Abstract
Background
Advances in Aeromonas taxonomy have led to the reclassification of aeromonads. Hereon, we aimed to re-evaluate the characteristics of Aeromonas bacteremia, including those of a novel species, Aeromonas dhakensis.
Methodology/Principal Findings
A retrospective study of monomicrobial Aeromonas bacteremia at a medical center in southern Taiwan from 2004–2011 was conducted. Species identification was based on rpoB sequencing. Of bacteremia of 153 eligible patients, A. veronii (50 isolates, 32.7%), A. dhakensis (48, 31.4%), A. caviae (43, 28.1%), and A. hydrophila (10, 6.5%) were the principal causative species. A. dhakensis and A. veronii bacteremia were mainly community-acquired and presented as primary bacteremia, spontaneous bacterial peritonitis, or skin and soft-tissue infection, whereas A. caviae was associated with hospital-onset bacteremia. The distribution of the AmpC β-lactamase and metallo-β-lactamase genes was species-specific: bla AQU-1, bla MOX, or bla CepH was present in A. dhakensis, A. caviae, or A. hydrophila, respectively, and bla CphA was present in A. veronii, A. dhakensis, and A. hydrophila. The cefotaxime resistance rates of the A. caviae, A. dhakensis, and A. hydrophila isolates were higher than that of A. veronii (39.5%%, 25.0%, and 30% vs. 2%, respectively). A. dhakensis bacteremia was linked to the highest 14-day sepsis-related mortality rate, followed by A. hydrophila, A. veronii, and A. caviae bacteremia (25.5%, 22.2%, 14.0%, and 4.7%, respectively; P = 0.048). Multivariate analysis revealed that A. dhakensis bacteremia, active malignancies, and a Pitt bacteremia score ≥ 4 was an independent mortality risk factor.
Conclusions/Significance
Characteristics of Aeromonas bacteremia vary between species. A. dhakensis prevalence and its associated poor outcomes suggest it an important human pathogen.
Introduction
Aeromonas species are aquatic gram-negative bacilli that are ubiquitously distributed in natural environments and implicated in a variety of human diseases [1]. Previous studies have indicated that three Aeromonas species, Aeromonas hydrophila, A. caviae, and A. veronii bv. sobria, accounted for > 95% of all Aeromonas blood-borne infections, and liver cirrhosis and malignancies are two well-known predisposing conditions associated with Aeromonas bacteremia [1,2]. Continuing advances in the field of Aeromonas taxonomy have led to the reclassification of aeromonads. Recent phylogenetic analyses indicated that A. aquariorum, a species first described in 2008, and A. hydrophila subsp. dhakensis are both incapable of fermenting arabinose and belong to the same taxon [3,4]. Therefore, a formal reclassification of both species as A. dhakensis sp. nov. comb nov. was proposed [3]. Although human A. dhakensis infections have been reported, their clinically relevant characteristics have not been thoroughly established [5–8]. A. dhakensis can be clearly differentiated from A. hydrophila by its gyrB, rpoB, or rpoD gene sequences, its inability to ferment arabinose, and the cluster analysis of spectra generated by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF) [6,7,9,10]. Our earlier work also showed that A. dhakensis exhibits higher pathogenicity than A. hydrophila, which justifies the need to further differentiate A. dhakensis from A. hydrophila [11,12].
To date, the clinically relevant characteristics of A. dhakensis bacteremia have not been well described. Due to the changing taxonomy, we re-evaluated these characteristics and antimicrobial resistance profiles of causative isolates associated with monomicrobial Aeromonas bacteremia, with consideration of the novel species A. dhakensis.
Methods
Patients and definition
A retrospective study of adults (age ≥ 18 years) with monomicrobial Aeromonas bacteremia at the National Cheng Kung University Hospital, a medical center in southern Taiwan, was conducted between 2004 and 2011 and was approved by the Institutional Review Board (B-ER-101-031) of the study hospital. Clinical information was retrieved from medical records. The patient information were anonymized and de-identified prior to analysis, and therefore the requirement for informed consent was waived by Institution Review Board.
Monomicrobial Aeromonas bacteremia was defined as the presence of an Aeromonas species in at least one blood culture from a patient with symptoms or signs of infection. Patients in which different species or multiple Aeromonas species were isolated from the blood were excluded. If a patient experienced more than one episode of Aeromonas bacteremia due to genetically related Aeromonas strains, all instances were counted as one episode. Community-onset infections were defined as those with the first positive blood culture collected within 48 hours after admission; the remaining infections were defined as hospital-onset infections. Those without apparent infection sites were defined as primary bacteremia. The severity of any underlying medical illness was determined as fatal, ultimately fatal, or nonfatal, according to the McCabe score [13]. The severity of the bacteremia on the day of onset was graded by the Pittsburgh (Pitt) bacteremia score, and critical illness was defined as a score of at least 4 points [14]. Steroid use was defined as the receipt of corticosteroid treatment (10 mg or an equivalent daily dosage) for more than 2 weeks. Recent antineoplastic chemotherapy or antimicrobial therapy was defined as the receipt of cancer chemotherapy or an oral or parenteral antimicrobial agent for > 48 hours within 2 weeks of the onset of bacteremia. Antimicrobial regimens given before the susceptibility results became available were defined as empirical therapy, whereas those subsequently adjusted accordingly were defined as definitive therapy. Appropriate drugs were those with demonstrable in vitro activity against the causative isolates. Breakthrough bacteremia was defined as a bacteremic episode occurring at least 48 hours after initiating antimicrobial therapy. Sepsis-related mortality was the death of a patient with a clinical course suggestive of persistently active infection without obvious other explanations for death.
Aeromonas species identification
Aeromonas blood isolates were stored at -70°C until use. The Aeromonas isolates were identified by a positive oxidase test, D-glucose fermentation, motility, the absence of growth in 6.5% sodium chloride, and resistance to the vibriostatic agent O/129 (150 μg), and by using the Vitek GNI Plus system (BioMérieux Marcy-l’Etoile, France). Species identification of each Aeromonas isolates was determined based on sequence analysis of partial rpoB with the PCR primers Pasrpob-L and Rpob-R and additional rpoD for A. dhakensis with the primers 70F and 70R [15,16]. The amplified sequences were compared with reference sequences from the GenBank database using a BLAST (http://www.ncbi.nlm.nih.gov/BLAST/) search of homologous sequences. Based on sequence analyses, the isolates with a dissimilarity value ≤ 2.0% for a given strain were identified as that strain [1]. L-arabinose fermentation was utilized in the species differentiation between A. dhakensis and A. hydrophila [3,7]. Genetic relatedness of isolates was examined by arbitrarily primed PCR with the primers ERIC-1R and ERIC-2R [17].
Antimicrobial drug susceptibility testing
The minimum inhibitory concentrations (MICs) of antimicrobial agents for Aeromonas isolates were determined by the broth microdilution method using the Trek Sensititre system (Trek Diagnostics, West Essex, England), and the results were interpreted following the Clinical and Laboratory Standards Institute (CLSI) recommendations for A. hydrophila complex [18]. The criteria for doxycycline and piperacillin susceptibility followed the CLSI recommendation for Enterobacteriaceae [19]. In isolates not susceptible to cefotaxime, ceftriaxone, or ceftazidime, the extended-spectrum β-lactamase (ESBL)-producing phenotypes were examined as previously described [20]. Resistance to broad-spectrum cephalosporins was defined as resistance to at least one third-generation cephalosporin, i.e., cefotaxime, ceftriaxone, or ceftazidime.
Detection of the AmpC and metallo-β-lactamase (MBL) genes
The study isolates were screened for the genes encoding AmpC β-lactamases (AQU-1, MOX-like, and CepH β-lactamases) and CphA (MBL) by a colony hybridization assay with digoxigenin-labeled probes (DIG DNA Labeling kit and Nucleic Acid Detection Kit, Roche, Germany). The probe for detection of bla AQU-1 consisted of bla AQU-1 amplified from A. dhakensis AAK1with the primers AQU-2F and AQU-2R [21]. The probe for bla MOX-like genes consisted of a gene sequence 98.0% identical to A. caviae bla MOX-6 (GenBank accession no. GQ152601) amplified from a blood isolate with the primers AcMOX-F and AcMOX-R. The probe for bla CepH-like genes consisted of a gene sequence 95.5% identical to A. hydrophila bla CepH (GenBank accession no. CP000462) amplified from a blood isolate with the primers CepH-F11 and CepH-R1129. The probe for bla CphA was previously described [22]. The DNA sequences of the primers used in this study are summarized in Table 1.
Table 1. PCR primers used in this study.
| Target gene | Primer | Primer sequence (5’–3’) | Reference or sources |
|---|---|---|---|
| rpoB | Pasrpob-L | F (5’-GCAGTGAAAGARTTCTTTGGTTC-3’) | [15] |
| Rpob-R | R (5’-GTTGCATGTTNGNACCCAT-3’) | ||
| rpoD | 70F | F (5’-ACGACTGACCCGGTACGCATGTAYATGMGNGARATGGGNACNGT-3’) | [16] |
| 70R | R (5’-ATAGAAATAACCAGACGTAAGTTNGCYTCNACCATYTCYTTYTT-3’) | ||
| AP-PCR a | ERIC-1R | F (5’-ATGTAAGCTCCTGGGGATTCAC-3’) | [17] |
| ERIC-2R | R (5’-AGTAAGTGACTGGGGTGAGCG-3’) | ||
| aqu-1 | AQU-2F | F (5’-GCTATGCTGGCGGCTTTCCAAC-3’) | [21] |
| AQU-2R | R (5’-TCAGGG AGCCAGCTTGCTCAG-3’) | ||
| bla MOX-like gene | AcMOX-F | F (5’-ATGCAACAACGACAATCCATCC-3’) | This study |
| AcMOX-R | R (5’-TTACCTGGCCAGTTGCGTCAG-3’) | ||
| bla CepH-like gene | CepH-F11 | F (5’-CCAGAKCCCTGCCACTGCTGGC-3’) | This study |
| CepH-R1129 | R (5’-AAATGGCATGGGCCGCGCTG-3’) | ||
| cphA | ANY-SSD/F | F (5’-GCTTAGAGCTCCTAAGGAGCAAGATGAAAGGTTGG-3’) | [22] |
| R (5’-GCATAGGTACCTTATGACTGGGGTGCGGCCTTG-3’) |
a Arbitrarily primed PCR.
Statistical analysis
All analyses were performed with the Statistical Package for the Social Sciences version 17.0 (SPSS, Chicago, IL, USA). Continuous variables are expressed as mean values ± standard deviation and were compared by the Analysis of Variance test. Categorical variables were compared by the Fisher exact test or chi-square test. All biologically plausible variables with a P value of ≤ 0.20 in the univariate analysis were considered for inclusion in the logistic regression model for multivariate analysis. The time to mortality among patients with bacteremia due to four Aeromonas species was analyzed using Kaplan–Meier survival analysis and the log-rank test. A P value < 0.05 was considered to be significant, and all tests were 2-tailed.
Results
There were 160 episodes of monomicrobial Aeromonas bacteremia between 2004 and 2011. Seven cases were excluded due to a lack of available causative isolates. Seven patients experienced two bacteremic episodes due to distinct Aeromonas isolates confirmed by AP-PCR (repeated bacteremia). Overall, 153 monomicrobial bacteremic episodes from 146 patients were included. For convenience, an episode was counted as one case.
Of 153 blood isolates, A. veronii (50 isolates, 32.7%), A. dhakensis (48, 31.4%), and A. caviae (43, 28.1%) were the three principal species involved in bacteremia according to rpoB sequencing, followed by A. hydrophila (10, 6.5%) and Aeromonas spp. (2, 1.3%). All 48 A. dhakensis isolates, also confirmed by rpoD sequencing, exhibited the inability to ferment arabinose, whereas all A. hydrophila isolates were able to ferment arabinose. Some A. veronii, A. dhakensis, A. caviae, and A. hydrophila isolates were first identified as A. veronii bv. sobria (41, 82%), A. hydrophila (46, 95.8%), A. caviae (41, 95.3%), and A. hydrophila (10, 100%), respectively, by the Vitek system. Detailed sequence information is described in the supplementary material (S1 Data).
Demographic data and clinical characteristics of the patients are shown in Table 2. Liver cirrhosis (43.8%), especially for A. veronii, A. dhakensis, and A. hydrophila bacteremia, and malignancies (42.5%) were two common underlying diseases. Of A. dhakensis bacteremia, 70.8% of the episodes were community-acquired infections, and primary bacteremia, spontaneous bacterial peritonitis (SBP), biliary tract infection (BTI), and skin and soft-tissue infection (SSTI) were the major presentations. Of 7 patients with repeated bacteremia (6 patients with liver cirrhosis and one with leukemia), 13 episodes were community-acquired, and 7 and 5 of these episodes were caused by A. dhakensis and A. veronii, respectively. A. caviae bacteremia was usually associated with hospital-onset infections, especially vascular catheter-related bacteremia, and less critical illness at the onset of bacteremia. Four patients with community-acquired bacteremia recalled contact histories: flame burn injury and frostbite (2 patients, SSTIs), ingestion of contaminated food (1, gastroenteritis) [23] and unboiled water (1, empyema). No clustering of hospital-onset infections was noted.
Table 2. Demographic data, underlying conditions, clinical presentations, and treatment outcomes of 153 patients with monomicrobial Aeromonas bacteremia from 2004 to 2011.
| Characteristic | Case no. (%) | P value | ||||
|---|---|---|---|---|---|---|
| All n = 153 | A. veronii n = 50 | A. dhakensis n = 48 | A. caviae n = 43 | A. hydrophila n = 10 | ||
| Age, mean ± standard deviation (years) | 59.8 ± 14.7 | 62.9 ± 14.3 | 55.7 ± 16.0 | 61.3 ± 12.5 | 57.7 ± 16.3 | 0.085 |
| Gender, male | 91 (59.5) | 28 (56) | 27 (56.3) | 26 (60.5) | 8 (80) | 0.533 |
| Hospital-onset bacteremia | 63 (41.2) | 10 (20) | 14 (29.2) | 33 (76.7) | 6 (60) | < 0.001 |
| Co-morbidity | ||||||
| Liver cirrhosis | 67 (43.8) | 24 (48) | 30 (62.5) | 7 (16.3) | 5 (50) | < 0.001 |
| Malignancies | 65 (42.5) | 25 (50) | 14 (29.2) | 19 (44.2) | 7 (70) | 0.052 |
| Leukemia/lymphoma/myeloma | 17 (11.1) | 11 (22) | 2 (4.2) | 2 (4.7) | 2 (20) | 0.013 |
| Myelodysplasia/aplastic anemia | 6 (3.9) | 3 (6) | 2 (4.2) | 1 (2.3) | 0 (0) | 0.738 |
| Hepatocellular carcinoma | 29 (19.0) | 11 (22) | 8 (16.7) | 6 (14.0) | 4 (40) | 0.261 |
| Pancreatobiliary cancer | 7 (4.6) | 1 (2) | 0 (0) | 6 (14.0) | 0 (0) | 0.007 |
| Solid cancer, other site | 13 (8.5) | 2 (4) | 4 (8.3) | 6 (14.0) | 1 (10) | 0.401 |
| Diabetes mellitus | 36 (23.5) | 11 (22) | 12 (25.0) | 10 (23.3) | 2 (20) | 0.980 |
| Obstructive biliary disease (stone or stricture) | 12 (7.8) | 3 (6) | 4 (8.3) | 3 (7.0) | 0 (0) | 0.807 |
| Steroid use | 9 (5.9) | 1 (2) | 1 (2.1) | 6 (14.0) | 1 (10) | 0.049 |
| Renal failure on dialysis | 5 (3.3) | 2 (4) | 3 (6.3) | 0 (0) | 0 (0) | 0.364 |
| Rapidly fatal underlying disease (McCabe classification) | 9 (5.9) | 8 (16) | 0 (0) | 0 (0) | 1 (10) | 0.002 |
| Previous procedures or conditions within 2 weeks of bacteremia onset | ||||||
| Prior antimicrobial therapy | 35 (22.9) | 8 (16) | 16 (33.3) | 9 (20.9) | 2 (20) | 0.221 |
| Endoscopic examination | 14 (9.2) | 4 (8) | 7 (14.6) | 2 (4.7) | 1 (10) | 0.423 |
| Surgery | 13 (8.5) | 4 (8) | 4 (8.3) | 5 (11.6) | 0 (0) | 0.690 |
| Neutropenia | 11 (7.2) | 8 (16) | 1 (2.1) | 0 (0) | 2 (20) | < 0.001 |
| Port-A catheter | 11 (7.2) | 2 (4) | 1 (2.1) | 7 (16.3) | 1 (10) | 0.046 |
| Indwelling central venous catheter other than Port-A | 6 (3.9) | 2 (4) | 3 (6.3) | 1 (2.3) | 0 (0) | 0.712 |
| Antineoplastic chemotherapy | 6 (3.9) | 3 (6) | 1 (2.1) | 2 (4.7) | 0 (0) | 0.693 |
| Intensive care unit care | 4 (2.6) | 1 (2) | 3 (6.3) | 0 (0) | 0 (0) | 0.268 |
| Sources of bacteremia | ||||||
| Primary bacteremia, non-neutropenic | 89 (58.2) | 28 (56) | 23 (47.9) | 31 (72.1) | 6 (60) | 0.132 |
| Primary bacteremia, neutropenic | 11 (7.2) | 8 (16) | 1 (2.1) | 0 (0) | 2 (20) | < 0.001 |
| Biliary tract infection | 15 (9.8) | 2 (4) | 5 (10.4) | 7 (16.3) | 0 (0) | 0.154 |
| Spontaneous bacterial peritonitis | 12 (7.8) | 3 (6) | 8 (16.7) | 0 (0) | 1 (10) | 0.029 |
| Skin and soft tissue infection | 10 (6.5) | 5 (10) | 5 (10.4) | 0 (0) | 0 (0) | 0.122 |
| Vascular-catheter related infection | 5 (3.3) | 0 (0) | 0 (0) | 4 (9.3) | 1 (10) | 0.023 |
| Enteritis | 3 (2.0) | 2 (4) | 2 (4.2) | 0 (0) | 0 (0) | 0.523 |
| Intra-abdominal infection | 3 (2.0) | 1 (2) | 2 (4.2) | 1 (2.3) | 0 (0) | 0.491 |
| Spontaneous bacterial empyema | 3 (2.0) | 2 (4) | 1 (2.1) | 0 (0) | 0 (0) | 0.548 |
| Pneumonia | 2 (1.3) | 1 (2) | 1 (2.1) | 0 (0) | 0 (0) | 0.778 |
| Pitt bacteremia score ≥ 4 | 31 (20.3) | 12 (24) | 13 (27.1) | 3 (7.0) | 3 (30) | 0.071 |
| Appropriate empirical antibiotics | 123 (80.4) | 48 (96) | 34 (70.8) | 31 (72.1) | 9 (90) | 0.004 |
| Appropriate definitive antibiotics | 121/134 (90.3) | 42/45 (93.3) | 35/38 (92.1) | 35/41 (85.4) | 8/8 (100) | 0.439 |
| Mortality rate | ||||||
| 14-day sepsis-related | 23/151 (15.2) | 7 (14) | 12/47 (25.5) | 2 (4.7) | 2/9 (22.2) | 0.048 |
| Crude in-hospital | 36/150 (24.0) | 12/49 (24.5) | 16/47 (34.0) | 6 (14.0) | 2/9 (22.2) | 0.176 |
Broad-spectrum cephalosporin resistance rates of A. caviae, A. dhakensis, and A. hydrophila were higher than that of A. veronii (39.5%, 29.2%, and 30% vs. 2%, respectively; P ≤ 0.001) (Table 3). A multivariate analysis revealed that preceding β-lactam therapy (≥ 48 hours) within 2 days before bacteremia onset was associated with broad-spectrum cephalosporin resistance (odds ratio [OR] 3.7; 95% confident interval [CI], 1.2–11.2; P = 0.022). One imipenem-resistant A. dhakensis isolate was isolated from a patient during ertapenem treatment for ESBL-producing E. coli BTI, as described previously [22]. Overall, A. veronii was susceptible to most drugs, whereas A. caviae was more likely to be resistant to the drugs tested.
Table 3. Distribution of β-lactamase genes detected by colony hybridization and antimicrobial resistance profiles of 153 Aeromonas blood isolates, 2004–2011.
| Isolate no. (%) | P values | |||||
|---|---|---|---|---|---|---|
| All isolatesn = 153 | A. veronii n = 50 | A. dhakensis n = 48 | A. caviae n = 43 | A. hydrophila n = 10 | ||
| Positive hybridization | ||||||
| Ambler class C β-lactamase | ||||||
| bla AQU-1 | 48 (31.4) | 0 (0) | 48 (100) | 0 (0) | 0 (0) | < 0.001 |
| bla MOX | 45 (29.4) | 0 (0) | 0 (0) | 43 (100) | 2 (20) | < 0.001 |
| bla CepH | 12 (7.8) | 0 (0) | 0 (0) | 2 (4.7) | 10 (100) | < 0.001 |
| Ambler class B β-lactamase | ||||||
| bla CphA | 109 (71.2) | 50 (100) | 48 (100) | 0 (0) | 10 (100) | < 0.001 |
| Antimicrobial resistance | ||||||
| Cefazolin | 149 (97.4) | 46 (92) | 48 (100) | 43 (100) | 10 (100) | 0.040 |
| Cefuroxime | 30 (19.6) | 1 (2) | 11 (22.9) | 15 (34.9) | 3 (30) | 0.001 |
| Cefoxitin | 76 (49.7) | 5 (10) | 44 (91.7) | 25 (58.1) | 2 (20) | < 0.001 |
| Cefotaxime | 33 (21.6) | 1 (2) | 12 (25.0) | 17 (39.5) | 3 (30) | <0.001 |
| Ceftazidime | 16 (10.5) | 1 (2) | 6 (12.5) | 7 (16.3) | 2 (20) | 0.090 |
| Ceftriaxone | 35 (22.9) | 1 (2) | 14 (29.2) | 17 (39.5) | 3 (30) | < 0.001 |
| Broad-spectrum cephalosporin a | 35 (22.9) | 1 (2) | 14 (29.2) | 17 (39.5) | 3 (30) | < 0.001 |
| Cefepime | 1 (0.7) | 0 (0) | 0 (0) | 1 (2.3) | 0 (0) | 0.470 |
| Aztreonam | 4 (2.6) | 1 (2) | 0 (0) | 3 (7.0) | 0 (0) | 0.188 |
| Ampicillin/sulbactam | 151 (98.7) | 50 (100) | 48 (100) | 42 (97.7) | 9 (90) | 0.060 |
| Piperacillin | 20 (13.1) | 1 (2) | 11 (22.9) | 8 (18.6) | 0 (0) | 0.007 |
| Piperacillin/tazobactam | 17 (11.1) | 2 (4) | 10 (20.8) | 5 (11.6) | 0 (0) | 0.040 |
| Imipenem | 2 (1.3) | 0 (0) | 2 (4.2) | 0 (0) | 0 (0) | 0.226 |
| Meropenem | 2 (1.3) | 0 (0) | 2 (4.2) | 0 (0) | 0 (0) | 0.226 |
| Doxycycline | 2 (1.3) | 0 (0) | 0 (0) | 2 (4.7) | 0 (0) | 0.165 |
| Gentamicin | 4 (2.6) | 0 (0) | 0 (0) | 3 (7.0) | 1 (10) | 0.048 |
| Amikacin | 1 (0.7) | 0 (0) | 0 (0) | 1 (2.3) | 0 (0) | 0.470 |
| Ciprofloxacin | 2 (1.3) | 0 (0) | 0 (0) | 2 (4.7) | 0 (0) | 0.165 |
| Levofloxacin | 1 (0.7) | 0 (0) | 0 (0) | 1 (2.3) | 0 (0) | 0.470 |
| Co-trimethoxazole | 24 (15.7) | 4 (8) | 7 (14.6) | 12 (27.9) | 0 (0) | 0.027 |
| ESBL phenotype b | 5 (3.3) | 0 (0) | 0 (0) | 4 (9.3) | 1 (10) | 0.023 |
a Resistance to at least one third-generation cephalosporin, i.e., cefotaxime, ceftriaxone, or ceftazidime.
b ESBL = extended-spectrum β-lactamase.
A colony hybridization assay revealed that bla AQU-1 was constantly present in A. dhakensis isolates but not in other species, whereas the bla MOX-like gene was present in all A. caviae isolates, and the bla CepH-like gene was present in all A. hydrophila isolates. The MBL bla CphA-like gene was present in all A. dhakensis, A. veronii, and A. hydrophila isolates but not in A. caviae isolates (Table 3).
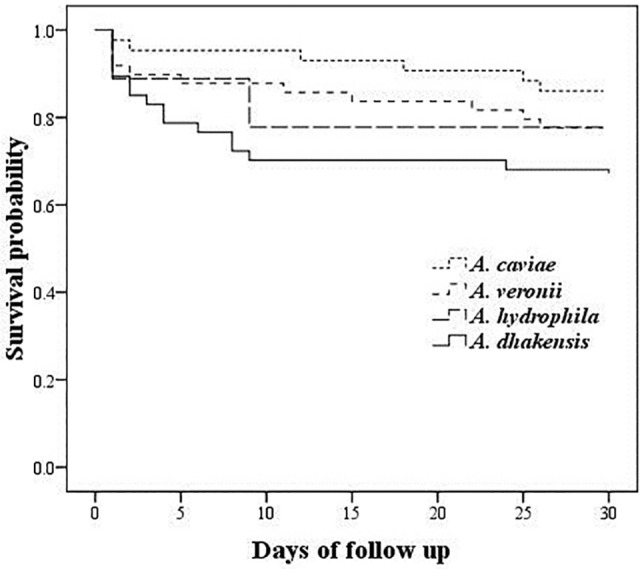
The 14-day and in-hospital clinical outcomes were assessed in 151 and 150 patients, respectively. The detailed antimicrobial treatments for these patients are provided in the supplementary material (S1 Data). Patients with A. dhakensis bacteremia had the highest 14-day sepsis-related mortality rate, followed by bacteremia due to A. hydrophila, A. veronii, or A. caviae (25.5%, 22.2%, 14.0%, and 4.7%, respectively; P = 0.048). Inappropriate empirical therapy or definite treatment was not associated with 14-day sepsis-related mortality. A multivariate analysis revealed that a Pitt bacteremia score ≥ 4 (OR 44.9; 95% CI 11.0–184.2; P < 0.001), A. dhakensis (OR 8.5; 95% CI 1.9–37.3; P = 0.005), and active malignancies (OR 9.1; 95% CI 1.9–43.7; P = 0.006) were independent risk factors for 14-day sepsis-related mortality. The Kaplan-Meier survival analysis revealed that A. dhakensis bacteremia heralded the worst clinical outcome (log-rank test, P = 0.020) (Fig. 1).
Fig 1. Kaplan-Meier survival curves for 148 patients with monomicrobial bacteremia caused by Aeromonas veronii, A. dhakensis, A. caviae, and A. hydrophila (Log-rank test, P = 0.02).
Despite receiving appropriate antibiotic treatment, one patient experienced breakthrough bacteremia, and two patients had relapsing bacteremia due to identical strains. Breakthrough A. dhakensis bacteremia occurred in a patient with a severe A. dhakensis burn wound infection, and cefotaxime resistance emerged after a 3-day ceftazidime therapy, as described previously [21]. Relapsing bacteremia after a 14-day treatment occurred in a cancer patient with Port A catheter-related A. caviae bacteremia treated by cefpirome without catheter removal and in a patient with unresectable pancreatic cancer with A. caviae BTI treated by ciprofloxacin. Overall, of 66 patients infected by cefotaxime-susceptible A. dhakensis, A. caviae, or A. hydrophila strains and treated by a β-lactam for at least 48 hours, only one (1.5%) was later colonized or infected with a resistant strain of the same species.
A subgroup analysis comparing the A. dhakensis and A. hydrophila groups revealed that co-morbidity with malignancy and cefoxitin susceptibility were more frequently associated with the A. hydrophila group (P = 0.027 and P < 0.001, respectively), and other clinical parameters were not significantly different.
Discussion
This study revealed that A. dhakensis, A. veronii and A. caviae were the three major species involved in Aeromonas bacteremia, while A. hydrophila played a minor role. Clinical characteristics, antimicrobial resistance profiles, and treatment outcomes of patients with bacteremia varied depending on the species. A. dhakensis and A. veronii bacteremia correlate with similar patient characteristics (liver cirrhosis), community acquisition, and infectious diseases (SBP and SSTIs). Manifestations of A. caviae bacteremia included less involvement of cirrhotic patients and frequent association with hospital-onset infections, as observed in an earlier report [24]. Oral ingestion, replacement of medical devices, or direct contact of abraded wounds with contaminated material can serve as the portals of entry of Aeromonas species [1]. We found that patients with either cirrhosis or leukemia experienced repeated episodes of Aeromonas bacteremia, which were mostly community-acquired and caused by either A. dhakensis or A. veronii. Because of the wide distribution of aeromonads in food products and A. dhakensis in the environment and the aquatic creatures [1,4,7], food safety and wound hygiene should be emphasized among susceptible hosts to reduce infections. Likewise, microbiological surveillance of hospital water and fluids for medical applications may be considered to prevent A. caviae-associated hospital-onset infections.
Differences in antimicrobial resistance phenotypes and genotypes are also associated with species variation. The distribution of chromosomal β-lactamases is species-specific among aeromonads, i.e., class B, C and D in A. hydrophila, class C and D in A. caviae, and class B and D in A. veronii [1,25]. The expression of the three chromosomal β-lactamases is often activated in the presence of inducers or due to the emergence of derepressed mutants [26]. We recently reported that A. dhakensis intrinsically carries class B, C and D β-lactamases, and AmpC bla AQU-1 is specific to A. dhakensis [21,22]. The colony hybridization assay that detected the AmpC and MBL genes in the present study yielded consistent results. Not unexpectedly, broad-spectrum cephalosporin resistance was associated with recent antibiotic selection pressure, as revealed by the multivariate analysis. Of note, 92% of the A. veronii isolates were resistant to cefazolin based on the current resistance criterion, i.e., MIC ≥ 4 μg/ml [18]. Such a result is different from the previous impression of cefazolin susceptibility of A. veronii [27], which was based on the earlier breakpoint, ≥ 32 μg/ml [28]. Only 2 (1.9%) of cphA-carrying A. dhakensis, A. hydrophila, and A. veronii isolates exhibited imipenem resistance, which is in concordance with the finding that CphA carbapenemase production is not easily detected by the conventional in vitro susceptibility test, unless using large inocula or a modified Hodge test [22,29].
As in our previous report [30], a low incidence (1.5%) of emergence of broad-spectrum cephalosporin-resistant Aeromonas isolates when treating Aeromonas bacteremia with a β-lactam. Notably, resistance mainly emerged in the cases of secondary bacteremia due to burn wound infections [21]. Therefore, the use of a broad-spectrum cephalosporin or carbapenem for bacteremia with a high tissue bacterial burden caused by AmpC-carrying or CphA-carrying species should be approached with caution [21,22].
Distinct survival curves were observed among patients with bacteremia due to different Aeromonas species. Although individual, phenotypically identified Aeromonas species were not shown to predict mortality in earlier reports [2,30], the present study demonstrated that A. dhakensis bacteremia had the highest mortality rate and was an independent risk factor for mortality, while A. caviae bacteremia led to the lowest fatality rate. This result is in agreement with the previous finding that A. caviae exhibits low pathogenic potential, as demonstrated by less toxicity to human cell lines and mice [23,27]. Additionally, higher pathogenicity of A. dhakensis was demonstrated by increased cytotoxicity to human cell lines and higher lethality to Caenorhabditis elegans and mice than A. hydrophila, A. veronii, or A. caviae [5,11,12,23]. Further studies to identify virulence determinants of A. dhakensis are warranted.
Our results are consistent with previous findings that A. dhakensis is widely distributed and often misidentified as A. hydrophila [7,8]. Collectively, the differences in microbiological characteristics, such as the rpoB, rpoD, or gyrB sequencing results, arabinose fermentation ability (negative for A. dhakensis and positive for A. hydrophila), MALDI/TOF spectra, and types of the AmpC β-lactamase gene presented herein (bla AQU-1 for A. dhakensis and bla CepH-like gene for A. hydrophila), support the concept that A. dhakensis and A. hydrophila are distinct species [6,7,9,10,21]. Although both in vitro and in vivo animal models have demonstrated higher pathogenicity of A. dhakensis than A. hydrophila [11,12], the comparison of clinical features and treatment outcomes between the cases of A. dhakensis and A. hydrophila bacteremia was inconclusive, likely owing to a limited number of the cases of the latter. Clinical studies enrolling more patients are warranted to clarify the issue.
In summary, A. dhakensis, A. veronii, and A. caviae are the three major species that cause Aeromonas bacteremia in southern Taiwan. Significant differences existed in their clinical characteristics and antimicrobial resistance profiles. The association with a poor clinical outcome suggests A. dhakensis an important human pathogen.
Supporting Information
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
CJW PLC PRH MCC PJT HIS HCW PHC WCK and CJW received grants from National Health Research Institutes (ID-100-PP-17 and IV-101-SP-13)(http://www.nhri.org.tw/NHRI_WEB/nhriw001Action.do). WCK received grants from National Science Council (NSC 102-2628-B-006-015-MY3) (http://www.most.gov.tw/) and Ministry of Health and Welfare (MOHW103-TDU-B-211-113002) (http://www.mohw.gov.tw/EN/Ministry/), Taiwan. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Janda JM, Abbott SL (2010) The genus Aeromonas: taxonomy, pathogenicity, and infection. Clin Microbiol Rev 23:35–73. 10.1128/CMR.00039-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ko WC, Chuang YC (1995) Aeromonas bacteremia: review of 59 episodes. Clin Infect Dis 20:1298–1304. [DOI] [PubMed] [Google Scholar]
- 3. Beaz-Hidalgo R, Martinez-Murcia A, Figueras MJ (2013) Reclassification of Aeromonas hydrophila subsp. dhakensis Huys et al. 2002 and Aeromonas aquariorum Martinez-Murcia et al. 2008 as Aeromonas dhakensis sp. nov. comb nov. and emendation of the species Aeromonas hydrophila . Syst Appl Microbiol 36:171–176. 10.1016/j.syapm.2012.12.007 [DOI] [PubMed] [Google Scholar]
- 4. Martinez-Murcia AJ, Saavedra MJ, Mota VR, Maier T, Stackebrandt E, et al. (2008) Aeromonas aquariorum sp. nov., isolated from aquaria of ornamental fish. Int J Syst Evol Microbiol 58:1169–1175. 10.1099/ijs.0.65352-0 [DOI] [PubMed] [Google Scholar]
- 5. Morinaga Y, Yanagihara K, Eugenin FL, Beaz-Hidalgo R, Kohno S, et al. (2013) Identification error of Aeromonas aquariorum: a causative agent of septicemia. Diagn Microbiol Infect Dis 76:106–109. 10.1016/j.diagmicrobio.2013.01.019 [DOI] [PubMed] [Google Scholar]
- 6. Puthucheary SD, Puah SM, Chua KH (2012) Molecular characterization of clinical isolates of Aeromonas species from Malaysia. PloS One 7:e30205 10.1371/journal.pone.0030205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Aravena-Roman M, Harnett GB, Riley TV, Inglis TJ, Chang BJ (2011) Aeromonas aquariorum is widely distributed in clinical and environmental specimens and can be misidentified as Aeromonas hydrophila . J Clin Microbiol 49:3006–3008. 10.1128/JCM.00472-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Figueras MJ, Alperi A, Saavedra MJ, Ko WC, Gonzalo N, et al. (2009) Clinical relevance of the recently described species Aeromonas aquariorum . J Clin Microbiol 47:3742–3746. 10.1128/JCM.02216-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen PL, Lee TF, Wu CJ, Teng SH, Teng LJ, et al. (2014) Matrix-assisted aser desorption ionization-time of flight mass spectrometry can accurately differentiate Aeromonas dhakensis from A. hydrophila, A. caviae, and A. veronii . J Clin Microbiol 52:2625–2628. 10.1128/JCM.01025-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lamy B, Laurent F, Kodjo A (2010) Validation of a partial rpoB gene sequence as a tool for phylogenetic identification of aeromonads isolated from environmental sources. Can J Microbiol 56:217–228. 10.1139/w10-006 [DOI] [PubMed] [Google Scholar]
- 11. Chen PL, Wu CJ, Chen CS, Tsai PJ, Tang HJ, et al. (2014) A comparative study of clinical Aeromonas dhakensis and Aeromonas hydrophila isolates in southern Taiwan: A. dhakensis is more predominant and virulent. Clin Microbiol Infect 20:O428–434. 10.1111/1469-0691.12456 [DOI] [PubMed] [Google Scholar]
- 12. Chen PL, Wu CJ, Tsai PJ, Tang HJ, Chuang YC, et al. (2014) Virulence diversity among bacteremic Aeromonas isolates: ex vivo, animal, and clinical evidences. PLoS One 9:e111213 10.1371/journal.pone.0111213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McCabe W, Jackson GG (1962) Gram-negative bacteremia, I: Etiology and ecology. Arch Intern Med 110:847–855. [Google Scholar]
- 14. Chow JW, Yu VL (1999) Combination antibiotic therapy versus monotherapy for gram-negative bacteraemia: a commentary. Int J Antimicrob Agents 11:7–12. [DOI] [PubMed] [Google Scholar]
- 15. Kupfer M, Kuhnert P, Korczak BM, Peduzzi R, Demarta A (2006) Genetic relationships of Aeromonas strains inferred from 16S rRNA, gyrB and rpoB gene sequences. Int J Syst Evol Microbiol 56:2743–2751. [DOI] [PubMed] [Google Scholar]
- 16. Soler L, Yanez MA, Chacon MR, Aguilera-Arreola MG, Catalan V, et al. (2004) Phylogenetic analysis of the genus Aeromonas based on two housekeeping genes. Int J Syst Evol Microbiol 54:1511–1519. [DOI] [PubMed] [Google Scholar]
- 17. Rice LB, Bonomo RA (1996) Genetic and biochemical mechanisms of bacterial resistance to antimicrobial agents, p 453–501. In Lorian V (ed), Antibiotics in laboratory medicine, 4th ed. Williams & Wilkins, Baltimore, Md. [Google Scholar]
- 18.Clinical and Laboratory Standards Institute (2010) Methods for antimicrobial dilution and disk susceptibility testing of infrequently isolated or fastidious bacteria; approved guideline-second edition. CLSI document M45-A2. Wayne, PA.
- 19.Clinical and Laboratory Standards Institute (2012) Performance standards for antimicrobial susceptibility testing; 22nd informational supplement. CLSI document M100-S22. Wayne, PA.
- 20. Wu CJ, Chuang YC, Lee MF, Lee CC, Lee HC, et al. (2011) Bacteremia due to extended-spectrum-beta-lactamase-producing Aeromonas spp. at a medical center in Southern Taiwan. Antimicrob Agents Chemother 55:5813–5818. 10.1128/AAC.00634-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wu CJ, Wang HC, Chen PL, Chang MC, Sun HS, et al. (2013) AQU-1, a chromosomal class C beta-lactamase, among clinical Aeromonas dhakensis isolates: Distribution and clinical significance. Int J Antimicrob Agents 42:456–461. 10.1016/j.ijantimicag.2013.08.002 [DOI] [PubMed] [Google Scholar]
- 22. Wu CJ, Chen PL, Wu JJ, Yan JJ, Lee CC, et al. (2012) Distribution and phenotypic and genotypic detection of a metallo-beta-lactamase, CphA, among bacteraemic Aeromonas isolates. J Med Microbiol 61:712–719. 10.1099/jmm.0.038323-0 [DOI] [PubMed] [Google Scholar]
- 23. Wu CJ, Tsai PJ, Chen PL, Wu IC, Lin YT, et al. (2012) Aeromonas aquariorum septicemia and enterocolitis in a cirrhotic patient. Diagn Microbiol Infect Dis 74:406–408. 10.1016/j.diagmicrobio.2012.08.005 [DOI] [PubMed] [Google Scholar]
- 24. Chuang HC, Ho YH, Lay CJ, Wang LS, Tsai YS, et al. (2011) Different clinical characteristics among Aeromonas hydrophila, Aeromonas veronii biovar sobria and Aeromonas caviae monomicrobial bacteremia. J Korean Med Sci 26:1415–1420. 10.3346/jkms.2011.26.11.1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fosse T, Giraud-Morin C, Madinier I (2003). Phenotypes of beta-lactam resistance in the genus Aeromonas . Pathol Biol (Paris). 51:290–296. [DOI] [PubMed] [Google Scholar]
- 26. Walsh TR, Stunt RA, Nabi JA, MacGowan AP, Bennett PM (1997) Distribution and expression of beta-lactamase genes among Aeromonas spp. J Antimicrob Chemoth 40:171–178. [DOI] [PubMed] [Google Scholar]
- 27. Janda JM, Guthertz LS, Kokka RP, Shimada T (1994) Aeromonas species in septicemia: laboratory characteristics and clinical observations. Clin Infect Dis 19:77–83. [DOI] [PubMed] [Google Scholar]
- 28.Clinical and Laboratory Standards Institute (2006) Methods for antimicrobial dilution and disk susceptibility testing of infrequently isolated or fastidious bacteria; approved guideline. CLSI document M45-A. Wayne, PA.
- 29. Rossolini GM, Zanchi A, Chiesurin A, Amicosante G, Satta G, et al. (1995) Distribution of cphA or related carbapenemase-encoding genes and production of carbapenemase activity in members of the genus Aeromonas . Antimicrob Agents Chemother 39:346–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ko WC, Lee HC, Chuang YC, Liu CC, Wu JJ (2000) Clinical features and therapeutic implications of 104 episodes of monomicrobial Aeromonas bacteraemia. J Infect 40:267–273. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.


