Abstract
Background
High mobility group box-1 (HMGB1) is a factor regulating malignant tumorigenesis, proliferation, and metastasis, and is associated with poor clinical pathology in various human cancers. We investigated the differential concentrations of HMGB1 in tissues and sera, and their clinical value for diagnosis in patients with breast cancer, benign breast disease, and healthy individuals.
Methods
HMGB1 levels in tumor tissues, adjacent normal tissues, and benign breast disease tissues was detected via immunohistochemistry. Serum HMGB1 was measured using an enzyme-linked immunosorbent assay in 56 patients with breast cancer, 25 patients with benign breast disease, and 30 healthy control subjects. The clinicopathological features of the patients were compared. Tissues were evaluated histopathologically by pathologists.
Results
HMGB1 levels in the tissues and sera of patients with breast cancer were significantly higher than those in patients with benign breast disease or normal individuals. The 56 cancer patients were classified as having high tissue HMGB1 levels (n=41) or low tissue HMGB1 levels (n=15), but the corresponsive serum HMGB1 in these two groups was not significantly different. HMGB1 levels in breast cancer tissues significantly correlated with differentiation grade, lymphatic metastasis, and tumor-node-metastasis stage, but not patient age, tumor size, or HER-2/neu expression; no association between serum HMGB1 levels and these clinicopathological parameters was found. The sensitivity and specificity of tissue HMGB1 levels for the diagnosis of breast cancer were 73.21% and 84.00%, respectively, while positive and negative predictive values were 91.11% and 58.33%.
Conclusion
HMGB1 might be involved in the development and progression of breast cancer and could be a supportive diagnostic marker for breast cancer. Serum HMGB1 could be a useful serological biomarker for diagnosis and screening of breast cancer.
Keywords: high mobility group box-1, breast cancer, serum biomarker
Introduction
Worldwide, breast cancer is the leading cause of death due to cancer in women.1 In the People’s Republic of China, the incidence of breast cancer has risen dramatically within the last 10 years; for example, rates in Beijing and Shanghai have increased by 23% and 31%, respectively.2 Although current comprehensive treatment for breast cancer prolongs survival, distant metastasis is still an important risk factor affecting prognosis.3 The majority of deaths associated with the disease are due to metastases in the advanced stages, making early detection crucial. Recently, cancer biomarkers have become attractive due to their potential application in the early diagnosis of cancer.4,5 Several useful and well established biomarkers, such as BRCA1/2, CA15.3, and CA27.29, are already used in the diagnosis of breast cancer.6,7 However, the sensitivity (BRCA1/2) and specificity (CA15.3, CA27.29) of the current tumor markers for breast cancer are not high enough.8,9 The US Food and Drug Administration recommended CA15.3 and CA27.29 as markers only for monitoring therapy or for recurrence of advanced breast cancer.10 Several novel and potential biomarkers, such as uPA, STAT3, PTEN, and lin28, are attractive but still far away from universal acceptance.6 Therefore, new biomarkers for cost-effective screening of breast cancer are urgently needed, to be used alone or in combination with an existing modality.
The process that transforms normal breast cells into highly malignant derivatives is complex, involving many genetic and epigenetic changes, and the molecular mechanisms for the initiation, progression, and metastasis of breast cancer are not fully understood. High mobility group box-1 (HMGB1) is a regulator of the tumorigenesis, expansion, and invasion of cancer cells and was identified relatively recently.11 HMGB1 was originally discovered in calf thymus 40 years ago, and named for its fast electrophoretic mobility in polyacrylamide gels.12 It is now known to be a non-histone architectural chromosomal protein ubiquitously present in all vertebrate nuclei, binding double-stranded DNA without sequence specificity.
HMGB1 protein is a ubiquitous widespread nuclear protein present in most cell types. It is typically localized in the nucleus and functions as a nuclear cofactor in regulation of transcription. However, HMGB1 can also be present in the cytoplasm and be released into the extracellular matrix, where it has crucial roles in carcinogenesis and inflammation.11 Once secreted, HMGB1 induces signal transduction through the receptor RAGE (receptor for advanced glycation end products), and perhaps the toll-like receptors (TLR2 and TLR4). Recent studies have shown that for a large variety of human neoplasms, levels of HMGB1 and its receptor RAGE are greater in tumors than in the normal surrounding epithelia. Examples include colorectal, prostate, and pancreatic cancers and hepatocellular carcinoma.13–18 Moreover, these studies show that overexpression of HMGB1 is strongly correlated with tumor invasiveness and a poor prognosis in cancer patients.19–21 Although the oncogenic role of HMGB1 in cancer is well recognized, its role in breast cancer remains poorly documented.22,23 Several studies have suggested that HMGB1 could promote growth of breast cancer cells in vitro, but its exact role in patients with breast cancer is still unclear.24,25 Furthermore, there is little information about the role of HMGB1 specifically in breast cancer.
In the present study, we investigated the differential levels of HMGB1 in the tissues and sera of patients with breast cancer, patients with benign breast disease, and healthy control subjects. We also assessed the associations between these levels and clinicopathological features of breast cancer, and their diagnostic value.
Materials and methods
The institutional ethics committee of Liaocheng People’s Hospital approved the study, and all patients provided their informed consent.
Patient characteristics
Fifty-six patients with breast cancer and 25 patients with benign breast disease were recruited, and the diagnoses were pathologically confirmed at the Liaocheng People’s Hospital, People’s Republic of China, between October 2012 and October 2013. The 56 cancer cases comprised 25 cases of axillary lymph node metastasis and 31 cases of non-metastasis, and 18, 24, and 14 cases were histological grades I, II, and III, respectively. Tumor-node-metastasis (TNM) staging was in accordance with the American Joint Committee on Cancer TNM classification system: five, ten, 22, 14, and five cases were in stage 0, I, II, III, and IV, respectively. The age range of the patients was from 24 years to 76 years, with a median of 46 years. The 25 patients with benign breast cancer included eight cases of hyperplasia, 14 cases of fibroadenoma, and three cases of intraductal papilloma.
To detect the expression of HMGB1 protein in tissues, tumor samples and corresponding adjacent normal tissues were collected from the 56 cancer patients undergoing modified radical mastectomy or breast conservation surgery, and tissue samples were obtained from the 25 patients with benign breast disease. To measure serum HMGB1 levels, blood samples were collected from the 56 cancer patients and 25 patients with benign breast disease, prior to any surgery, radiotherapy, chemotherapy, or molecular-targeted therapy. All patients had normal liver, kidney, heart function and without evidence of malignant cancers from other tissues except breast. In addition, blood samples were collected from 30 healthy volunteers as the control group.
Immunohistochemistry
HMGB1 protein in tissues was detected using a HMGB1 immunohistochemistry kit (Beijing Bioss Biotechnology, People’s Republic of China). The protocol was performed in accordance with the manufacturer’s instructions (a streptavidin peroxidase-conjugated method). Briefly, tissues were formalin-fixed, dehydrated, and paraffin-embedded for sectioning at 4 μm. The sections were dewaxed and rehydrated, treated with 3% hydrogen peroxide to block endogenous peroxidase activity, sealed with non-specific serum, and incubated with primary antibodies at 4°C overnight. The sections were placed at room temperature and incubated with biotin-labeled secondary antibody for 30 minutes at 37°C, then mixed with streptococcus avidin-peroxidase-labeled solution. After washing with phosphate-buffered saline, 3,3′-diaminobenzidine tetrahydrochloride chromogen was applied to develop color. The sections were counterstained with hematoxylin and dehydrated. As a negative control, a parallel experiment was performed by substituted the primary antibodies with phosphate-buffered saline.
The sections were examined under a light microscope. HMGB1 exists mainly in the nucleus.11 To assess protein expression, five visual fields were randomly selected at high power, and 200 cells per visual field were counted. All samples were reviewed and diagnosed by two independent pathologists. If an inconsistency occurred, a third experienced pathologist was consulted to achieve consensus. Staining frequency was divided into four classes and scored according to the proportion of positively-stained cancer cells among all the tumor cells: 0, <25%; 1, 25%–50%; 2, 51%–75%; and 3, >76%. Additionally, staining intensity was divided into four classes and scored as follows: 0, uncolored; 1, pale yellow; 2, yellow; 3, brown-yellow. For each subject, scores for staining intensity and staining frequency were summed. Total scores of 0 to 2 were considered low for HMGB1, and scores >2 were considered high.
Enzyme-linked immunosorbent assay
Serum concentrations of HMGB1 were evaluated using an HMGB1 enzyme-linked immunosorbent assay kit (R&D Systems, Minneapolis, MN, USA). The serum fraction was obtained by centrifugation within the first 3 hours after blood sample collection, and stored at −80°C until assayed. The enzyme-linked immunosorbent assay was performed in accordance with the manufacturer’s instructions. Briefly, 10 μL of serum sample was mixed with diluent (1:10 dilution) and added to microtiter plate wells. Standards and controls were also added to the wells. The microtiter plates were incubated for 30 minutes at 37°C. After washing, 50 μL of enzyme reagent was added to each well and incubated for 30 minutes at 37°C. After washing, substrate solution was added to each well and incubated for 15 minutes at 37°C, and the reaction was stopped with 50 μL of stop solution. Absorbance was immediately read at 450 nm. The concentrations of HMGB1 were based on a calibration curve prepared from standards.
Statistical analysis
All data were analyzed using Statistical Package for the Social Sciences version 19.0 software (SPSS Inc, Chicago, IL, USA). A t-test or a t′-test was used to compare the means of two samples. A chi-squared test (χ2) was used to assess the difference between the ratios. Calculation of sensitivity, specificity, positive predictive value, and negative predictive value was based on the following formula: sensitivity (SEN) = a/(a + c); specificity (SPE) = d/(b + d); positive predictive value = a/(a + b); and negative predictive value = d/(c + d), where a is the true positive number, b is the false positive number, c is the false negative number, and d is the true negative number. P<0.05 was considered to indicate a statistically significant difference.
Results
HMGB1 levels in tissues and sera
Immunohistochemistry was used to detect evidence of HMGB1 in tissues, including tumor tissues and corresponding adjacent normal tissues, from 56 patients with breast cancer and samples from 25 patients with benign breast disease (Figure 1). In cancer patients, 73% (41/56) were classified as having high HMGB1 levels. However, of the 56 samples of adjacent normal tissue and the 25 samples of benign breast disease tissue, only nine (16%) and four (16%), respectively, had high HMGB1 levels. Statistical analysis revealed that HMGB1 expression was significantly higher in breast cancer tissues than in the corresponding adjacent normal tissues and benign breast disease tissues (P<0.05). However, there were no statistically significant differences in HMGB1 levels between normal tissue in breast cancer patients and benign breast disease tissue (P=0.9935).
Figure 1.
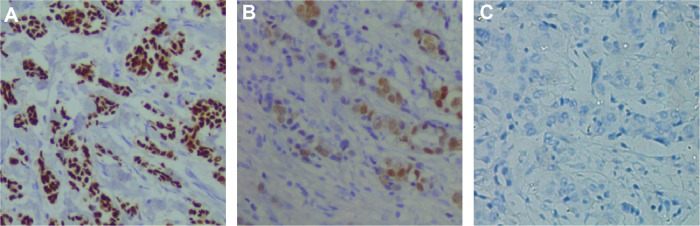
Representative high mobility group box-1 immunohistochemistry staining in breast cancer tissues (A), patients with benign breast disease (B) and healthy subjects (C).
We measured serum HMGB1 levels in the 56 patients with breast cancer, 25 patients with benign breast disease, and 30 healthy subjects who had no evidence of carcinoma (Figure 2). The mean serum HMGB1 concentration in breast cancer patients was 4.64±2.50 ng/mL, which was significantly higher than in those with benign breast disease [1.32±0.68 ng/mL; t′=9.2044, t′(0.05)=2.0125, P<0.05] and in healthy subjects [1.36±0.75 ng/mL; t′=9.0846, t′(0.05)=0.0100, P<0.05]. Mean serum HMGB1 levels in the patients with benign breast disease and the control subjects were similar (t=0.2054, P=0.8380).
Figure 2.

Serum HMGB1 levels in (A) 56 patients with breast cancer, (B) 25 patients with benign breast disease, (C) 30 healthy subjects, (D) 41 patients with breast cancer and high levels of HMGB1 in tissues, and (E) 15 patients with breast cancer and low levels of HMGB1 in tissues. *P<0.05.
Notes: The mean serum HMGB1 concentration in breast cancer patients was significantly higher than in those with benign breast disease and in healthy subjects. Mean serum HMGB1 levels in the patients with benign breast disease and the control subjects were similar. Similarly, no statistically significant difference was found between 41 breast cancer patients who tested high for HMGB1 levels and 15 patients who having low HMGB1 tissue levels.
Abbreviation: HMGB1, high mobility group box-1.
We compared the mean serum HMGB1 concentration in the 41 breast cancer patients who tested high for HMGB1 levels (4.84±2.43 ng/mL) with that of the 15 patients having low HMGB1 tissue levels (4.00±2.69 ng/mL), and found no statistically significant difference between the two (t=1.1186, P=0.2683; Figure 2).
Correlation between HMGB1 levels and clinicopathological features in breast cancer
To assess the clinical value of HMGB1 from our collected tissues and blood sample sets, we stratified the 56 breast cancer cases according to age, tumor size, HER-2/neu levels, differentiation grade, lymphatic metastasis, and TNM stage (Table 1). Tissue HMGB1 levels correlated significantly with the differentiation grade (P=0.0008), lymphatic metastasis (P=0.0248), and TNM stage (P=0.0490), but were not associated with patient age (P=0.7938), tumor size (P=0.2346), or HER-2/neu levels (P=0.4879). No association was found between serum HMGB1 levels and any of the clinicopathological parameters examined.
Table 1.
HMGB1 levels and clinicopathological parameters in patients with breast cancer
| n | Tissue HMGB1, n
|
Serum HMGB1
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Low | High | χ2 | P-value | ng/mL | t-value | P-value | |||
| Age, years | ≤50 | 32 | 9 | 23 | 0.0683 | 0.7938 | 4.84±2.31 | 0.7160 | 0.4771 |
| >50 | 24 | 6 | 18 | 4.36±2.76 | |||||
| Menopause | Yes | 25 | 8 | 17 | 0.6261 | 0.4288 | 4.32±2.57 | 0.8452 | 0.4017 |
| No | 31 | 7 | 24 | 4.89±2.46 | |||||
| HER-2/neu | Negative | 41 | 12 | 29 | 0.4810 | 0.4879 | 4.42±2.40 | 0.9916 | 0.3258 |
| Positive | 15 | 3 | 12 | 5.12±2.80 | |||||
| Tumor diameter | ≤2 cm | 30 | 10 | 20 | 1.4125 | 0.2346 | 4.51±2.55 | 0.4114 | 0.6824 |
| >2 cm | 26 | 5 | 21 | 4.79±2.49 | |||||
| Axillary lymph node metastasis | Negative | 31 | 12 | 19 | 5.0344 | 0.0248* | 4.34±2.36 | 0.9878 | 0.3277 |
| Positive | 25 | 3 | 22 | 5.00±2.67 | |||||
| Differentiation grade | High | 18 | 10 | 8 | 11.1957 | 0.0008* | 4.17±2.51 | 0.9148 | 0.3644 |
| Intermediate/low | 38 | 5 | 33 | 4.83±2.51 | |||||
| TNM stage | 0, I, II | 37 | 13 | 24 | 3.8766 | 0.0490* | 4.55±2.41 | 0.3596 | 0.7206 |
| III, IV | 19 | 2 | 17 | 4.81±2.73 | |||||
Note:
P<0.05.
Abbreviations: TNM, tumor-node-metastasis; HMGB1, high mobility group box-1.
Correlation between tissue HMGB1 levels and diagnosis in breast cancer
We assessed the diagnostic value of HMGB1 levels in breast cancer tissues (Table 2). The sensitivity, specificity, positive predictive value, and negative predictive value of HMGB1 levels in tissues for the diagnosis of breast cancer were 73.21%, 84.00%, 91.11%, and 58.33%, respectively. With regard to differentiation grade and lymph node metastasis, the sensitivity and positive and negative predictive values of HMGB1 levels in tissues were high, but specificity was low, while the sensitivity and negative predictive value were high but specificity and positive predictive value were low for TNM stage.
Table 2.
Tissue HMGB1 and diagnostic and clinicopathological features of breast cancer
| Statistic | Breast cancer | Differentiation grade | Lymph node metastasis | TNM stage |
|---|---|---|---|---|
| Sensitivity | 73.21% (41/56) | 86.84% (33/38) | 88.00% (22/25) | 89.47% (17/19) |
| Specificity | 84.00% (21/25) | 55.55% (10/18) | 38.71% (12/31) | 35.14% (13/37) |
| Positive predictive value | 91.11% (41/45) | 80.49% (33/41) | 53.66% (22/41) | 41.46% (17/41) |
| Negative predictive value | 58.33% (21/36) | 66.67% (10/15) | 80.00% (12/15) | 86.67% (13/15) |
Abbreviations: TNM, tumor-node-metastasis; HMGB1, high mobility group box-1.
Discussion
In this study, we investigated levels of HMGB1 in tissues and sera in breast cancer, and evaluated the diagnostic significance of these levels. We found that HMGB1 levels were significantly higher in breast cancer tissue than in either adjacent normal tissue or benign breast disease tissue. This finding suggests that HMGB1 may have a role in tumorigenesis. Our findings are consistent with previous reports of overexpression of HMGB1 in human breast cancer.26–29 The mechanisms of HMGB1 in the initiation and development of cancer may involve three aspects. First, overexpression of HMGB1 may cause certain gene disorders, leading to certain tumor phenotypes. In addition, HMGB1 may participate in the regulation of apoptosis, and high levels may promote tumor growth through antiapoptotic effects.30 Finally, HMGB1 and heat shock protein 60 may contribute to the growth of mammary carcinoma cells, at least in part through Myd88-dependent signaling, and these damage-associated molecular pattern molecules are expressed in a Myd88-dependent manner.25
Our results imply that tissue HMGB1 levels are closely correlated with the differentiation, lymphatic metastasis, and TNM stage in breast cancer, and HMGB1 might be involved in invasion and metastasis of breast cancer. The data indicate that HMGB1 might be involved in invasion and metastasis of breast cancer and could be a new target for molecular therapy. Dolde et al31 reported that HMGB1 levels were higher in metastatic cell lines (Hs5787 and MDA-MB-231) than in less aggressive MCF-7 cells. Decreasing HMGB1 proteins using an antisense ribozyme approach inhibited transformation in human breast cancer cells. In contrast, increased expression of HMGB1 in normal human breast cells led to transformation. Many studies have explored the possible mechanisms of HMGB1 in tumor metastases. For example, HMGB1 may be involved in tumor metastasis by binding cell surface receptors including RAGE. The RAGE-HMGB1 interaction promotes tumor growth and metastasis by activation of mitogen-activated protein kinases,32 Rac1, nuclear factor kappa B,33 extracellular signal regulated kinase 1/2 (ERK 1/2), and the protein kinase B pathway.34 This in turn results in the expression of matrix metalloproteinases (MMPs), such as MMP-2 and MMP-9, to degrade extracellular matrix proteins and induce tumor invasion and metastasis.35,36 HMGB1 may also induce tumor angiogenesis and metastasis by upregulating the expression of vascular endothelial growth factor-C via activation of the phosphatidylinositol-4 5-bisphosphate 3-kinase/Akt signaling pathway.37 Finally, HMGB1 enhances endothelial growth factor activity by inducing intracellular Ca2+ influx, changing the actin cytoskeleton and enhancing cell motility by activating RK, which leads to tumor invasion and metastasis.38
In the present study, we also found that serum HMGB1 levels were significantly higher in patients with breast cancer than in patients with benign breast disease and normal healthy subjects. Since HMGB1 is translocated to the cytoplasm and secreted by cancer cells, we measured serum HMGB1 levels to evaluate the diagnostic significance of serum HMGB1 in breast cancer. Our findings suggest that serum HMGB1 levels can be used as a novel diagnostic marker. Previous studies have shown that serum HMGB1 levels are elevated in patients with various types of cancer, including gastric, non-small cell lung, and cervical cancers, and hepatocellular carcinoma.39–42 However, in contrast with our finding that tissue HMGB1 levels closely correlated with the differentiation, lymphatic metastasis, and TNM stage of breast cancer, there was no correlation between the corresponding serum HMGB1 levels and these clinicopathological parameters. This may be because very low amounts of HMGB1 are secreted from cancer cells. In addition, the relatively small number of samples is a limitation of our study. Our preliminary results justify a larger-scale study to confirm the diagnostic value of HMGB1.
Conclusion
In summary, we found that HMGB1 protein levels were elevated in human breast cancer tissue over that of normal adjacent tissues or benign breast disease tissue, and were closely related to TNM stage, differentiation, and metastasis. This suggests that HMGB1 has an important role in the initiation, progression, and metastasis of breast cancer. We also found that serum HMGB1 levels were significantly higher in patients with breast cancer than in patients with benign breast disease or normal healthy subjects. This implies that serum HMGB1 levels may be useful as a diagnostic marker. Thus, HMGB1 warrants further study for targeted biological treatment of breast cancer.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Fang Q, Qiong W, Zhang L, et al. Analysis of breast cancer epidemic. Chin J Soc Med. 2012;29:333–335. [Google Scholar]
- 3.Mantovani A. Cancer: Inflaming metastasis. Nature. 2009;457(7225):36–37. doi: 10.1038/457036b. [DOI] [PubMed] [Google Scholar]
- 4.Salto-Tellez M, James JA, Hamilton PW. Molecular pathology – the value of an integrative approach. Mol Oncol. 2014;8(7):1163–1168. doi: 10.1016/j.molonc.2014.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Y, Jiao J, Yang P, Lu H. Mass spectrometry-based N-glycoproteomics for cancer biomarker discovery. Clin Proteomics. 2014;11(1):18. doi: 10.1186/1559-0275-11-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dos Anjos Pultz B, da Luz FA, de Faria PR, Oliveira AP, de Araujo RA, Silva MJ. Far beyond the usual biomarkers in breast cancer: a review. J Cancer. 2014;5(7):559–571. doi: 10.7150/jca.8925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friebel TM, Domchek SM, Rebbeck TR. Modifiers of cancer risk in BRCA1 and BRCA2 mutation carriers: systematic review and meta-analysis. J Natl Cancer Inst. 2014;106(6):dju091. doi: 10.1093/jnci/dju091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gion M, Mione R, Leon AE, et al. Comparison of the diagnostic accuracy of CA27.29 and CA15.3 in primary breast cancer. Clin Chem. 1999;45(5):630–637. [PubMed] [Google Scholar]
- 9.Hou MF, Chen YL, Tseng TF, et al. Evaluation of serum CA27.29, CA15-3 and CEA in patients with breast cancer. Kaohsiung J Med Sci. 1999;15(9):520–528. [PubMed] [Google Scholar]
- 10.Li J, Zhang Z, Rosenzweig J, et al. Proteomics and bioinformatics approaches for identification of serum biomarkers to detect breast cancer. Clin Chem. 2002;48(8):1296–1304. [PubMed] [Google Scholar]
- 11.Tang D, Kang R, Zeh HJ, 3rd, et al. High-mobility group box 1 and cancer. Biochim Biophys Acta. 2010;1799(1–2):131–140. doi: 10.1016/j.bbagrm.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang J, Zhu JS, Zhou Z, et al. Inhibitory effects of ethyl pyruvate administration on human gastric cancer growth via regulation of the HMGB1-RAGE and Akt pathways in vitro and in vivo. Oncol Rep. 2012;27(5):1511–1519. doi: 10.3892/or.2012.1623. [DOI] [PubMed] [Google Scholar]
- 13.Völp K, Brezniceanu ML, Bösser S, et al. Increased expression of high mobility group box 1 (HMGB1) is associated with an elevated level of the antiapoptotic c-IAP2 protein in human colon carcinomas. Gut. 2006;55(2):234–242. doi: 10.1136/gut.2004.062729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pardo M, García A, Thomas B, et al. The characterization of the invasion phenotype of uveal melanoma tumour cells shows the presence of MUC18 and HMG-1 metastasis markers and leads to the identification of DJ-1 as a potential serum biomarker. Int J Cancer. 2006;119(5):1014–1022. doi: 10.1002/ijc.21942. [DOI] [PubMed] [Google Scholar]
- 15.Ishiguro H, Nakaigawa N, Miyoshi Y, et al. Receptor for advanced glycation end products (RAGE) and its ligand, amphoterin are over expressed and associated with prostate cancer development. Prostate. 2005;64(1):92–100. doi: 10.1002/pros.20219. [DOI] [PubMed] [Google Scholar]
- 16.Choi YR, Kim H, Kang HJ, et al. Overexpression of high mobility group box 1 in gastrointestinal stromal tumors with KIT mutation. Cancer Res. 2003;63(9):2188–2193. [PubMed] [Google Scholar]
- 17.Hirata K, Takada M, Suzuki Y, et al. Expression of receptor for advanced glycation end products (RAGE) in human biliary cancer cells. Hepatogastroenterology. 2003;50(53):1205–1207. [PubMed] [Google Scholar]
- 18.Evans A, Lennard TW, Davies BR. High-mobility group protein 1(Y): metastasis-associated or metastasis-inducing? J Surg Oncol. 2004;88(2):86–99. doi: 10.1002/jso.20136. [DOI] [PubMed] [Google Scholar]
- 19.Sparvero LJ, Asafu-Adjei D, Kang R, et al. RAGE (receptor for advanced glycation endproducts), RAGE ligands, and their role in cancer and inflammation. J Transl Med. 2009;7:17. doi: 10.1186/1479-5876-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kostova N, Zlateva S, Ugrinova I, Pasheva E. The expression of HMGB1 protein and its receptor RAGE in human malignant tumors. Mol Cell Biochem. 2010;337(1–2):251–258. doi: 10.1007/s11010-009-0305-0. [DOI] [PubMed] [Google Scholar]
- 21.Yang GL, Zhang LH, Bo JJ, et al. Increased expression of HMGB1 is associated with poor prognosis in human bladder cancer. J Surg Oncol. 2012;106(1):57–61. doi: 10.1002/jso.23040. [DOI] [PubMed] [Google Scholar]
- 22.Kang R, Chen R, Zhang Q, et al. HMGB1 in health and disease. Mol Aspects Med. 2014;40C:1–116. doi: 10.1016/j.mam.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang R, Zhang Q, Zeh HJ, 3rd, Lotze MT, Tang D. HMGB1 in cancer: good, bad, or both? Clin Cancer Res. 2013;19(15):4046–4057. doi: 10.1158/1078-0432.CCR-13-0495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiao Y, Wang HC, Fan SJ. Growth suppression and radiosensitivity increase by HMGB1 in breast cancer. Acta Pharmacol Sin. 2007;28(12):1957–1967. doi: 10.1111/j.1745-7254.2007.00669.x. [DOI] [PubMed] [Google Scholar]
- 25.Chalmers SA, Eidelman AS, Ewer JC, et al. A role for HMGB1, HSP60 and Myd88 in growth of murine mammary carcinoma in vitro. Cell Immunol. 2013;282(2):136–145. doi: 10.1016/j.cellimm.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flohr AM, Rogalla P, Meiboom M, et al. Variation of HMGB1 expression in breast cancer. Anticancer Res. 2001;21(6A):3881–3885. [PubMed] [Google Scholar]
- 27.Stoetzer OJ, Fersching DM, Salat C, et al. Circulating immunogenic cell death biomarkers HMGB1 and RAGE in breast cancer patients during neoadjuvant chemotherapy. Tumour Biol. 2013;34(1):81–90. doi: 10.1007/s13277-012-0513-1. [DOI] [PubMed] [Google Scholar]
- 28.Apetoh L, Tesniere A, Ghiringhelli F, et al. Molecular interactions between dying tumor cells and the innate immune system determine the efficacy of conventional anticancer therapies. Cancer Res. 2008;68(11):4026–4030. doi: 10.1158/0008-5472.CAN-08-0427. [DOI] [PubMed] [Google Scholar]
- 29.Brezniceanu ML, Völp K, Bosse S, et al. HMGB1 inhibits cell death in yeast and mammalian cells and is abundantly expressed in human breast carcinoma. FASEB J. 2003;17(8):1295–1297. doi: 10.1096/fj.02-0621fje. [DOI] [PubMed] [Google Scholar]
- 30.Müller S, Ronfani L, Bianchi ME. Regulated expression and subcellular localization of HMGB1, a chromatin protein with a cytokine function. J Intern Med. 2004;255(3):332–343. doi: 10.1111/j.1365-2796.2003.01296.x. [DOI] [PubMed] [Google Scholar]
- 31.Dolde CE, Mukherjee M, Cho C, et al. HMG-1/Y in human breast cancer cell lines. Breast Cancer Res Treat. 2002;71(3):181–191. doi: 10.1023/a:1014444114804. [DOI] [PubMed] [Google Scholar]
- 32.Ghavami S, Rashedi I, Dattilo BM, et al. S100A8/A9 at low concentration promotes tumor cell growth via RAGE ligation and MAP kinase-dependent pathway. J Leukoc Biol. 2008;83(6):1484–1492. doi: 10.1189/jlb.0607397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palumbo R, Galvez BG, Pusterla T, et al. Cells migrating to sites of tissue damage in response to the danger signal HMGB1 require NF-kappaB activation. J Cell Biol. 2007;179(1):33–40. doi: 10.1083/jcb.200704015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arumugam T, Simeone DM, Schmidt AM, Logsdon CD. S100P stimulates cell proliferation and survival via receptor for activated glycation end products (RAGE) J Biol Chem. 2004;279(7):5059–5065. doi: 10.1074/jbc.M310124200. [DOI] [PubMed] [Google Scholar]
- 35.Ohmori H, Luo Y, Kuniyasu H. Non-histone nuclear factor HMGB1 as a therapeutic target in colorectal cancer. Expert Opin Ther Targets. 2011;15(2):183–193. doi: 10.1517/14728222.2011.546785. [DOI] [PubMed] [Google Scholar]
- 36.Taguchi A, Blood DC, del Toro G, et al. Blockade of RAGE-amphoterin signalling suppresses tumour growth and metastases. Nature. 2000;405(6784):354–360. doi: 10.1038/35012626. [DOI] [PubMed] [Google Scholar]
- 37.Schlueter C, Weber H, Meyer B, et al. Angiogenetic signaling through hypoxia: HMGB1: an angiogenetic switch molecule. Am J Pathol. 2005;166(4):1259–1263. doi: 10.1016/S0002-9440(10)62344-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sparatore B, Patrone M, Passalacqua M, et al. Activation of A431 human carcinoma cell motility by extracellular high-mobility group box 1 protein and epidermal growth factor stimuli. Biochem J. 2005;389(Pt 1):215–221. doi: 10.1042/BJ20050053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chung HW, Lee SG, Kim H, et al. Serum high mobility group box-1 (HMGB1) is closely associated with the clinical and pathologic features of gastric cancer. J Transl Med. 2009;7:38. doi: 10.1186/1479-5876-7-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng BQ, Jia CQ, Liu CT, et al. Serum high mobility group box chromosomal protein 1 is associated with clinicopathologic features in patients with hepatocellular carcinoma. Dig Liver Dis. 2008;40(6):446–452. doi: 10.1016/j.dld.2007.11.024. [DOI] [PubMed] [Google Scholar]
- 41.Shang GH, Jia CQ, Tian H, et al. Serum high mobility group box protein 1 as a clinical marker for non-small cell lung cancer. Respir Med. 2009;103(12):1949–1953. doi: 10.1016/j.rmed.2009.05.019. [DOI] [PubMed] [Google Scholar]
- 42.Sheng X, Du X, Zhang X, et al. Clinical value of serum HMGB1 levels in early detection of recurrent squamous cell carcinoma of uterine cervix: comparison with serum SCCA, CYFRA21-1, and CEA levels. Croat Med J. 2009;50(5):455–464. doi: 10.3325/cmj.2009.50.455. [DOI] [PMC free article] [PubMed] [Google Scholar]


