SUMMARY
Due to the importance of human immunodeficiency virus type 1 (HIV-1) integrase as a drug target, the biochemistry and structural aspects of retroviral DNA integration have been the focus of intensive research during the past three decades. The retroviral integrase enzyme acts on the linear double-stranded viral DNA product of reverse transcription. Integrase cleaves specific phosphodiester bonds near the viral DNA ends during the 3′ processing reaction. The enzyme then uses the resulting viral DNA 3′-OH groups during strand transfer to cut chromosomal target DNA, which simultaneously joins both viral DNA ends to target DNA 5′-phosphates. Both reactions proceed via direct transesterification of scissile phosphodiester bonds by attacking nucleophiles: a water molecule for 3′ processing, and the viral DNA 3′-OH for strand transfer. X-ray crystal structures of prototype foamy virus integrase-DNA complexes revealed the architectures of the key nucleoprotein complexes that form sequentially during the integration process and explained the roles of active site metal ions in catalysis. X-ray crystallography furthermore elucidated the mechanism of action of HIV-1 integrase strand transfer inhibitors, which are currently used to treat AIDS patients, and provided valuable insights into the mechanisms of viral drug resistance.
INTRODUCTION
Retroviruses are the only animal viruses that require the stable integration of genetic information into the genome of the host cell as an obligate step in replication. All members of the virus family Retroviridae accordingly carry with them integrase, which is a specialized DNA recombination enzyme. Integration is required for efficient expression of retroviral genes by the host transcriptional machinery and hence productive virus replication. The integrase encoded by human immunodeficiency virus type 1 (HIV-1) is thus an important antiviral target in the fight against HIV/AIDS (1). Integration additionally ensures replication and segregation of viral genes to daughter cells during cell division. Stable association of HIV-1 with cellular DNA underlies the notorious incurable nature of AIDS despite highly active antiretroviral therapy (HAART) (2).
Retroviridae is composed of two virus subfamilies, Orthoretrovirinae and Spumaretrovirinae. While six viral genera, including alpha through epsilon and lenti, belong to Orthoretrovirinae, the spumaviruses solely comprise Spumaretrovirinae. Spumavirus biology accordingly differs somewhat from the other retroviruses. For brevity, this chapter describes generalities that apply to most, if not all, retroviruses; specific viruses or genera are discussed as applicable. Viruses pathogenic to humans include the lentiviruses HIV-1 and HIV-2 and the deltaretroviruses human T-lymphotropic virus type 1 (HTLV-1) and HTLV-2.
Retroviruses carry two copies of plus-sense genomic RNA. Integrase is encoded at the 3′ end of the pol gene (3-5), which also encodes for the RNA-dependent DNA polymerase reverse transcriptase (RT) enzyme that converts the genomic RNA into linear, double-stranded DNA. Integrase and RT are translated as part of a Gag-Pol precursor polyprotein in virus producer cells, which is cleaved by the viral protease as particles bud from the plasma membrane and mature into infectious virions. Integrase and RT, together with viral RNA in association with the viral nucleocapsid protein, are situated within the viral capsid core during particle maturation.
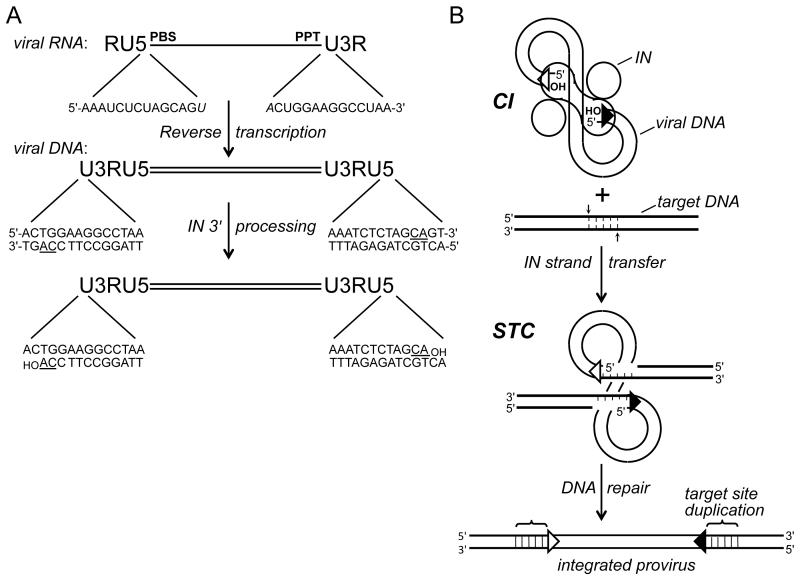
Viral DNA synthesis occurs within the context of the reverse transcription complex, which is a large subviral complex derived from the virus core (6, 7). Retroviral RNAs harbor a terminal repeat (R) element that adjoins unique sequences at the 5′ and 3′ ends (U5 and U3, respectively). U3 and U5 become duplicated during reverse transcription such that the DNA contains a copy of 5′-U3RU5-3′, dubbed the long terminal repeat (LTR), at both ends. Integrase engages approximately 16-20 bp of the LTR termini to integrate the viral DNA substrate into host DNA (8-12) (Fig. 1A).
Figure 1.
Integration substrate and integrase activities. (A) Reverse transcription yields linear, double-stranded DNA with U3RU5 long terminal repeats. The 14 terminal bp of HIV-1 DNA are compared with their originating positions in viral RNA (italicized bases form part of the primer binding site (PBS) and polypurine tract (PPT) that are important for DNA synthesis). The positions of the invariant CA dinucleotides (underlined in the DNA) relative to the 3′ ends of the PBS and PPT determines whether 3′ processing is required to yield a CAOH 3′ terminus; 3′ processing by HIV-1 integrase liberates a pGpTOH dinucleotide from each viral DNA end. (B) Two monomers (oblong shape) of an HIV-1 integrase tetramer within the cleaved intasome (CI) use the viral DNA 3′-hydroxyl groups to cut the target DNA (thick bold lines) with a 5 bp stagger, which concomitantly joins the LTR ends to target DNA 5′ phosphates. Repair of the strand transfer complex (STC) yields a 5 bp duplication of target DNA flanking the integrated HIV-1 provirus. Open and filled triangles, U3 and U5 termini. Integrase was omitted from the drawing of the STC for simplicity. IN, integrase.
INTEGRASE ACTIVITIES
Two integrase activities, 3′ processing and strand transfer, are required for productive virus replication. The associated DNA cutting and joining steps were initially deciphered using extracts of acutely infected cells. All retroviruses harbor an invariant CA dinucleotide in the immediate proximity to each 3′ end of the unintegrated viral DNA (Fig. 1A, underline), and downstream sequences (up to 3 nucleotides depending on the viral species) are removed during 3′ processing (13-17). A key breakthrough came with the demonstration that the DNA-containing nucleoprotein complex, termed the preintegration complex (PIC), can integrate the endogenous reverse transcript into heterologous target DNA in vitro (18). The processed viral DNA CA-3′ ends are joined to target DNA during strand transfer, while the unprocessed viral DNA 5′ ends remain unjoined (14, 15, 17) (Fig. 1B). Cellular enzymes remove the two unpaired nucleotides from the viral DNA 5′ ends and repair the single-stranded gaps in the recombination intermediate, which yields a short (4-6 bp, depending on the virus) duplication of target DNA flanking the integrated provirus (Fig. 1B). PIC activity requires divalent metal ions such as Mn2+ or Mg2+, but does not require a high-energy cofactor (14, 18-21).
Purified recombinant integrase proteins display 3′ processing and strand transfer activities in vitro (22-30). Simplified assay systems utilized relatively short (~17-22 bp) double-stranded oligonucleotides that model the ends of the LTRs for 3′ processing reaction substrates (22, 24) and also for acceptor target DNA during strand transfer (23, 26, 27, 29, 30). Because pre-processed substrates that lacked sequences normally removed during 3′ processing supported strand transfer activity, the integration of retroviral DNA ends is mechanistically separable from dinucleotide cleavage (23, 26, 27, 29, 30). Concordantly, 3′ processing of HIV-1 DNA ends can occur soon after the LTRs are synthesized (31, 32), which precedes integration into chromosomal DNA minimally by several hours (33, 34).
Purified integrase proteins display two additional endonucleolytic activities in vitro, disintegration (35) and alcoholysis (36, 37). Though it seems unlikely that either of these activities occurs in the context of virus infection, their study has nevertheless yielded valuable information on integrase domain organization and reaction mechanism.
3′ Processing and Strand Transfer Reaction Mechanisms
Oligonucleotide-based biochemical assays enabled relatively rapid assessment of DNA substrate and integrase protein requirements for 3′ processing and strand transfer activities, as well as the mechanism of DNA recombination. Because integrase activity does not require a high-energy cofactor, the energy to drive the formation of the new viral-target DNA bond during strand transfer must come from the pre-existing target DNA phosphodiester bond. Two different mechanisms can be considered: the viral DNA could attack the target DNA directly, which through isoenergetic transesterification would concomitantly drive the formation of the viral-target DNA phosphodiester bond, or the bond energy could be temporarily stored in the form of an integrase-DNA covalent intermediate prior to viral-target DNA bond formation (38). Essentially all enzyme-mediated phosphoryl transfer reactions proceed by bimolecular nucleophilic substitution (SN2) displacement (39); as the hallmark of SN2 chemistry is inversion of chirality, monitoring the stereochemical course of the strand transfer reaction afforded a means by which to address its mechanism. The phosphodiester group can be made chiral by substituting one of its non-bridging oxygen atoms (Pro-Sp or Pro-Rp) with sulfur. Finding the retention of phosphorothioate chirality in strand transfer reaction products would be consistent with a protein-DNA covalent intermediate reaction mechanism, as formation of the integrase-DNA covalent bond would invert chirality, and its subsequent resolution would invert chirality a second time to overall retention (40). Because phosphorothioate chirality was inverted in HIV-1 DNA reaction products, strand transfer proceeds via an odd number of transesterification reactions (41). Though convoluted models that consider 3 or 5 separate chemical reactions could be entertained, it was evident that strand transfer proceeds via a single transesterification reaction (41).
HIV-1 integrase uses a water molecule under Mg2+-dependent conditions to hydrolyze CA\GT (backslash denotes scissile phosphodiester bond throughout the chapter) during 3′ processing (42). Reaction specificity is loosened somewhat in the presence of Mn2+, such that two or three carbon-containing diols, or the 3′ end of the viral DNA itself, can additionally serve as the nucleophile (36, 41). Determining phosphorothioate chirality in the cyclic dinucleotide reaction product formed by DNA end-mediated processing provided a means to monitor the stereochemical course of the reaction. Because chirality was inverted, the HIV-1 integrase 3′ processing reaction, like strand transfer, proceeds via a single SN2 transesterification (41).
Similarities with Other DNA Recombination Systems
Retroviral and LTR retrotransposon integrase proteins are evolutionarily related to a large variety of metal ion-dependent polynucleotidyl transferase enzymes that include bacterial transposases, RuvC resolvase, RNase H (43), the Argonaute component of RISC (RNA-induced silencing complex) (44), the RAG1 component of the RAG1/2 recombinase that catalyzes V(D)J recombination (45, 46), and RT (47). The key similarities include the enzyme active sites and reaction mechanisms.
The bacteriophage MuA transposase protein catalyzes DNA cutting and joining reactions that are analogous to retroviral integrase 3′ processing and strand transfer activities (38). Monitoring the stereochemical course of the phage Mu strand transfer reaction revealed inversion of phosphorothioate chirality (40). The utilization of H2 18O afforded the monitoring of the stereochemical course of Mu DNA end hydrolysis. Similar to the results obtained for DNA-mediated 3′ processing of HIV-1 DNA, MuA transposase-mediated hydrolysis yielded inversion of phosphorothioate chirality (48).
The RAG1/2 recombinase yields a DNA hairpin product during the first step of V(D)J recombination (see the chapter by David Roth in this monograph). Monitoring the stereochemical course of the reaction revealed the inversion of phosphorothioate chirality, which is consistent with a single step transesterification (49). The phosphorothioate stereoselectivity of the various reactions catalyzed by the Tn10 transposase protein were moreover similar to those catalyzed by MuA transposase, HIV-1 integrase, and RAG1/2 recombinase (50, 51). Thus, similar mechanisms underlie the DNA cutting and joining reactions that are catalyzed by elements as seemingly disparate as HIV-1 integrase, MuA transposase, and RAG1/2 recombinase.
Challenges of Working with Recombinant Integrase Proteins
Integration proceeds through the pairwise or concerted integration of both ends of linear viral DNA into chromosomal DNA (Fig. 1B). However, purified integrase proteins vary greatly in their ability to catalyze the concerted integration of substrate DNA in vitro. Heterologous circular target DNA is used to distinguish strand transfer reaction products that result from the integration of single ends of oligonucleotide duplexes from those that result from the concerted integration of two independent duplex oligonucleotide ends: single end integration yields a nicked circular DNA product, while concerted integration yields a linearized product after deproteinization (23). Many integrase proteins yield mixtures of the two types of reaction products (23, 52-55) whereas others, such as those derived from the spumavirus prototype foamy virus (PFV) (56) and lentivirus equine infectious leukemia virus (54), predominantly display concerted integration activity. HIV-1 integrase by contrast predominantly catalyzes the integration of single oligonucleotide DNA ends in vitro (26). The reason behind the relatively poor behavior of HIV-1 integrase in concerted integration reactions remains unclear. Modifications that included relatively long viral DNA substrates (57-59) and/or the addition of viral nucleocapsid (60) or host lens epithelium-derived growth factor (LEDGF)/p75 protein (61) have enhanced concerted integration activity. Purification under conditions that disfavor protein aggregation has also been reported to enhance the ability of the HIV-1 enzyme to integrate oligonucleotide substrate DNA in concerted fashion (62).
Retroviral integrase proteins display a range of solubility properties. Epsilonretrovirus integrase was reportedly insoluble following its expression in Escherichia coli under a variety of conditions (55). HIV-1 integrase purified following its expression in bacteria can attain concentrations of 1 mg/ml or greater, however this strictly depends on non-physiological concentrations of salt (e.g., 1 M NaCl). PFV integrase is by contrast highly soluble, with concentrations in excess of 10 mg/ml achieved in buffer containing 200 mM NaCl (56, 63, 64). The favorable solubility of PFV integrase is at least partially responsible for its utility as a crystallography substrate with DNA (see below).
Various modifications, most notably the use of solubilizing mutations, can improve the solubility of HIV-1 integrase (42, 65, 66). The lentiviral integration cofactor LEDGF/p75 possesses favorable solubility, and binding to HIV-1 integrase yields a protein complex that displays generally favorable solubility properties (67, 68). Such observations have sparked interest in the structural biology of lentiviral integrase-LEDGF/p75 complexes (61, 68-70).
INTEGRASE PROTEIN STRUCTURES
Domain Organization of Integrase Proteins
Retroviral integrase proteins harbor three common domains, the N-terminal domain (NTD), catalytic core domain (CCD), and C-terminal domain (CTD) (71-76) (Fig. 2A). All evidence suggests that the three-dimensional structures of the individual domains are preserved across the different integrase proteins (77). At the level of amino acid sequence, the CCD and CTD display the greatest and least extents of conservation, respectively.
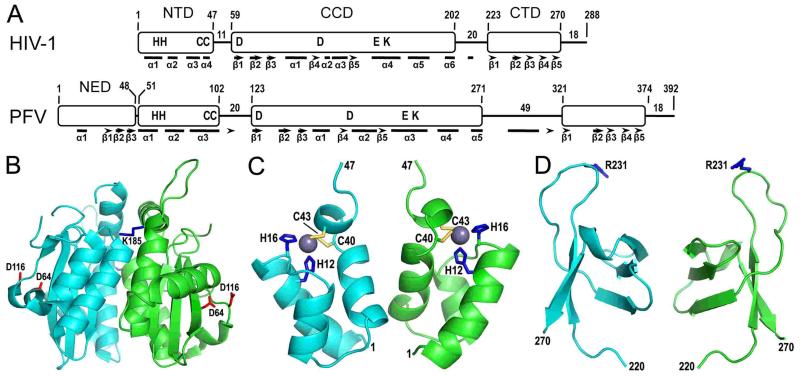
Figure 2.
Retroviral integrase domain organization and HIV-1 integrase domain structures. (A) The N-terminal domain (NTD), catalytic core domain (CCD), and C-terminal domain (CTD) are common among all retroviral integrase proteins, whereas sequence analysis indicates that gammaretrovirus and epsilonretrovirus in addition to spumavirus proteins harbor an N-terminal extension domain (NED) (55, 94). HIV-1 and PFV integrases were aligned by NTD N-termini, with positions of domain boundaries and lengths of interdomain linker and C-terminal tail regions indicated. Residues conserved across all retroviral integrase proteins are shown in single letter code. Bars and arrows indicate alpha helix and beta strand secondary elements as determined by X-ray crystallography for PFV integrase (94) and by a combination of X-ray crystallography (66, 149) and molecular modeling (105) for HIV-1 integrase. (B) The X-ray crystal structure of the HIV-1 integrase CCD [protein database (pdb) code 1ITG] (43) highlights in red sticks the aspartate residues of the DDE catalytic triad (the glutamic acid was not visualized in this structure) and in blue sticks the Lys185 substitution that enhanced protein solubility and enabled protein crystallization (the Lys residue at the rear face of the dimer is barely visible in this projection). (C) The NMR structure of the integrase NTD (pdb code 1WJC) highlights the His (blue sticks) and Cys (yellow sticks) residues that coordinate a single zinc atom (grey sphere) (99). (D) The structure of the HIV-1 integrase CTD as determined by NMR (pdb code 1IHV) (142) highlights Arg231, which has been implicated in target DNA binding (108, 166).
The CCD harbors the enzyme active site, at the heart of which are the invariant amino acid residues of the DDE catalytic triad (73, 78-80) that coordinate a pair of magnesium ions during catalysis (see below). Initially recognized as a DX39-58DX35E motif conserved among retroviral and retrotransposon integrases and bacterial IS3 insertion sequences (74, 78, 81), the advent of genomic sequencing has since expanded the DD(E/D) superfamily of polynucleotidyl transferases to include the transposase proteins of numerous prokaryotic and eukaryotic transposable elements (47, 82-84). To date, the roles of the active site residues during integrative recombination are most thoroughly understood from X-ray crystallographic analyses of active PFV integrase-DNA complexes (85).
The CCD harbors additional conserved residues that contact viral DNA (86-94) and target DNA (90, 95-98) during integration. The lysine residue that engages the phosphate backbone of the invariant CA dinucleotide in viral DNA (86, 94) is conserved across retroviral integrase proteins and some bacterial insertion elements (Fig. 2A), although not among the retrotransposon integrase proteins despite the conservation of the terminal CA sequence (78, 86).
Retroviral and retrotransposon integrase NTDs harbor a conserved HHCC motif that coordinates the binding of a single zinc atom (72, 74, 99, 100). Metal binding stabilizes the native fold of the NTD and concordantly stimulates HIV-1 integrase catalytic activity (61, 101, 102). The NTD is involved in integrase multimerization and interacts with viral DNA during integration (94, 102-105).
Although the extent of amino acid sequence conservation among integrase CTDs is less prevalent than for the other common domains, certain conserved amino acid motifs are recognizable across subsets of retroviral integrase proteins (106). The CTD binds DNA in a sequence non-specific fashion (75, 107-109) and contributes to the functional multimerization of full-length integrase proteins (42, 108, 110).
The NTD and CTD play important roles in integrase 3′ processing and DNA strand transfer activities in vitro (74, 76, 111-116). The isolated CCD can catalyze disintegration activity (74, 113, 114), which was consistent with the notion that it housed the enzyme active site (73, 74). The integrase proteins from three of the viral genera, including the gammaretroviruses, epsilonretroviruses, and spumaviruses, carry a fourth domain termed the N-terminal extension domain (NED) based on sequence and X-ray crystallographic analyses (55, 94) (Fig. 2A).
Integrase Functions as a Multimer
Kinetic measures of integrase activities (117, 118) and chemical cross-linking (58, 111, 119-121) provided initial evidence that integrase functions as a multimer. The discovery that mixtures of defective integrase deletion mutant proteins restored as much as 50% of wild type activity provided a means to probe the functional organization of the different integrase domains within the multimer (111, 115, 116, 122, 123). For example, the function of the NTD was required in trans to the catalytically active protomer (111, 116, 122).
Mutagenesis experiments revealed that the invariant CA/TG bp is the most critical sequence for oligonucleotide-based integrase activity in vitro (29, 124-126). Titers of viruses that contained one mutated LTR end were modestly reduced, whereas double end-mutant viruses were essentially dead (11, 12, 127). Sequence analysis revealed large insertions or deletions of cellular DNA associated with the integration of mutant LTR ends (128, 129). Though virus replication normally proceeds through the pairwise insertion of both LTRs (Fig. 1B), integrase-mediated insertion of one end can apparently template the integration of the second, mutated end via cell-mediated DNA recombination.
Retroviral integration and DNA transposition proceed through a series of stable nucleoprotein complexes (85) (for reviews, see 130 and 131). In the case of integration, the two ends of viral DNA are bridged together by a tetramer of integrase in a complex that is called the intasome or stable synaptic complex (SSC) (31, 58, 94, 120, 121, 132-135). Processing of the viral DNA ends converts the SSC to the cleaved intasome (CI) (85) (Fig. 1B), which is referred to as the cleaved donor complex or type 1 transpososome in the transposition literature (130). The CI subsequently morphs into the target capture complex (TCC) upon target DNA binding. Covalent joining of the viral DNA ends to target DNA yields the strand transfer complex (STC) (85, 98).
Whereas an integrase tetramer catalyzes strand transfer activity (58, 94, 118, 120), the form of the enzyme that catalyzes 3′ processing activity during virus infection has been debated. Although an integrase dimer appears to suffice to process single LTR ends in vitro (58, 136), a variety of information needs to be taken into account when considering whether 3′ processing is mediated by an integrase dimer bound to a single viral DNA end or by the integrase tetramer during infection. DNA end processing prior to SSC formation could account for the different apparent rates of U5 versus U3 cleavage during HIV-1 infection (31) and is consistent with integrase-mediated integration of the sole wild type ends of single LTR end mutant viruses (128, 129). However, 3′ processing and strand transfer of single viral DNA ends can occur in the context of the SSC in vitro (120). Crystallized PFV intasomes are functional (see below), and the integrase tetramer catalyzed 3′ processing activity in crystallo (85). The PFV integrase NTD moreover functioned in trans with the CCD during 3′ processing (85). Therefore, if an integrase dimer were active for 3′ processing, it would need to assume a cis-trans conformation that is different from the active form observed in the crystals. While not impossible, we favor the straightforward interpretation that 3′ processing activity is catalyzed by the integrase tetramer in the context of the SSC during viral infection. Of note, results of small angle X-ray scattering (SAXS) have yielded novel “reaching dimer” solution conformations for full-length HIV-1 and ASLV integrase in the absence of DNA (137, 138). Although extensive CTD-CTD interactions preclude these structures from functionally engaging DNA, they nevertheless could represent intermediates on the pathway to SSC formation (see the chapter by Anna Marie Skalka).
Integrase Domain Structures
A high-resolution structure of a full-length retroviral integrase protein in the absence of DNA has yet to be reported, which is likely due to the inherent flexibility of integrase interdomain linkers (Fig. 2A) (64, 104). Over the years, structures of isolated integrase domains and 2-domain constructs have been elucidated. Because the integrase CCD could catalyze disintegration activity (74, 113, 114), it was earmarked early on as a target of interest. The HIV-1 integrase CCD was however insoluble following its expression in E. coli (74, 139). An important advance came from the identification of amino acid substitutions, including F185K, which significantly increased the solubility of HIV-1 integrase CCD protein (65) and afforded its crystallization and structural determination by X-ray diffraction (43). The CCD harbors an RNase H fold, which situates the active site residues of the DDE catalytic triad in close proximity to one another (Fig. 2B). Retroviral integrase CCDs in the vast majority of cases crystallize as a homodimer with a large dimeric interface (43, 140) (reviewed in 77), and the Lys185 side-chain composed part of the HIV-1 integrase CCD/F185K interface (Fig. 2B). The ~35 Å distance between the active sites within the homodimer was incompatible with pairwise insertion of two ends of HIV-1 DNA across a major groove (43), which is ~17 Å in canonical B-form DNA. More recent studies of PFV intasomes have helped to clarify the role of the CCD dimer during integration (see below).
The F185K change also enhanced the solubility of full-length HIV-1 integrase, and the mutant enzyme retained integrase 3′ processing and strand transfer activities in vitro (42). The mutation however rendered HIV-1 replication-defective, which was attributed to pleiotropic defects at the steps of virus particle assembly and reverse transcription (42). The CCD from the alpharetrovirus avian-sarcoma leukosis virus (ASLV) integrase was sufficiently soluble to permit its crystallization in the absence of solubility-enhancing mutations (140).
The NTD adopts a helix-turn-helix fold around a single zinc atom, which is chelated by the side chains of the conserved residues of the HHCC motif (99, 100, 141) (Fig. 2C). The CTD folds into a 5-stranded beta barrel with homology to Src homology 3 (SH3) domains (142-144) (Fig. 2D). Though SH3 domains in general interact with Pro-rich regions of proteins (145), some, such as Sso7d from Sulfolobus solfatarius, also mediate DNA binding (146). Each integrase domain notably engages DNA in the context of the active PFV intasome (94).
Integrase Two-Domain Structures
Structures of 2-domain integrase constructs provided initial insight into the organization of the different protein domains during integration. The HIV-1 integrase CCD-CTD fragment containing five solubilizing mutations (including F185K) crystallized as a homodimer, with the CTDs positioned at the ends of alpha helical extensions that emanated from the common CCD dimer (66) (Fig. 3A). The structures of two other integrase CCD-CTD proteins that were solved at around the same time, including those from simian immunodeficiency virus (147) and ASLV (148), revealed different positions of these CTDs relative to the integrase CCD dimer (reviewed in 77). Protein contacts made during crystallization therefore likely influenced the positions of the CTDs relative to the CCD dimer in each of the 2-domain CCD-CTD structures (77). Consistent with this interpretation, the CCD-CTD linker region of PFV integrase adopts an extended conformation that is largely devoid of secondary structure in the presence of viral DNA (see below).
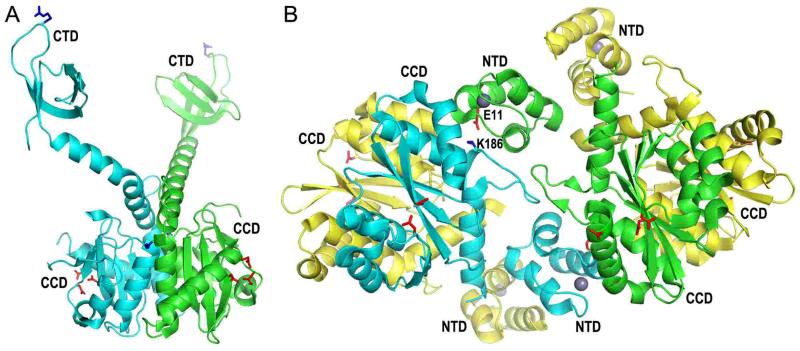
Figure 3.
Structures of 2-domain HIV-1 integrase constructs. (A) The X-ray crystal structure of the HIV-1 integrase CCD-CTD dimer (pdb code 1EX4) (66), highlighting the CCD and CTD side-chains that were shown in Fig. 2. (B) The crystal structure of the NTD-CCD asymmetric unit (pdb code 1K6Y) (149) highlights NTD residue Glu11 and CCD residue Lys186 of the green and cyan molecules, respectively, which play important roles in integrase concerted integration activity and HIV-1 infection (70). The other pair of interacting residues (Glu11 from the cyan NTD and Lys186 from the green CCD) is not visible in this projection. The side chains of the DDE catalytic triad (red sticks) and NTD-coordinated zinc atoms (grey spheres) are also shown.
Perhaps the most interesting of the partial integrase structures was that of the HIV-1 NTD-CCD construct, where the asymmetric unit harbored four protein molecules (Fig. 3B) (149). Although each CCD participated in canonical dimer interface formation (yellow-cyan and green-yellow dimers in the figure), a novel interface was observed between two “inner” molecules of the tetramer (Fig. 3B, green and cyan). The NTDs of the cyan and green integrase protomers were moreover seemingly positioned to work in trans with the apposing CCD, though incomplete electron density maps precluded assignments of interdomain NTD-CCD connectors in this structure. The active sites of the two inner monomers additionally seemed too far apart to catalyze pairwise insertion of HIV-1 DNA ends (149).
LEDGF/p75-Integrase Structures
LEDGF/p75 is a lentiviral-specific integrase-binding protein that helps to guide integration to active genes (reviewed in 150 and 151; also see the chapter by Craigie and Bushman in this monograph). LEDGF/p75 harbors a conserved domain, called the integrase-binding domain (IBD), which is necessary and sufficient for binding to integrase (152). On the integrase side, the CCD is minimally required for LEDGF/p75 binding, with the NTD contributing to high affinity binding (61, 153).
The LEDGF/p75 IBD is a compact alpha helical domain, with the hairpins from two helix-hairpin-helix folds situated on one end of the elongated structure (154). A crystal structure of the HIV-1 integrase CCD-IBD complex revealed that the host factor predominantly utilizes hairpin residues, most notably Ile365 and Asp366 at the tip of the first hairpin, to nestle into the CCD-CCD dimer interface (69) (Fig. 4A). A series of quinoline-based antiviral compounds has been developed that mimics the amino acid contacts of the IBD-CCD interaction, and the small molecules accordingly compete for the binding of LEDGF/p75 to HIV-1 integrase in vitro (155) (reviewed in 156). It is however currently unclear to what extent this inhibition contributes to the antiviral activities of the compounds (see Craigie and Bushman chapter).
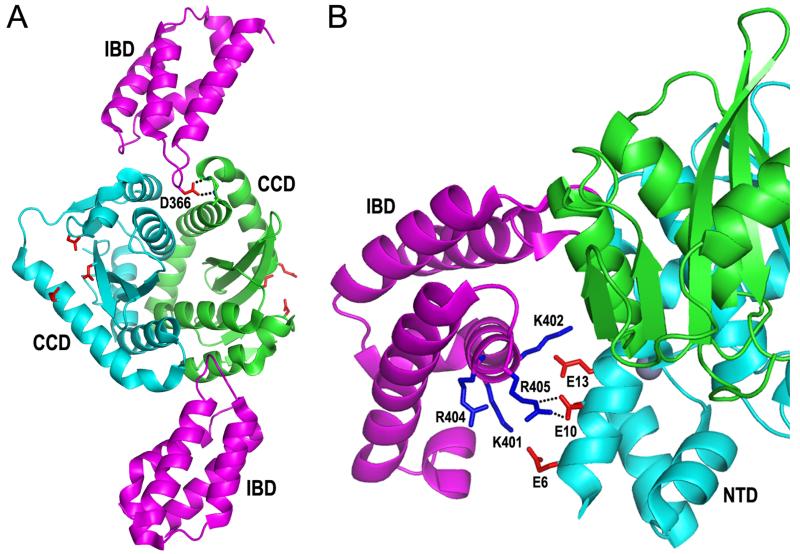
Figure 4.
Lentiviral integrase-LEDGF/p75 IBD structures. (A) X-ray crystal structure of the IBD (magenta cartoon)-HIV-1 integrase CCD/F185K (cyan/green dimer) complex (pdb code 2B4J) (69). Highlighted is the Asp366 side-chain from the upper IBD molecule (red stick) hydrogen bonding (dashed lines) to backbone amides of integrase residues within the linker between CCD α helices 4 and 5. The extent to which integrase CCD α4/5 connector regions contribute to forming analogous CCD-CCD dimer interface pockets at least in part accounts for the lentiviral specificity of the LEDGF/p75-integrase interaction (54, 69). The Asp and Glu side chains of the catalytic DDE triad are also painted red. (B) The crystal structure of the HIV-2 integrase NTD-CCD–IBD complex highlights the electrostatic interaction between the integrase NTD and the second helix-hairpin-helix unit of the IBD (pdb code 3F9K, chains A, B, and C) (61). Salt bridges between IBD residue Arg405 and integrase residue Glu10 are indicated by dashed lines; IBD residue Asp366 is behind the green CCD, hidden from view.
Structures of lentiviral integrase NTD-CCD 2-domain constructs with the LEDGF/p75 IBD elucidated the structural basis of the NTD-IBD interaction (61) and clarified the organization of the NTD and CCD within the active tetramer (70). Owing to the favorable solubility of LEDGF/p75 protein, crystallization in these cases proceeded in the absence of solubility enhancing mutations. Electronegative side chains on one face of the HIV-2 integrase NTD alpha 1 helix apposed electropositive side chains from the second helix-hairpin-helix repeat of the IBD (Fig. 4B). The electrostatic interaction is mutagenetically reversible, as HIV-1 integrase containing Lys residues in place of Glu10 and Glu13 was active only in the presence of reverse-charge LEDGF/p75 mutant protein (61). Such mutant co-dependent function can in theory guide the customized integration of defective integrase mutant viruses in cells that express complementary mutant LEDGF/p75 protein (61, 157).
Similar to the HIV-1 integrase 2-domain construct (149), the maedi-visna virus integrase NTD-CCD fragment crystallized as a dimer of dimers in the presence of the LEDGF/p75 IBD (70). Because an NTD-CCD interdomain linker was resolved in one of two crystal forms, all four NTD-CCD linkers could be unambiguously assigned in this structure. Within the inner dimer, CCD residue Lys188 formed an intermolecular salt bridge with NTD residue Glu11. Mutating the analogous HIV-1 integrase residues (Lys186 and Glu11; Fig. 3B) revealed that the salt bridge played an important role in integrase concerted integration activity and the establishment of HIV-1 infection (70). Thus, although there was limited information on the positioning of the CTDs or viral DNA strands within the intasome, these studies established the basic geometry of the CCD and NTD within a functional integrase tetramer.
FOAMY VIRUS INTASOME STRUCTURES
A key breakthrough in the field of retroviral integrase structural biology came with the crystallization and structural determination of functional PFV integrase-DNA complexes. As alluded to above, PFV integrase is well behaved in solution, and the intasome complex moreover retained its structural and functional integrity upon challenge with relatively high concentrations of salt (94).
Dimer-of-Dimers Architecture
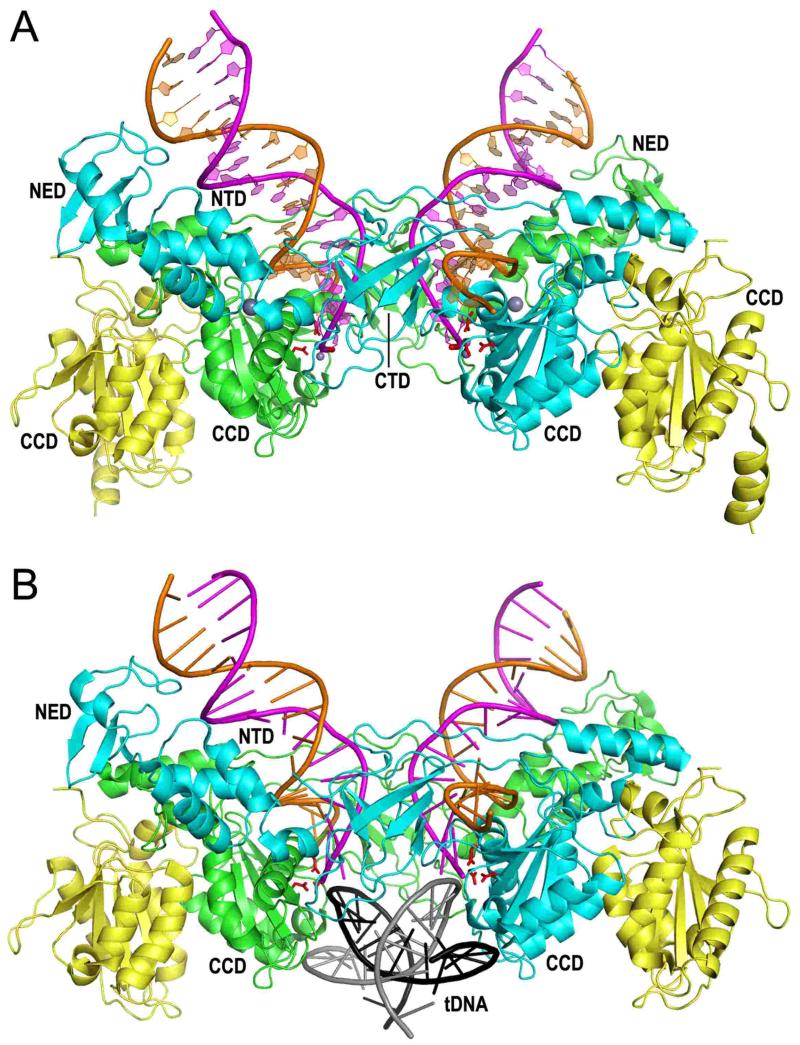
As hinted at from prior NTD-CCD structures that were solved in the absence of DNA (70, 149), the PFV intasome contains a tetramer of integrase with the dimer-of-dimers architecture (Fig. 5A) (94). Each dimer harbors the canonical CCD dimer interface (green-yellow and cyan-yellow dimers in the figure). The inner integrase protomers (green and cyan) within the tetramer make all the contacts with the viral DNA ends and contribute the active sites that process and integrate the viral DNA (85, 94, 98). The NED and NTD of each inner monomer span the structure to interact with the apposing DNA duplex, and, in the case of the NTD, the apposing CCD (Fig. 5A). The CTD from each inner monomer positions in between each NTD and CCD, poised to interact with target DNA (94) (Fig. 5).
Figure 5.
PFV intasome structures. (A) Structure containing 19 bp pre-cleaved U5 DNA end (94, 207) (pdb code 3OY9). The inner integrase monomers of the tetramer, which contact the viral DNA, are painted cyan and green; the outer integrase molecules are yellow. The transferred DNA strands with CAOH 3′ ends are painted magenta whereas the non-transferred strands are orange. The large grey spheres are NTD-coordinated zinc; small grey spheres are Mn atoms coordinated by the DDE active site residues (red sticks) and viral DNA end. (B) Structure of the TCC (98) (pdb code 3OS1). Although a 30 bp target DNA (tDNA) was utilized during crystallization, only 18 bp (grey plus strand sequence -7GCACGTG\CTAGCACGTGC10) was resolved in the electron density maps.
TCC and STC Structures
Sequence analysis of retroviral integration sites revealed weakly conserved palindromes that center on the staggered cut in target DNA, indicating that an integrase multimer with dyad symmetry prefers particular nucleotides at the sites of viral DNA joining (158-162). PFV preferentially integrates at sites that on average harbor the sequence -3KWK\VYRBMWM6 (written using International Union of Biochemistry base codes; the backslash indicates the position of U5 DNA joining, which occurs at the −1\0 position; the target DNA sequence that is duplicated after integration is underline) (56). PFV integrase catalyzed the integration of pre-cleaved U5 DNA into a 30 bp symmetric target DNA duplex during intasome crystallogenesis, which allowed the structural determination of the STC (98). TCC structures were determined by omitting divalent metal ion, or by using viral DNA that terminated in dideoxy adenylate and therefore lacked reactive CAOH−3′ ends (98) (Fig. 5B).
The overall geometry of the PFV integrase-viral DNA complex does not change during target DNA binding and integration (98) (Fig. 5). Approximately 26.5 Å separates the two active sites of the inner monomers; the intasome accordingly accommodates target DNA in a severely bent conformation to enable integration across the expanded major groove. The deformation of the target DNA duplex is localized at the central base pair step, which incurs a negative roll of approximately 55° (98) (Fig. 5B). The severe kink impressively occurs in the absence of direct protein-DNA stacking interactions.
As anticipated from the relatively weak nature of target DNA palindrome sequence conservation, the majority of PFV integrase-target DNA interactions are mediated through the phosphodiester backbone (98). Two amino acids, Ala188 in the integrase CCD and Arg329 in the CTD, by contrast make base-specific contacts: Ala188 interacts with cytosine at position 6 through van der Waals interaction whereas Arg329 hydrogen bonds with guanosine 3, guanosine −1, and thymine −2 (see Fig. 5B for target DNA sequence). Thus, within the tetramer, Ala188 and Arg329 interact with all −3KWK\VYRBMWM6 consensus bases aside from the central dinucleotide (YR). The sequencing of in vitro concerted integration products accordingly revealed that R329S, R329E, and A188S mutant integrases displayed novel target DNA nucleotide preferences (98). The Arg329 mutants additionally preferred sites that harbored relatively flexible dinucleotides at the center.
Depending on their composition, DNA dinucleotides differ in their base-stacking propensity, and hence vary in their ability to accommodate a distortion of a DNA duplex. Pyrimidine-purine (YR) and RY are the most and least flexible, respectively, with intermediate flexibilities for YY and RR dinucleotides (163). Thus, the consensus target DNA palindrome for PFV integrase underscores features of DNA bendability, including bases that register with interacting amino acids Arg329 and Ala188 surrounding a central, flexible YR step (98). Bendability may moreover underlie a general property of retroviral integration and DNA transposition sites (164, 165). The consensus HIV-1 integrase target DNA sequence −3TDG\(G/V)TWA(C/B)CHA7 harbors the central nucleotide signature 0RYXRY4 (166). Though seemingly enriched for rigid RY dinucleotides, the pattern actually ensures for relatively flexible sequences at the center of the integration site: Y at the center X position yields YY and YR at nucleotide positions 1 and 2 and at positions 2 and 3, respectively, whereas R at the center X yields YR and RR at these respective positions. Due to the lack of HIV-1 intasome structures, less is known about HIV-1 integrase-target DNA interactions than is known for PFV integrase. Nevertheless, mutagenesis experiments suggest that Ser119 of HIV-1 integrase, a residue that is structurally equivalent to Ala188 in PFV integrase, also interacts with bases that lie 3 positions upstream from the positions of viral DNA joining (166). Altered patterns of strand transfer reaction products on sequencing gels notably first highlighted a role for Ser119 in HIV-1 integrase and the analogous residue Ser124 in ASLV integrase in target DNA binding (96, 97, 167).
Structural Basis for Integrase Enzymatic Activities
The chemistry of transesterification starts with deprotonation of an attacking nucleophile and concludes with the protonation of a leaving group (168-170). The key role of the integrase active site residues in this process is to coordinate the binding of divalent metal ions; the metal ions in turn orchestrate the chemistry. One metal ion accordingly positions and deprotonates the attacking nucleophile, which is water for 3′ processing and the 3′-OH of cleaved viral DNA for strand transfer, whereas the other metal ion helps to destabilize the scissile phosphodiester bond and promote the formation of the pentavalent phosphorane reaction intermediate (168, 169, 171).
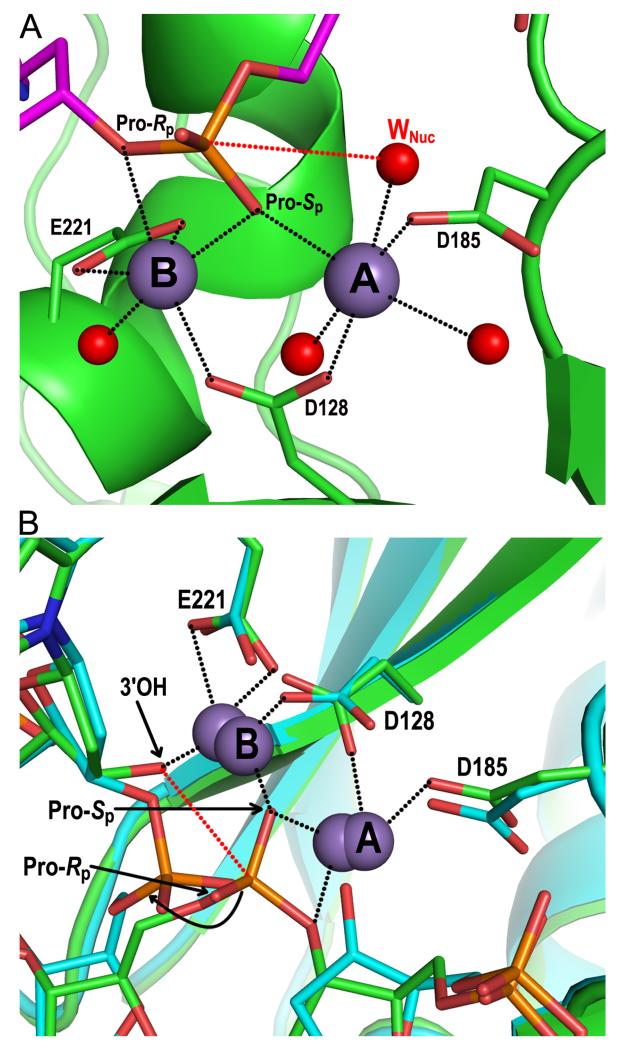
The co-crystallization of PFV integrase with uncleaved U5 DNA led to the structural determination of the SSC (85). Soaking the SSC crystals with divalent metal ions (Mg2+ or Mn2+) supported integrase 3′ processing activity in crystallo (85). A brief exposure of the crystals to manganese chloride allowed freezing out the active form of the SSC just prior to viral DNA cleavage and elucidating the roles of the divalent metal ions in 3′ processing activity. Metal ion A is in near perfect octahedral coordination though its engagement of active site residues Asp128 and Asp185, the non-bridging Pro-Sp oxygen of the scissile CA\AT phosphodiester bond, and three water molecules, including the nucleophilic water (Fig. 6A). The distance between the attacking water and scissile phosphodiester bond, 3.3 Å (red dashed line in the figure), was notably identical to that observed in a metal-bound structure of the RNase H enzyme (172). Metal ion B, coordinated through active site residues Asp128 and Glu221, a water molecule, and a bridging oxygen atom in addition to Pro-Sp, is in a less ideal environment, which may aid scissile phosphodiester bond destabilization (Fig. 6A) (85). The structure is consistent with the stereoselectivity of the HIV-1 integrase 3′ processing reaction, as the substitution of the Pro-Rp oxygen with sulfur was tolerated by integrase to a much greater extent than was the substitution of the Pro-Sp position (50).
Figure 6.
Structural basis of integrase 3′ processing and strand transfer activities. (A) Structure of the manganese-bound SSC (pdb code 4E7I). The DNA and integrase backbones are colored magenta and green, respectively; red and orange sticks are oxygen and phosphorus atoms, respectively. Gray and red spheres are manganese ions and water molecules, respectively, with the nucleophilic water labeled WNuc. Black dashed lines indicate metal ion interactions; the red dashed line connects the nucleophile and scissile phosphodiester bond. (B) Overlay of metal ion-bound TCC (pdb code 4E7K; DNA and protein in green) and STC (pdb code 4E7L; elements painted in cyan) structures. Both sets of metal ions are shown; the 3.8 Å spacing between ions in the TCC contracts to 3.2 Å in the STC (85). The curved black line indicates the displacement of the viral-target DNA phosphodiester bond after strand transfer relative to the scissile bond in target DNA. Other labeling is the same as in panel A.
The crystallized PFV TCC was also catalytically proficient. Due to the relatively rapid kinetics of the strand transfer reaction, numerous crystals were surveyed at early times post-metal ion exposure to determine manganese-bound TCC and STC structures (85). The overlay of both structures clarified the roles of the metal ions during strand transfer as well as the irreversible nature of the reaction (Fig. 6B). The roles of the metal ions in 3′ processing activity reverse during strand transfer, even though the majority of coordination contacts, including those with active site residues and Pro-Sp oxygen, remain. Thus, reaction specificity is in large part determined by the positions of attacking nucleophile and scissile bond relative to the metals. Metal ion B accordingly activates the 3′-OH of viral DNA for nucleophilic attack, whereas metal ion A helps to destabilize the scissile bond through its contact with a bridging oxygen atom (Fig. 6B). The structure is again consistent with the stereoselectivity of the HIV-1 integrase enzyme, as the substitution of sulfur for the Pro-Sp as compared to Pro-Rp oxygen preferentially inhibited strand transfer activity (50).
The phosphodiester bond that is formed between the viral and target DNA during strand transfer is displaced from the active site relative to the position of the scissile bond in target DNA prior to catalysis (Fig. 6B, curved arrow). Because isoenergetic reactions like strand transfer are in theory reversible, the displacement ensures that virus integration into chromosomal DNA is a largely irreversible process. The torsional stress that is applied to the target DNA by integrase (Fig. 5B) is the likely driving force behind the displacement (85, 98).
Future Directions in Integrase Structural Biology
The NED, NTD, and CTD of the outer integrase protomers were not resolved in the PFV intasome X-ray crystal structures (Fig. 5) (85, 94, 98). The interaction of these domains with the inner integrase dimer and/or target DNA therefore seems dispensable for integrase 3′ processing and strand transfer activities. We accordingly infer that the main role of the outer two molecules in PFV integrase catalysis in vitro is architectural, to frame the critical inner integrase dimer and viral DNA ends together.
Retroviral integration prefers nucleosomal DNA both in vitro (173-176) and during virus infection (177-179). One potential role for the outer integrase protomers may be to orchestrate interactions with nucleoprotein targets as compared to the naked target DNA utilized during intasome crystallogenesis (85, 98). SAXS and small angle neutron scattering studies have yielded models for the outer integrase NED, NTD, and CTD in the PFV intasome (64), and it could accordingly prove useful to conduct similar experiments with nucleoprotein targets such as reconstituted nucleosomes.
Four integrase molecules comprise the intasome, yet retrovirus particles harbor ~100-200 copies of integrase (180). It is unclear what fraction of this population may associate with the viral DNA as the PIC forms and traffics through the cell. Retroviral integrase proteins have been reported to bind to numerous cellular proteins (181-183) and some of these, like LEDGF/p75, have been confirmed to play an important role in integration. Other potential roles for integrase binding partners include the facilitation of reverse transcription (184) and PIC nuclear import (185-189), though the biological relevance of some of these interactions, for example that between HIV-1 integrase and the beta-karyopherin transportin 3, has been brought into question (190, 191). Complexes of integrase and cellular binding proteins that are verified to play important roles in virus replication are obvious candidates for structural biology studies moving forward.
Despite the advances afforded from recent success with PFV integrase structural biology, the spumaviruses are but one of seven retroviral genera. The PFV integrase interdomain linkers are relatively long (Fig. 2A), bringing into question as to whether integrase proteins that harbor shorter linker regions, like those derived from HIV-1 (Fig. 2A) or ASLV (77), will support the formation of the dimer-of-dimers architecture observed with the PFV intasome. Numerous groups have assembled HIV-1 models based on the PFV structure (105, 135, 192, 193), indicating reasonable potential for concordance (64). With ASLV integrase NTD-CCD and CCD-CTD linkers as seemingly short as 13 and 8 amino acid residues, respectively (77), the structure of the ASLV intasome should shed light on the potential universality of the PFV intasome architecture as a virus family-wide model.
THE INTEGRASE ACTIVE SITE AS A TARGET FOR ANTI-HIV DRUGS
The critical requirement of integrase for productive HIV-1 replication highlighted the enzyme as a target for drug development (194, 195). Initial attempts however yielded compounds with limited antiviral specificity that predominantly inhibited the assembly of active integrase-DNA complexes, often via electrostatic interactions (1). Screening assays staged with integrase pre-bound to immobilized viral DNA turned out to be key for the discovery and the development of the first clinically useful HIV-1 integrase inhibitors (1, 196, 197).
Integrase Strand Transfer Inhibitors (INSTIs)
Whereas 3′ processing of HIV-1 LTR ends occurs soon after reverse transcription (31, 32), strand transfer is delayed until the HIV-1 TCC is formed in the nucleus. Depending on the activation state of the infected cell, retroviral PICs are accordingly vulnerable to small molecule inhibitors of integrase strand transfer activity for a period of several hours to days (33, 34, 198). The initial compounds in this drug class, which contained a common diketo acid moiety (199), chelated divalent metal ions in the active site and competed with target DNA for binding to the intasome in vitro (200, 201).
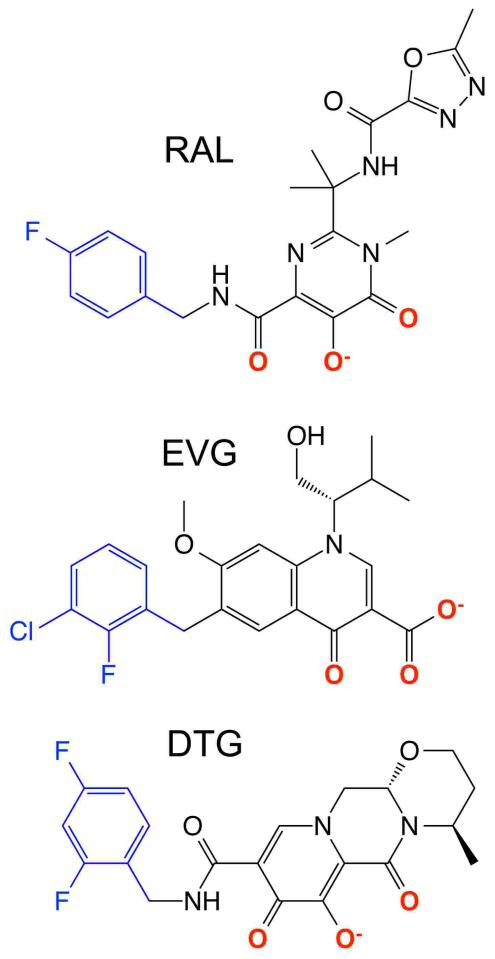
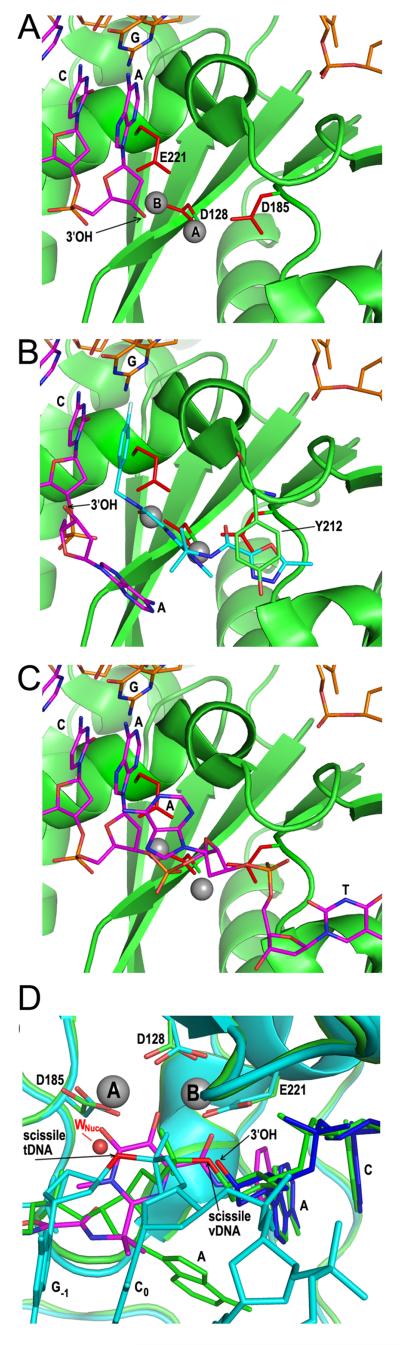
There are currently three INSTIs approved for the treatment of HIV/AIDS, raltegravir (202), elvitegravir (203), and dolutegravir (204) (Fig. 7). Commonalities among the molecules define aspects of the pharmacophore critical for antiviral activity. Each compound harbors coplanar oxygen atoms (highlighted in red in the figure) that resemble the diketo acid moiety of progenitor INSTIs. The compounds additionally harbor a halogenated benzyl group on a flexible linker (Fig. 7, blue). INSTIs display generally broad-spectrum antiretroviral activity (56, 205, 206), and co-crystallization with the PFV CI accordingly revealed critical aspects of the mechanism of INSTI action (94, 207, 208). As predicted from prior solution-based measures (201), the co-planar oxygen atoms engage the divalent metal ions at the integrase active site. Through stacking interactions with the cytosine of the invariant CA dinucleotide and its guanosine partner on the non-transferred DNA strand, the INSTI halobenzyl group supplants the invariant adenosine base and accordingly ejects the 3′ deoxyadenylate from the active site (compare Fig. 8B with 8A) (94, 207, 208). Ejection of the 3′-OH strand transfer nucleophile from the active site underscores the mechanism of INSTI action. The structure of the uncleaved dinucleotide in the active site of the integrase SSC accounts for the mechanistic selectivity of INSTIs, as the binding sites of the DNA and compounds coincide (Fig. 8B and 8C). Inhibition of 3′ processing activity would accordingly require the ejection of a trinucleotide from the integrase active site (85).
Figure 7.
Chemical structures of INSTIs raltegravir (RAL), elvitegravir (EVG), and dolutegravir (DTG).
Figure 8.
Structural basis of INSTI mechanism of action. (A) The active site from pdb code 3OY9 highlights PFV integrase DDE residues, Mn2+ ions A and B, and the 3′-OH of the terminal deoxyadenylate. (B) Structure of raltegravir (cyan)-bound integrase active site (pdb code 3OYA) highlighting positions of supplanted deoxyadenylate 3′-OH, magnesium ions (grey), and integrase residue Tyr212. (C) The integrase active site in the context of the PFV SSC highlights the position of the unprocessed AT dinucleotide. Additional panel A-C coloring: green, integrase; magenta, transferred DNA strand; orange, non-transferred strand. (D) Overlaid structures of the PFV SSC (pdb code 4E7I; integrase and viral DNA in green), TCC (pdb code 4E7K; integrase and target DNA in cyan and viral DNA in blue), and raltegravir-bound CI (pdb code 3OYA; raltegravir in magenta) highlights the common positioning of raltegravir oxygen atoms with strand transfer and 3′ processing attacking and leaving groups. Subscript numbers denote target DNA bases. tDNA, target DNA; vDNA, viral DNA.
Overlaying drug-bound structures with those of the metal-bound SSC and TCC revealed additional insight into the function of INSTI co-planar oxygen atoms. The position of the raltegravir oxygen atom that is distal from the halobenzyl group mimics the positions of the nucleophilic water molecule for 3′ processing activity and a bridging oxygen atom of the scissile phosphodiester bond in target DNA. Vice versa, the position of the raltegravir oxygen atom proximal to the halobenzyl group mimics those of the 3′-OH strand transfer nucleophile and a bridging oxygen atom of the viral DNA scissile phosphodiester bond (Fig. 8D). Therefore, the INSTIs are in fact substrate mimics of the 3′ processing and strand transfer reactions (85).
Mechanisms of Viral Resistance to INSTIs
The PFV intasome model has additionally proved useful in probing the structural basis of INSTI drug resistance. Raltegravir, elvitegravir, and dolutegravir as of this writing have been in the clinic for roughly 7, 2, and 1 years, respectively, and the mechanism of raltegravir resistance has accordingly been investigated most thoroughly. Three resistance pathways have been described, including those involving changes of HIV-1 integrase residues Gln148, Asn155, and Tyr143 (209); PFV integrase harbors Ser217, Asn224, and Tyr212 at these respective positions. Tyr212 stacked against a unique 5-member oxadiazole ring in raltegravir (Fig. 8B), so loss of a direct drug binding contact likely accounts for the Tyr143 resistance pathway (94). Perturbation of local active site structure as compared to loss of direct drug contact is potentially the basis for Gln148 and Asn155 resistance pathways. In particular, INSTI binding to the PFV integrase mutant S217H, which mimicked the HIV-1 integrase resistance mutation Q148H, correlated with an approximate 1-Å shift in the conformation of the active site. INSTI binding requires the mutant active site to adopt a wild type-like conformation, and the associated energy cost should reduce drug binding affinity (207).
The capacity of a small molecule to trap the HIV-1 PIC in a long-lived inhibited state correlates with its antiviral activity, and extended dissociative half-time (td1/2) emerged as an important benchmark in INSTI development (210, 211). The second-generation INSTIs dolutegravir and MK-2048 (an experimental compound), which possess td1/2s of roughly 71 and 32 hours, respectively, dissociate much more slowly from wild type-integrase DNA complexes in vitro than either raltegravir (with a td1/2 of 8.8 hours) or elvitegravir (2.7 hours) (210, 211) and are much more potent against viruses with Q148H or N155H resistance mutations (212, 213). Plausibly, the tighter binding INSTIs are less affected by the energetic penalty associated with the need for the active site to reconfigure to a wild type-like conformation (207). In addition, the higher flexibility of dolutegravir was suggested to play a role in its reduced susceptibility to canonical INSTI resistance mutations (208).
Future Directions in Integrase Active Site Drug Development
Though INSTIs are ineffective 3′ processing inhibitors, designs that side step the requirement for nucleotide ejection could be entertained. For example, small molecules that engage the viral DNA-integrase interface in the SSC could insert a chemically inert moiety at the position for the nucleophilic water molecule. Molecular dynamic simulations of the PFV integrase strand transfer reaction could moreover suggest improved positions for INSTI metal chelating groups, which in turn could improve INSTI potency (85). Toward these ends, we suspect that the pharmaceutical industry will employ the PFV model system to help develop novel inhibitors of the integrase active site and HIV-1 DNA integration.
ACKNOWLEDGEMENTS
We are grateful to Robert Craigie and Frederic Bushman for critically reading the manuscript. This work was supported by U.S. National Institutes of Health grants AI039394 and AI070042 (to A.E.) and by Medical Research Council UK grants G0900116 and G1000917 (to P.C.).
REFERENCES
- 1.Métifiot M, Marchand C, Pommier Y. HIV integrase inhibitors: 20-year landmark and challenges. Adv. Pharmacol. 2013;67:75–105. doi: 10.1016/B978-0-12-405880-4.00003-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ruelas DS, Greene WC. An integrated overview of HIV-1 latency. Cell. 2013;155:519–529. doi: 10.1016/j.cell.2013.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Donehower LA, Varmus HE. A mutant murine leukemia virus with a single missense codon in pol is defective in a function affecting integration. Proc. Natl. Acad. Sci. USA. 1984;81:6461–6465. doi: 10.1073/pnas.81.20.6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Panganiban AT, Temin HM. The retrovirus pol gene encodes a product required for DNA integration: Identification of a retrovirus int locus. Proc. Natl. Acad. Sci. USA. 1984;81:7885–7889. doi: 10.1073/pnas.81.24.7885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwartzberg P, Colicelli J, Goff SP. Construction and analysis of deletion mutations in the pol gene of moloney murine leukemia virus: A new viral function required for productive infection. Cell. 1984;37:1043–1052. doi: 10.1016/0092-8674(84)90439-2. [DOI] [PubMed] [Google Scholar]
- 6.Fassati A, Goff SP. Characterization of intracellular reverse transcription complexes of Moloney murine leukemia virus. J. Virol. 1999;73:8919–8925. doi: 10.1128/jvi.73.11.8919-8925.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fassati A, Goff SP. Characterization of intracellular reverse transcription complexes of human immunodeficiency virus type 1. J. Virol. 2001;75:3626–3635. doi: 10.1128/JVI.75.8.3626-3635.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Panganiban AT, Temin HM. The terminal nucleotides of retrovirus DNA are required for integration but not virus production. Nature. 1983;306:155–160. doi: 10.1038/306155a0. [DOI] [PubMed] [Google Scholar]
- 9.Colicelli J, Goff SP. Mutants and pseudorevertants of Moloney murine leukemia virus with alterations at the integration site. Cell. 1985;42:573–580. doi: 10.1016/0092-8674(85)90114-x. [DOI] [PubMed] [Google Scholar]
- 10.Colicelli J, Goff SP. Sequence and spacing requirements of a retrovirus integration site. J. Mol. Biol. 1988;199:47–59. doi: 10.1016/0022-2836(88)90378-6. [DOI] [PubMed] [Google Scholar]
- 11.Masuda T, Kuroda MJ, Harada S. Specific and independent recognition of U3 and U5 att sites by human immunodeficiency virus type 1 integrase in vivo. J. Virol. 1998;72:8396–8402. doi: 10.1128/jvi.72.10.8396-8402.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown HEV, Chen H, Engelman A. Structure-based mutagenesis of the human immunodeficiency virus type 1 DNA attachment site: effects on integration and cDNA synthesis. J. Virol. 1999;73:9011–9020. doi: 10.1128/jvi.73.11.9011-9020.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roth MJ, Schwartzberg PL, Goff SP. Structure of the termini of DNA intermediates in the integration of retroviral DNA: dependence on IN function and terminal DNA sequence. Cell. 1989;58:47–54. doi: 10.1016/0092-8674(89)90401-7. [DOI] [PubMed] [Google Scholar]
- 14.Fujiwara T, Mizuuchi K. Retroviral DNA integration: structure of an integration intermediate. Cell. 1988;54:497–504. doi: 10.1016/0092-8674(88)90071-2. [DOI] [PubMed] [Google Scholar]
- 15.Brown PO, Bowerman B, Varmus HE, Bishop JM. Retroviral integration: structure of the initial covalent product and its precursor, and a role for the viral IN protein. Proc. Natl. Acad. Sci. USA. 1989;86:2525–2529. doi: 10.1073/pnas.86.8.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pauza CD. Two bases are deleted from the termini of HIV-1 linear DNA during integrative recombination. Virology. 1990;179:886–889. doi: 10.1016/0042-6822(90)90161-j. [DOI] [PubMed] [Google Scholar]
- 17.Lee YM, Coffin JM. Relationship of avian retrovirus DNA synthesis to integration in vitro. Mol. Cell Biol. 1991;11:1419–1430. doi: 10.1128/mcb.11.3.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown PO, Bowerman B, Varmus HE, Bishop JM. Correct integration of retroviral DNA in vitro. Cell. 1987;49:347–356. doi: 10.1016/0092-8674(87)90287-x. [DOI] [PubMed] [Google Scholar]
- 19.Lee YM, Coffin JM. Efficient autointegration of avian retrovirus DNA in vitro. J. Virol. 1990;64:5958–5965. doi: 10.1128/jvi.64.12.5958-5965.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ellison V, Abrams H, Roe T, Lifson J, Brown P. Human immunodeficiency virus integration in a cell-free system. J. Virol. 1990;64:2711–2715. doi: 10.1128/jvi.64.6.2711-2715.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farnet CM, Haseltine WA. Integration of human immunodeficiency virus type 1 DNA in vitro. Proc. Natl. Acad. Sci. USA. 1990;87:4164–4168. doi: 10.1073/pnas.87.11.4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katzman M, Katz RA, Skalka AM, Leis J. The avian retroviral integration protein cleaves the terminal sequences of linear viral DNA at the in vivo sites of integration. J. Virol. 1989;63:5319–5327. doi: 10.1128/jvi.63.12.5319-5327.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Craigie R, Fujiwara T, Bushman F. The IN protein of Moloney murine leukemia virus processes the viral DNA ends and accomplishes their integration in vitro. Cell. 1990;62:829–837. doi: 10.1016/0092-8674(90)90126-y. [DOI] [PubMed] [Google Scholar]
- 24.Sherman PA, Fyfe JA. Human immunodeficiency virus integration protein expressed in Escherichia coli possesses selective DNA cleaving activity. Proc. Natl. Acad. Sci. USA. 1990;87:5119–5123. doi: 10.1073/pnas.87.13.5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bushman FD, Fujiwara T, Craigie R. Retroviral DNA integration directed by HIV integration protein in vitro. Science. 1990;249:1555–1558. doi: 10.1126/science.2171144. [DOI] [PubMed] [Google Scholar]
- 26.Bushman FD, Craigie R. Activities of human immunodeficiency virus (HIV) integration protein in vitro: Specific cleavage and integration of HIV DNA. Proc. Natl. Acad. Sci. USA. 1991;88:1339–1343. doi: 10.1073/pnas.88.4.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katz RA, Merkel G, Kulkosky J, Leis J, Skalka AM. The avian retroviral IN protein is both necessary and sufficient for integrative recombination in vitro. Cell. 1990;63:87–95. doi: 10.1016/0092-8674(90)90290-u. [DOI] [PubMed] [Google Scholar]
- 28.Vora AC, Fitzgerald ML, Grandgenett DP. Removal of 3′-OH-terminal nucleotides from blunt-ended long terminal repeat termini by the avian retrovirus integration protein. J. Virol. 1990;64:5656–5659. doi: 10.1128/jvi.64.11.5656-5659.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.LaFemina RL, Callahan PL, Cordingley MG. Substrate specificity of recombinant human immunodeficiency virus integrase protein. J. Virol. 1991;65:5624–5630. doi: 10.1128/jvi.65.10.5624-5630.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pahl A, Flugel RM. Endonucleolytic cleavages and DNA-joining activities of the integration protein of human foamy virus. J. Virol. 1993;67:5426–5434. doi: 10.1128/jvi.67.9.5426-5434.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller M, Farnet C, Bushman F. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J. Virol. 1997;71:5382–5390. doi: 10.1128/jvi.71.7.5382-5390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Munir S, Thierry S, Subra F, Deprez E, Delelis O. Quantitative analysis of the time-course of viral DNA forms during the HIV-1 life cycle. Retrovirology. 2013;10:87. doi: 10.1186/1742-4690-10-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Butler SL, Hansen MS, Bushman FD. A quantitative assay for HIV DNA integration in vivo. Nat. Med. 2001;7:631–634. doi: 10.1038/87979. [DOI] [PubMed] [Google Scholar]
- 34.Vandegraaff N, Kumar R, Burrell CJ, Li P. Kinetics of human immunodeficiency virus type 1 (HIV) DNA integration in acutely infected cells as determined using a novel assay for detection of integrated HIV DNA. J. Virol. 2001;75:11253–11260. doi: 10.1128/JVI.75.22.11253-11260.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chow SA, Vincent KA, Ellison V, Brown PO. Reversal of integration and DNA splicing mediated by integrase of human immunodeficiency virus. Science. 1992;255:723–726. doi: 10.1126/science.1738845. [DOI] [PubMed] [Google Scholar]
- 36.Vink C, Yeheskiely E, van der Marel GA, Van Boom JH, Plasterk RHA. Site-specific hydrolysis and alcoholysis of human immunodeficiency virus DNA termini mediated by the viral integrase protein. Nucleic Acids Res. 1991;19:6691–6698. doi: 10.1093/nar/19.24.6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Katzman M, Sudol M. Mapping domains of retroviral integrase responsible for viral DNA specificity and target site selection by analysis of chimeras between human immunodeficiency virus type 1 and visna virus integrases. J. Virol. 1995;69:5687–5696. doi: 10.1128/jvi.69.9.5687-5696.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mizuuchi K. Transpositional recombination: Mechanistic insights from studies of Mu and other elements. Annu. Rev. Biochem. 1992;61:1011–1051. doi: 10.1146/annurev.bi.61.070192.005051. [DOI] [PubMed] [Google Scholar]
- 39.Knowles JR. Enzyme-catalyzed phosphoryl transfer reactions. Annu. Rev. Biochem. 1980;49:877–919. doi: 10.1146/annurev.bi.49.070180.004305. [DOI] [PubMed] [Google Scholar]
- 40.Mizuuchi K, Adzuma K. Inversion of the phosphate chirality at the target site of Mu DNA strand transfer: evidence for a one-step transesterification mechanism. Cell. 1991;66:129–140. doi: 10.1016/0092-8674(91)90145-o. [DOI] [PubMed] [Google Scholar]
- 41.Engelman A, Mizuuchi K, Craigie R. HIV-1 DNA integration: mechanism of viral DNA cleavage and DNA strand transfer. Cell. 1991;67:1211–1221. doi: 10.1016/0092-8674(91)90297-c. [DOI] [PubMed] [Google Scholar]
- 42.Jenkins TM, Engelman A, Ghirlando R, Craigie R. A soluble active mutant of HIV-1 integrase: involvement of both the core and the C-terminal domains in multimerization. J. Biol. Chem. 1996;271:7712–7718. doi: 10.1074/jbc.271.13.7712. [DOI] [PubMed] [Google Scholar]
- 43.Dyda F, Hickman AB, Jenkins TM, Engelman A, Craigie R, Davies DR. Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science. 1994;266:1981–1986. doi: 10.1126/science.7801124. [DOI] [PubMed] [Google Scholar]
- 44.Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- 45.Landree MA, Wibbenmeyer JA, Roth DB. Mutational analysis of RAG1 and RAG2 identifies three catalytic amino acids in RAG1 critical for both cleavage steps of V(D)J recombination. Genes Dev. 1999;13:3059–3069. doi: 10.1101/gad.13.23.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim DR, Dai Y, Mundy CL, Yang W, Oettinger MA. Mutations of acidic residues in RAG1 define the active site of the V(D)J recombinase. Genes Dev. 1999;13:3070–3080. doi: 10.1101/gad.13.23.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nowotny M. Retroviral integrase superfamily: the structural perspective. EMBO Rep. 2009;10:144–151. doi: 10.1038/embor.2008.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mizuuchi K, Nobbs TJ, Halford SE, Adzuma K, Qin J. A new method for determining the stereochemistry of DNA cleavage reactions: application to the SfiI and HpaII restriction endonucleases and to the MuA transposase. Biochemistry. 1999;38:4640–4648. doi: 10.1021/bi990054p. [DOI] [PubMed] [Google Scholar]
- 49.van Gent DC, Mizuuchi K, Gellert M. Similarities between initiation of V(D)J recombination and retroviral integration. Science. 1996;271:1592–1594. doi: 10.1126/science.271.5255.1592. [DOI] [PubMed] [Google Scholar]
- 50.Gerton JL, Herschlag D, Brown PO. Stereospecificity of reactions catalyzed by HIV-1 integrase. J. Biol. Chem. 1999;274:33480–33487. doi: 10.1074/jbc.274.47.33480. [DOI] [PubMed] [Google Scholar]
- 51.Kennedy AK, Haniford DB, Mizuuchi K. Single active site catalysis of the successive phosphoryl transfer steps by DNA transposases: insights from phosphorothioate stereoselectivity. Cell. 2000;101:295–305. doi: 10.1016/s0092-8674(00)80839-9. [DOI] [PubMed] [Google Scholar]
- 52.Vora AC, McCord M, Fitzgerald ML, Inman RB, Grandgenett DP. Efficient concerted integration of retrovirus-like DNA in vitro by avian myeloblastosis virus integrase. Nucleic Acids Res. 1994;22:4454–4461. doi: 10.1093/nar/22.21.4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang F, Roth MJ. Assembly and catalysis of concerted two-end integration events by Moloney murine leukemia virus integrase. J. Virol. 2001;75:9561–9670. doi: 10.1128/JVI.75.20.9561-9570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cherepanov P. LEDGF/p75 interacts with divergent lentiviral integrases and modulates their enzymatic activity in vitro. Nucleic Acids Res. 2007;35:113–124. doi: 10.1093/nar/gkl885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ballandras-Colas A, Naraharisetty H, Li X, Serrao E, Engelman A. Biochemical characterization of novel retroviral integrase proteins. PLoS One. 2013;8:e76638. doi: 10.1371/journal.pone.0076638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Valkov E, Gupta SS, Hare S, Helander A, Roversi P, McClure M, Cherepanov P. Functional and structural characterization of the integrase from the prototype foamy virus. Nucleic Acids Res. 2009;37:243–255. doi: 10.1093/nar/gkn938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sinha S, Pursley MH, Grandgenett DP. Efficient concerted integration by recombinant human immunodeficiency virus type 1 integrase without cellular or viral cofactors. J. Virol. 2002;76:3105–3113. doi: 10.1128/JVI.76.7.3105-3113.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Faure A, Calmels C, Desjobert C, Castroviejo M, Caumont-Sarcos A, Tarrago-Litvak L, Litvak S, Parissi V. HIV-1 integrase crosslinked oligomers are active in vitro. Nucleic Acids Res. 2005;33:977–986. doi: 10.1093/nar/gki241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li M, Craigie R. Processing of viral DNA ends channels the HIV-1 integration reaction to concerted integration. J. Biol. Chem. 2005;280:29334–29339. doi: 10.1074/jbc.M505367200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Carteau S, Gorelick RJ, Bushman FD. Coupled integration of human immunodeficiency virus type 1 cDNA ends by purified integrase in vitro: Stimulation by the viral nucleocapsid protein. J. Virol. 1999;73:6670–6679. doi: 10.1128/jvi.73.8.6670-6679.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hare S, Shun MC, Gupta SS, Valkov E, Engelman A, Cherepanov P. A novel co-crystal structure affords the design of gain-of-function lentiviral integrase mutants in the presence of modified PSIP1/LEDGF/p75. PLoS Pathog. 2009;5:e1000259. doi: 10.1371/journal.ppat.1000259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pandey KK, Bera S, Grandgenett DP. The HIV-1 integrase monomer induces a specific interaction with LTR DNA for concerted integration. Biochemistry. 2011;50:9788–9796. doi: 10.1021/bi201247f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Delelis O, Carayon K, Guiot E, Leh H, Tauc P, Brochon JC, Mouscadet JF, Deprez E. Insight into the integrase-DNA recognition mechanism. A specific DNA-binding mode revealed by an enzymatically labeled integrase. J. Biol. Chem. 2008;283:27838–27849. doi: 10.1074/jbc.M803257200. [DOI] [PubMed] [Google Scholar]
- 64.Gupta K, Curtis JE, Krueger S, Hwang Y, Cherepanov P, Bushman FD, Van Duyne GD. Solution conformations of prototype foamy virus integrase and its stable synaptic complex with U5 viral DNA. Structure. 2012;20:1918–1928. doi: 10.1016/j.str.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jenkins T, Hickman A, Dyda F, Ghirlando R, Davies D, Craigie R. Catalytic domain of human immunodeficiency virus type 1 integrase: Identification of a soluble mutant by systematic replacement of hydrophobic residues. Proc. Natl. Acad. Sci. USA. 1995;92:6057–6061. doi: 10.1073/pnas.92.13.6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen JC-H, Krucinski J, Miercke LJW, Finer-Moore JS, Tang AH, Leavitt AD, Stroud RM. Crystal structure of the HIV-1 integrase catalytic core and C-terminal domains: A model for viral DNA binding. Proc. Natl. Acad. Sci. USA. 2000;97:8233–8238. doi: 10.1073/pnas.150220297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Busschots K, Vercammen J, Emiliani S, Benarous R, Engelborghs Y, Christ F, Debyser Z. The interaction of LEDGF/p75 with integrase is lentivirus-specific and promotes DNA binding. J. Biol. Chem. 2005;280:17841–17847. doi: 10.1074/jbc.M411681200. [DOI] [PubMed] [Google Scholar]
- 68.Michel F, Crucifix C, Granger F, Eiler S, Mouscadet JF, Korolev S, Agapkina J, Ziganshin R, Gottikh M, Nazabal A, Emiliani S, Benarous R, Moras D, Schultz P, Ruff M. Structural basis for HIV-1 DNA integration in the human genome, role of the LEDGF/P75 cofactor. EMBO J. 2009;28:980–991. doi: 10.1038/emboj.2009.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cherepanov P, Ambrosio ALB, Rahman S, Ellenberger T, Engelman A. From the Cover: Structural basis for the recognition between HIV-1 integrase and transcriptional coactivator p75. Proc. Natl. Acad. Sci. USA. 2005;102:17308–17313. doi: 10.1073/pnas.0506924102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hare S, Di Nunzio F, Labeja A, Wang J, Engelman A, Cherepanov P. Structural basis for functional tetramerization of lentiviral integrase. PLoS Pathog. 2009;5:e1000515. doi: 10.1371/journal.ppat.1000515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Khan E, Mack JPG, Katz RA, Kulkosky J, Skalka AM. Retroviral integrase domains: DNA binding and the recognition of LTR sequences. Nucleic Acids Res. 1991;19:851–860. doi: 10.1093/nar/19.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Burke CJ, Sanyal G, Bruner MW, Ryan JA, LaFemina RL, Robbins HL, Zeft AS, Middaugh CR, Cordingley MG. Structural implications of spectroscopic characterization of a putative zinc finger peptide from HIV-1 integrase. J. Biol. Chem. 1992;267:9639–9644. [PubMed] [Google Scholar]
- 73.Engelman A, Craigie R. Identification of conserved amino acid residues critical for human immunodeficiency virus type 1 integrase function in vitro. J. Virol. 1992;66:6361–6369. doi: 10.1128/jvi.66.11.6361-6369.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bushman FD, Engelman A, Palmer I, Wingfield P, Craigie R. Domains of the integrase protein of human immunodeficiency virus type 1 responsible for polynucleotidyl transfer and zinc binding. Proc. Natl. Acad. Sci. USA. 1993;90:3428–3432. doi: 10.1073/pnas.90.8.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Woerner AM, Klutch M, Levin JG, Marcus-Sekura CJ. Localization of DNA binding activity of HIV-1 integrase to the C-terminal half of the protein. AIDS Res. Hum. Retroviruses. 1992;8:297–304. doi: 10.1089/aid.1992.8.297. [DOI] [PubMed] [Google Scholar]
- 76.Vink C, Oude Groeneger AM, Plasterk RHA. Identification of the catalytic and DNA-binding region of the human immunodeficiency virus type I integrase protein. Nucleic Acids Res. 1993;21:1419–1425. doi: 10.1093/nar/21.6.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li X, Krishnan L, Cherepanov P, Engelman A. Structural biology of retroviral DNA integration. Virology. 2011;411:194–205. doi: 10.1016/j.virol.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kulkosky J, Jones KS, Katz RA, Mack JP, Skalka AM. Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol. Cell. Biol. 1992;12:2331–2338. doi: 10.1128/mcb.12.5.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Drelich M, Wilhelm R, Mous J. Identification of amino acid residues critical for endonuclease and integration activities of HIV-1 IN protein in vitro. Virology. 1992;188:459–468. doi: 10.1016/0042-6822(92)90499-f. [DOI] [PubMed] [Google Scholar]
- 80.Leavitt AD, Shiue L, Varmus HE. Site-directed mutagenesis of HIV-1 integrase demonstrates differential effects on integrase functions in vitro. J. Biol. Chem. 1993;268:2113–2119. [PubMed] [Google Scholar]
- 81.Fayet O, Ramond P, Polard P, Prère MF, Chandler M. Functional similarities between retroviruses and the IS3 family of bacterial insertion sequences? Mol. Microbiol. 1990;4:1771–1777. doi: 10.1111/j.1365-2958.1990.tb00555.x. [DOI] [PubMed] [Google Scholar]
- 82.Yuan Y-W, Wessler SR. The catalytic domain of all eukaryotic cut-and-paste transposase superfamilies. Proc. Natl. Acad. Sci. USA. 2011;108:7884–7889. doi: 10.1073/pnas.1104208108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kojima KK, Jurka J. A superfamily of DNA transposons targeting multicopy small RNA genes. PLoS One. 2013;8:e68260. doi: 10.1371/journal.pone.0068260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Guérillot R, Siguier P, Gourbeyre E, Chandler M, Glaser P. The diversity of prokaryotic DDE transposases of the mutator superfamily, iInsertion specificity, and association with conjugation machineries. Genome Biol. Evol. 2014;6:260–272. doi: 10.1093/gbe/evu010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hare S, Maertens GN, Cherepanov P. 3′-Processing and strand transfer catalysed by retroviral integrase in crystallo. EMBO J. 2012;31:3020–3028. doi: 10.1038/emboj.2012.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jenkins TM, Esposito D, Engelman A, Craigie R. Critical contacts between HIV-1 integrase and viral DNA identified by structure-based analysis and photo-crosslinking. EMBO J. 1997;16:6849–6859. doi: 10.1093/emboj/16.22.6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Esposito D, Craigie R. Sequence specificity of viral end DNA binding by HIV-1 integrase reveals critical regions for protein-DNA interaction. EMBO J. 1998;17:5832–5843. doi: 10.1093/emboj/17.19.5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gerton JL, Brown PO. The core domain of HIV-1 integrase recognizes key features of its DNA substrates. J. Biol. Chem. 1997;272:25809–25815. doi: 10.1074/jbc.272.41.25809. [DOI] [PubMed] [Google Scholar]
- 89.Gerton JL, Ohgi S, Olsen M, DeRisi J, Brown PO. Effects of mutations in residues near the active site of human immunodeficiency virus type 1 integrase on specific enzyme-substrate interactions. J. Virol. 1998;72:5046–5055. doi: 10.1128/jvi.72.6.5046-5055.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Heuer TS, Brown PO. Mapping features of HIV-1 integrase near selected sites on viral and target DNA molecules in an active enzyme-DNA complex by photo-cross-linking. Biochemistry. 1997;36:10655–10665. doi: 10.1021/bi970782h. [DOI] [PubMed] [Google Scholar]
- 91.Chen A, Weber IT, Harrison RW, Leis J. Identification of amino acids in HIV-1 and avian sarcoma virus integrase subsites required for specific recognition of the long terminal repeat ends. J. Biol. Chem. 2006;281:4173–4182. doi: 10.1074/jbc.M510628200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Johnson AA, Santos W, Pais GCG, Marchand C, Amin R, Burke TR, Jr, Verdine G, Pommier Y. Integration requires a specific interaction of the donor DNA terminal 5′-cytosine with glutamine 148 of the HIV-1 integrase flexible loop. J. Biol. Chem. 2006;281:461–467. doi: 10.1074/jbc.M511348200. [DOI] [PubMed] [Google Scholar]
- 93.Hobaika Z, Zargarian L, Boulard Y, Maroun RG, Mauffret O, Fermandjian S. Specificity of LTR DNA recognition by a peptide mimicking the HIV-1 integrase α4 helix. Nucleic Acids Res. 2009;37:7691–7700. doi: 10.1093/nar/gkp824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hare S, Gupta SS, Valkov E, Engelman A, Cherepanov P. Retroviral intasome assembly and inhibition of DNA strand transfer. Nature. 2010;464:232–236. doi: 10.1038/nature08784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Appa RS, Shin C-G, Lee P, Chow SA. Role of the nonspecific DNA-binding region and alpha helices within the core domain of retroviral integrase in selecting target DNA sites for integration. J. Biol. Chem. 2001;276:45848–45855. doi: 10.1074/jbc.M107365200. [DOI] [PubMed] [Google Scholar]
- 96.Harper AL, Skinner LM, Sudol M, Katzman M. Use of patient-derived human immunodeficiency virus type 1 integrases to identify a protein residue that affects target site selection. J. Virol. 2001;75:7756–7762. doi: 10.1128/JVI.75.16.7756-7762.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Harper AL, Sudol M, Katzman M. An amino acid in the central catalytic domain of three retroviral integrases that affects target site selection in nonviral DNA. J. Virol. 2003;77:3838–3845. doi: 10.1128/JVI.77.6.3838-3845.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Maertens GN, Hare S, Cherepanov P. The mechanism of retroviral integration through X-ray structures of its key intermediates. Nature. 2010;468:326–329. doi: 10.1038/nature09517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cai M, Zheng R, Caffrey M, Craigie R, Clore GM, Gronenborn AM. Solution structure of the N-terminal zinc binding domain of HIV-1 integrase. Nat. Struct. Biol. 1997;4:567–577. doi: 10.1038/nsb0797-567. [DOI] [PubMed] [Google Scholar]
- 100.Eijkelenboom AP, van den Ent FM, Vos A, Doreleijers JF, Hard K, Tullius TD, Plasterk RH, Kaptein R, Boelens R. The solution structure of the amino-terminal HHCC domain of HIV-2 integrase: a three-helix bundle stabilized by zinc. Curr. Biol. 1997;7:739–746. doi: 10.1016/s0960-9822(06)00332-0. [DOI] [PubMed] [Google Scholar]
- 101.Lee SP, Han MK. Zinc stimulates Mg2+-dependent 3′-processing activity of human immunodeficiency virus type 1 integrase in vitro. Biochemistry. 1996;35:3837–3844. doi: 10.1021/bi952056p. [DOI] [PubMed] [Google Scholar]
- 102.Zheng R, Jenkins TM, Craigie R. Zinc folds the N-terminal domain of HIV-1 integrase, promotes multimerization, and enhances catalytic activity. Proc. Natl. Acad. Sci. USA. 1996;93:13659–13664. doi: 10.1073/pnas.93.24.13659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.van den Ent FMI, Vos A, Plasterk RHA. Dissecting the role of the N-terminal domain of human immunodeficiency virus integrase by trans-complementation analysis. J. Virol. 1999;73:3176–3183. doi: 10.1128/jvi.73.4.3176-3183.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhao Z, McKee CJ, Kessl JJ, Santos WL, Daigle JE, Engelman A, Verdine G, Kvaratskhelia M. Subunit-specific protein footprinting reveals significant structural rearrangements and a role for N-terminal Lys-14 of HIV-1 integrase during viral DNA binding. J. Biol. Chem. 2008;283:5632–5641. doi: 10.1074/jbc.M705241200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Krishnan L, Li X, Naraharisetty HL, Hare S, Cherepanov P, Engelman A. Structure-based modeling of the functional HIV-1 intasome and its inhibition. Proc. Natl. Acad. Sci. USA. 2010;107:15010–15915. doi: 10.1073/pnas.1002346107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cannon PM, Byles ED, Kingsman SM, Kingsman AJ. Conserved sequences in the carboxyl terminus of integrase that are essential for human immunodeficiency virus type 1 replication. J. Virol. 1996;70:651–657. doi: 10.1128/jvi.70.1.651-657.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Engelman A, Hickman AB, Craigie R. The core and carboxyl-terminal domains of the integrase protein of human immunodeficiency virus type 1 each contribute to nonspecific DNA binding. J. Virol. 1994;68:5911–5917. doi: 10.1128/jvi.68.9.5911-5917.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Puras Lutzke RA, Plasterk RHA. Structure-based mutational analysis of the C-terminal DNA-binding domain of human immunodeficiency virus type 1 integrase: critical residues for protein oligomerization and DNA binding. J. Virol. 1998;72:4841–4848. doi: 10.1128/jvi.72.6.4841-4848.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Puras Lutzke RA, Vink C, Plasterk RHA. Characterization of the minimal DNA-binding domain of the HIV integrase protein. Nucleic Acids Res. 1994;22:4125–4131. doi: 10.1093/nar/22.20.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Andrake MD, Skalka AM. Multimerization determinants reside in both the catalytic core and C terminus of avian sarcoma virus integrase. J. Biol. Chem. 1995;270:29299–29306. doi: 10.1074/jbc.270.49.29299. [DOI] [PubMed] [Google Scholar]
- 111.Engelman A, Bushman FD, Craigie R. Identification of discrete functional domains of HIV-1 integrase and their organization within an active multimeric complex. EMBO J. 1993;12:3269–3275. doi: 10.1002/j.1460-2075.1993.tb05996.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.van Gent DC, Oude Groeneger AAM, Plasterk RHA. Identification of amino acids in HIV-2 integrase involved in site-specific hydrolysis and alcoholysis of viral DNA termini. Nucleic Acids Res. 1993;21:3373–3377. doi: 10.1093/nar/21.15.3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bushman FD, Wang B. Rous sarcoma virus integrase protein: mapping functions for catalysis and substrate binding. J. Virol. 1994;68:2215–2223. doi: 10.1128/jvi.68.4.2215-2223.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kulkosky J, Katz RA, Merkel G, Skalka AM. Activities and substrate specificity of the evolutionarily conserved central domain of retroviral integrase. Virology. 1995;206:448–456. doi: 10.1016/s0042-6822(95)80060-3. [DOI] [PubMed] [Google Scholar]
- 115.Pahl A, Flügel RM. Characterization of the human spuma retrovirus integrase by site-directed mutagenesis, by complementation analysis, and by swapping the zinc finger domain of HIV-1. J. Biol. Chem. 1995;270:2957–2966. doi: 10.1074/jbc.270.7.2957. [DOI] [PubMed] [Google Scholar]
- 116.Jonsson C, Donzella G, Gaucan E, Smith C, Roth M. Functional domains of Moloney murine leukemia virus integrase defined by mutation and complementation analysis. J. Virol. 1996;70:4585–4597. doi: 10.1128/jvi.70.7.4585-4597.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jones KS, Coleman J, Merkel GW, Laue TM, Skalka AM. Retroviral integrase functions as a multimer and can turn over catalytically. J. Biol. Chem. 1992;267:16037–16040. [PubMed] [Google Scholar]
- 118.Bao KK, Wang H, Miller JK, Erie DA, Skalka AM, Wong I. Functional oligomeric state of avian sarcoma virus integrase. J. Biol. Chem. 2003;278:1323–1327. doi: 10.1074/jbc.C200550200. [DOI] [PubMed] [Google Scholar]
- 119.Cherepanov P, Maertens G, Proost P, Devreese B, Van Beeumen J, Engelborghs Y, De Clercq E, Debyser Z. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J. Biol. Chem. 2003;278:372–381. doi: 10.1074/jbc.M209278200. [DOI] [PubMed] [Google Scholar]
- 120.Li M, Mizuuchi M, Burke TRJ, Craigie R. Retroviral DNA integration: reaction pathway and critical intermediates. EMBO J. 2006;25:1295–1304. doi: 10.1038/sj.emboj.7601005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bera S, Pandey KK, Vora AC, Grandgenett DP. Molecular Interactions between HIV-1 integrase and the two viral DNA ends within the synaptic complex that mediates concerted integration. J. Mol. Biol. 2009;389:183–198. doi: 10.1016/j.jmb.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.van Gent DC, Vink C, Groeneger AAMO, Plasterk RHA. Complementation between HIV integrase proteins mutated in different domains. EMBO J. 1993;12:3261–3267. doi: 10.1002/j.1460-2075.1993.tb05995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ellison V, Gerton J, Vincent KA, Brown PO. An essential interaction between distinct domains of HIV-1 integrase mediates assembly of the active multimer. J. Biol. Chem. 1995;270:3320–3326. doi: 10.1074/jbc.270.7.3320. [DOI] [PubMed] [Google Scholar]
- 124.Vink C, van Gent DC, Elgersma Y, Plasterk RH. Human immunodeficiency virus integrase protein requires a subterminal position of its viral DNA recognition sequence for efficient cleavage. J. Virol. 1991;65:4636–4644. doi: 10.1128/jvi.65.9.4636-4644.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Leavitt AD, Rose RB, Varmus HE. Both substrate and target oligonucleotide sequences affect in vitro integration mediated by human immunodeficiency virus type 1 integrase protein produced in Saccharomyces cerevisiae. J. Virol. 1992;66:2359–2368. doi: 10.1128/jvi.66.4.2359-2368.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sherman PA, Dickson ML, Fyfe JA. Human immunodeficiency virus type 1 integration protein: DNA sequence requirements for cleaving and joining reactions. J. Virol. 1992;66:3593–3601. doi: 10.1128/jvi.66.6.3593-3601.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Masuda T, Planelles V, Krogstad P, Chen IS. Genetic analysis of human immunodeficiency virus type 1 integrase and the U3 att site: unusual phenotype of mutants in the zinc finger-like domain. J. Virol. 1995;69:6687–6696. doi: 10.1128/jvi.69.11.6687-6696.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Oh J, Chang KW, Alvord WG, Hughes SH. Alternate polypurine tracts affect Rous sarcoma virus integration in vivo. J. Virol. 2006;80:10281–10284. doi: 10.1128/JVI.00361-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Oh J, Chang KW, Hughes SH. Mutations in the U5 sequences adjacent to the primer binding site do not affect tRNA cleavage by Rous sarcoma virus RNase H but do cause aberrant integrations in vivo. J. Virol. 2006;80:451–459. doi: 10.1128/JVI.80.1.451-459.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chaconas G. Studies on a “jumping gene machine”: Higher-order nucleoprotein complexes in Mu DNA transposition. Biochem. Cell Biol. 1999;77:487–491. [PubMed] [Google Scholar]
- 131.Krishnan L, Engelman A. Retroviral integrase proteins and HIV-1 DNA integration. J. Biol. Chem. 2012;287:40858–40866. doi: 10.1074/jbc.R112.397760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wei S-Q, Mizuuchi K, Craigie R. A large nucleoprotein assembly at the ends of the viral DNA mediates retroviral DNA integration. EMBO J. 1997;16:7511–7520. doi: 10.1093/emboj/16.24.7511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Chen H, Wei S-Q, Engelman A. Multiple integrase functions are required to form the native structure of the human immunodeficiency virus type I intasome. J. Biol. Chem. 1999;274:17358–17364. doi: 10.1074/jbc.274.24.17358. [DOI] [PubMed] [Google Scholar]
- 134.Kotova S, Li M, Dimitriadis EK, Craigie R. Nucleoprotein intermediates in HIV-1 DNA integration visualized by atomic force microscopy. J. Mol. Biol. 2010;399:491–500. doi: 10.1016/j.jmb.2010.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kessl JJ, Li M, Ignatov M, Shkriabai N, Eidahl JO, Feng L, Musier-Forsyth K, Craigie R, Kvaratskhelia M. FRET analysis reveals distinct conformations of IN tetramers in the presence of viral DNA or LEDGF/p75. Nucleic Acids Res. 2011;39:9009–9022. doi: 10.1093/nar/gkr581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Guiot E, Carayon K, Delelis O, Simon F, Tauc P, Zubin E, Gottikh M, Mouscadet J-F, Brochon J-C, Deprez E. Relationship between the oligomeric status of HIV-1 integrase on DNA and enzymatic activity. J. Biol. Chem. 2006;281:22707–22719. doi: 10.1074/jbc.M602198200. [DOI] [PubMed] [Google Scholar]
- 137.Bojja RS, Andrake MD, Weigand S, Merkel G, Yarychkivska O, Henderson A, Kummerling M, Skalka AM. Architecture of a full-length retroviral integrase monomer and dimer, revealed by small angle X-ray scattering and chemical cross-linking. J. Biol. Chem. 2011;286:17047–17059. doi: 10.1074/jbc.M110.212571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Bojja RS, Andrake MD, Merkel G, Weigand S, Dunbrack RLJ, Skalka AM. Architecture and assembly of HIV integrase multimers in the absence of DNA substrates. J. Biol. Chem. 2013;288:7373–7386. doi: 10.1074/jbc.M112.434431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hickman AB, Palmer I, Engelman A, Craigie R, Wingfield P. Biophysical and enzymatic properties of the catalytic domain of HIV-1 integrase. J. Biol. Chem. 1994;269:29279–29287. [PubMed] [Google Scholar]
- 140.Bujacz G, Jaskolski M, Alexandratos J, Wlodawer A, Merkel G, Katz RA, Skalka AM. High-resolution structure of the catalytic domain of avian sarcoma virus integrase. J. Mol. Biol. 1995;253:333–346. doi: 10.1006/jmbi.1995.0556. [DOI] [PubMed] [Google Scholar]
- 141.Eijkelenboom AP, van den Ent FM, Wechselberger R, Plasterk RH, Kaptein R, Boelens R. Refined solution structure of the dimeric N-terminal HHCC domain of HIV-2 integrase. J. Biomol. NMR. 2000;18:119–128. doi: 10.1023/a:1008342312269. [DOI] [PubMed] [Google Scholar]
- 142.Lodi PJ, Ernst JA, Kuszewski J, Hickman AB, Engelman A, Craigie R, Clore GM, Gronenborn AM. Solution structure of the DNA binding domain of HIV-1 integrase. Biochemistry. 1995;34:9826–9833. doi: 10.1021/bi00031a002. [DOI] [PubMed] [Google Scholar]
- 143.Eijkelenboom APAM, Puras Lutzke RA, Boelens R, Plasterk RHA, Kaptein R, Hård K. The DNA-binding domain of HIV-1 integrase has an SH3-like fold. Nat. Struct. Biol. 1995;2:807–810. doi: 10.1038/nsb0995-807. [DOI] [PubMed] [Google Scholar]
- 144.Eijkelenboom APAM, Sprangers R, Hård K, Puras Lutzke RA, Plasterk RHA, Boelens R, Kaptein R. Refined solution structure of the C-terminal DNA-binding domain of human immunovirus-1 integrase. Proteins. 1999;36:556–564. doi: 10.1002/(sici)1097-0134(19990901)36:4<556::aid-prot18>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 145.Kaneko T, Li L, Li SS. The SH3 domain--a family of versatile peptide- and protein-recognition module. Front. Biosci. 2008;13:4938–4952. doi: 10.2741/3053. [DOI] [PubMed] [Google Scholar]
- 146.Baumann H, Knapp S, Lundbäck T, Ladenstein R, Härd T. Solution structure and DNA-binding properties of a thermostable protein from the archaeon Sulfolobus solfataricus. Nat. Struct. Biol. 1994;1:808–819. doi: 10.1038/nsb1194-808. [DOI] [PubMed] [Google Scholar]
- 147.Chen Z, Yan Y, Munshi S, Li Y, Zugay-Murphy J, Xu B, Witmer M, Felock P, Wolfe A, Sardana V. X-ray structure of simian immunodeficiency virus integrase containing the core and C-terminal domain (residues 50-293) - an initial glance of the viral DNA binding platform. J. Mol. Biol. 2000;296:521–533. doi: 10.1006/jmbi.1999.3451. [DOI] [PubMed] [Google Scholar]
- 148.Yang Z-N, Mueser TC, Bushman FD, Hyde CC. Crystal structure of an active two-domain derivative of rous sarcoma virus integrase. J. Mol. Biol. 2000;296:535–548. doi: 10.1006/jmbi.1999.3463. [DOI] [PubMed] [Google Scholar]
- 149.Wang J-Y, Ling H, Yang W, Craigie R. Structure of a two-domain fragment of HIV-1 integrase: implications for domain organization in the intact protein. EMBO J. 2001;20:7333–7343. doi: 10.1093/emboj/20.24.7333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Engelman A, Cherepanov P. The lentiviral integrase binding protein LEDGF/p75 and HIV-1 replication. PLoS Pathog. 2008;4:e1000046. doi: 10.1371/journal.ppat.1000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Poeschla EM. Integrase, LEDGF/p75 and HIV replication. Cell. Mol. Life Sci. 2008;65:1403–1424. doi: 10.1007/s00018-008-7540-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Cherepanov P, Devroe E, Silver PA, Engelman A. Identification of an evolutionarily-conserved domain in LEDGF/p75 that binds HIV-1 integrase. J. Biol. Chem. 2004;279:48883–48892. doi: 10.1074/jbc.M406307200. [DOI] [PubMed] [Google Scholar]
- 153.Maertens G, Cherepanov P, Pluymers W, Busschots K, De Clercq E, Debyser Z, Engelborghs Y. LEDGF/p75 Is essential for nuclear and chromosomal targeting of HIV-1 integrase in human cells. J. Biol. Chem. 2003;278:33528–33539. doi: 10.1074/jbc.M303594200. [DOI] [PubMed] [Google Scholar]
- 154.Cherepanov P, Sun Z-YJ, Rahman S, Maertens G, Wagner G, Engelman A. Solution structure of the HIV-1 integrase-binding domain in LEDGF/p75. Nat. Struct. Mol. Biol. 2005;12:526–532. doi: 10.1038/nsmb937. [DOI] [PubMed] [Google Scholar]
- 155.Christ F, Voet A, Marchand A, Nicolet S, Desimmie BA, Marchand D, Bardiot D, Van der Veken NJ, Van Remoortel B, Strelkov SV, De Maeyer M, Chaltin P, Debyser Z. Rational design of small-molecule inhibitors of the LEDGF/p75-integrase interaction and HIV replication. Nat. Chem. Biol. 2010;6:442–448. doi: 10.1038/nchembio.370. [DOI] [PubMed] [Google Scholar]
- 156.Jurado KA, Engelman A. Multimodal mechanism of action of allosteric HIV-1 integrase inhibitors. Expert Rev. Mol. Med. 2013;15:e14. doi: 10.1017/erm.2013.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Wang H, Shun MC, Li X, Di Nunzio F, Hare S, Cherepanov P, Engelman A. Efficient transduction of LEDGF/p75 mutant cells by complementary gain-of-function HIV-1 integrase mutant viruses. Mol. Ther. Methods Clin. Dev. 2014;1:2. doi: 10.1038/mtm.2013.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Stevens SW, Griffith JD. Sequence analysis of the human DNA flanking sites of human immunodeficiency virus type 1 integration. J. Virol. 1996;70:6459–6462. doi: 10.1128/jvi.70.9.6459-6462.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Carteau S, Hoffmann C, Bushman F. Chromosome structure and human immunodeficiency virus type 1 cDNA integration: Centromeric alphoid repeats are a disfavored target. J. Virol. 1998;72:4005–4014. doi: 10.1128/jvi.72.5.4005-4014.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Holman AG, Coffin JM. Symmetrical base preferences surrounding HIV-1, avian sarcoma/leukosis virus, and murine leukemia virus integration sites. Proc. Natl. Acad. Sci. USA. 2005;102:6103–6107. doi: 10.1073/pnas.0501646102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Wu X, Li Y, Crise B, Burgess SM, Munroe DJ. Weak palindromic consensus sequences are a common feature found at the integration target sites of many retroviruses. J. Virol. 2005;79:5211–5214. doi: 10.1128/JVI.79.8.5211-5214.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Berry C, Hannenhalli S, Leipzig J, Bushman FD. Selection of target sites for mobile DNA integration in the human genome. PLoS Comput. Biol. 2006;2:e157. doi: 10.1371/journal.pcbi.0020157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Johnson RC, Stella S, Heiss JK. Bending and compaction of DNA by proteins. In: Rice PA, Correll CC, editors. Protein-Nucleic Acid Interactions: Structural Biology. RSC Publishing; London: 2008. pp. 176–220. [Google Scholar]
- 164.Liao G-c, Rehm EJ, Rubin GM. Insertion site preferences of the P transposable element in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA. 2000;97:3347–3351. doi: 10.1073/pnas.050017397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Haapa-Paananen S, Rita H, Savilahti H. DNA transposition of bacteriophage Mu: A quantitative analysis of target site selection in vitro. J. Biol. Chem. 2002;277:2843–2851. doi: 10.1074/jbc.M108044200. [DOI] [PubMed] [Google Scholar]
- 166.Serrao E, Krishnan L, Shun M-C, Li X, Cherepanov P, Engelman A, Maertens GN. Integrase residues that determine nucleotide preferences at sites of HIV-1 integration: implications for the mechanism of target DNA binding. Nucleic Acids Res. 2014;42:5146–5176. doi: 10.1093/nar/gku136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Nowak MG, Sudol M, Lee NE, Konsavage WMJ, Katzman M. Identifying amino acid residues that contribute to the cellular-DNA binding site on retroviral integrase. Virology. 2009;389:141–148. doi: 10.1016/j.virol.2009.04.014. [DOI] [PubMed] [Google Scholar]
- 168.Mizuuchi K. Polynucleotidyl transfer reactions in transpositional DNA recombination. J. Biol. Chem. 1992;267:21273–21276. [PubMed] [Google Scholar]
- 169.Yang W, Lee JY, Nowotny M. Making and breaking nucleic acids: two-Mg2+-ion catalysis and substrate specificity. Mol. Cell. 2006;22:5–13. doi: 10.1016/j.molcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 170.Rosta E, Nowotny M, Yang W, Hummer G. Catalytic mechanism of RNA backbone cleavage by ribonuclease H from quantum mechanics/molecular mechanics simulations. J. Am. Chem. Soc. 2011;133:8934–8941. doi: 10.1021/ja200173a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Nowotny M, Yang W. Stepwise analyses of metal ions in RNase H catalysis from substrate destabilization to product release. EMBO J. 2006;25:1924–1933. doi: 10.1038/sj.emboj.7601076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Nowotny M, Gaidamakov SA, Crouch RJ, Yang W. Crystal structures of RNase H bound to an RNA/DNA hybrid: substrate specificity and metal-dependent catalysis. Cell. 2005;121:1005–1016. doi: 10.1016/j.cell.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 173.Pryciak PM, Varmus HE. Nucleosomes, DNA-binding proteins, and DNA sequence modulate retroviral integration target site selection. Cell. 1992;69:769–780. doi: 10.1016/0092-8674(92)90289-o. [DOI] [PubMed] [Google Scholar]
- 174.Pryciak PM, Sil A, Varmus HE. Retroviral integration into minichromosomes in vitro. EMBO J. 1992;11:291–303. doi: 10.1002/j.1460-2075.1992.tb05052.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Pruss D, Bushman F, Wolffe A. Human immunodeficiency virus integrase directs integration to sites of severe DNA distortion within the nucleosome core. Proc. Natl. Acad. Sci. USA. 1994;91:5913–5917. doi: 10.1073/pnas.91.13.5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Pruss D, Reeves R, Bushman FD, Wolffe AP. The influence of DNA and nucleosome structure on integration events directed by HIV integrase. J. Biol. Chem. 1994;269:25031–25041. [PubMed] [Google Scholar]
- 177.Pryciak PM, Müller HP, Varmus HE. Simian virus 40 minichromosomes as targets for retroviral integration in vivo. Proc. Natl. Acad. Sci. USA. 1992;89:9237–9241. doi: 10.1073/pnas.89.19.9237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Wang GP, Ciuffi A, Leipzig J, Berry CC, Bushman FD. HIV integration site selection: Analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res. 2007;17:1186–1194. doi: 10.1101/gr.6286907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Roth SL, Malani N, Bushman FD. Gammaretroviral integration into nucleosomal target DNA in vivo. J. Virol. 2011;85:7393–7401. doi: 10.1128/JVI.00635-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Coffin JM, Hughes SH, Varmus HE. Retroviruses. Cold Spring Harbor Laboratory Press; Plainville, N.Y.: 1997. [PubMed] [Google Scholar]
- 181.Turlure F, Devroe E, Silver PA, Engelman A. Human cell proteins and human immunodeficiency virus DNA integration. Front. Biosci. 2004;9:3187–3208. doi: 10.2741/1472. [DOI] [PubMed] [Google Scholar]
- 182.Van Maele B, Busschots K, Vandekerckhove L, Christ F, Debyser Z. Cellular co-factors of HIV-1 integration. Trends Biochem. Sci. 2006;31:98–105. doi: 10.1016/j.tibs.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 183.Engelman A. Host cell factors and HIV-1 integration. Future HIV Ther. 2007;1:415–426. [Google Scholar]
- 184.Nishitsuji H, Hayashi T, Takahashi T, Miyano M, Kannagi M, Masuda T. Augmentation of reverse transcription by integrase through an interaction with host factor, SIP1/Gemin2 Is critical for HIV-1 infection. PLoS One. 2009;4:e7825. doi: 10.1371/journal.pone.0007825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Gallay P, Hope T, Chin D, Trono D. HIV-1 infection of nondividing cells through the recognition of integrase by the importin/karyopherin pathway. Proc. Natl. Acad. Sci. USA. 1997;94:9825–9830. doi: 10.1073/pnas.94.18.9825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Fassati A, Gorlich D, Harrison I, Zaytseva L, Mingot JM. Nuclear import of HIV-1 intracellular reverse transcription complexes is mediated by importin 7. EMBO J. 2003;22:3675–3685. doi: 10.1093/emboj/cdg357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Ao Z, Danappa Jayappa K, Wang B, Zheng Y, Kung S, Rassart E, Depping R, Kohler M, Cohen EA, Yao X. Importin alpha3 interacts with HIV-1 integrase and contributes to HIV-1 nuclear import and replication. J. Virol. 2010;84:8650–8663. doi: 10.1128/JVI.00508-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Ao Z, Huang G, Yao H, Xu Z, Labine M, Cochrane AW, Yao X. Interaction of human immunodeficiency virus type 1 integrase with cellular nuclear import receptor importin 7 and its impact on viral replication. J. Biol. Chem. 2007;282:13456–13467. doi: 10.1074/jbc.M610546200. [DOI] [PubMed] [Google Scholar]
- 189.Christ F, Thys W, De Rijck J, Gijsbers R, Albanese A, Arosio D, Emiliani S, Rain JC, Benarous R, Cereseto A, Debyser Z. Transportin-SR2 imports HIV into the nucleus. Curr. Biol. 2008;18:1192–1202. doi: 10.1016/j.cub.2008.07.079. [DOI] [PubMed] [Google Scholar]
- 190.Krishnan L, Matreyek KA, Oztop I, Lee K, Tipper CH, Li X, Dar MJ, Kewalramani VN, Engelman A. The requirement for cellular transportin 3 (TNPO3 or TRN-SR2) during infection maps to human immunodeficiency virus type 1 capsid and not integrase. J. Virol. 2010;84:397–406. doi: 10.1128/JVI.01899-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Maertens GN, Cook NJ, Wang W, Hare S, Gupta SS, Öztop I, Lee K, Pye VE, Cosnefroy O, Snijders AP, KewalRamani VN, Fassati A, Engelman A, Cherepanov P. Structural basis for nuclear import of splicing factors by human Transportin 3. Proc. Natl. Acad. Sci. USA. 2014;111:2728–2733. doi: 10.1073/pnas.1320755111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Quashie PK, Mesplède T, Han Y-S, Oliveira M, Singhroy DN, Fujiwara T, Underwood MR, Wainberg MA. Characterization of the R263K mutation in HIV-1 integrase that confers low-level resistance to the second-generation integrase strand transfer inhibitor dolutegravir. J. Virol. 2012;86:2696–2705. doi: 10.1128/JVI.06591-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Johnson BC, Métifiot M, Ferris A, Pommier Y, Hughes SH. A homology model of HIV-1 integrase and analysis of mutations designed to test the model. J. Mol. Biol. 2013;425:2133–2146. doi: 10.1016/j.jmb.2013.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.LaFemina R, Schneider C, Robbins H, Callahan P, LeGrow K, Roth E, Schleif W, Emini E. Requirement of active human immunodeficiency virus type 1 integrase enzyme for productive infection of human T-lymphoid cells. J Virol. 1992;66:7414–7419. doi: 10.1128/jvi.66.12.7414-7419.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Sakai H, Kawamura M, Sakuragi J, Sakuragi S, Shibata R, Ishimoto A, Ono N, Ueda S, Adachi A. Integration is essential for efficient gene expression of human immunodeficiency virus type 1. J. Virol. 1993;67:1169–1174. doi: 10.1128/jvi.67.3.1169-1174.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Wolfe AL, Felock PJ, Hastings JC, Blau CU, Hazuda DJ. The role of manganese in promoting multimerization and assembly of human immunodeficiency virus type 1 integrase as a catalytically active complex on immobilized long terminal repeat substrates. J. Virol. 1996;70:1424–1432. doi: 10.1128/jvi.70.3.1424-1432.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Hazuda D, Felock PJ, Hastings JC, Pramanik B, Wolfe AL. Discovery and analysis of inhibitors of the human immunodeficiency integrase. Drug Des. Discov. 1997;15:17–24. [PubMed] [Google Scholar]
- 198.Pierson TC, Zhou Y, Kieffer TL, Ruff CT, Buck C, Siliciano RF. Molecular characterization of preintegration latency in human immunodeficiency virus type 1 infection. J. Virol. 2002;76:8518–8531. doi: 10.1128/JVI.76.17.8518-8531.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Hazuda DJ, Felock P, Witmer M, Wolfe A, Stillmock K, Grobler JA, Espeseth A, Gabryelski L, Schleif W, Blau C, Miller MD. Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science. 2000;287:646–650. doi: 10.1126/science.287.5453.646. [DOI] [PubMed] [Google Scholar]
- 200.Espeseth AS, Felock P, Wolfe A, Witmer M, Grobler J, Anthony N, Egbertson M, Melamed JY, Young S, Hamill T, Cole JL, Hazuda DJ. HIV-1 integrase inhibitors that compete with the target DNA substrate define a unique strand transfer conformation for integrase. Proc. Natl. Acad. Sci. USA. 2000;97:11244–11249. doi: 10.1073/pnas.200139397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Grobler JA, Stillmock K, Hu B, Witmer M, Felock P, Espeseth AS, Wolfe A, Egbertson M, Bourgeois M, Melamed J, Wai JS, Young S, Vacca J, Hazuda DJ. Diketo acid inhibitor mechanism and HIV-1 integrase: Implications for metal binding in the active site of phosphotransferase enzymes. Proc. Natl. Acad. Sci. USA. 2002;99:6661–6666. doi: 10.1073/pnas.092056199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Summa V, Petrocchi A, Bonelli F, Crescenzi B, Donghi M, Ferrara M, Fiore F, Gardelli C, Gonzalez Paz O, Hazuda DJ, Jones P, Kinzel O, Laufer R, Monteagudo E, Muraglia E, Nizi E, Orvieto F, Pace P, Pescatore G, Scarpelli R, Stillmock K, Witmer MV, Rowley M. Discovery of raltegravir, a potent, selective orally bioavailable HIV-integrase inhibitor for the treatment of HIV-AIDS infection. J. Med. Chem. 2008;51:5843–5855. doi: 10.1021/jm800245z. [DOI] [PubMed] [Google Scholar]
- 203.Sato M, Motomura T, Aramaki H, Matsuda T, Yamashita M, Ito Y, Kawakami H, Matsuzaki Y, Watanabe W, Yamataka K, Ikeda S, Kodama E, Matsuoka M, Shinkai H. Novel HIV-1 integrase inhibitors derived from quinolone antibiotics. J. Med. Chem. 2006;49:1506–1508. doi: 10.1021/jm0600139. [DOI] [PubMed] [Google Scholar]
- 204.Johns BA, Kawasuji T, Weatherhead JG, Taishi T, Temelkoff DP, Yoshida H, Akiyama T, Taoda Y, Murai H, Kiyama R, Fuji M, Tanimoto N, Jeffrey J, Foster SA, Yoshinaga T, Seki T, Kobayashi M, Sato A, Johnson MN, Garvey EP, Fujiwara T. Carbamoyl pyridone HIV-1 integrase inhibitors 3. A diastereomeric approach to chiral nonracemic tricyclic ring systems and the discovery of dolutegravir (S/GSK1349572) and (S/GSK1265744) J. Med. Chem. 2013;56:5901–5916. doi: 10.1021/jm400645w. [DOI] [PubMed] [Google Scholar]
- 205.Shimura K, Kodama E, Sakagami Y, Matsuzaki Y, Watanabe W, Yamataka K, Watanabe Y, Ohata Y, Doi S, Sato M, Kano M, Ikeda S, Matsuoka M. Broad antiretroviral activity and resistance profile of the novel human immunodeficiency virus integrase inhibitor elvitegravir (JTK-303/GS-9137) J. Virol. 2008;82:764–774. doi: 10.1128/JVI.01534-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Koh Y, Matreyek KA, Engelman A. Differential sensitivities of retroviruses to integrase strand transfer inhibitors. J. Virol. 2011;85:3677–3682. doi: 10.1128/JVI.02541-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Hare S, Vos AM, Clayton RF, Thuring JW, Cummings MD, Cherepanov P. Molecular mechanisms of retroviral integrase inhibition and the evolution of viral resistance. Proc. Natl. Acad. Sci. USA. 2010;107:20057–20062. doi: 10.1073/pnas.1010246107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Hare S, Smith SJ, Métifiot M, Jaxa-Chamiec A, Pommier Y, Hughes SH, Cherepanov P. Structural and functional analyses of the second-generation integrase strand transfer inhibitor dolutegravir (S/GSK1349572) Mol. Pharmacol. 2011;80:565–572. doi: 10.1124/mol.111.073189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Cooper DA, Steigbigel RT, Gatell JM, Rockstroh JK, Katlama C, Yeni P, Lazzarin A, Clotet B, Kumar PN, Eron JE, Schechter M, Markowitz M, Loutfy MR, Lennox JL, Zhao J, Chen J, Ryan DM, Rhodes RR, Killar JA, Gilde LR, Strohmaier KM, Meibohm AR, Miller MD, Hazuda DJ, Nessly ML, DiNubile MJ, Isaacs RD, Teppler H, Nguyen B-Y, the BENCHMRK Study Teams Subgroup and resistance analyses of raltegravir for resistant HIV-1 infection. N. Engl. J. Med. 2008;359:355–365. doi: 10.1056/NEJMoa0708978. [DOI] [PubMed] [Google Scholar]
- 210.Grobler JA, McKenna PM, Ly S, Stillmock KA, Bahnck CM, Danovich RM, Dornadula G, Hazuda DJ, Miller MD. HIV integrase inhibitor dissociation rates correlate with efficacy in vitro. Antiviral Ther. 2009;14(Suppl 1):A27. [Google Scholar]
- 211.Hightower KE, Wang R, Deanda F, Johns BA, Weaver K, Shen Y, Tomberlin GH, Carter HLr, Broderick T, Sigethy S, Seki T, Kobayashi M, Underwood MR. Dolutegravir (S/GSK1349572) exhibits significantly slower dissociation than raltegravir and elvitegravir from wild-type and integrase inhibitor-resistant HIV-1 integrase-DNA complexes. Antimicrob. Agents Chemother. 2011;55:4552–4559. doi: 10.1128/AAC.00157-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Goethals O, Vos A, Van Ginderen M, Geluykens P, Smits V, Schols D, Hertogs K, Clayton R. Primary mutations selected in vitro with raltegravir confer large fold changes in susceptibility to first-generation integrase inhibitors, but minor fold changes to inhibitors with second-generation resistance profiles. Virology. 2010;402:338–346. doi: 10.1016/j.virol.2010.03.034. [DOI] [PubMed] [Google Scholar]
- 213.Canducci F, Ceresola ER, Boeri E, Spagnuolo V, Cossarini F, Castagna A, Lazzarin A, Clementi M. Cross-resistance profile of the novel integrase inhibitor dolutegravir (S/GSK1349572) using clonal viral variants selected in patients failing raltegravir. J. Infect. Dis. 2011;204:1811–1815. doi: 10.1093/infdis/jir636. [DOI] [PubMed] [Google Scholar]