Abstract
Background
The fractional concentration of nitric oxide in exhaled air (FeNO) is a biomarker of eosinophilic airway inflammation and associated with childhood asthma. Identification of common genetic variants associated with childhood FeNO may help to define biological mechanisms related to specific asthma phenotypes.
Objective
To identify genetic variants associated with childhood FeNO, and their relation with asthma.
Methods
FeNO was measured in children aged 5 to 15 years. In 14 genome-wide association (GWA) studies (N = 8,858), we examined the associations of ~2.5 million single nucleotide polymorphisms (SNPs) with FeNO. Subsequently, we assessed whether significant SNPs were expression quantitative trait loci (eQTLs) in genome-wide expression datasets of lymphoblastoid cell lines (N = 1,830), and were related with asthma in a previously published GWA dataset (cases: n=10,365; controls: n=16,110).
Results
We identified 3 SNPs associated with FeNO: rs3751972 in LYR motif containing 9 (LYRM9) (P = 1.97×10−10) and rs944722 in inducible nitric oxide synthase 2 (NOS2) (P = 1.28×10−9) both located at 17q11.2-q12, and rs8069176 near gasdermin B (GSDMB) (P = 1.88×10−8) at 17q12-q21. We found a cis eQTL for the transcript soluble galactoside-binding lectin 9 (LGALS9) that is in linkage disequilibrium with rs944722. Rs8069176 was associated with GSDMB and ORM1-like 3 (ORMDL3) expression. Rs8069176 at 17q12-q21, and not rs3751972 and rs944722 at 17q11.2-q12, were associated with physician-diagnosed asthma.
Conclusion
This study identified 3 variants associated with FeNO, explaining 0.95% of the variance. Identification of functional SNPs and haplotypes in these regions might provide novel insight in the regulation of FeNO. This study highlights that both shared and distinct genetic factors affect FeNO and childhood asthma.
Keywords: airway inflammation, asthma phenotypes, biomarker, genetics, genome-wide association study
INTRODUCTION
Asthma is a complex disease with different phenotypes, influenced by many genetic and environmental factors1. Why children develop specific asthma phenotypes is still poorly understood2, 3. Genetic association studies may help to identify biological pathways underlying the clinical expression of asthma. Recent genome-wide association (GWA) studies provided evidence that different common genetic variants are associated with specific asthma-related outcomes such as childhood onset asthma4-6, adult asthma5-7, impaired lung function8-11, and atopy12-14.
The fractional concentration of nitric oxide in exhaled air (FeNO) is a noninvasive biomarker of eosinophilic airway inflammation15-17. Higher FeNO is associated with childhood asthma symptoms18, exacerbations19, physician-diagnosed asthma15-17 and atopy20. Nitric oxide is a reactive free-radical gas generated in the airway epithelium when L-arginine is oxidized to L-citrulline17. This reaction is catalyzed by nitric oxide synthases (NOS), that are upregulated in the presence of pro-inflammatory cytokines and inflammatory mediators17. Nitric oxide regulates airway and blood vessel tone and high concentrations have antimicrobial effects17. Although 60% of the variance in FeNO in adults can be explained by heritability21, the genetic loci that influence FeNO are largely unknown. Identification of common genetic variants associated with childhood FeNO may help to define biological mechanisms related to specific asthma phenotypes2, 3, 22, 23.
To identify common genetic variants associated with childhood FeNO, we examined the association of ~2.5 million directly genotyped and imputed single nucleotide polymorphisms (SNPs) with FeNO in 14 independent pediatric discovery GWA studies (N = 8,858).
METHODS
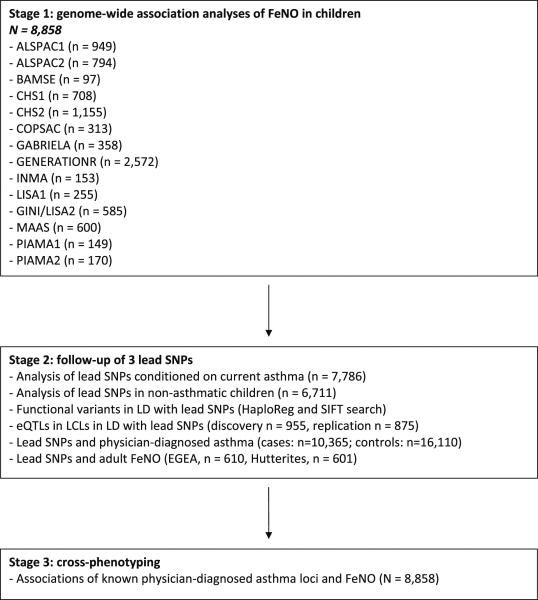
FeNO was measured online in children aged 5 to 15 years according to European Respiratory Society (ERS) and American Thoracic Society (ATS) guidelines16. FeNO was natural-log transformed to obtain a normal distribution. We applied linear regression between allele dosages obtained from imputations and natural-log FeNO adjusted for sex and age at time of measurement. Details on the SNP discovery analysis and additional analyses, including the analysis to determine independent SNP effects, explained variance analyses and stratified analysis for current asthma, are presented in the Online Repository Materials methods section, and an overview of our study design is outlined in Figure I. Details on individual study characteristics, SNP genotyping platforms and study association analyses are provided in Repository Table E1.
Figure I. Study design.
SNPs, single nucleotide polymorphisms; LD, linkage disequilibrium; eQTLs, expression quantitative trait loci; LCLs, lymphoblastoid cell lines.
We assessed whether significant SNPs or SNPs in linkage disequilibrium (LD, a measure of correlation between SNPs) with our lead SNPs were functional annotated SNPs using HaploReg24 and SIFT (http://sift.jcvi.org/), and were situated in genomic loci that are involved in the regulation of messenger RNA expression (the so-called ‘expression quantitative trait loci’ or eQTLs). For the second purpose we used available genome-wide expression datasets of human lymphoblastoid cell lines (N = 1,830)25, 26.
We tested the relation of significant SNPs with asthma using a previously published GWA dataset of physician-diagnosed asthma (cases: n=10,365; controls: n=16,110)5. We explored whether the SNPs identified in the present GWA study were related with FeNO in adults in the Epidemiological study on the Genetics and Environment of Asthma (EGEA) and in Hutterites (N = 1,211).
Finally, we explored whether common genetic variants known to be associated with physician-diagnosed asthma5 were related with childhood FeNO.
The institutional review boards for human studies approved the protocols and written consent was obtained from the participating subjects or their caregivers if required by the institutional review board.
RESULTS
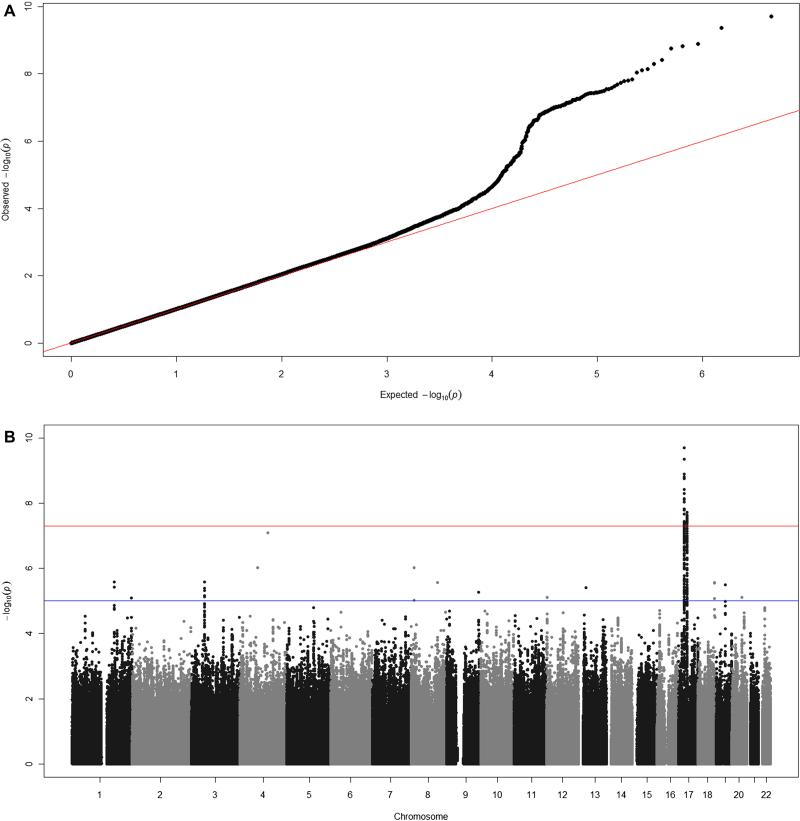
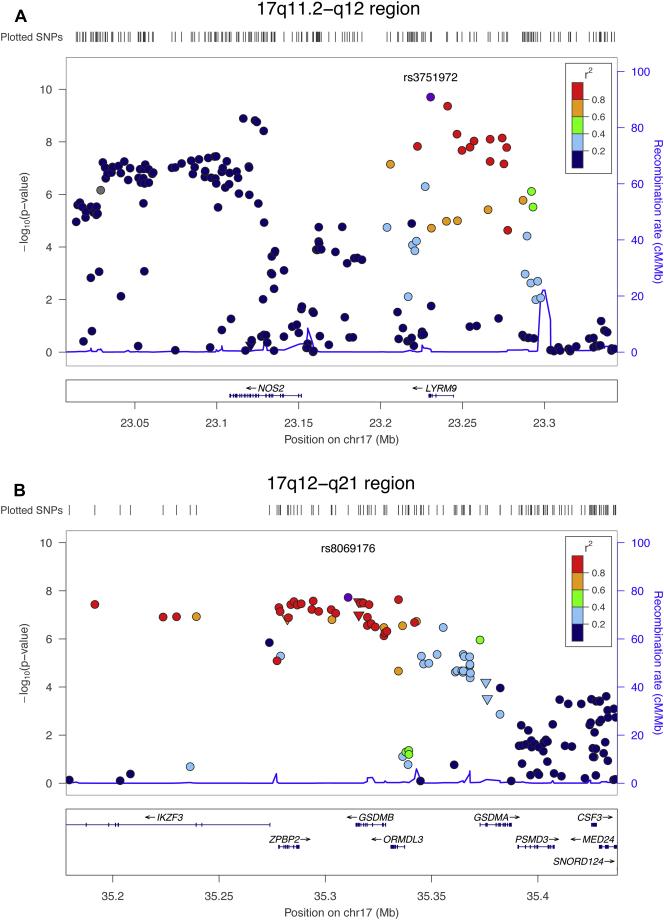
We identified genome-wide significant (P < 5×10−8) association of childhood FeNO and SNPs at 3 genetic loci. Two SNPs were located at chromosome 17q11.2-q12: the SNP rs3751972 in the LYR motif containing 9 (LYRM9) gene and rs944722 in the NOS2 gene (Table I). Each C allele of rs3751972 was associated with higher ln(FeNO) (ß = 0.09 ppb; S.E. = 0.014; P = 1.97×10−10; explained variance = 0.23%), and each C allele of rs944722 was associated with lower ln(FeNO) (ß = -0.07 ppb; S.E. = 0.012; P = 1.28×10−9; explained variance = 0.30%). Rs3751972 and rs944722 are in neighboring loci with low LD, indicating that the two SNPs might not represent the same genetic variation (HapMap pairwise LD, phase II release 22 CEU; D’ = 0.237, r2 = 0.014). A third SNP, rs8069176 near the gasdermin B (GSDMB) gene at 17q12-q21 was also associated with childhood FeNO. Each A allele of rs8069176 was associated with lower ln(FeNO) (ß = -0.07 ppb; S.E. = 0.012; P = 1.88×10−8; explained variance = 0.41%). Figure II-IV show the QQ-, Manhattan-, regional association- and forest plots of the 3 signals.
Table I.
Summary statistics of the 3 SNPs at P < 5×10−8.
| Marker | MAF | β | S.E. | P | I2 | HetP |
|---|---|---|---|---|---|---|
| rs3751972[C] at 17q11.2 (LYRM9) | 0.25 | 0.086 | 0.014 | 1.97×10−10 | 27.4 | 0.161 |
| rs944722[C] at 17q11.2-q12 (NOS2) | 0.38 | −0.073 | 0.012 | 1.28×10−09 | 37.8 | 0.075 |
| rs8069176[A] at 17q12-q21 (nearest genes ZPBP2-GSDMB) | 0.43 | −0.066 | 0.012 | 1.88×10−08 | 0.0 | 0.668 |
Single nucleotide polymorphisms (SNPs) markers are identified according to their standard rs numbers (NCBI build 36). Independent SNPs with a genome-wide significant effect on FeNO levels in children are shown (P < 5×10−8). The total sample includes data of 14 independent GWA datasets (N = 8,858). MAF, minor allele frequency; S.E., standard error. β reflects differences in natural log-transformed FeNO per minor allele. P values are obtained from linear regression of each SNP against natural log-transformed FeNO adjusted for sex and age at time of measurement (fixed-effect additive genetic model). Derived inconsistency statistic I2 and HetP values reflect heterogeneity across studies with the use of Cochran's Q tests.
Figure II. QQ and Manhattan plots of 2,253,077 SNPs of 14 GWA studies (N = 8,858).
QQ plot of 2,253,077 SNPs of 14 GWA studies. The black dots represent observed P values and the red line represents the expected P values under the null distribution. Manhattan plot showing the association P values of FeNO of the 14 studies. The –log10 of the P value for each of 2,253,077 SNPs (y-axis) is plotted against the genomic position (x-axis).
Figure IV. Forest plots of the associations between FeNO and the 3 SNPs associated with FeNO at P < 5 × 10−8.
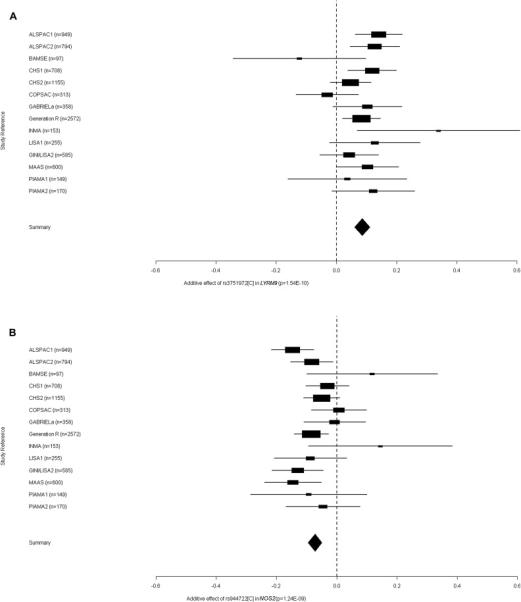
Forest plots of the associations between FeNO and the SNPs in LYRM9 (a), NOS2 (b) and near ZPBP2-GSDMB (c) at P < 5×10−8. In each plot, the triangle indicates the effect size and the confidence interval in the 14 studies. The P values in the plots are without genomic control correction.
We used the genome-wide complex trait analysis (GCTA) tool to determine if SNP effects were independent. We conditioned on all SNPs of the meta-analysis27, and showed that rs3751972 and rs944722 were indeed independent signals and did not represent the same genetic variation (Repository Table E2). After conditioning on all SNPs of the meta-analysis, rs3751972 and rs2274894 showed the strongest association in the LYRM9 gene (P = 2.06×10−9) and in the NOS2 gene (P = 1.50×10−8, rs2274894 not rs944722 is the strongest signal using GCTA) respectively. Using the same approach, rs8069176 showed the strongest association at 17q12-q21 (P = 2.14×10−8).
The 3 genome-wide significant SNPs showed low heterogeneity between studies (all P ≥ 0.075, I2 = 0 – 37.8%). The 3 SNPs together explained 0.95% of the variance in FeNO. Other suggestive loci that were associated with FeNO, but did not reach genome-wide significance (P < 1×10−5), are given in Repository Tables E3 and E4. The associations of genetic variants in the nitric oxide synthases or arginase genes might be different among asthmatic versus non-asthmatic children28. Therefore, we performed a sensitivity analysis adjusting for current asthma and this produced comparable results for the SNPs in LYRM9 and NOS2 and a slightly lower effect for the SNP in the 17q12-q21 locus (Repository Table E5). In addition, we showed that the 3 SNPs were also associated with FeNO in non-asthmatic children (Repository Table E6).
We assessed whether there were common non-synonymous variants with deleterious functional implications in LD (r2 > 0.80) with our 3 genome-wide significant SNPs using HaploReg24, a data base for functional annotation of SNPs. We found 3 variants, rs11557467, rs2305480 and rs2305479 that were in high LD with rs8069176 at 17q12-q21. Rs11557467 is located in the zona pellucida binding protein 2 (ZPBP2) gene, holding a high risk deleterious effect consisting of a missense variation resulting in a non-conservative amino acid change. Rs2305480 and rs2305479 in the GSDMB gene are both variations with a high risk of deleterious effect resulting from a missense change leading to abolishment of a protein domain. We did not find functional implications for rs3751972 and rs944722 at 17q11.2-q12. The nature of the amino-acid changes, and predicted functional significances using SIFT (http://sift.jcvi.org/), as well as the frequencies, LD with the index SNP at 17q12-q21 and P values for FeNO association are depicted in Repository Table E7.
Subsequently, we assessed whether the identified 3 loci were eQTLs in genome-wide expression datasets of lymphoblastoid cell lines (N = 1,830)25, 26. We found a cis eQTL for the transcript soluble galactoside-binding lectin 9 (LGALS9) in LD with rs944722 in two independent datasets (Repository Tables E8 and E9). LGALS9 is downstream of the NOS2 gene. Rs8069176 was associated with both GSDMB- and ORM1-like 3 (ORMDL3) gene expression. We did not find eQTLs for rs3751972.
We tested the associations of the 3 FeNO-associated SNPs with physician-diagnosed asthma in a previously published GWA dataset (cases: n=10,365; controls: n=16,110)5. The SNP rs8069176 was not available and we used rs2305480 as a proxy. The rs2305480[A] minor allele at the 17q12-q21 locus was associated with a decreased risk of asthma (odds ratio (OR) 0.85; 95% CI 0.81 - 0.88; P = 7.93×10−17; Table II). This is in line with the association with lower FeNO that we found for rs8069176[A]. The SNPs rs3751972 and rs944722 were not associated with an asthma diagnosis (P ≥ 0.3). The 3 childhood FeNO-associated SNPs were not associated with adult FeNO (N = 1,211, Table II).
Table II.
Association of the 3 SNPs related to childhood FeNO with physician-diagnosed asthma and adult FeNO.
| Physician-diagnosed asthma (cases = 10,365 : controls = 16,110)5 | ||
|---|---|---|
| Marker | OR (95% CI) | P |
| Proxy for rs3751972: rs4796222[A] (r2=1.000; D′=1.000) at 17q11.2 (LYRM9) | 0.98 (0.93-1.02) | 0.303 |
| Proxy for rs944722: rs2274894[T] (r2=0.967; D′=1.000) at 17q11.2-q12 (NOS2) | 1.00 (0.96-1.04) | 0.983 |
| Proxy for rs8069176: rs2305480[A] (r2=1.000; D′=1.000) at 17q12-q21 (nearest genes ZPBP2-GSDMB) | 0.85 (0.81-0.88) | 7.93×10−17 |
| Adult FeNO | |||
|---|---|---|---|
| Marker (EGEA, n = 610) | β | S.E. | P |
| rs3751972[C] at 17q11.2 (LYRM9) | 0.125 | 0.065 | 0.057 |
| rs944722[C] at 17q11.2-q12 (NOS2) | −0.015 | 0.061 | 0.802 |
| rs8069176[A] at 17q12-q21 (nearest genes ZPBP2-GSDMB) | −0.113 | 0.062 | 0.067 |
| Marker (Hutterites, n = 601) | Z score | P |
|---|---|---|
| Proxy for rs3751972: rs4796228[G] (r2=0.659; D′=1.000) at 17q11.2 (LYRM9) | −1.536 | 0.125 |
| Proxy for rs944722: rs2314809[T] (r2=0.967; D′=1.000) at 17q11.2-q12 (NOS2) | −2.322 | 0.020 |
| Proxy for rs8069176: rs11078927[T] (r2=1.000; D′=1.000) at 17q12-q21 (nearest genes ZPBP2-GSDMB) | 0.505 | 0.613 |
Single nucleotide polymorphisms (SNPs) markers are identified according to their standard rs numbers (NCBI build 36). Independent SNPs with a genome-wide significant effect on FeNO levels in children are shown (P < 5×10−8) in relation to physician-diagnosed asthma5 and adult FeNO. S.E., standard error. Odds ratios (OR) with 95% confidence interval (CI) for physician-diagnosed asthma5. β reflects differences in natural log-transformed FeNO per minor allele for adult FeNO in EGEA. Z-score reflects the strength of the association between SNP and natural log-transformed FeNO and the direction of the effect of the minor allele in Hutterites.
Finally, we explored whether common genetic variants known to be associated with physician-diagnosed asthma5 were related with childhood FeNO. We found that known asthma SNPs rs2305480 at 17q12 (GSDMB), rs3894194 at 17q21.1 (GSDMA), rs744910 at 15q22.33 (SMAD3) and rs1295686 at 5q31 (IL13) were indeed associated with childhood FeNO (all P ≤ 0.005, after Bonferroni correction; Table III). The directions of the SNP effects were as expected. The asthma SNPs together explained 0.32% of the variance in FeNO.
Table III.
Association of known physician-diagnosed asthma loci, from a previous GWA study5 with childhood FeNO.
| Physician-diagnosed asthma5 | ||||||
|---|---|---|---|---|---|---|
| Marker | MAF | β | S.E. | P | I2 | HetP |
| rs2305480[A] decreasing risk-allele at 17q12 (GSDMB) | 0.42 | −0.065 | 0.012 | 2.83×10−08 | 0.0 | 0.731 |
| rs3894194[A] increasing risk-allele at 17q21.1 (GSDMA) | 0.47 | 0.048 | 0.012 | 6.35×10−05 | 9.5 | 0.349 |
| rs744910[A] decreasing risk-allele at 15q22.33 (SMAD3) | 0.49 | −0.039 | 0.012 | 8.41×10−04 | 0.0 | 0.491 |
| rs1295686[T] increasing risk-allele at 5q31 (IL13) | 0.27 | 0.044 | 0.014 | 1.25×10−03 | 4.6 | 0.401 |
| rs1342326[C] increasing risk-allele at 9p24.1 (IL33) | 0.17 | 0.025 | 0.016 | 0.119 | 0.0 | 0.515 |
| rs9273349[T] decreasing risk-allele at 6p21.3 (HLA-DQ) | 0.37 | −0.022 | 0.022 | 0.310 | 0.0 | 0.802 |
| rs11071559[T] decreasing risk-allele at 15q22.2 (RORA) | 0.14 | −0.014 | 0.017 | 0.415 | 0.0 | 0.651 |
| rs3771166[A] decreasing risk-allele at 2q12 (IL18R1) | 0.35 | −0.009 | 0.012 | 0.463 | 7.4 | 0.371 |
| rs2284033[A] decreasing risk-allele at 22q13.1 (IL2RB) | 0.42 | 0.005 | 0.012 | 0.705 | 0.0 | 0.633 |
| rs2073643[T] increasing risk-allele at 5q23.3 (SLC22A5) | 0.47 | 0.000 | 0.012 | 0.993 | 0.0 | 0.590 |
Single nucleotide polymorphisms (SNPs) markers are identified according to their standard rs numbers (NCBI build 36). We explored whether common genetic variants known to be related with physician-diagnosed asthma5 were associated with childhood FeNO. The total sample includes data of 14 independent GWA datasets (N = 8,858). MAF, minor allele frequency; S.E., standard error. β reflects differences in natural log-transformed FeNO per minor allele. P values are obtained from linear regression of each SNP against natural log-transformed FeNO adjusted for sex and age at time of measurement (fixed-effect additive genetic model). Derived inconsistency statistic I2 and HetP values reflect heterogeneity across studies with the use of Cochran's Q tests.
DISCUSSION
We identified associations between FeNO and genetic variants at 3 loci. The common variants in and near the LYRM9 and NOS2 genes were located at 17q11.2-q12, the third signal was at 17q12-q21, harboring the ZPBP2, GSDMB, and ORMDL3 genes. The three independently associated genetic variants at the 3 loci explained 0.95% of the total variance in FeNO.
The function of the LYRM9 gene is unknown; variants in the nitric oxide synthases and arginase genes jointly contributed to differences in FeNO in previous studies28-31, and variation in arginase genes to asthma severity32. We did not find associations between the NOS2 and LYRM9 SNPs and asthma. It has been shown previously that the inducible NOS2 protein is higher in adults with severe asthma33. Unfortunately, we do not have data of the two SNPs and severe asthma cases. Inducible NOS2 is expressed in airway epithelium and is synthesized in response to pro-inflammatory cytokines and mediators. Expression of inducible NOS2 may be beneficial in host defense and in modulating the immune response17, 34. In our study genetic variants in inducible NOS2, but not in neuronal NOS1 and constitutive NOS3, were robustly associated with childhood FeNO. A previous study suggested that DNA methylation in promotor regions of arginase genes were associated with FeNO in children with asthma29. Thus, DNA methylation could also play an important role in epigenetic regulation of other genes for NO production.
We found a cis eQTL for the transcript LGALS9 in LD with rs944722, downstream of NOS2, and this suggests that the protein Gal-9 may be involved in the regulation of FeNO. Gal-9 plays a crucial role in immune responses, including allergic inflammation. Gal-9 was shown to inhibit allergic airway inflammation, and airway hyperresponsiveness by modulating CD44-dependent leukocyte recognition of the extracellular matrix in mice35. Results in guinea pigs showed that Gal-9 might be involved in prolonged eosinophil accumulation in the lung36. A recent study suggested a novel function of Gal-9 in mast cells and suggested that Gal-9 might be an interesting new target for the treatment of allergic disorders including asthma37.
The 17q12-q21 asthma locus, harboring the ZPBP2, GSDMB, and ORMDL3 ‘asthma genes’, is a complex region with high LD4, 5, 38, 39. GSDMB may be involved in the regulation of the growth and differentiation of epithelial cells40, 41. The function of the upstream ORMDL3 gene in humans is not clear. The ORMDL family genes encode for transmembrane proteins located in the endoplasmic reticulum membrane. In mice, double knockout of the ORMDL genes leads to slower growth and higher sensitivity to toxic compounds in mice42. The function of the downstream ZPBP2 gene is not known. Hence, the mechanisms by which 17q12-q21 variants may regulate FeNO remains to be elucidated.
The three genetic variants identified in the present study explained only a small proportion of the total variance in FeNO, while earlier work on twins indicated that most of FeNO variation is genetically determined. One explanation could be that the heritability of FeNO was overestimated. Lund et al estimated the heritability but did not adjust for body height, a determinant of adult FeNO31. Furthermore, atopic adults were excluded from their analysis21. In the present study we did not exclude atopic children. Most GWA studies are underpowered to detect a large fraction of the variance conferred by polygenic traits. Big consortia showed consistent genetic architecture of > 1000 alleles for the average polygenic trait43, 44. We determined the genetic variance explained at the whole genome SNP level using a GCTA analysis27, which was 21.3% (P = 0.100) in the largest cohort (Generation R Study, Caucasians only, n = 1,332). The missing heritability in our study is most likely explained by other genetic mechanisms, including missing information on causal (rare) variants, interaction between genes, between environmental factors and genes, and by epigenetic mechanisms45. It has also been suggested that the association between asthma and FeNO may be entirely explained by atopy46. We found an association between the 17q12-q21 childhood asthma locus and FeNO. This suggests that FeNO is related with asthma independent of allergy, as variants at the 17q12-q21 locus are not associated with specific atopic outcomes. The signals in NOS2 and LYRM9 were not associated with asthma, which conflicts with a possible causal effect of FeNO on asthma. One explanation could be that FeNO and asthma are not directly related but may have mechanisms in common. Unfortunately, we were not able to assess haplotypes or other types of genetic variation in the NOS2 and LYRM9 regions that could play a role in the development of asthma in our in silico database of patients with childhood- and adult-onset asthma.
In summary, we identified 3 independent signals that were associated with childhood FeNO in the LYRM9 and NOS2 genes, which are both located at 17q11.2-q12, and near the GSDMB gene at 17q12-q21. The 3 SNPs together explained 0.95% of the variance in FeNO. Identification of functional SNPs and haplotypes in these regions might provide novel insight in the regulation of FeNO. This study highlights that both shared and distinct genetic factors affect FeNO and childhood asthma.
Supplementary Material
Key messages.
■ We identified 3 independent genetic variants associated with childhood FeNO, one of the variants was also associated with physician-diagnosed asthma.
■ Future studies are needed to unravel the mechanisms by which the variants regulate childhood FeNO and asthma.
Capsule summary.
This study highlights that both shared and distinct genetic factors affect childhood FeNO and asthma.
Figure III. Association plots of the 17q11.2-q12 and 17q12-q21 regions.
For both the 17q11.2-q12 and 17q12-q21 regions, SNPs are plotted with their P values (as –log10 values; left y-axis) as a function of genomic position (x-axis). Estimated recombination rates (right y-axis) taken from HapMap are plotted to reflect the local LD structure around the top associated SNP (purple circle) and their correlated proxies (according to a blue to red scale from r2 = 0 to 1). Triangles represent nonsynonymous SNPs.
ACKNOWLEDGEMENTS
The acknowledgements per study can be found in the online supplement.
Funding
DE is supported by UK Medical Research Council Centre (G0600705); GS is supported by UK Medical Research Council Centre (G0600705); JK is funded by a Wellcome Trust 4-year PhD studentship in molecular, genetic, and life course epidemiology (WT083431MA); LD is supported by a European Respiratory Society/Marie Curie Joint Research Fellowship of the European Respiratory Society and the European Community's Seventh Framework Programme FP7/2007-2013 - Marie Curie Actions under grant agreement RESPIRE, PCOFUND-GA-2008-229571 (nr MC 1226-2009); NT is supported by UK Medical Research Council Centre (G0600705); RV was partly supported by an unrestricted personal grant from GlaxoSmithKline, NL; VJ is supported by the Netherlands Organization for Health Research and Development (ZonMw 90700303, 916.10159).
Abbreviations
- ATS
American Thoracic Society
- EGEA
Epidemiological study on the Genetics and Environment of Asthma
- eQTLs
expression quantitative trait loci
- ERS
European Respiratory Society
- FeNO
fractional concentration of nitric oxide in exhaled air
- GCTA
genome-wide complex trait analysis
- GSDMB
gasdermin B
- GWA
genome-wide association
- LD
linkage disequilibrium
- LGALS9
soluble galactoside-binding lectin 9
- LYRM9
LYR motif containing 9
- NOS
nitric oxide synthases
- ORMDL3
ORM1-like 3
- SNPs
single nucleotide polymorphisms
- ZPBP2
zona pellucida binding protein 2
Footnotes
Author's contributions
All authors participated substantially in the design of the study, data acquisition or analysis, and writing or revising the manuscript. The manuscript has been reviewed and approved by all authors.
None of the authors have conflict of interest.
REFERENCES
- 1.Vercelli D. Discovering susceptibility genes for asthma and allergy. Nat Rev Immunol. 2008;8:169–82. doi: 10.1038/nri2257. [DOI] [PubMed] [Google Scholar]
- 2.A plea to abandon asthma as a disease concept. Lancet. 2006;368:705. doi: 10.1016/S0140-6736(06)69257-X. [DOI] [PubMed] [Google Scholar]
- 3.Anderson GP. Endotyping asthma: new insights into key pathogenic mechanisms in a complex, heterogeneous disease. Lancet. 2008;372:1107–19. doi: 10.1016/S0140-6736(08)61452-X. [DOI] [PubMed] [Google Scholar]
- 4.Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–3. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 5.Moffatt MF, Gut IG, Demenais F, Strachan DP, Bouzigon E, Heath S, et al. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 2010;363:1211–21. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Torgerson DG, Ampleford EJ, Chiu GY, Gauderman WJ, Gignoux CR, Graves PE, et al. Meta-analysis of genome-wide association studies of asthma in ethnically diverse North American populations. Nat Genet. 2011;43:887–92. doi: 10.1038/ng.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirota T, Takahashi A, Kubo M, Tsunoda T, Tomita K, Doi S, et al. Genome-wide association study identifies three new susceptibility loci for adult asthma in the Japanese population. Nat Genet. 2011;43:893–6. doi: 10.1038/ng.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Repapi E, Sayers I, Wain LV, Burton PR, Johnson T, Obeidat M, et al. Genome-wide association study identifies five loci associated with lung function. Nat Genet. 2010;42:36–44. doi: 10.1038/ng.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hancock DB, Eijgelsheim M, Wilk JB, Gharib SA, Loehr LR, Marciante KD, et al. Meta-analyses of genome-wide association studies identify multiple loci associated with pulmonary function. Nat Genet. 2010;42:45–52. doi: 10.1038/ng.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilk JB, Chen TH, Gottlieb DJ, Walter RE, Nagle MW, Brandler BJ, et al. A genome-wide association study of pulmonary function measures in the Framingham Heart Study. PLoS Genet. 2009;5:e1000429. doi: 10.1371/journal.pgen.1000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Imboden M, Bouzigon E, Curjuric I, Ramasamy A, Kumar A, Hancock DB, et al. Genome-wide association study of lung function decline in adults with and without asthma. J Allergy Clin Immunol. 2012;129:1218–28. doi: 10.1016/j.jaci.2012.01.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gudbjartsson DF, Bjornsdottir US, Halapi E, Helgadottir A, Sulem P, Jonsdottir GM, et al. Sequence variants affecting eosinophil numbers associate with asthma and myocardial infarction. Nat Genet. 2009;41:342–7. doi: 10.1038/ng.323. [DOI] [PubMed] [Google Scholar]
- 13.Weidinger S, Gieger C, Rodriguez E, Baurecht H, Mempel M, Klopp N, et al. Genome-wide scan on total serum IgE levels identifies FCER1A as novel susceptibility locus. PLoS Genet. 2008;4:e1000166. doi: 10.1371/journal.pgen.1000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castro-Giner F, Bustamante M, Ramon Gonzalez J, Kogevinas M, Jarvis D, Heinrich J, et al. A pooling-based genome-wide analysis identifies new potential candidate genes for atopy in the European Community Respiratory Health Survey (ECRHS). BMC Med Genet. 2009;10:128. doi: 10.1186/1471-2350-10-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith AD, Cowan JO, Brassett KP, Herbison GP, Taylor DR. Use of exhaled nitric oxide measurements to guide treatment in chronic asthma. N Engl J Med. 2005;352:2163–73. doi: 10.1056/NEJMoa043596. [DOI] [PubMed] [Google Scholar]
- 16.ATS/ERS recommendations for standardized procedures for the online and offline measurement of exhaled lower respiratory nitric oxide and nasal nitric oxide, 2005. Am J Respir Crit Care Med. 2005;171:912–30. doi: 10.1164/rccm.200406-710ST. [DOI] [PubMed] [Google Scholar]
- 17.Pijnenburg MW, De Jongste JC. Exhaled nitric oxide in childhood asthma: a review. Clin Exp Allergy. 2008;38:246–59. doi: 10.1111/j.1365-2222.2007.02897.x. [DOI] [PubMed] [Google Scholar]
- 18.Stern G, de Jongste J, van der Valk R, Baraldi E, Carraro S, Thamrin C, et al. Fluctuation phenotyping based on daily fraction of exhaled nitric oxide values in asthmatic children. J Allergy Clin Immunol. 2011;128:293–300. doi: 10.1016/j.jaci.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 19.van der Valk RJ, Baraldi E, Stern G, Frey U, de Jongste JC. Daily exhaled nitric oxide measurements and asthma exacerbations in children. Allergy. 2012;67:265–71. doi: 10.1111/j.1398-9995.2011.02734.x. [DOI] [PubMed] [Google Scholar]
- 20.van der Valk RJ, Caudri D, Savenije O, Koppelman GH, Smit HA, Wijga AH, et al. Childhood wheezing phenotypes and FeNO in atopic children at age 8. Clin Exp Allergy. 2012;42:1329–36. doi: 10.1111/j.1365-2222.2012.04010.x. [DOI] [PubMed] [Google Scholar]
- 21.Lund MB, Kongerud J, Nystad W, Boe J, Harris JR. Genetic and environmental effects on exhaled nitric oxide and airway responsiveness in a population-based sample of twins. Eur Respir J. 2007;29:292–8. doi: 10.1183/09031936.00044805. [DOI] [PubMed] [Google Scholar]
- 22.McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–69. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 23.McCarthy MI, Hirschhorn JN. Genome-wide association studies: potential next steps on a genetic journey. Hum Mol Genet. 2008;17:R156–65. doi: 10.1093/hmg/ddn289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–4. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.L Liang NM, Dixon A, Lathrop G, Abecasis G, Moffatt M, Cookson W. A cross-platform catalogue of 14,177 expression quantitative trait loci derived from lymphoblastoid cell lines (Submitted) 2012 doi: 10.1101/gr.142521.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Granell R, Henderson AJ, Timpson N, St Pourcain B, Kemp JP, Ring SM, et al. Examination of the relationship between variation at 17q21 and childhood wheeze phenotypes. J Allergy Clin Immunol. 2012 doi: 10.1016/j.jaci.2012.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang J, Ferreira T, Morris AP, Medland SE, Madden PA, Heath AC, et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat Genet. 2012;44:369–75. doi: 10.1038/ng.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salam MT, Bastain TM, Rappaport EB, Islam T, Berhane K, Gauderman WJ, et al. Genetic variations in nitric oxide synthase and arginase influence exhaled nitric oxide levels in children. Allergy. 2011;66:412–9. doi: 10.1111/j.1398-9995.2010.02492.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Breton CV, Byun HM, Wang X, Salam MT, Siegmund K, Gilliland FD. DNA methylation in the arginase-nitric oxide synthase pathway is associated with exhaled nitric oxide in children with asthma. Am J Respir Crit Care Med. 2011;184:191–7. doi: 10.1164/rccm.201012-2029OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dahgam S, Nyberg F, Modig L, Naluai AT, Olin AC. Single nucleotide polymorphisms in the NOS2 and NOS3 genes are associated with exhaled nitric oxide. J Med Genet. 2012;49:200–5. doi: 10.1136/jmedgenet-2011-100584. [DOI] [PubMed] [Google Scholar]
- 31.Bouzigon E, Monier F, Boussaha M, Le Moual N, Huyvaert H, Matran R, et al. Associations between nitric oxide synthase genes and exhaled NO-related phenotypes according to asthma status. PLoS One. 2012;7:e36672. doi: 10.1371/journal.pone.0036672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vonk JM, Postma DS, Maarsingh H, Bruinenberg M, Koppelman GH, Meurs H. Arginase 1 and arginase 2 variations associate with asthma, asthma severity and beta2 agonist and steroid response. Pharmacogenet Genomics. 2010;20:179–86. doi: 10.1097/FPC.0b013e328336c7fd. [DOI] [PubMed] [Google Scholar]
- 33.Yamamoto M, Tochino Y, Chibana K, Trudeau JB, Holguin F, Wenzel SE. Nitric oxide and related enzymes in asthma: relation to severity, enzyme function and inflammation. Clin Exp Allergy. 2012;42:760–8. doi: 10.1111/j.1365-2222.2011.03860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stuehr DJ. Mammalian nitric oxide synthases. Biochim Biophys Acta. 1999;1411:217–30. doi: 10.1016/s0005-2728(99)00016-x. [DOI] [PubMed] [Google Scholar]
- 35.Katoh S, Ishii N, Nobumoto A, Takeshita K, Dai SY, Shinonaga R, et al. Galectin-9 inhibits CD44-hyaluronan interaction and suppresses a murine model of allergic asthma. Am J Respir Crit Care Med. 2007;176:27–35. doi: 10.1164/rccm.200608-1243OC. [DOI] [PubMed] [Google Scholar]
- 36.Yamamoto H, Kashio Y, Shoji H, Shinonaga R, Yoshimura T, Nishi N, et al. Involvement of galectin-9 in guinea pig allergic airway inflammation. Int Arch Allergy Immunol. 2007;143(Suppl 1):95–105. doi: 10.1159/000101414. [DOI] [PubMed] [Google Scholar]
- 37.Niki T, Tsutsui S, Hirose S, Aradono S, Sugimoto Y, Takeshita K, et al. Galectin-9 is a high affinity IgE-binding lectin with anti-allergic effect by blocking IgE-antigen complex formation. J Biol Chem. 2009;284:32344–52. doi: 10.1074/jbc.M109.035196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holloway JW, Koppelman GH. 17q21 variants and asthma - questions and answers. N Engl J Med. 2008;359:2043–5. doi: 10.1056/NEJMe0807576. [DOI] [PubMed] [Google Scholar]
- 39.Verlaan DJ, Berlivet S, Hunninghake GM, Madore AM, Lariviere M, Moussette S, et al. Allele-specific chromatin remodeling in the ZPBP2/GSDMB/ORMDL3 locus associated with the risk of asthma and autoimmune disease. Am J Hum Genet. 2009;85:377–93. doi: 10.1016/j.ajhg.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tamura M, Tanaka S, Fujii T, Aoki A, Komiyama H, Ezawa K, et al. Members of a novel gene family, Gsdm, are expressed exclusively in the epithelium of the skin and gastrointestinal tract in a highly tissue-specific manner. Genomics. 2007;89:618–29. doi: 10.1016/j.ygeno.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 41.Fujii T, Tamura M, Tanaka S, Kato Y, Yamamoto H, Mizushina Y, et al. Gasdermin D (Gsdmd) is dispensable for mouse intestinal epithelium development. Genesis. 2008;46:418–23. doi: 10.1002/dvg.20412. [DOI] [PubMed] [Google Scholar]
- 42.Hjelmqvist L, Tuson M, Marfany G, Herrero E, Balcells S, Gonzalez-Duarte R. ORMDL proteins are a conserved new family of endoplasmic reticulum membrane proteins. Genome Biol. 2002;3:RESEARCH0027. doi: 10.1186/gb-2002-3-6-research0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lango Allen H, Estrada K, Lettre G, Berndt SI, Weedon MN, Rivadeneira F, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–8. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42:565–9. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maher B. Personal genomes: The case of the missing heritability. Nature. 2008;456:18–21. doi: 10.1038/456018a. [DOI] [PubMed] [Google Scholar]
- 46.Franklin PJ, Stick SM. The value of FeNO measurement in asthma management: the motion against FeNO to help manage childhood asthma--reality bites. Paediatr Respir Rev. 2008;9:122–6. doi: 10.1016/j.prrv.2007.12.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.