Abstract
Numerous accessory factors modulate RNA polymerase response to regulatory signals and cellular cues and establish communications with co-transcriptional RNA processing. Transcription regulators are astonishingly diverse, with similar mechanisms arising via convergent evolution. NusG/Spt5 elongation factors comprise the only universally conserved and ancient family of regulators. They bind to the conserved clamp helices domain of RNA polymerase, which also interacts with non-homologous initiation factors in all domains of life, and reach across the DNA channel to form processivity clamps that enable uninterrupted RNA chain synthesis. In addition to this ubiquitous function, NusG homologs exert diverse, and sometimes opposite, effects on gene expression by competing with each other and other regulators for binding to the clamp helices and by recruiting auxiliary factors that facilitate termination, antitermination, splicing, translation, etc. This surprisingly diverse range of activities and the underlying unprecedented structural changes make studies of these “transformer” proteins both challenging and rewarding.
Keywords: antitermination, NusG, refolding, RfaH, transcription, translation
Introduction
Transcription, the first step of gene expression, is elaborately controlled at every step, from initiation to termination, and is tightly coordinated with other cellular processes, such as translation in prokaryotes [1,2] and RNA capping and splicing in eukaryotes [3], creating a dynamic regulatory network of many interlinked factors. In bacteria and archaea, a single RNA polymerase (RNAP) enzyme is responsible for all transcription, whereas in eukaryotes, distinct classes of genes are transcribed by multiple different RNAP species. Despite differences in subunit composition, with five subunits in bacterial (α2ββ′ω) and more than 12 in archaeal and eukaryotic RNAPs, these enzymes share an overall shape, make similar contacts to the nucleic acid chains, and have highly conserved active centers [4]. However, they differ significantly with respect to regulation, with many more accessory factors modulating every step of transcription in eukaryotes. In bacteria, specific initiation at promoters requires only one accessory factor called sigma (σ) that, when bound to RNAP, recognizes and melts promoter DNA [5]. Eukaryotic RNAPs use two key basal factors, TBP and TFIIB, which bind to DNA and subsequently recruit RNAP and additional factors to form a core initiation complex [4,6]; underscoring this regulatory complexity is Mediator, a 25-subunit 1.4 MDa complex (in yeast) that bridges gene-specific initiation factors and the RNAP II and is required for transcription of most, if not all, protein-coding genes [7]. Requirements for the termination of transcription are also quite distinct. In bacteria, a simple signal composed of an RNA hairpin followed by a run of U residues is sufficient to trigger RNA release [8]. Although similar RNA elements may induce termination in archaea and eukaryotes [9,10], this step is regulated very differently across the three kingdoms [8,11-13]. Among myriads of different accessory factors that modulate transcription, the NusG/Spt5 transcription elongation factors stand out as the only example of the universally conserved regulators that coevolved with the core RNAP since the last universal common ancestor [14].
The sequence and core structure are well conserved among all NusG homologs [14,15] (Fig. 1). These proteins contain a single NGN (NusG-like N-terminal) domain [16], connected via a flexible linker to one or more C-terminal domains (CTDs) containing a KOW motif [17]. The N-terminal domain (NTD) binds to RNAP and nucleic acids to mediate effects on RNA synthesis, while the CTD functions as a tether bridging the transcription apparatus and other cellular machineries [18]. Eukaryotic proteins contain multiple KOW domains as well as additional regulatory domains that serve as docking sites for other proteins [19]. In bacteria, two separate groups of NusG homologs are present [20], the first including proteins closely related to NusG and the second comprising specialized NusG proteins (NusGSP) that cluster with Escherichia coli RfaH, the best studied operon-specific NusG paralog [15].
Figure 1.
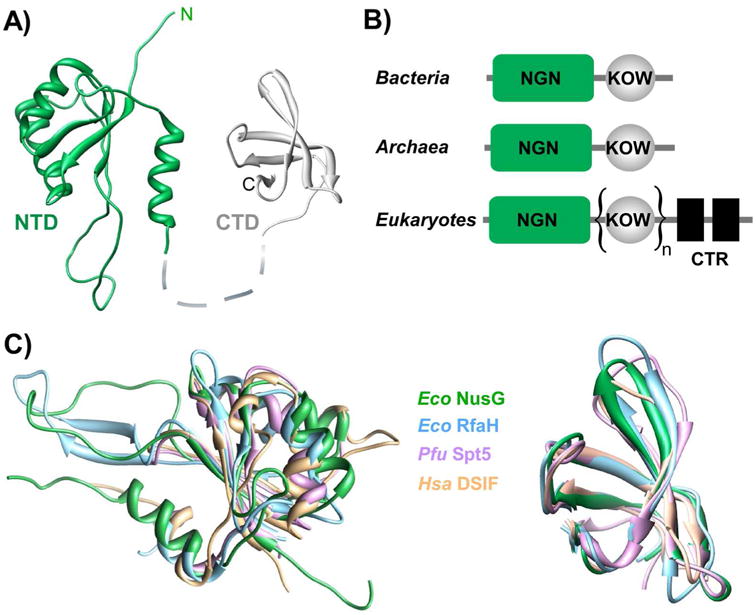
Structural conservation among NusG family members. A: E. coli NusG (PDB: 2K06, 2JVV) consists of two domains connected by a flexible linker; the NTD (green) has an NGN fold [16], the CTD has a KOW motif [17]. B: Domain organization of NusG homologs in bacteria, archaea, and eukaryotes. The NGNs bind to RNAP and the DNA [18,51,63]; the KOW domains tether transcribing RNAP to other proteins (e.g. Rho and ribosome in bacteria [86,87,107]) the C-terminal repeats (CTR) present in eukaryotes are regulated by phosphorylation and interact with diverse cellular partners [19,108-111]. C: Structural superposition of NGN (left) and KOW (right) domains prepared using Chimera. NGN domain PDB IDs: E. coli (Eco) NusG: 2KO6; E. coli RfaH: 2OUG; P. furiosus (Pfu) Spt5: 3P8B; Human (Hsa) DSIF: 3H7H; KOW PDB IDs: E. coli NusG: 2JVV; E. coli RfaH: 2LCL; P. furiosus Spt5: 3P8B; Human DSIF: 2E70.
The founding member of this family, Escherichia coli NusG, and all its characterized homologs share an ability to increase the rate of RNA chain elongation, which is frequently referred to as antitermination (AT) modification [21,22], when assayed in vitro [14,15]. Together with the absolute requirement for RNAP processivity (uninterrupted RNA synthesis between a promoter and a terminator), particularly in eukaryotes where mRNAs can be hundreds of thousands of nucleotides long and take hours to synthesize, this property was thought to be universally important, underscoring the ubiquity of NusG-like factors in all kingdoms [4]. However, recent studies painted a much more complex picture in which these proteins have context-dependent effects on RNA chain elongation, have different and sometimes opposite effects on transcription termination, and enable communications between the transcribing RNAP and diverse co-transcriptional processes that ultimately determine the gene expression program in all living cells. Studies of NusG and its homologs in bacteria were punctuated by many twists and turns, which, somewhat unexpectedly, facilitated unraveling of the molecular mechanism and revealed regulatory plasticity displayed by members of this fascinating family of regulators. In this review, we will describe the key points of these studies, point out similarities to other, more complex transcription machineries, and discuss how factors with the identical binding site on RNAP and the ubiquitous AT mechanism may have apparently opposite effects on gene expression.
E. coli NusG - the founding member of the family
NusG was identified as part of the phage λ N AT complex along with other host Nus (N-utilization substance) proteins [23]. The λ N protein binds to the nut site on the phage RNA and, together with the Nus factors, forms an AT complex that modifies RNAP into a state able to resist RNA release induced by nascent RNA structures or a termination factor Rho [22]. This modification is essential for the completion of the phage lytic cycle. NusG was also found in multi-component complexes required for efficient synthesis of rRNAs [24]. rRNAs are not translated and are thus potential targets for Rho that prematurely terminates transcription which is uncoupled from translation [8,25], e.g. at a premature stop codon [26]. rRNAs comprise the bulk of cellular RNA and their synthesis is tightly controlled to match the cell's metabolic status [27]; the maximal rate of rRNA synthesis observed at high growth rates was proposed to depend on AT [28]. This constraint could explain why NusG is essential in wild-type E. coli [29] and, consistent with its role in AT, E. coli NusG increases the RNA synthesis rate in vitro [30]. However, in both the λ and rRNA AT complexes NusG plays an auxiliary role, with another protein, λ N [31] or ribosomal protein S4 [32], acting as a principal player, respectively.
Strikingly, the essential role of NusG in E. coli turned out to be exactly the opposite, i.e. gene silencing by assisting Rho-dependent termination. Genome-wide analyses revealed that Rho and NusG jointly inhibit expression of foreign DNA elements, in particular the toxic kil gene in the rac prophage [33], and pervasive antisense transcription [25]. Consistently, NusG becomes partially dispensable when the rac prophage is deleted [33]. The enhancement of Rho-dependent termination by E. coli NusG is well-documented in vitro and is mediated by a direct contact between NusG and Rho [18,34] but may not be a common property of all NusGs. Interestingly, although Rho is essential and responsible for termination of ∼20% of transcripts in E. coli [35], NusG and Rho are dispensable in some bacteria, such as Bacillus subtilis [36] and Staphylococcus aureus [37].
By contrast, the AT modification of RNAP by NusG-like proteins appears to be universally conserved, consistent with a view that a requirement for high processivity of RNAP, which cannot rebind a prematurely released RNA, arose early in evolution [14]. The principal details of this mechanism have been elucidated, owing in equal parts to recent progress in structural analysis of transcription and an amenable model system [14,15].
A new nucleic acid target of RfaH — a key to the AT mechanism
Knowing the target site of a regulator is crucial for understanding its mechanism of action. Based on the cellular AT function and a consistent pattern of effects on elongation observed in vitro, NusG and its homologs have been annotated in genomes as AT factors, but the underlying mechanism remained elusive. Even though E. coli NusG was shown to bind to RNAP in 1992 [23], its binding site was unknown until 2007, in part because NusG is essential in wild-type E. coli and is bound to RNAP transcribing almost all genes [38]. E. coli RfaH, a sequence-specific nonessential NusG paralog that controls just a handful of genes [39], was used instead to elucidate where these factors bind on, and how they modify the properties of, the transcription elongation complex (TEC).
Genetic analysis demonstrated that a 12-nt ops sequence located in leader regions of operons under RfaH control was required for the RfaH action [39]. This sequence could be recognized by RfaH either as DNA or as RNA. RNA is a logical target for an AT factor because pulling on the nascent RNA, either by an RNA hairpin or by Rho, leads to termination [8]. The majority of characterized processive AT complexes assemble on the nascent RNA (e.g. λ N or rRNA complexes [40,41]) or act as free-standing RNA elements (e.g. put RNA [42]). The idea that RfaH could bind RNA was consistent with the presence of KOW motifs, which are found in ribosomal proteins and RNA helicases and bind RNA directly [43], in all NusG paralogs [17]. Furthermore, the ops element was shown to induce RNAP pausing in vivo [44], a regulatory property commonly attributed to RNA secondary structures [45]. However, the ops element is too short to encode a multicomponent RNA hairpin-dependent pause [46], and was shown to mediate pausing by an alternative mechanism [47] that is related to RNAP's ability to backtrack along the RNA chain [48,49]. Biochemical studies demonstrated that RfaH did not bind to the nascent RNA and instead recognized the non-template DNA strand exposed on the surface of RNAP paused at the ops site [50] (Fig. 2).
Figure 2.
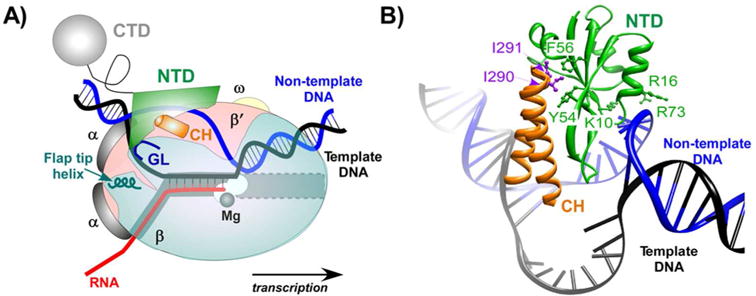
RfaH binding site on the transcription complex. A: A TEC cartoon with the template (black) and non-template (blue) DNA strands forming a transcription bubble that encompasses the nucleotides in the active site (near the catalytic Mg2+ ion; black sphere). RNAP subunits and RfaH domains are colored as follows: β (light cyan), β′ (light pink), α (grey), ω (light yellow), RfaH NTD (green), and RfaH CTD (cyan). The nascent RNA (red) pairs to the template DNA to form the RNA:DNA hybrid, and exits the complex under the β flap domain; the helix at the tip of the flap domain is shown. The same color scheme is used in other figures. The NTD directly crosslinks to the non-template DNA strand in the TEC and interacts with the β and β′ subunits with the gate loop (GL; deep cyan) and the β′ clamp helices (CH; orange) elements. B: A close-up of a structural model of RfaH/TEC complex [51]. A hydrophobic cavity on NTD interacts with two hydrophobic residues at the tip of the β′ CH (purple; Ile290 and Ile291 in E. coli), the polar and charged residues on the opposite side of the NTD interact with DNA. This model is supported by biochemical analysis of mutationally-altered RNAP and RfaH variants [51,60]. RfaH residues making the key contacts to the β′ CH (Tyr54, Phe56) and DNA (Lys10, Arg16, and Arg73) are shown.
This finding of non-template DNA interaction was the sole experimental lead that allowed us to identify the RfaH binding site on the RNAP and to propose its mode of AT action [51]. The position of the ops element is equivalent to that of the -10 element recognized by a transcription initiation factor σ in an open promoter complex [52]. Using the interaction with the non-template DNA as a constraint, RfaH was modeled to interact with the β′ clamp helices (CH) [51], the main σ binding site on the core RNAP [53]. The β′ CH appeared to be the least likely target for RfaH because NusA, which competes with σ [54] but not NusG [30] for binding to RNAP, was proposed to interact with the β′ CH based on structural modeling [55]. However, subsequent studies demonstrated that RfaH, NusG and their archaeal homolog Spt5 bind to the CH and compete with initiation factors for binding to RNAP [18,56-58], whereas NusA binds to the β flap domain [59], another contact site for the σ factor.
The position of RfaH on the TEC (Fig. 3) also suggested a plausible mechanism of AT. In addition to the β′CH, RfaH was within an interaction distance from the β gate loop (β GL), a flexible part of the β lobe located near the β′CH on the opposite side of the DNA channel. This interaction is essential for the AT activity of RfaH but not for its binding to RNAP [60,61]. By making simultaneous contacts to the β and β′ pincers, RfaH would enable the RNAP to clamp down on the DNA and inhibit isomerization into a paused state, thereby increasing the rate of transcription, as observed in vitro. Despite significant differences between bacterial and archaeal transcription complexes, archaeal Spt5 interacts with the RNAP in an identical fashion [62,63]. These results show that RNAPs utilize NusG homologs as dissociable clamps which encircle the DNA to enhance enzyme processivity, a universal strategy that is also common among DNA polymerases and helicases [64].
Figure 3.

The β′ CH as a universally conserved target site for the initiation and elongation factors. A: Crystal structure of the E.coli RNAP (PDB ID: 4IGC) with the β and β′ (cyan and light pink, respectively), α (grey) and ω (light yellow) subunits shown as ribbons. The active site Mg2+ ion (black sphere), the β GL (residues 368 to 376), and the β′ CH (residues 265 to 310) motifs are highlighted. The β′CH serves as a docking site for many initiation and elongation factors; σ forms direct polar interactions largely with the C-terminal part (red) and many indirect interactions covering nearly all of the exposed CH surface in both the E. coli and T. thermophilus RNAP holoenzymes [112,113], whereas RfaH NTD makes van der Waals contacts with the tip of the β′ CH [51]. Archaeal Spt5 proteins make identical contacts to the β′ CH [62,63]. Basal transcription initiation factors Methanocaldococcus jannaschii TFE [57] and Saccharomyces cerevisiae TFIIB [114] also interact with the CH domain, setting up an orchestrated transition from the initiation to the elongation phase through competition for the CH [115]. B: Sequence alignment of the CH domain prepared using DNAStar MegAlign module; identical residues are boxed, regions are color-coded as in panel A. Eco: E. coli; Pfu: P. furiosus; Sce: S. cerevisiae.
The non-template DNA strand – a sliding target for diverse regulators
While the nascent RNA and double-stranded DNA are recognized by a wide array of transcription factors, the non-template DNA may appear to be an odd target. It is a moving target, because the rapidly elongating RNAP spends just a fraction of a second at a given template register. Such a sliding window may present a challenge for a protein that recognizes a specific sequence, and RNAP stalling at the target site may be required to facilitate recruitment, e.g. at the ops pause site (RfaH; [50]), at a promoter-proximal pause site (Spt5; [65]), or at a lesion in the transcribed DNA (UvrD; [66]).
Base-specific interactions with the non-template DNA could play diverse roles in transcription. This specificity is well established in the case of σ [67] and RfaH [60]. A model of yeast TEC bound by Spt5 revealed an interaction between NGN and the non-template DNA [63], suggesting that NGN-DNA contacts are broadly conserved among the NusG homologs in all three domains of life. It could be expected that a nucleic acid-binding protein would exhibit some sequence preference. By analogy to promoter escape, wherein interactions between σ and promoter elements hold the RNAP back to hinder promoter clearance [68], persistent interactions with the non-template DNA would be expected to inhibit RNAP translocation. Indeed, RfaH significantly delays RNAP escape from the ops site, while reducing pausing at other signals [50], and decelerates pause-deficient RNAPs [69]. B. subtilis and Thermus thermophilus NusGs slow their cognate enzymes down [70,71], possibly due to inhibiting contacts with the non-template DNA. B. subtilis NusG stimulates pausing at U-rich sequences and protects three residues on the non-template DNA in the paused complexes [71,72], consistent with sequence-specific recognition of T-rich sequences [73]. Recent data also demonstrate the importance of direct interactions between the core RNAP and the non-template DNA. This contact was first observed in a promoter initiation complex, with a key Asp446 residue in a β subunit pocket specifically interacting with a G residue at +1 position, one nucleotide ahead of the active site [52]. This interaction appears to modulate transcription initiation and pausing in vivo [74].
Specific contacts with the non-template DNA could mediate differential recruitment of NusG homologs to RNAP transcribing different genes, control timing/position of their binding to the TEC, provide time for subsequent binding of auxiliary factors to the quaternary RNAP/NusG complex, and induce conformational changes in the TEC. Binding to the transcription bubble places a factor into a strategic position that offers unique advantages. A factor could push the RNAP forward, by favoring the strand reannealing at the rear end of the bubble, thereby aiding translocation and decreasing pausing, or push RNAP back to induce arrest. Intimate interactions between the non-template strand and the RNAP may ensure that a factor that stably associates with the non-template DNA essentially becomes an integral part of the TEC able to scan the transcribed region for a specific sequence or a DNA lesion. Recent studies show that non-template DNA is targeted by proteins that couple transcription to DNA repair and recombination. UvrD, a DNA helicase which plays a key role in nucleotide excision DNA repair pathway [75], has been proposed to contact the non-template DNA and push the RNAP stalled at a DNA lesion backwards, thus exposing the lesion to the nucleotide excision repair complex [66]. Activation-induced cytidine deaminase (AID), which induces targeted DNA breaks that give rise to antibody class switching by recombination and somatic hypermutation, also binds to the non-template DNA [76].
Pervasive association of NusG [38] and Spt5 [77] with the transcribing RNAP suggests that other regulators would cooperate or compete with these general factors to gain access to the non-template strand. For example, since σ and TFE bind to overlapping sites on the bacterial and archaeal RNAP, respectively [56,57], factor swapping downstream from a promoter could assist RNAP during promoter escape [78]. Such an effect has been documented for Pyrococcus furiosus TFE [57] which binds nonspecifically to single-stranded non-template DNA in early elongation complexes [79]. Conversely, a productive interaction may guide a factor to the non-template DNA (e.g. AID associates with RNAP via Spt5 [80]).
Binding to the non-template strand could also facilitate recruitment of other auxiliary factors to the nascent RNA, which emerges from the enzyme's exit channel nearby [81]. Spt5 interactions with the exosome may underlie exosome-mediated pre-mRNA surveillance [82], whereas Spt5-dependent recruitment of pre-mRNA cleavage factor I (CFI) [83] and mRNA capping enzymes [84] would coordinate mRNA processing and synthesis. Sub1, a factor implicated in activation of transcription, termination, and 3′-end formation, modulates the RNA elongation rate by binding to Spt5 [85]. In prokaryotic cells, NusG homologs could coordinate RNA synthesis and processing with translation. E. coli RfaH and NusG have been shown to interact with the ribosomal protein S10 [86,87] and could couple transcription to the leading round of translation [88]. Since the interaction surfaces are conserved, similar contacts could be established in archaea.
Do all NusG-like proteins work similarly?
The presence of more than one NusG-like factor in the cell implies different targets/functions or expression under specific conditions. E. coli NusG is not known to be regulated and is present in nearly all transcription units [38], suggesting that NusGSPs are required for the expression of some “special needs” genes. All characterized NusGSPs activate transcription of their targets, akin to NusG acting within large AT complexes. While mechanistic details are lacking for most of these factors, unlike NusG, RfaH functions as a stand-alone AT factor that is only effective against Rho [50,89,90].
RfaH was discovered as a factor required for lipopolysaccharide biosynthesis[91] and the expression of F plasmid tra operon [92] and shown to increase expression of the distal parts of several ops-containing operons [39], including rfb. Although the ubiquitous AT modification of RNAP could explain this effect, by helping RNAP to escape Rho kinetically [93], RfaH effects on gene expression in vivo are dramatically larger than those observed on model templates in vitro, even in the presence of cell extracts [50]. This discrepancy suggests that some features of the RfaH-controlled operons underlie these differences. In vivo analysis demonstrated that Rho and RfaH are associated with the RNAP transcribing the rfb operon, whereas NusG is not [20,38], and that Rho reduces rfb RNA synthesis ∼800-fold in the absence and only ∼2-fold in the presence of RfaH [61]. Rho-dependent termination is enhanced by NusG and inhibited by an increase in RNAP elongation rate or activation of translation [94]. Thus, such a dramatic anti-Rho effect could be due to a combination of (i) competition with NusG; (ii) antipausing activity; or (iii) increase in mRNA translation.
The first scenario is supported by the absence of NusG from all operons associated with RfaH in vivo and by RfaH's ability to exclude NusG from RNAP in vitro [20]. The second scenario is consistent with the accepted mode of action of all NusG-like factors, which is readily observed on a variety of templates in vitro [69] but appears too modest to confer dramatic effects observed in vivo. Identification of the β gate loop as an AT determinant for RfaH and NusG provided means to probe the importance of AT in transcription activation by RfaH. Deletion of the gate loop eliminated the ability of RfaH to reduce pausing in vitro, but made only a small contribution to the rfb expression in vivo [61], implying that direct effects on RNAP, even though undeniably conserved, are not sufficient for RfaH regulation. An idea that all regulation can be explained by RfaH competition with NusG is at odds with observations that NusG effects are rather modest and are exhibited only at some Rho-dependent sites [25]. What are we missing?
Complete silencing of the rfb operon by Rho suggested that this operon may be poorly translated. In fact, all experimentally verified RfaH targets lack canonical Shine-Dalgarno elements and have many rare codons, potentially hindering both the initial binding and translocation of the ribosome, and making the nascent RNA an easy target for Rho. The ribosome recruitment appears to be the major step targeted by RfaH since deletion of a Shine-Dalgarno element is sufficient to confer the dependence of a heterologous lux operon on RfaH [86]. Several lines of evidence are consistent with the direct contacts between the ribosome and RfaH, including the solution NMR structure of the RfaH CTD bound to S10 [86], and support a model in which RfaH acts in lieu of a Shine-Dalgarno element, anchoring the 30S subunit to the transcribing RNAP to increase its local concentration in the vicinity of the nascent mRNA.
Thus, RfaH may function primarily as a translation initiation factor dedicated to a small set of horizontally transferred operons. This activity requires that RfaH is recruited to RNAP at the ops site and then establishes a contact with the ribosome while being bound to RNAP. If these bridging interactions are maintained throughout the entire operon, RfaH could couple transcription to translation, thereby offering additional protection against Rho. An analogous mechanism has been proposed for NusG [95], and S10 makes essentially identical contacts to RfaH and NusG [86]. However, unlike NusG, RfaH must undergo dramatic conformational changes in order to bind to RNAP and S10.
RfaH has to transform to bind to RNAP and ribosome
In free RfaH, the interaction surfaces for RNAP and S10 are hidden (Fig. 4). The RNAP-binding site on the NTD is masked by an α-helical CTD, and binding of an exposed surface on the NTD to the ops bases is proposed to trigger the separation of the RfaH domains and permit RNAP binding. Consistently, the isolated NTD does not require ops for recruitment to the TEC [51]. This auto-inhibitory mechanism limits RfaH action to a small set of targets [20], avoiding the competition with NusG and expression of potentially harmful genes. A much more striking transformation occurs in the released CTD, which completely refolds into a β barrel that is nearly identical to that of NusG and is able to bind to S10 [86]. The CTD folds into a β-barrel when expressed alone or separated from the NTD upon proteolytic digestion of the interdomain linker. Surprisingly, the presence of the NTD is sufficient to chaperone CTD into the α-helix, even starting from a denatured chain [96]. Thus, the two alternative states of the CTD play key roles during recruitment of both the RNAP and the ribosome, prompting to coin the term “transformer protein” for RfaH [97]. The structural plasticity of this domain is astounding, particularly since the SH3 β-barrel is a rather common structural motif, and suggests that other NusG homologs may undergo similar transformations, enabling them to make different functional interactions. How fast this transition occurs, whether it is reversible at the end of the transcription cycle, whether there could be a protein that naturally exists in equilibrium between the two states, and many other questions remain open. From a practical perspective, it may not be possible to predict whether an uncharacterized NusG homolog will fold as RfaH or as NusG (or be able to change folds) because the primary sequence will likely be compatible with either fold, as is the case of RfaH. Yet the protein fold determines whether these proteins require activation by DNA binding, the extreme form of sequence dependence.
Figure 4.
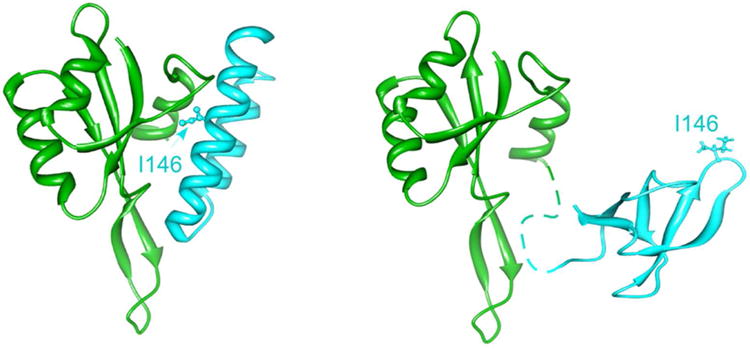
RfaH activation by transformation. In the crystal structure of free RfaH (left; 2OUG), the two domains form a large hydrophobic interface that masks the RNAP-binding site on the NTD (green) and captures the CTD cyan) in an α-helical state, in which the key residues interacting with S10 (e.g. Ile146) face the domain interface. By contrast, an isolated CTD folds as a β-barrel (right; 2LCL) in which Ile146 is exposed to bind S10. This dramatic transformation can be induced by artificial separation of the two domains in vitro [86] but requires the recognition of the ops DNA element in the course of transcription. Direct NTD/ops contacts are thought to transduce a signal to the domain interface that weakens the interactions with the CTD, allowing for domain dissociation and subsequent CTD refolding. The freed NTD and CTD can establish the contacts with RNAP and ribosome, respectively, to modulate transcription, translation, and coupling of these processes.
Alternative transcription elongation factors
Sigma competition is a paradigm for transcriptional control by sequence-specific accessory factors, which compete for the pool of free RNAP core molecules and direct them to dedicated subsets of promoters, thereby reprogramming gene expression in response to cellular triggers [5]. The most abundant primary σA (σ70 in E. coli) recognizes housekeeping promoters and is the best studied. It shares the regulatory space with several alternative σs (6 in E. coli) and binds to many regions of transcription complex, among which the non-template strand and the β′CH play the key roles during both initiation and elongation [53]. NusG and its paralogs comprise an analogous family of alternative factors that bind to an overlapping target on the TEC (Fig. 5). Similarly to σ, analysis of NusG/RfaH competition revealed clear partitioning of the transcriptome, with “housekeeping” NusG associated with most transcribed genes and RfaH - just a few. However, even in the well-studied E. coli this family includes additional members, ActX and TraB, encoded on conjugative plasmids R6K and F, respectively [98,99]; these proteins phylogenetically cluster with RfaH but nothing is known about their mechanisms.
Figure 5.

Families of alternative transcription factors. In E. coli, alternative σ factors recognize specific sequences (in double-stranded DNA and on the non-template stand in the transcription bubble) and the β′CH domain to direct RNAP to specific promoters; σ70 is the most abundant and mediates transcription of most genes. Following escape from the promoter and release of σ, alternative elongation factors bind to distinct but overlapping elements on the non-template DNA stand and modulate RNA chain elongation and translation; among them, the most abundant NusG mediates transcription of most genes. The well-established competition among the factors from each group controls initiation and elongation phases of transcription. In addition, transition between these phases, the promoter escape and core RNAP recycling after termination, may be modulated by competition between an initiation and an elongation factor for their β′CH/non-template DNA target.
How do NusGSPs find their targets? While σs achieve specificity through recognition of different DNA sequences, the mechanisms used by NusG homologs could be more multifaceted. In E. coli NusG, the two domains do not interact [100] and the RNAP-binding site is always available. In contrast, free RfaH is in autoinhibited state, and contacts to ops are required to trigger domain dissociation and expose the RNAP-binding site [51]. Masking interdomain interactions may occur even in a housekeeping NusG. In Thermotoga maritima NusG, the two domains form a stable and dynamic interface which masks the RNAP-binding site on the NTD and the hypothetical Rho- and S10-binding surfaces on the β-barrel CTD [101]. It is possible that other NusGs may establish autoinhibitory contacts that can be subject to regulation by cellular cues or specific DNA binding [73]. A NusG homolog from Streptomyces virginiae has an N-terminal extension proposed to bind butyrolactone [102]; interestingly, addition of a tag to the N-terminus of RfaH reduces dependence on the ops element, presumably due to the domain interface destabilization. Recognition of a specific DNA sequence is a common targeting mechanism, and it could be used by NusGSP regulators such as E. coli ActX and TraB; however, their gene context suggests an alternative in cis recruitment mechanism (Fig. 6) wherein the newly synthesized NTD loads onto RNAP as soon as it emerges from the trailing ribosome [1]. The cis mechanism could represent a first step in evolution of a dedicated regulator occurring early after the duplication of nusG but before acquisition of an ops-like sequence [20].
Figure 6.

Recruitment strategies of NusG-like proteins. A: A protein in an open conformation can bind to RNAP at any site, although some sequence specificity could be present. B: A protein in a closed, autoinhibited conformation requires a specific target for recruitment. C: A protein which is encoded by the first gene of the controlled operon can be recruited to RNAP as soon as it emerges from the ribosome, if translation and transcription are closely coupled. Examples of RfaH-like regulators which could be recruited in cis and are known to control their resident operons are TAA [104], AnfA1 [103], and Ubx [116].
A specific sequence may be unnecessary if a regulator has just one target. Many RfaH-like proteins are encoded as a first gene within an operon whose expression they activate, such as Serratia entomophila AnfA1 from a 30 kb prophage thought to encode a transport system for toxin delivery into insect larvae [103] and Myxococcus xanthus TAA from a ∼80 kb polyketide antibiotic gene synthesis cluster [104]. Importantly, in cis recruitment does not preclude the protein from functioning in trans, particularly if produced in large quantities, but it would allow a protein made in a single copy (relative to its target) to protect the mRNA against Rho during the pioneer round of translation. Once the complete transcript is made, it is immune to Rho, making an AT dispensable.
Having a built-in AT factor could help to evade Rho-mediated silencing upon arrival into a new host, enabling efficient horizontal transfer. ActX and TraB are encoded on transmissible plasmids in front of type IV pilus synthesis operons [98,99], which are necessary for conjugation but are likely targets for Rho. It remains to be determined if these putative regulators act via AT and require specific sequences for activation, but an idea of a self-sufficient conjugation apparatus that is immune to Rho action appears attractive.
Conclusions
In all organisms, NusG-like proteins bind to the transcribing RNAP and increase its processivity and establish communications with enzymes that carry out processing of the nascent RNA. These communications can be facilitated by physical contacts with the CTD or by transcriptional pausing induced by sequence-specific DNA contacts to the NTD; by delaying RNAP escape from a regulatory site, B. subtilis NusG [72] and RfaH [15] are thought to control the ribosome binding to the mRNA. In bacteria, NusG paralogs comprise a family of alternative transcription elongation factors that compete for binding to RNAP. Despite similar effects of these proteins on transcription in vitro [69,105], the cellular outcomes of their action are astonishingly different, including silencing of foreign and antisense RNAs by potentiating Rho-mediated transcription termination [25,33], activation of horizontally acquired genes by blocking Rho-mediated termination [61], and activation of mRNA translation, presumably by direct recruitment of the 30S ribosomal subunit [86]. Even their structures can change dramatically, enabling sequence-specific recruitment to RNAP and acquisition of new interaction surfaces. We hypothesize that alternative NusGSP proteins have evolved to work against the housekeeping NusG to allow expression of dedicated subsets of horizontally transferred operons which encode virulence and fertility determinants. While some NusGSPs reside on the chromosome and control several scattered genes, others appear to travel with their target operons on transmissible plasmids, likely contributing to the rapid spread of antibiotic resistance in clinical populations [106]. While these factors share the binding site on RNAP, structural and regulatory complexity precludes simple sequence-based prediction of their target sites and molecular mechanisms of action, making their analysis both challenging and exciting.
Acknowledgments
This project was supported by the National Institutes of Health GM67153 grant.
Abbreviations
- AT
antitermination
- CH
clamp helix
- CTD
C-terminal domain
- NGN domain
NusG-like N-terminal domain
- NTD
N-terminal domain
- NusGSP
specialized NusG proteins
- RNAP
RNA polymerase
- TEC
transcription elongation complex
Footnotes
Conflict of interest: none declared
References
- 1.Proshkin S, Rahmouni AR, Mironov A, Nudler E. Cooperation between translating ribosomes and RNA polymerase in transcription elongation. Science. 2010;328:504–8. doi: 10.1126/science.1184939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.French SL, Santangelo TJ, Beyer AL, Reeve JN. Transcription and translation are coupled in Archaea. Mol Biol Evol. 2007;24:893–5. doi: 10.1093/molbev/msm007. [DOI] [PubMed] [Google Scholar]
- 3.Braun KA, Young ET. Coupling mRNA synthesis and decay. Mol Cell Biol. 2014 doi: 10.1128/MCB.00535-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Werner F, Grohmann D. Evolution of multisubunit RNA polymerases in the three domains of life. Nat Rev Microbiol. 2011;9:85–98. doi: 10.1038/nrmicro2507. [DOI] [PubMed] [Google Scholar]
- 5.Feklistov A, Sharon BD, Darst SA, Gross CA. Bacterial Sigma Factors: A Historical, Structural, and Genomic Perspective. Annu Rev Microbiol. 2014 doi: 10.1146/annurev-micro-092412-155737. [DOI] [PubMed] [Google Scholar]
- 6.Muhlbacher W, Sainsbury S, Hemann M, Hantsche M, et al. Conserved architecture of the core RNA polymerase II initiation complex. Nature communications. 2014;5:4310. doi: 10.1038/ncomms5310. [DOI] [PubMed] [Google Scholar]
- 7.Lariviere L, Plaschka C, Seizl M, Petrotchenko EV, et al. Model of the Mediator middle module based on protein cross-linking. Nucleic Acids Res. 2013;41:9266–73. doi: 10.1093/nar/gkt704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peters JM, Vangeloff AD, Landick R. Bacterial transcription terminators: the RNA 3′-end chronicles. J Mol Biol. 2011;412:793–813. doi: 10.1016/j.jmb.2011.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nielsen S, Yuzenkova Y, Zenkin N. Mechanism of eukaryotic RNA polymerase III transcription termination. Science. 2013;340:1577–80. doi: 10.1126/science.1237934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Santangelo TJ, Reeve JN. Archaeal RNA polymerase is sensitive to intrinsic termination directed by transcribed and remote sequences. J Mol Biol. 2006;355:196–210. doi: 10.1016/j.jmb.2005.10.062. [DOI] [PubMed] [Google Scholar]
- 11.Kaplan CD. Basic mechanisms of RNA polymerase II activity and alteration of gene expression in Saccharomyces cerevisiae. Biochim Biophys Acta. 2013;1829:39–54. doi: 10.1016/j.bbagrm.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arimbasseri AG, Rijal K, Maraia RJ. Comparative overview of RNA polymerase II and III transcription cycles, with focus on RNA polymerase III termination and reinitiation. Transcription. 2013;4 doi: 10.4161/trns.27369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grohmann D, Werner F. Recent advances in the understanding of archaeal transcription. Curr Opin Microbiol. 2011;14:328–34. doi: 10.1016/j.mib.2011.04.012. [DOI] [PubMed] [Google Scholar]
- 14.Werner F. A nexus for gene expression-molecular mechanisms of Spt5 and NusG in the three domains of life. J Mol Biol. 2012;417:13–27. doi: 10.1016/j.jmb.2012.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tomar SK, Artsimovitch I. NusG-Spt5 proteins-Universal tools for transcription modification and communication. Chem Rev. 2013;113:8604–19. doi: 10.1021/cr400064k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ponting CP. Novel domains and orthologues of eukaryotic transcription elongation factors. Nucleic Acids Res. 2002;30:3643–52. doi: 10.1093/nar/gkf498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kyrpides NC, Woese CR, Ouzounis CA. KOW: a novel motif linking a bacterial transcription factor with ribosomal proteins. Trends Biochem Sci. 1996;21:425–6. doi: 10.1016/s0968-0004(96)30036-4. [DOI] [PubMed] [Google Scholar]
- 18.Mooney RA, Schweimer K, Rosch P, Gottesman M, et al. Two structurally independent domains of E. coli NusG create regulatory plasticity via distinct interactions with RNA polymerase and regulators. J Mol Biol. 2009;391:341–58. doi: 10.1016/j.jmb.2009.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hartzog GA, Fu J. The Spt4-Spt5 complex: a multi-faceted regulator of transcription elongation. Biochim Biophys Acta. 2013;1829:105–15. doi: 10.1016/j.bbagrm.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Belogurov GA, Mooney RA, Svetlov V, Landick R, et al. Functional specialization of transcription elongation factors. Embo J. 2009;28:112–22. doi: 10.1038/emboj.2008.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Santangelo TJ, Artsimovitch I. Termination and antitermination: RNA polymerase runs a stop sign. Nat Rev Microbiol. 2011;9:319–29. doi: 10.1038/nrmicro2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weisberg RA, Gottesman ME. Processive antitermination. J Bacteriol. 1999;181:359–67. doi: 10.1128/jb.181.2.359-367.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J, Horwitz R, McCracken S, Greenblatt J. NusG, a new Escherichia coli elongation factor involved in transcriptional antitermination by the N protein of phage lambda. J Biol Chem. 1992;267:6012–9. [PubMed] [Google Scholar]
- 24.Squires CL, Greenblatt J, Li J, Condon C. Ribosomal RNA antitermination in vitro: requirement for Nus factors and one or more unidentified cellular components. Proc Natl Acad Sci U S A. 1993;90:970–4. doi: 10.1073/pnas.90.3.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peters JM, Mooney RA, Grass JA, Jessen ED, et al. Rho and NusG suppress pervasive antisense transcription in Escherichia coli. Genes Dev. 2012;26:2621–33. doi: 10.1101/gad.196741.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richardson JP, Grimley C, Lowery C. Transcription termination factor rho activity is altered in Escherichia coli with suA gene mutations. Proc Natl Acad Sci U S A. 1975;72:1725–8. doi: 10.1073/pnas.72.5.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Condon C, Squires C, Squires CL. Control of rRNA transcription in Escherichia coli. Microbiological reviews. 1995;59:623–45. doi: 10.1128/mr.59.4.623-645.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klumpp S, Hwa T. Stochasticity and traffic jams in the transcription of ribosomal RNA: Intriguing role of termination and antitermination. Proc Natl Acad Sci U S A. 2008;105:18159–64. doi: 10.1073/pnas.0806084105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sullivan SL, Gottesman ME. Requirement for E. coli NusG protein in factor-dependent transcription termination. Cell. 1992;68:989–94. doi: 10.1016/0092-8674(92)90041-a. [DOI] [PubMed] [Google Scholar]
- 30.Burns CM, Richardson LV, Richardson JP. Combinatorial effects of NusA and NusG on transcription elongation and Rho-dependent termination in Escherichia coli. J Mol Biol. 1998;278:307–16. doi: 10.1006/jmbi.1998.1691. [DOI] [PubMed] [Google Scholar]
- 31.Rees WA, Weitzel SE, Yager TD, Das A, et al. Bacteriophage lambda N protein alone can induce transcription antitermination in vitro. Proc Natl Acad Sci U S A. 1996;93:342–6. doi: 10.1073/pnas.93.1.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Torres M, Condon C, Balada JM, Squires C, et al. Ribosomal protein S4 is a transcription factor with properties remarkably similar to NusA, a protein involved in both non-ribosomal and ribosomal RNA antitermination. Embo J. 2001;20:3811–20. doi: 10.1093/emboj/20.14.3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cardinale CJ, Washburn RS, Tadigotla VR, Brown LM, et al. Termination factor Rho and its cofactors NusA and NusG silence foreign DNA in E. coli. Science. 2008;320:935–8. doi: 10.1126/science.1152763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burns CM, Nowatzke WL, Richardson JP. Activation of Rho-dependent transcription termination by NusG. Dependence on terminator location and acceleration of RNA release. J Biol Chem. 1999;274:5245–51. doi: 10.1074/jbc.274.8.5245. [DOI] [PubMed] [Google Scholar]
- 35.Peters JM, Mooney RA, Kuan PF, Rowland JL, et al. Rho directs widespread termination of intragenic and stable RNA transcription. Proc Natl Acad Sci U S A. 2009;106:15406–11. doi: 10.1073/pnas.0903846106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ingham CJ, Dennis J, Furneaux PA. Autogenous regulation of transcription termination factor Rho and the requirement for Nus factors in Bacillus subtilis. Mol Microbiol. 1999;31:651–63. doi: 10.1046/j.1365-2958.1999.01205.x. [DOI] [PubMed] [Google Scholar]
- 37.Washburn RS, Marra A, Bryant AP, Rosenberg M, et al. rho is not essential for viability or virulence in Staphylococcus aureus. Antimicrob Agents Chemother. 2001;45:1099–103. doi: 10.1128/AAC.45.4.1099-1103.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mooney RA, Davis SE, Peters JM, Rowland JL, et al. Regulator trafficking on bacterial transcription units in vivo. Mol Cell. 2009;33:97–108. doi: 10.1016/j.molcel.2008.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bailey MJ, Hughes C, Koronakis V. RfaH and the ops element, components of a novel system controlling bacterial transcription elongation. Mol Microbiol. 1997;26:845–51. doi: 10.1046/j.1365-2958.1997.6432014.x. [DOI] [PubMed] [Google Scholar]
- 40.Barik S, Ghosh B, Whalen W, Lazinski D, et al. An antitermination protein engages the elongating transcription apparatus at a promoter-proximal recognition site. Cell. 1987;50:885–99. doi: 10.1016/0092-8674(87)90515-0. [DOI] [PubMed] [Google Scholar]
- 41.Li SC, Squires CL, Squires C. Antitermination of E. coli rRNA transcription is caused by a control region segment containing lambda nut-like sequences. Cell. 1984;38:851–60. doi: 10.1016/0092-8674(84)90280-0. [DOI] [PubMed] [Google Scholar]
- 42.King RA, Banik-Maiti S, Jin DJ, Weisberg RA. Transcripts that increase the processivity and elongation rate of RNA polymerase. Cell. 1996;87:893–903. doi: 10.1016/s0092-8674(00)81996-0. [DOI] [PubMed] [Google Scholar]
- 43.Ban N, Nissen P, Hansen J, Moore PB, et al. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–20. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 44.Leeds JA, Welch RA. Enhancing transcription through the Escherichia coli hemolysin operon, hlyCABD: RfaH and upstream JUMPStart DNA sequences function together via a postinitiation mechanism. J Bacteriol. 1997;179:3519–27. doi: 10.1128/jb.179.11.3519-3527.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Landick R, Yanofsky C. Stability of an RNA secondary structure affects in vitro transcription pausing in the trp operon leader region. J Biol Chem. 1984;259:11550–5. [PubMed] [Google Scholar]
- 46.Chan CL, Wang D, Landick R. Multiple interactions stabilize a single paused transcription intermediate in which hairpin to 3′ end spacing distinguishes pause and termination pathways. J Mol Biol. 1997;268:54–68. doi: 10.1006/jmbi.1997.0935. [DOI] [PubMed] [Google Scholar]
- 47.Artsimovitch I, Landick R. Pausing by bacterial RNA polymerase is mediated by mechanistically distinct classes of signals. Proc Natl Acad Sci U S A. 2000;97:7090–5. doi: 10.1073/pnas.97.13.7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Komissarova N, Kashlev M. Transcriptional arrest: Escherichia coli RNA polymerase translocates backward, leaving the 3′ end of the RNA intact and extruded. Proc Natl Acad Sci U S A. 1997;94:1755–60. doi: 10.1073/pnas.94.5.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nudler E. RNA polymerase backtracking in gene regulation and genome instability. Cell. 2012;149:1438–45. doi: 10.1016/j.cell.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Artsimovitch I, Landick R. The transcriptional regulator RfaH stimulates RNA chain synthesis after recruitment to elongation complexes by the exposed nontemplate DNA strand. Cell. 2002;109:193–203. doi: 10.1016/s0092-8674(02)00724-9. [DOI] [PubMed] [Google Scholar]
- 51.Belogurov GA, Vassylyeva MN, Svetlov V, Klyuyev S, et al. Structural basis for converting a general transcription factor into an operon-specific virulence regulator. Mol Cell. 2007;26:117–29. doi: 10.1016/j.molcel.2007.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Y, Feng Y, Chatterjee S, Tuske S, et al. Structural basis of transcription initiation. Science. 2012;338:1076–80. doi: 10.1126/science.1227786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mooney RA, Darst SA, Landick R. Sigma and RNA polymerase: an on-again, off-again relationship? Mol Cell. 2005;20:335–45. doi: 10.1016/j.molcel.2005.10.015. [DOI] [PubMed] [Google Scholar]
- 54.Gill SC, Weitzel SE, von Hippel PH. Escherichia coli sigma 70 and NusA proteins. I. Binding interactions with core RNA polymerase in solution and within the transcription complex. J Mol Biol. 1991;220:307–24. doi: 10.1016/0022-2836(91)90015-x. [DOI] [PubMed] [Google Scholar]
- 55.Borukhov S, Lee J, Laptenko O. Bacterial transcription elongation factors: new insights into molecular mechanism of action. Mol Microbiol. 2005;55:1315–24. doi: 10.1111/j.1365-2958.2004.04481.x. [DOI] [PubMed] [Google Scholar]
- 56.Sevostyanova A, Svetlov V, Vassylyev DG, Artsimovitch I. The elongation factor RfaH and the initiation factor sigma bind to the same site on the transcription elongation complex. Proc Natl Acad Sci U S A. 2008;105:865–70. doi: 10.1073/pnas.0708432105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grohmann D, Nagy J, Chakraborty A, Klose D, et al. The initiation factor TFE and the elongation factor Spt4/5 compete for the RNAP clamp during transcription initiation and elongation. Mol Cell. 2011;43:263–74. doi: 10.1016/j.molcel.2011.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hirtreiter A, Damsma GE, Cheung AC, Klose D, et al. Spt4/5 stimulates transcription elongation through the RNA polymerase clamp coiled-coil motif. Nucleic Acids Res. 2010;38:4040–51. doi: 10.1093/nar/gkq135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang X, Lewis PJ. The interaction between RNA polymerase and the elongation factor NusA. RNA Biol. 2010;7:272–5. doi: 10.4161/rna.7.3.12021. [DOI] [PubMed] [Google Scholar]
- 60.Belogurov GA, Sevostyanova A, Svetlov V, Artsimovitch I. Functional regions of the N-terminal domain of the antiterminator RfaH. Mol Microbiol. 2010;76:286–301. doi: 10.1111/j.1365-2958.2010.07056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sevostyanova A, Belogurov GA, Mooney RA, Landick R, et al. The beta subunit gate loop is required for RNA polymerase modification by RfaH and NusG. Mol Cell. 2011;43:253–62. doi: 10.1016/j.molcel.2011.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Klein BJ, Bose D, Baker KJ, Yusoff ZM, et al. RNA polymerase and transcription elongation factor Spt4/5 complex structure. Proc Natl Acad Sci U S A. 2011;108:546–50. doi: 10.1073/pnas.1013828108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Martinez-Rucobo FW, Sainsbury S, Cheung AC, Cramer P. Architecture of the RNA polymerase-Spt4/5 complex and basis of universal transcription processivity. Embo J. 2011;30:1302–10. doi: 10.1038/emboj.2011.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Svetlov V, Nudler E. Clamping the clamp of RNA polymerase. Embo J. 2011;30:1190–1. doi: 10.1038/emboj.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Andrulis ED, Guzman E, Doring P, Werner J, et al. High-resolution localization of Drosophila Spt5 and Spt6 at heat shock genes in vivo: roles in promoter proximal pausing and transcription elongation. Genes Dev. 2000;14:2635–49. doi: 10.1101/gad.844200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Epshtein V, Kamarthapu V, McGary K, Svetlov V, et al. UvrD facilitates DNA repair by pulling RNA polymerase backwards. Nature. 2014;505:372–7. doi: 10.1038/nature12928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Feklistov A, Darst SA. Structural basis for promoter-10 element recognition by the bacterial RNA polymerase sigma subunit. Cell. 2011;147:1257–69. doi: 10.1016/j.cell.2011.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Revyakin A, Liu C, Ebright RH, Strick TR. Abortive initiation and productive initiation by RNA polymerase involve DNA scrunching. Science. 2006;314:1139–43. doi: 10.1126/science.1131398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Svetlov V, Belogurov GA, Shabrova E, Vassylyev DG, et al. Allosteric control of the RNA polymerase by the elongation factor RfaH. Nucleic Acids Res. 2007;35:5694–705. doi: 10.1093/nar/gkm600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sevostyanova A, Artsimovitch I. Functional analysis of Thermus thermophilus transcription factor NusG. Nucleic Acids Res. 2010;38:7432–45. doi: 10.1093/nar/gkq623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yakhnin AV, Yakhnin H, Babitzke P. Function of the Bacillus subtilis transcription elongation factor NusG in hairpin-dependent RNA polymerase pausing in the trp leader. Proc Natl Acad Sci U S A. 2008;105:16131–6. doi: 10.1073/pnas.0808842105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yakhnin AV, Babitzke P. Mechanism of NusG-stimulated pausing, hairpin-dependent pause site selection and intrinsic termination at overlapping pause and termination sites in the Bacillus subtilis trp leader. Mol Microbiol. 2010;76:690–705. doi: 10.1111/j.1365-2958.2010.07126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yakhnin AV, Babitzke P. NusG/Spt5: are there common functions of this ubiquitous transcription elongation factor? Curr Opin Microbiol. 2014;18:68–71. doi: 10.1016/j.mib.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vvedenskaya IO, Vahedian-Movahed H, Bird JG, Knoblauch JG, et al. Transcription. Interactions between RNA polymerase and the “core recognition element” counteract pausing. Science. 2014;344:1285–9. doi: 10.1126/science.1253458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Spivak G, Ganesan AK. The complex choreography of transcription-coupled repair. DNA Repair (Amst) 2014;19:64–70. doi: 10.1016/j.dnarep.2014.03.025. [DOI] [PubMed] [Google Scholar]
- 76.Daniel JA, Nussenzweig A. The AID-induced DNA damage response in chromatin. Mol Cell. 2013;50:309–21. doi: 10.1016/j.molcel.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mayer A, Lidschreiber M, Siebert M, Leike K, et al. Uniform transitions of the general RNA polymerase II transcription complex. Nat Struct Mol Biol. 2010;17:1272–8. doi: 10.1038/nsmb.1903. [DOI] [PubMed] [Google Scholar]
- 78.Blombach F, Daviter T, Fielden D, Grohmann D, et al. Archaeology of RNA polymerase: factor swapping during the transcription cycle. Biochem Soc Trans. 2013;41:362–7. doi: 10.1042/BST20120274. [DOI] [PubMed] [Google Scholar]
- 79.Grunberg S, Bartlett MS, Naji S, Thomm M. Transcription factor E is a part of transcription elongation complexes. J Biol Chem. 2007;282:35482–90. doi: 10.1074/jbc.M707371200. [DOI] [PubMed] [Google Scholar]
- 80.Pavri R, Gazumyan A, Jankovic M, Di Virgilio M, et al. Activation-induced cytidine deaminase targets DNA at sites of RNA polymerase II stalling by interaction with Spt5. Cell. 2010;143:122–33. doi: 10.1016/j.cell.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vassylyev DG, Vassylyeva MN, Perederina A, Tahirov TH, et al. Structural basis for transcription elongation by bacterial RNA polymerase. Nature. 2007;448:157–62. doi: 10.1038/nature05932. [DOI] [PubMed] [Google Scholar]
- 82.Andrulis ED, Werner J, Nazarian A, Erdjument-Bromage H, et al. The RNA processing exosome is linked to elongating RNA polymerase II in Drosophila. Nature. 2002;420:837–41. doi: 10.1038/nature01181. [DOI] [PubMed] [Google Scholar]
- 83.Mayer A, Schreieck A, Lidschreiber M, Leike K, et al. The spt5 C-terminal region recruits yeast 3′ RNA cleavage factor I. Mol Cell Biol. 2012;32:1321–31. doi: 10.1128/MCB.06310-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lidschreiber M, Leike K, Cramer P. Cap completion and C-terminal repeat domain kinase recruitment underlie the initiation-elongation transition of RNA polymerase II. Mol Cell Biol. 2013;33:3805–16. doi: 10.1128/MCB.00361-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Garcia A, Collin A, Calvo O. Sub1 associates with Spt5 and influences RNA polymerase II transcription elongation rate. Molecular biology of the cell. 2012;23:4297–312. doi: 10.1091/mbc.E12-04-0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Burmann BM, Knauer SH, Sevostyanova A, Schweimer K, et al. An alpha helix to beta barrel domain switch transforms the transcription factor RfaH into a translation factor. Cell. 2012;150:291–303. doi: 10.1016/j.cell.2012.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Burmann BM, Schweimer K, Luo X, Wahl MC, et al. A NusE:NusG complex links transcription and translation. Science. 2010;328:501–4. doi: 10.1126/science.1184953. [DOI] [PubMed] [Google Scholar]
- 88.Roberts JW. Molecular biology. Syntheses that stay together. Science. 2010;328:436–7. doi: 10.1126/science.1189971. [DOI] [PubMed] [Google Scholar]
- 89.Farewell A, Brazas R, Davie E, Mason J, et al. Suppression of the abnormal phenotype of Salmonella typhimurium rfaH mutants by mutations in the gene for transcription termination factor Rho. J Bacteriol. 1991;173:5188–93. doi: 10.1128/jb.173.16.5188-5193.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Stevens MP, Clarke BR, Roberts IS. Regulation of the Escherichia coli K5 capsule gene cluster by transcription antitermination. Mol Microbiol. 1997;24:1001–12. doi: 10.1046/j.1365-2958.1997.4241780.x. [DOI] [PubMed] [Google Scholar]
- 91.Wilkinson RG, Gemski P, Jr, Stocker BA. Non-smooth mutants of Salmonella typhimurium: differentiation by phage sensitivity and genetic mapping. J Gen Microbiol. 1972;70:527–54. doi: 10.1099/00221287-70-3-527. [DOI] [PubMed] [Google Scholar]
- 92.Beutin L, Achtman M. Two Escherichia coli chromosomal cistrons, sfrA and sfrB, which are needed for expression of F factor tra functions. J Bacteriol. 1979;139:730–7. doi: 10.1128/jb.139.3.730-737.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Burns CM, Richardson JP. NusG is required to overcome a kinetic limitation to Rho function at an intragenic terminator. Proc Natl Acad Sci U S A. 1995;92:4738–42. doi: 10.1073/pnas.92.11.4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Boudvillain M, Figueroa-Bossi N, Bossi L. Terminator still moving forward: expanding roles for Rho factor. Curr Opin Microbiol. 2013;16:118–24. doi: 10.1016/j.mib.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 95.Burmann BM, Rosch P. The role of E. coli Nus-factors in transcription regulation and transcription:translation coupling: From structure to mechanism. Transcription. 2011;2:130–4. doi: 10.4161/trns.2.3.15671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tomar SK, Knauer SH, Nandymazumdar M, Rosch P, et al. Interdomain contacts control folding of transcription factor RfaH. Nucleic Acids Res. 2013;41:10077–85. doi: 10.1093/nar/gkt779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Knauer SH, Artsimovitch I, Rosch P. Transformer proteins. Cell Cycle. 2012;11:4289–90. doi: 10.4161/cc.22468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Arutyunov D, Frost LS. F conjugation: back to the beginning. Plasmid. 2013;70:18–32. doi: 10.1016/j.plasmid.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 99.Nunez B, Avila P, de la Cruz F. Genes involved in conjugative DNA processing of plasmid R6K. Mol Microbiol. 1997;24:1157–68. doi: 10.1046/j.1365-2958.1997.4111778.x. [DOI] [PubMed] [Google Scholar]
- 100.Burmann BM, Scheckenhofer U, Schweimer K, Rosch P. Domain interactions of the transcription-translation coupling factor Escherichia coli NusG are intermolecular and transient. Biochem J. 2011;435:783–9. doi: 10.1042/BJ20101679. [DOI] [PubMed] [Google Scholar]
- 101.Drogemuller J, Stegmann CM, Mandal A, Steiner T, et al. An autoinhibited state in the structure of Thermotoga maritima NusG. Structure. 2013;21:365–75. doi: 10.1016/j.str.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Okamoto S, Nihira T, Kataoka H, Suzuki A, et al. Purification and molecular cloning of a butyrolactone autoregulator receptor from Streptomyces virginiae. J Biol Chem. 1992;267:1093–8. [PubMed] [Google Scholar]
- 103.Hurst MR, Beard SS, Jackson TA, Jones SM. Isolation and characterization of the Serratia entomophila antifeeding prophage. FEMS Microbiol Lett. 2007;270:42–8. doi: 10.1111/j.1574-6968.2007.00645.x. [DOI] [PubMed] [Google Scholar]
- 104.Paitan Y, Orr E, Ron EZ, Rosenberg E. A NusG-like transcription anti-terminator is involved in the biosynthesis of the polyketide antibiotic TA of Myxococcus xanthus. FEMS Microbiol Lett. 1999;170:221–7. doi: 10.1111/j.1574-6968.1999.tb13377.x. [DOI] [PubMed] [Google Scholar]
- 105.Herbert KM, Zhou J, Mooney RA, Porta AL, et al. E. coli NusG inhibits backtracking and accelerates pause-free transcription by promoting forward translocation of RNA polymerase. J Mol Biol. 2010;399:17–30. doi: 10.1016/j.jmb.2010.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chen L, Chavda KD, Fraimow HS, Mediavilla JR, et al. Complete nucleotide sequences of blaKPC-4- and blaKPC-5-harboring IncN and IncX plasmids from Klebsiella pneumoniae strains isolated in New Jersey. Antimicrob Agents Chemother. 2013;57:269–76. doi: 10.1128/AAC.01648-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chalissery J, Muteeb G, Kalarickal NC, Mohan S, et al. Interaction surface of the transcription terminator Rho required to form a complex with the C-terminal domain of the antiterminator NusG. J Mol Biol. 2011;405:49–64. doi: 10.1016/j.jmb.2010.10.044. [DOI] [PubMed] [Google Scholar]
- 108.Doamekpor SK, Sanchez AM, Schwer B, Shuman S, et al. How an mRNA capping enzyme reads distinct RNA polymerase II and Spt5 CTD phosphorylation codes. Genes Dev. 2014;28:1323–36. doi: 10.1101/gad.242768.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chen Y, Yamaguchi Y, Tsugeno Y, Yamamoto J, et al. DSIF, the Paf1 complex, and Tat-SF1 have nonredundant, cooperative roles in RNA polymerase II elongation. Genes Dev. 2009;23:2765–77. doi: 10.1101/gad.1834709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ding B, LeJeune D, Li S. The C-terminal repeat domain of Spt5 plays an important role in suppression of Rad26-independent transcription coupled repair. J Biol Chem. 2010;285:5317–26. doi: 10.1074/jbc.M109.082818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhou K, Kuo WH, Fillingham J, Greenblatt JF. Control of transcriptional elongation and cotranscriptional histone modification by the yeast BUR kinase substrate Spt5. Proc Natl Acad Sci U S A. 2009;106:6956–61. doi: 10.1073/pnas.0806302106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Murakami KS. X-ray crystal structure of Escherichia coli RNA polymerase sigma70 holoenzyme. J Biol Chem. 2013;288:9126–34. doi: 10.1074/jbc.M112.430900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Vassylyev DG, Sekine S, Laptenko O, Lee J, et al. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 A resolution. Nature. 2002;417:712–9. doi: 10.1038/nature752. [DOI] [PubMed] [Google Scholar]
- 114.Sainsbury S, Niesser J, Cramer P. Structure and function of the initially transcribing RNA polymerase II-TFIIB complex. Nature. 2013;493:437–40. doi: 10.1038/nature11715. [DOI] [PubMed] [Google Scholar]
- 115.Hartzog GA, Kaplan CD. Competing for the clamp: promoting RNA polymerase processivity and managing the transition from initiation to elongation. Mol Cell. 2011;43:161–3. doi: 10.1016/j.molcel.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 116.Chatzidaki-Livanis M, Coyne MJ, Comstock LE. A family of transcriptional antitermination factors necessary for synthesis of the capsular polysaccharides of Bacteroides fragilis. J Bacteriol. 2009;191:7288–95. doi: 10.1128/JB.00500-09. [DOI] [PMC free article] [PubMed] [Google Scholar]


