression distributions could be characterized by a few informative parameters. For example, when the gene expression data are normally distributed, they can simply be described by their mean and variance. In our case, we fitted a normal distribution (i.e., a unimodal distribution) or two normal distributions (i.e., a bimodal distribution) to our data using the maximum likelihood method to estimate the best-fitting statistical parameters. This was done for all images individually and for the collective data distributions corresponding to the clusters identified in the previous section. The fitted probability distributions were superimposed on the distribution data shown in Fig. 3B. The best-fitting normal distributions are shown in blue, and the two normal distributions are shown in red.
For the first data set, we observed only a bimodal distribution in tapA expression in clusters 1 and 2 (i.e., a bimodality of CFP fluorescence intensity distributions), although the bimodal distribution in cluster 2 was less apparent, because only a few cells expressed tapA. In both clusters, only a minority of cells expressed tapA. Cluster 1 showed more cells expressing tapA than cluster 2, and in cluster 3 there were no cells that expressed tapA (in cluster 4 there was no expression, because the strain was unlabeled). In contrast to tapA, srfA did not show a bimodal distribution; in all clusters, except for the unlabeled strain (cluster 4), cells expressed srfA (i.e., the pixel fluorescence intensities of clusters 1 to 3 were higher than the pixel fluorescence intensity of cluster 4, which resulted from higher transcription rates). It is therefore not a question of whether cells expressed srfA but, rather, how strongly they did so. Interestingly, for srfA the probability distributions did not always fit the data perfectly (e.g., cluster 1), which indicates that the data are distributed according to a probability distribution different from the distributions tested here. For the second data set, tapA was expressed by all cells in cluster 1 and by very few cells in cluster 2. Interestingly, in cluster 1, tapA expression followed a bimodal distribution. These results indicate that there are (at least) three phenotypic states: cells that do not express tapA (cluster 2), cells that weakly express tapA (the left part of the bimodal distribution of cluster 1), and cells that strongly express tapA (the right part of the bimodal distribution of cluster 1). The bimodal distribution of eps expression in cluster 1 was less apparent. Furthermore, in contrast to tapA expression, all cells expressed eps in both cluster 1 and cluster 2. This indicates that tapA expression and eps expression are not identical.
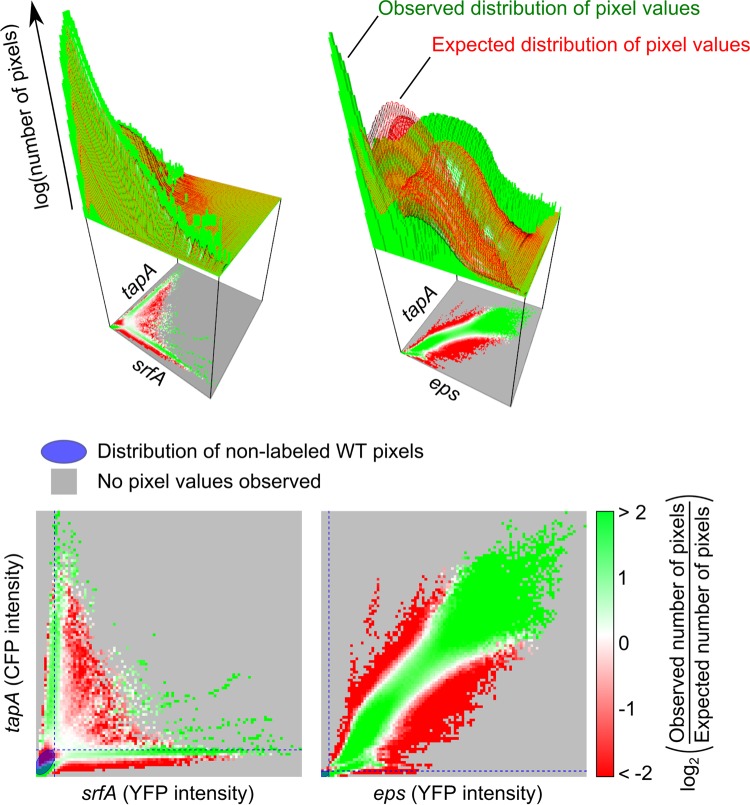
The distance metric that we used to construct the dendrograms (Fig. 3A) ignores the covariance of genes, since the Cramér-von Mises statistics can be applied only to one-dimensional distributions. The question of if and how genes are coexpressed therefore remained: how does the expression of one gene depend on that of the other? To examine this, we studied the two-dimensional distribution of pixel fluorescence intensities. We again used the pixel fluorescence intensity as a proxy for gene expression. This distribution can be used to determine if genes are expressed independently and, if not, how the expression of a gene depends on that of another other gene. Figure 4 shows the two-dimensional distribution for the first (left) and second (right) data sets. For each combination, we plotted the observed (green histogram bars) and expected (red grid surface) distribution of pixel values. The expected number of pixels was calculated by assuming that genes are expressed independently. For example, say that gene A has expression level x with a chance Px and gene B has expression level y with a chance Py. If genes A and B are expressed independently, then the chance of having expression x and y, Pxy, is equal to Px · Py.
FIG 4.
Coexpression of genes in (PtapA-cfp PsrfA-yfp) strains and (PtapA-cfp Peps-yfp) strains. (Top) Three-dimensional plots show the observed (green histogram) and expected (red surface) distribution of the fluorescence intensity (the number of pixels that belong to each fluorescence intensity combination is log transformed); (bottom) for each combination, the log-transformed ratio of the observed versus the expected number of pixels is shown. When the number of pixels observed was more (or less) than expected, the fluorescent combination is colored green (or red). When there was no difference, the fluorescent combination is colored white. When no pixels were observed, the fluorescent combination was colored gray. The blue ellipse shows the bivariate normal density contour containing all unlabeled wild-type (WT) pixels. The corresponding blue dotted lines show the maximal fluorescence intensity measured in the unlabeled wild type for each fluorescence channel.
In most studies, one uses only the observed gene expression distribution to conclude whether and how genes are coexpressed. For example, for the first data set, one did not observe many cells that express both tapA and srfA (see the green histogram bars in Fig. 4). Therefore, one would conclude that the expression of tapA and srfA is mutually exclusive. However, if cells are unlikely to express tapA and they are unlikely to express srfA, then they are even less likely to express both. Not observing cells that expressed both tapA and srfA might therefore be a sampling error.
A more accurate method to examine the coexpression of genes is by comparing the observed distribution of the fluorescence intensity level with the expected distribution. The difference between the two distributions was mapped and is shown below the two-dimensional distributions in the lower graphs of Fig. 4 (see also Fig. S8 in the supplemental material). In these graphs we show results in green if more pixels than expected were observed (for a given combination of gene expression values), in red if we observed fewer pixels than expected, and in white if we observed as many pixels as expected (results in gray indicate that we did not observe any pixels). For both the first and second data sets, there was a difference between the observed and expected distributions, which means that the expression of tapA and srfA and the expression of tapA and eps are not independent of each other. The interdependence in gene expression might result from the discrete environmental conditions that cells experience or from the regulatory control of gene expression. For the first data set, cells that strongly expressed tapA (i.e., high CFP intensity) and weakly expressed srfA (i.e., weak YFP intensity) or vice versa were more abundant than the abundance expected by chance. Thus, even though some cells weakly expressed both tapA and srfA, the expression of tapA and srfA was indeed largely mutually exclusive. For the second data set, high levels of both tapA and eps expression were observed more often than the frequency expected by chance. Thus, despite the differences between the gene expression distributions of tapA and eps in clusters 1 and 2 (Fig. 3), these results show that tapA expression and eps expression are strongly correlated. The strength of this correlation differed, however, between the clusters: it was stronger for images from cluster 1 than those from cluster 2 (see Fig. S9 in the supplemental material).
Differential gene expression in Bacillus subtilis colonies.
In this section, we examine the actual phase-contrast and fluorescence images. Since we performed a cluster analysis, the representative images can simply be chosen by randomly selecting one image from each cluster in the dendrogram. Since the images within each cluster are more or less the same (i.e., they have similar fluorescence intensity distributions), we are confident that the representative images cover the variety of microscopy images present in our data sets. In the previous section we used pixel fluorescence intensities as a proxy for gene expression. In this section we can compare these results with the actual microscopy images.
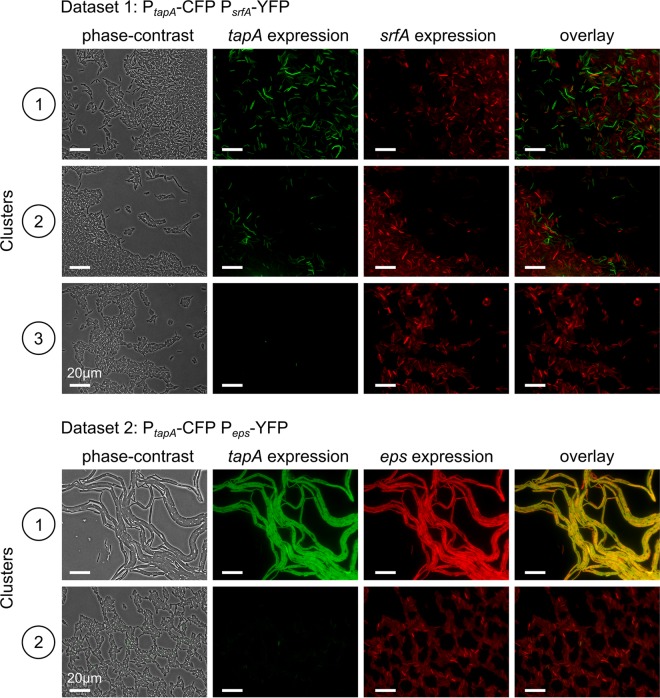
Figure 5 shows representative images. In agreement with the findings of the quantitative analysis (Fig. 3 and 4), the images from clusters 1, 2, and 3 of the first data set are more similar to each other than the images from clusters 1 and 2 of the second data set. In the first data set, the fraction of tapA-expressing cells is the highest for cluster 1 and less high for cluster 2, and tapA-expressing cells are completely absent in cluster 3. This finding is also in agreement with the results of the quantitative analysis presented in the previous section. What is not immediately obvious from the microscopy images is that tapA shows a bimodal distribution, while srfA does not (compare Fig. 3 and 5). In addition, one cannot immediately tell from the images if cells that express both tapA and srfA are less or more common than expected by chance (compare Fig. 4 and 5). Thus, the microscopy images and the quantitative analysis complement each other. Microscopy images of the second data set also show some expected results: the expression of tapA and eps is stronger and more alike in cluster 1 than the expression of tapA and eps in cluster 2. Interestingly, the cell morphology is different between clusters 1 and 2: cluster 1 contains mostly chains of elongated cells, and cluster 2 contains mostly single cells. If the colony had been dissected and analyzed by flow cytometry, such details would have been lost. Thus, one can conclude that, even though, on average, there is a positive correlation between tapA and eps expression, the expression of tapA and eps is not identical and differs in space (i.e., images were taken at different spatial positions at the colony edge).
FIG 5.
Representative microscopy images for each of the phenotypic clusters for cells expressing PtapA-cfp-PsrfA-yfp and PtapA-cfp-Peps-yfp. From left to right, the phase-contrast, cyan fluorescent (falsely colored green), yellow fluorescent (falsely colored red), and overlay images are shown. For PtapA-cfp-PsrfA-yfp-expressing cells, the first three phenotypic clusters are shown (clusters 1 to 3 in Fig. 3). For PtapA-cfp-Peps-yfp-expressing cells, the first two clusters are shown (clusters 1 and 2 in Fig. 3).
DISCUSSION
In this study, we demonstrate how one can analyze and compare fluorescence microscopy images. Our methods consist of two steps. First, we selected our raw data from the microscopy images through a process of image segmentation. For this purpose, we developed simple image-analysis software that, like other software (23, 47, 48), can segment microscopy images. In contrast to other programs, our image-analysis software is based on a pixel-dependent proxy of gene expression. This proxy is less accurate than a cell-based measurement but can be executed faster (in the supplemental material, we compare the results obtained using the pixel-based proxy with those obtained using a cell-based proxy). Second, we performed data analysis by converting the raw pixel data to distribution data. The distribution data that are associated with each image can subsequently be used to compare images via a cluster analysis. In addition, we used the distribution data to characterize gene expression in the image clusters. To illustrate the simplicity and additive value of our methods, we scrutinized two data sets. For the first data set, microscopy images were taken from the colony edge of a B. subtilis strain doubly labeled for the expression of tapA and srfA, genes expressed in matrix-producing and surfactin-producing cells, respectively. For the second data set, microscopy images were taken from the colony edge of a B. subtilis strain doubly labeled for the expression of tapA and eps, both of which are expressed in matrix-producing cells.
Although our primary goal was to introduce some simple methods to compare microscopy images, our study also resulted in some new insights that partly contradict the results from previous studies on surfactin-producing and matrix-producing cells. For example, for the first data set, we show that srfA expression does not show a bimodal distribution. Instead, all cells express srfA. In contrast, tapA expression does show a bimodal distribution. Thus, all cells that express tapA also express srfA, yet the expression of tapA and the expression of srfA are largely mutually exclusive, because cells that strongly express tapA only weakly express srfA and cells that strongly express srfA do not or only weakly express tapA. López and colleagues (32) studied tapA and srfA expression using flow cytometry data and showed that both srfA expression and tapA expression have a bimodal distribution. Furthermore, they showed that none of the tapA-expressing cells express srfA and vice versa. Since surfactin acts as a communication signal that triggers matrix production (38), the authors concluded that surfactin-producing cells induce matrix production in those cells that do not express surfactin, a form of paracrine communication. However, under our conditions, all cells expressed srfA to some extent, including those that expressed tapA. It is therefore plausible that under our growth conditions srfA-expressing cells can be triggered to produce matrix. Alternatively, there could be a threshold level of srfA gene expression that is required for the actual production of the surfactin molecule. The contrasting results are likely to be the consequence of different culturing conditions and the times at which the samples were analyzed. Only a time-lapse experiment could conclusively show if surfactin-producing cells can and do differentiate to produce matrix.
For the second data set, we examined the expression of tapA and eps. One typically assumes that both genes are coexpressed in matrix-producing cells, because they are necessary for the production of TasA and extracellular polysaccharides (41). Here we show that even though there was a strong positive correlation, there were some differences between tapA and eps expression. The differences were most pronounced in cluster 2, where single cells weakly expressed eps but did not express tapA (this was apparent both from our pixel-based image analysis and from the fluorescence microscopy images). Interestingly, in many studies on matrix-producing cells in B. subtilis (27, 49), tapA expression is used to determine which cells produce matrix and which ones do not, thereby implicitly assuming that tapA and eps are coexpressed. tapA expression and eps expression are in part regulated by the same proteins: SinR, AbrB, and RemA (50–56) (see Fig. S10 in the supplemental material). SinR and AbrB repress the transcription of both the tapA and eps operons, while RemA activates the transcription of both operons. SinR negatively regulates the eps operon by inhibiting the activation of RemA (56), thereby functioning as an antiactivator. Once cells enter the stationary phase, tapA and eps get derepressed, due to the Spo0A-mediated repression of both SinR and AbrB (57–60). Spo0A is a key regulatory protein that controls biofilm formation and sporulation (1, 61, 62). Even though both the tapA and eps operons are repressed by the same regulatory proteins, the repressive effects of SinR and AbrB are not equivalent (54): SinR is more efficient in repressing the eps operon, while AbrB is more efficient in repressing the tapA operon. These differences might partly explain the differences between eps and tapA expression that we observed. In addition, tapA expression is also regulated by SlrR, a regulatory protein that forms a double negative-feedback loop with SinR; SinR respresses slrR expression, while SlrR sequesters SinR by direct protein-protein binding (54, 63, 64). In contrast to tapA expression, eps expression is not directly regulated by SlrR (54). Upon starvation, SinR-mediated repression of slrR is released, due to which SlrR subsequently stimulates tapA expression. In addition to SlrR, there is another regulator, LutR, that directly regulates tapA expression, while it only indirectly affects eps expression (65). Thus, even though tapA expression and eps expression are in part regulated by the same regulatory proteins, there are many small regulatory differences that could explain our results (see Fig. S10 in the supplemental material). Future studies are necessary to see which of the regulatory differences is responsible for the differences in gene expression observed in this study.
The simplicity of our methods allows their broad applicability. For example, instead of examining doubly labeled strains, one can examine singly labeled strains. One can also compare microscopy images of colonies that are grown under different culturing conditions and for different time periods. In this way, one can study the generality of certain cell behaviors or compare colony development in different environments and at different times. Another advantage of performing a quantitative analysis on microscopy data is that they can be compared to quantitative data from other sources, e.g., flow cytometry data. In those cases, one should be aware of the types of data that are actually compared. For example, we used a pixel-based proxy for gene expression, while in flow cytometry, a cell-based measure is used. These are different types of data, even though they both relate to gene expression. It is therefore impossible to make a dendrogram that contains both microscopy data and flow cytometry data. Instead, one can make a dendrogram of each data type separately and examine if the overall structure between samples is the same. In other words, samples can be compared with respect to each other. All in all, we hope that our study stimulates microbiologists to further analyze their microscopy data and thereby acquire a better understanding of bacterial behavior.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grant GM58213 to R.K. J.V.G. was subsidized by grants of the Graduate Programme of the Netherlands Science Foundation (NWO), the Ubbo Emmius Programme of the University of Groningen, and the Gratama Foundation.
J.V.G. thanks Paula Montero Llopis, Linda van Zomeren, and Franjo Weissing for useful discussions and help.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02501-14.
REFERENCES
- 1.Piggot PJ, Hilbert DW. 2004. Sporulation of Bacillus subtilis. Curr Opin Microbiol 7:579–586. doi: 10.1016/j.mib.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 2.López D, Vlamakis H, Kolter R. 2009. Generation of multiple cell types in Bacillus subtilis. FEMS Microbiol Rev 33:152–163. doi: 10.1111/j.1574-6976.2008.00148.x. [DOI] [PubMed] [Google Scholar]
- 3.Flores E, Herrero A. 2010. Compartmentalized function through cell differentiation in filamentous cyanobacteria. Nat Rev Microbiol 8:39–50. doi: 10.1038/nrmicro2242. [DOI] [PubMed] [Google Scholar]
- 4.Higgs PI, Hartzell PL, Holkenbrink C, Hoiczyk E. 2014. Myxococcus xanthus vegetative and developmental cell differentiation. In Myxobacteria: genomics and molecular biology. Horizon Scientific Press, Norfolk, United Kingdom. [Google Scholar]
- 5.Avery SV. 2006. Microbial cell individuality and the underlying sources of heterogeneity. Nat Rev Microbiol 4:577–587. doi: 10.1038/nrmicro1460. [DOI] [PubMed] [Google Scholar]
- 6.Smits WK, Kuipers OP, Veening J-W. 2006. Phenotypic variation in bacteria: the role of feedback regulation. Nat Rev Microbiol 4:259–271. doi: 10.1038/nrmicro1381. [DOI] [PubMed] [Google Scholar]
- 7.Raj A, van Oudenaarden A. 2008. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell 135:216–226. doi: 10.1016/j.cell.2008.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Veening J-W, Smits WK, Kuipers OP. 2008. Bistability, epigenetics, and bet-hedging in bacteria. Annu Rev Microbiol 62:193–210. doi: 10.1146/annurev.micro.62.081307.163002. [DOI] [PubMed] [Google Scholar]
- 9.Parkinson JS. 1993. Signal transduction schemes of bacteria. Cell 73:857–871. doi: 10.1016/0092-8674(93)90267-T. [DOI] [PubMed] [Google Scholar]
- 10.Stock AM, Robinson VL, Goudreau PN. 2000. Two-component signal transduction. Annu Rev Biochem 69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 11.Stewart PS, Franklin MJ. 2008. Physiological heterogeneity in biofilms. Nat Rev Microbiol 6:199–210. doi: 10.1038/nrmicro1838. [DOI] [PubMed] [Google Scholar]
- 12.Konovalova A, Petters T, Søgaard-Andersen L. 2010. Extracellular biology of Myxococcus xanthus. FEMS Microbiol Rev 34:89–106. doi: 10.1111/j.1574-6976.2009.00194.x. [DOI] [PubMed] [Google Scholar]
- 13.Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM. 1995. Microbial biofilms. Annu Rev Microbiol 49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- 14.O'Toole G, Kaplan HB, Kolter R. 2000. Biofilm formation as microbial development. Annu Rev Microbiol 54:49–79. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- 15.Nadell CD, Xavier JB, Foster KR. 2009. The sociobiology of biofilms. FEMS Microbiol Rev 33:206–224. doi: 10.1111/j.1574-6976.2008.00150.x. [DOI] [PubMed] [Google Scholar]
- 16.Monds RD, O'Toole GA. 2009. The developmental model of microbial biofilms: ten years of a paradigm up for review. Trends Microbiol 17:73–87. doi: 10.1016/j.tim.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 17.Vlamakis H, Chai Y, Beauregard P, Losick R, Kolter R. 2013. Sticking together: building a biofilm the Bacillus subtilis way. Nat Rev Microbiol 11:157–168. doi: 10.1038/nrmicro2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Locke JCW, Elowitz MB. 2009. Using movies to analyse gene circuit dynamics in single cells. Nat Rev Microbiol 7:383–392. doi: 10.1038/nrmicro2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Süel GM, Garcia-Ojalvo J, Liberman LM, Elowitz MB. 2006. An excitable gene regulatory circuit induces transient cellular differentiation. Nature 440:545–550. doi: 10.1038/nature04588. [DOI] [PubMed] [Google Scholar]
- 20.Garcia-Betancur JC, Yepes A, Schneider J, López D. 2012. Single-cell analysis of Bacillus subtilis biofilms using fluorescence microscopy and flow cytometry. J Vis Exp 2012:3796. doi: 10.3791/3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Norman TM, Lord ND, Paulsson J, Losick R. 2013. Memory and modularity in cell-fate decision making. Nature 503:481–486. doi: 10.1038/nature12804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ljosa V, Carpenter AE. 2009. Introduction to the quantitative analysis of two-dimensional fluorescence microscopy images for cell-based screening. PLoS Comput Biol 5:e1000603. doi: 10.1371/journal.pcbi.1000603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sliusarenko O, Heinritz J, Emonet T, Jacobs-Wagner C. 2011. High-throughput, subpixel precision analysis of bacterial morphogenesis and intracellular spatio-temporal dynamics. Mol Microbiol 80:612–627. doi: 10.1111/j.1365-2958.2011.07579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Veening J-W, Stewart EJ, Berngruber TW, Taddei F, Kuipers OP, Hamoen LW. 2008. Bet-hedging and epigenetic inheritance in bacterial cell development. Proc Natl Acad Sci U S A 105:4393–4398. doi: 10.1073/pnas.0700463105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levine JH, Fontes ME, Dworkin J, Elowitz MB. 2012. Pulsed feedback defers cellular differentiation. PLoS Biol 10:e1001252. doi: 10.1371/journal.pbio.1001252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Jong IG, Beilharz K, Kuipers OP, Veening J-W. 2011. Live cell imaging of Bacillus subtilis and Streptococcus pneumoniae using automated time-lapse microscopy. J Vis Exp 2011:3145. doi: 10.3791/3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vlamakis H, Aguilar C, Losick R, Kolter R. 2008. Control of cell fate by the formation of an architecturally complex bacterial community. Genes Dev 22:945–953. doi: 10.1101/gad.1645008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marlow VL, Cianfanelli FR, Porter M, Cairns LS, Dale JK, Stanley-Wall NR. 2014. The prevalence and origin of exoprotease-producing cells in the Bacillus subtilis biofilm. Microbiology 160:56–66. doi: 10.1099/mic.0.072389-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fall R, Kearns DB, Nguyen T. 2006. A defined medium to investigate sliding motility in a Bacillus subtilis flagella-less mutant. BMC Microbiol 6:31. doi: 10.1186/1471-2180-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kobayashi K. 2007. Bacillus subtilis pellicle formation proceeds through genetically defined morphological changes. J Bacteriol 189:4920–4931. doi: 10.1128/JB.00157-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hamze K, Julkowska D, Autret S, Hinc K, Nagorska K, Sekowska A, Holland IB, Séror SJ. 2009. Identification of genes required for different stages of dendritic swarming in Bacillus subtilis, with a novel role for phrC. Microbiology 155:398–412. doi: 10.1099/mic.0.021477-0. [DOI] [PubMed] [Google Scholar]
- 32.López D, Vlamakis H, Losick R, Kolter R. 2009. Paracrine signaling in a bacterium. Genes Dev 23:1631–1638. doi: 10.1101/gad.1813709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hamze K, Autret S, Hinc K, Laalami S, Julkowska D, Briandet R, Renault M, Absalon C, Holland IB, Putzer H, Séror SJ. 2011. Single-cell analysis in situ in a Bacillus subtilis swarming community identifies distinct spatially separated subpopulations differentially expressing hag (flagellin), including specialized swarmers. Microbiology 157:2456–2469. doi: 10.1099/mic.0.047159-0. [DOI] [PubMed] [Google Scholar]
- 34.Nakano MM, Magnuson R, Myers A, Curry J, Grossman AD, Zuber P. 1991. srfA is an operon required for surfactin production, competence development, and efficient sporulation in Bacillus subtilis. J Bacteriol 173:1770–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Branda SS, González-Pastor JE, Ben-Yehuda S, Losick R, Kolter R. 2001. Fruiting body formation by Bacillus subtilis. Proc Natl Acad Sci U S A 98:11621–11626. doi: 10.1073/pnas.191384198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kinsinger RF, Shirk MC, Fall R. 2003. Rapid surface motility in Bacillus subtilis is dependent on extracellular surfactin and potassium ion. J Bacteriol 185:5627–5631. doi: 10.1128/JB.185.18.5627-5631.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kearns DB, Chu F, Rudner R, Losick R. 2004. Genes governing swarming in Bacillus subtilis and evidence for a phase variation mechanism controlling surface motility. Mol Microbiol 52:357–369. doi: 10.1111/j.1365-2958.2004.03996.x. [DOI] [PubMed] [Google Scholar]
- 38.López D, Fischbach MA, Chu F, Losick R, Kolter R. 2009. Structurally diverse natural products that cause potassium leakage trigger multicellularity in Bacillus subtilis. Proc Natl Acad Sci U S A 106:280–285. doi: 10.1073/pnas.0810940106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bais HP, Fall R, Vivanco JM. 2004. Biocontrol of Bacillus subtilis against infection of Arabidopsis roots by Pseudomonas syringae is facilitated by biofilm formation and surfactin production. Plant Physiol 134:307–319. doi: 10.1104/pp.103.028712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Branda SS, González-Pastor JE, Dervyn E, Ehrlich SD, Losick R, Kolter R. 2004. Genes involved in formation of structured multicellular communities by Bacillus subtilis. J Bacteriol 186:3970–3979. doi: 10.1128/JB.186.12.3970-3979.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Branda SS, Chu F, Kearns DB, Losick R, Kolter R. 2006. A major protein component of the Bacillus subtilis biofilm matrix. Mol Microbiol 59:1229–1238. doi: 10.1111/j.1365-2958.2005.05020.x. [DOI] [PubMed] [Google Scholar]
- 42.Branda SS, Vik S, Friedman L, Kolter R. 2005. Biofilms: the matrix revisited. Trends Microbiol 13:20–26. doi: 10.1016/j.tim.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 43.Marvasi M, Visscher PT, Casillas Martinez L. 2010. Exopolymeric substances (EPS) from Bacillus subtilis: polymers and genes encoding their synthesis. FEMS Microbiol Lett 313:1–9. doi: 10.1111/j.1574-6968.2010.02085.x. [DOI] [PubMed] [Google Scholar]
- 44.Romero D, Aguilar C, Losick R, Kolter R. 2010. Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc Natl Acad Sci U S A 107:2230–2234. doi: 10.1073/pnas.0910560107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Romero D, Vlamakis H, Losick R, Kolter R. 2011. An accessory protein required for anchoring and assembly of amyloid fibres in B. subtilis biofilms. Mol Microbiol 80:1155–1168. doi: 10.1111/j.1365-2958.2011.07653.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Asally M, Kittisopikul M, Rué P, Du Y, Hu Z, Çağatay T, Robinson AB, Lu H, Garcia-Ojalvo J, Süel GM. 2012. Localized cell death focuses mechanical forces during 3D patterning in a biofilm. Proc Natl Acad Sci U S A 109:18891–18896. doi: 10.1073/pnas.1212429109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garner EC. 2011. MicrobeTracker: quantitative image analysis designed for the smallest organisms. Mol Microbiol 80:577–579. doi: 10.1111/j.1365-2958.2011.07580.x. [DOI] [PubMed] [Google Scholar]
- 48.Young JW, Locke JCW, Altinok A, Rosenfeld N, Bacarian T, Swain PS, Mjolsness E, Elowitz MB. 2012. Measuring single-cell gene expression dynamics in bacteria using fluorescence time-lapse microscopy. Nat Protoc 7:80–88. doi: 10.1038/nprot.2011.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beauregard PB, Chai Y, Vlamakis H, Losick R, Kolter R. 2013. Bacillus subtilis biofilm induction by plant polysaccharides. Proc Natl Acad Sci U S A 110:E1621–E1630. doi: 10.1073/pnas.1218984110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hamon MA, Stanley NR, Britton RA, Grossman AD, Lazazzera BA. 2004. Identification of AbrB-regulated genes involved in biofilm formation by Bacillus subtilis. Mol Microbiol 52:847–860. doi: 10.1111/j.1365-2958.2004.04023.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kearns DB, Chu F, Branda SS, Kolter R, Losick R. 2005. A master regulator for biofilm formation by Bacillus subtilis. Mol Microbiol 55:739–749. doi: 10.1111/j.1365-2958.2004.04440.x. [DOI] [PubMed] [Google Scholar]
- 52.Chu F, Kearns DB, Branda SS, Kolter R, Losick R. 2006. Targets of the master regulator of biofilm formation in Bacillus subtilis. Mol Microbiol 59:1216–1228. doi: 10.1111/j.1365-2958.2005.05019.x. [DOI] [PubMed] [Google Scholar]
- 53.Strauch MA, Bobay BG, Cavanagh J, Yao F, Wilson A, Breton YL. 2007. Abh and AbrB control of Bacillus subtilis antimicrobial gene expression. J Bacteriol 189:7720–7732. doi: 10.1128/JB.01081-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chu F, Kearns DB, McLoon A, Chai Y, Kolter R, Losick R. 2008. A novel regulatory protein governing biofilm formation in Bacillus subtilis. Mol Microbiol 68:1117–1127. doi: 10.1111/j.1365-2958.2008.06201.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Winkelman JT, Blair KM, Kearns DB. 2009. RemA (YlzA) and RemB (YaaB) regulate extracellular matrix operon expression and biofilm formation in Bacillus subtilis. J Bacteriol 191:3981–3991. doi: 10.1128/JB.00278-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Winkelman JT, Bree AC, Bate AR, Eichenberger P, Gourse RL, Kearns DB. 2013. RemA is a DNA-binding protein that activates biofilm matrix gene expression in Bacillus subtilis. Mol Microbiol 88:984–997. doi: 10.1111/mmi.12235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Strauch M, Webb V, Spiegelman G, Hoch JA. 1990. The Spo0A protein of Bacillus subtilis is a repressor of the abrB gene. Proc Natl Acad Sci U S A 87:1801–1805. doi: 10.1073/pnas.87.5.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bai U, Mandic-Mulec I, Smith I. 1993. SinI modulates the activity of SinR, a developmental switch protein of Bacillus subtilis, by protein-protein interaction. Genes Dev 7:139–148. doi: 10.1101/gad.7.1.139. [DOI] [PubMed] [Google Scholar]
- 59.Lewis RJ, Brannigana JA, Smith I, Wilkinson AJ. 1996. Crystallisation of the Bacillus subtilis sporulation inhibitor SinR, complexed with its antagonist, SinI. FEBS Lett 378:98–100. doi: 10.1016/0014-5793(95)01432-2. [DOI] [PubMed] [Google Scholar]
- 60.Chai Y, Chu F, Kolter R, Losick R. 2008. Bistability and biofilm formation in Bacillus subtilis. Mol Microbiol 67:254–263. doi: 10.1111/j.1365-2958.2007.06040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hamon MA, Lazazzera BA. 2001. The sporulation transcription factor Spo0A is required for biofilm development in Bacillus subtilis. Mol Microbiol 42:1199–1209. doi: 10.1046/j.1365-2958.2001.02709.x. [DOI] [PubMed] [Google Scholar]
- 62.Fujita M, González-Pastor JE, Losick R. 2005. High- and low-threshold genes in the Spo0A regulon of Bacillus subtilis. J Bacteriol 187:1357–1368. doi: 10.1128/JB.187.4.1357-1368.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chai Y, Kolter R, Losick R. 2010. Reversal of an epigenetic switch governing cell chaining in Bacillus subtilis by protein instability. Mol Microbiol 78:218–229. doi: 10.1111/j.1365-2958.2010.07335.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chai Y, Norman T, Kolter R, Losick R. 2010. An epigenetic switch governing daughter cell separation in Bacillus subtilis. Genes Dev 24:754–765. doi: 10.1101/gad.1915010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Irigül-Sonmez Ö, Köroğlu TE, Öztürk B, Kovács ÁT, Kuipers OP, Yazgan-Karataş A. 2014. In Bacillus subtilis LutR is part of the global complex regulatory network governing the adaptation to the transition from exponential growth to stationary phase. Microbiology 160:243–260. doi: 10.1099/mic.0.064675-0. [DOI] [PubMed] [Google Scholar]