Abstract
Mutations in the BCR-ABL1 kinase domain are an established mechanism of tyrosine kinase inhibitor (TKI) resistance in Philadelphia chromosome-positive leukemia, but fail to explain many cases of clinical TKI failure. In contrast, it is largely unknown why some patients fail TKI therapy despite continued suppression of BCR-ABL1 kinase activity, a situation termed BCRABL1 kinase-independent TKI resistance. Here, we identified activation of signal transducer and activator of transcription 3 (STAT3) by extrinsic or intrinsic mechanisms as an essential feature of BCR-ABL1 kinase-independent TKI resistance. By combining synthetic chemistry, in vitro reporter assays, and molecular dynamics-guided rational inhibitor design and high-throughput screening, we discovered BP-5-087, a potent and selective STAT3 SH2 domain inhibitor that reduces STAT3 phosphorylation and nuclear transactivation. Computational simulations, fluorescence polarization assays, and hydrogen-deuterium exchange assays establish direct engagement of STAT3 by BP-5-087 and provide a high-resolution view of the STAT3 SH2 domain/BP-5-087 interface. In primary cells from CML patients with BCR-ABL1 kinase-independent TKI resistance, BP-5-087 (1.0 μM) restored TKI sensitivity to therapy-resistant CML progenitor cells, including leukemic stem cells (LSCs). Our findings implicate STAT3 as a critical signaling node in BCR-ABL1 kinase-independent TKI resistance, and suggest that BP-5-087 has clinical utility for treating malignancies characterized by STAT3 activation.
INTRODUCTION
Chronic myeloid leukemia (CML) is caused by the BCR-ABL1 tyrosine kinase, the result of the t(9;22)(q34;q11) translocation, which is cytogenetically visible as the Philadelphia chromosome (Ph). Targeting BCR-ABL1 with tyrosine kinase inhibitors (TKIs) such as imatinib induces complete cytogenetic responses in many patients with chronic phase CML (CP-CML)1. However, ~20-30% of CP-CML patients fail imatinib due to primary or acquired resistance2, and TKI responses in patients with blastic phase CML (BP-CML) are not durable.
Point mutations in the BCR-ABL1 kinase domain are the most commonly cited mechanism of TKI resistance3, 4. Beyond imatinib, the regulatory approval of four additional TKIs with differing point mutation susceptibilities renders this mechanism of resistance clinically addressable5. However, BCR-ABL1 point mutations fail to explain many cases of clinical TKI failure, as many patients with resistance express exclusively native BCR-ABL1. In these cases, BCR-ABL1 kinase-independent mechanisms activate alternative signaling pathways that maintain survival despite BCR-ABL1 inhibition6. BCR-ABL1 kinase-independent resistance likely plays a key role in preventing disease eradication in patients responding to therapy, as imatinib inhibits BCR-ABL1 kinase activity but does not trigger cell death in primitive CML cells cultured ex vivo7, 8.
Activation of signal transducer and activator of transcription 3 (STAT3) by bone marrow (BM)-derived factors protects CML cells upon TKI-mediated BCR-ABL1 inhibition9, 10. We now demonstrate that, in CML patients with BCR-ABL1 kinase-independent resistance, STAT3 is activated without a requirement for BM-derived factors, and represents a major signaling node conferring TKI resistance. We hypothesized that targeting STAT3 in addition to BCR-ABL1 would resensitize CML cells with kinase-independent resistance to TKI therapy. Using structure-activity relationship (SAR) studies and compound library screens, we identified BP-5-087, a potent and selective STAT3 inhibitor. Computational simulations and hydrogen-deuterium exchange assays confirmed binding of BP-5-087 to the STAT3 SH2 domain. Experiments on TKI-resistant CML cell lines and primary CML stem and progenitor cells reveal that BP-5-087 restores TKI sensitivity in vitro and ex vivo, with no toxicity to normal hematopoietic stem or progenitor cells. We conclude that targeting STAT3 with BP-5-087 may be useful for the treatment of kinase-independent TKI resistance and for other malignancies driven by STAT3 activation.
MATERIALS AND METHODS
Cell cultures and primary cells
Imatinib-resistant K562R and AR230R cells were derived by long-term culture in the presence of low-dose imatinib, followed by incremental increases in concentration (0.1-1.0 μM imatinib), and maintenance of a single clone in continual 1.0 μM imatinib (isogenic TKI-sensitive K562S and AR230S cells were used as controls). Mononuclear cells (MNCs) from peripheral blood (PB) of CP-CML patients or healthy donors were CD34+ selected to >90% purity and kept overnight without cytokines prior to use in 96 hr inhibitor treatment assays. All donors gave informed consent and all studies were approved by The University of Utah Institutional Review Board (IRB). For additional information see Supplementary Materials and Methods.
Clonogenic assays
Methylcellulose colony assays were performed by plating CML cell lines or patient samples in 0.9% MethoCult (H4230; Stem Cell Technologies). For cell lines, 103 cells were plated in cytokine-free conditions +/− imatinib (1.0 μM) and/or the indicated STAT3 inhibitors. For patient samples, cells were treated for 96 hr in regular medium (RM) or HS-5 conditioned medium (CM), without additional cytokines, +/− the indicated inhibitors. Following culture, 103 viable CD34+ cells were plated in the presence of rhIL-3 (20 ng/mL), rhIL-6 (20 ng/mL), rhFlt-3 ligand (100 ng/mL), and rhSCF (100 ng/mL).
Immunoblot analysis
For CML cell lines, 1.5×105 cells/mL were cultured in an equal volume of either RM or HS-5 CM and treated with TKI for 24-36 hr. For CMLCD34+ cells, assays were performed with 106 cells/mL in RM or HS-5 CM containing 10% BIT9500 instead of FBS, in the absence of cytokines, and treated with TKI for 24 hr. For additional information see Supplementary Materials and Methods.
Pharmacologic inhibitors
Imatinib was a gift from Novartis. Dasatinib and nilotinib were purchased from Selleck. STAT3 inhibitors were produced as described in Chemical Methods. Proliferation was assessed by methanethiosulfonate (MTS)-based viability assay (CellTiter 96 AQueous One; Promega).
Fluorescence polarization (FP) assay
To assess STAT3 SH2 domain binding, a high-throughput FP assay was used as described11.
Chemical Methods
Synthesis and characterization of STAT3 inhibitors was performed as described in Supplementary Materials and Methods. Briefly, all compounds were purified by silica gel column chromatography, with final molecules characterized by both 1H and 13C NMR and high resolution mass spectrometry. Inhibitor purity was evaluated by analytical reversed-phase HPLC. Prior to biological testing, all compounds were subject to fluorescence polarization and/or luciferase inhibitor screening as described.
Luciferase inhibitor screen
To detect endogenous STAT3 activity, AR230R cells were transduced with the pGreenFire Lenti-Reporter system (pGF1; System Biosciences) harboring two sequential STAT3-inducible elements (SIE) or mutated negative control (NEG) sequences (Supplementary Figure 3b). AR230R cells (3×105) expressing either pGF1-SIE (AR230R-SIE) or pGF1-NEG (AR230R-NEG) were exposed to imatinib (1.0 μM) and/or STAT3 inhibitors (5-10 μM) for 6 hr, followed by detection of luciferase reporter activity. See Statistical Analyses and Supplementary Materials and Methods for details.
Docking simulations
The crystal structure of the STAT3B-DNA complex (PDB entry 1BG1) utilized for docking was prepared with Protein Preparation Wizard. Initial docking simulation was performed using Glide Extra Precision (GlideXP module, version 5.7) (Suite 2012: LigPrep, version 2.5, Schrödinger, LLC, New York, NY, 2012), followed by induced fit docking simulation12. Residues within 7 Å of the initial binding pose were optimized by side chain reorientations in Prime module. Receptor and ligand van der Waals spheres were scaled by a factor of 0.5 to allow unusual contacts, then refined by Prime module to readjust orientation. See Supplementary Materials and Methods.
STAT3 analysis by site-specific TRESI-MS/HDX
STAT3 residues 127-688 (pET15b_STAT3, provided by Dr. Rob Laister) was subcloned into pMAL-c5X (New England Biolabs) to generate an N-terminal MBP-tagged fusion. MBPSTAT3(127-688) was expressed in E. coli BL21(DE3) and purified by amylose-affinity chromatography. MBP-STAT3(127-688) samples were prepared for mass spectrometry (MS) by buffer exchange into 100 mM ammonium acetate (pH 7.5) on a Vivaspin 20 (GE Healthcare). BP-5-087 (200 mM) was dissolved in DMSO. MBP-STAT3(127-688) (80 μM) was incubated with or without BP-5-087 (600 μM) for 2 hr on ice. Site-specific time-resolved electrospray ionization mass spectrometry (TRESI-MS) and hydrogen-deuterium exchange (HDX) was conducted on a microfluidic device13 as described in Supplementary Materials and Methods.
Long-term culture-initiating cell (LTC-IC) assays
Following 96 hr culture +/− imatinib (2.5 μM) and/or BP-5-087 (1 μM), in the absence of cytokines, 5x103 viable CD34+ cells were plated in MyeloCult (H5100; Stem Cell Technologies) on top of irradiated (80 Gy) M210B4 cells in duplicate LTC-IC assays as described14, 15. Following 6 weeks of culture, cells were trypsinized, plated into methylcellulose colony assays (H4435; Stem Cell Technologies), and scored after 18 days. Colony numbers were adjusted to reflect the total number of viable LTC-ICs present following the 96 hr culture. BCR-ABL1+ colonies were identified by qRT-PCR for BCR-ABL1 mRNA16.
Cytospin and immunofluorescence
CMLCD34+ cells were cultured for 24 hr in the indicated conditions prior to cytospin. Cells were fixed, permeabilized, and incubated with rabbit anti-pSTAT3Y705 (Cell Signaling Technologies), followed by detection using an AlexaFluor 594-conjugated goat anti-rabbit IgG (Invitrogen). Slides were examined using a Nikon Eclipse E600 equipped with a CRI Nuance multispectral imaging system (model N-MSI-420-FL).
Statistical analyses
A two-tailed Student's t test was used for assays with identical cell lines and for immunoblot densitometry. Luminescence of SIE and NEG constructs were assessed in triplicate for 74 inhibitors and standardized to 6 measures of luciferase control for a given construct in each run. A total of three such runs were independently performed. Luciferase controls were assessed for normality in each construct/run. One construct in the third run had a wide bimodal distribution, and was hence excluded from analyses based on non-uniformity of controls. Average values for each inhibitor's effects on SIE and NEG constructs were calculated and plotted to identify those with the most potent (assessed by a high negative SIE luminescence value) and selective (assessed by a high NEG value) luciferase inhibition. Patient CMLCD34+ colony data was analyzed using Welch's t-test for unequal variances. Data were considered statistically different when p values were <0.05. For MTS assays, three distinct runs each with 4 replicates per concentration were performed on unique plates with untreated controls. Median values for each concentration were calculated as a percentage of the plate's control. IC50 values were calculated from a 4-parameter variable-slope logistic equation: and fit by Prism Software. Significant differences in IC50 run values between inhibitors was calculated by Welch's t-test.
RESULTS
STAT3 is activated in BCR-ABL1 kinase-independent TKI resistance
TKI resistance in CML occurs through reactivation of BCR-ABL1 by kinase domain mutations, or through mechanisms allowing survival despite continued BCR-ABL1 inhibition, known as kinase-independent resistance. The latter may be caused through activation of alternative signaling pathways by extrinsic, BM-derived factors, or through intrinsic, cell-autonomous mechanisms. To model extrinsic resistance, we cultured CMLCD34+ cells from newly diagnosed patients or parental TKI-sensitive cell lines (K562S and AR230S) in HS-5 CM9, 10. To model intrinsic resistance without BM-derived factors, we used CMLCD34+ cells from patients with treatment failure on two or more TKIs, as well as TKI-resistant CML cell lines adapted for growth in the presence of 1.0 μM imatinib (K562R and AR230R). These cells demonstrate cross-resistance to nilotinib and dasatinib, thereby modeling resistance to multiple TKIs as observed in our patient samples (Supplementary Figures 1 and 2). All cells express exclusively native BCRABL1 and therefore harbor no detectable kinase domain mutations. For details on primary samples see Supplementary Table 1.
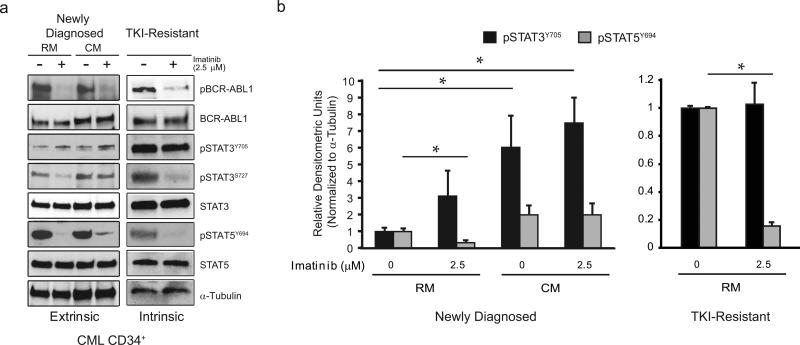
TKI-sensitive or TKI-resistant CML cell lines and primary CMLCD34+ cells were cultured in RM or HS-5 CM +/− imatinib. To identify active signaling pathways common to BCR-ABL1 kinase-independent resistance, phosphorylated versions of canonical signaling proteins were examined by immunoblot analyses. Regardless of TKI sensitivity, imatinib suppressed BCRABL1 kinase activity (Figure 1a; Supplementary Figure 3a), verifying kinase-independent resistance. As expected, in extrinsic, BM-derived TKI resistance, pSTAT3Y705 levels were elevated in parental CMLCD34+ cells from newly diagnosed CML patients, as well as K562S and AR230S cells, when cultured in HS-5 CM compared to RM in the presence of imatinib (Figures 1a and 1b; Supplementary Figures 3b and 3c). In contrast, imatinib markedly reduced pSTAT5Y694 and pSTAT3S727 levels in both RM and HS-5 CM.
Figure 1. pSTAT3Y705 is activated in TKI-resistant CML cells in the presence of imatinib.
(a) CML CD34+ cells from newly diagnosed or TKI-resistant patients lacking BCR-ABL1 kinase domain mutations were cultured in RM or HS-5 CM with or without 2.5 μM imatinib for 24 hr followed by immunoblot with the specified antibodies. Activated pBCR-ABL1 was detected using a phosphotyrosine-specific antibody. The dose of imatinib was chosen to achieve near complete suppression of BCR-ABL1 kinase activity. pSTAT3Y705 was elevated in CD34+ cells from newly diagnosed patients when cultured in HS-5 CM (n=5), and in CD34+ cells from TKI-resistant patients (n=5) in the presence of imatinib. TKI-sensitive CD34+ cells cultured in RM (n=5) were examined as controls. (b) Data presented in panel a are quantified by densitometry for pSTAT3Y705 and pSTAT5Y694 for both newly diagnosed (n=4) and TKI-resistant (n=5) patients. Error bars represent SEM. *p<0.05.
Similar to extrinsic resistance, pSTAT3Y705 levels were elevated in CD34+ cells from TKI-resistant compared to treatment-naïve newly diagnosed CML patients, in the absence of exogenous cytokines (Figures 1a and 1b). Three of five samples from newly diagnosed patients had no detectable levels of pSTAT3Y705 when cultured in RM, whereas pSTAT3S727 and total STAT3 were readily detectable. In contrast, pSTAT3Y705 was highly detectable in all five TKI-resistant samples analyzed, in both the presence and absence of imatinib. However, not all samples were run on the same blot, precluding direct quantitative comparisons between newly diagnosed and TKI-resistant samples. In contrast to pSTAT3Y705, imatinib reduced the levels of pSTAT5Y694 and pSTAT3S727 in both newly diagnosed and TKI-resistant CML cells, implying that these sites remain under the control of BCR-ABL1 (Figures 1a and 1b). In intrinsically TKI-resistant K562R and AR230R cell lines, imatinib suppression of BCR-ABL1 kinase activity also correlated with increased pSTAT3Y705 levels compared to parental K562S and AR230S controls under the same conditions (Supplementary Figure 3a). Importantly, pSTAT3Y705 activation was maintained in K562R and AR230R cells treated with imatinib, nilotinib, or dasatinib (Supplementary Figure 2). K562R and AR230R cells demonstrated increased levels of the SRC family kinases, pSRCY416 and pLYNY507, which decreased with dasatinib in a concentration-dependent manner (Supplementary Figure 2). In both cell lines, dasatinib resulted in a partial reduction of pSTAT3Y705 that was not further reduced with escalating dasatinib concentrations, consistent with partial but not full dependence on SRC family kinases (Supplementary Figure 2). pJAK2Y1007/1008 was also elevated in K562R and AR230R cells in the absence of TKIs, but were reduced to low levels in the presence of TKIs (Supplementary Figure 2), implying that JAK2 is not directly involved in STAT3 activation.
Altogether, we conclude that CML cells with intrinsic TKI resistance activate pSTAT3Y705 upon BCR-ABL1 inhibition in the absence of BM-derived factors, suggesting that pSTAT3Y705 represents a point of convergence for extrinsic and intrinsic resistance pathways promoting kinase-independent resistance.
STAT3 inhibition reduces survival of TKI-resistant CML cell lines and primary CMLCD34+ progenitor cells
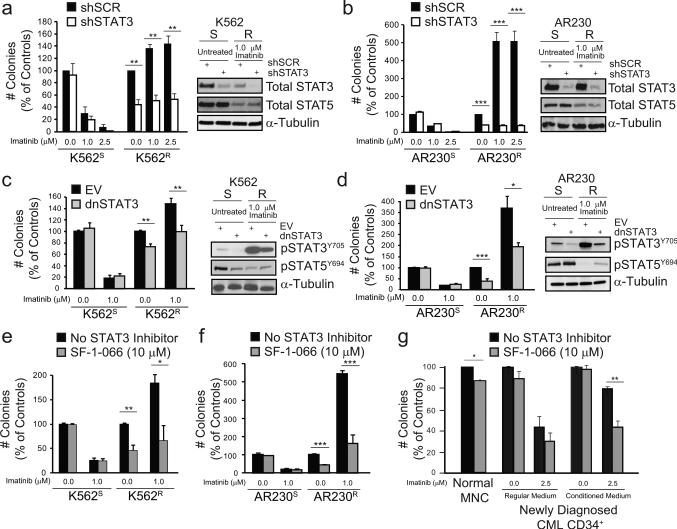
To determine whether STAT3 confers TKI resistance, we used retroviral or lentiviral delivery of shRNA targeting STAT3 (shSTAT3). shRNA-mediated knockdown of STAT3 and not STAT5 was confirmed by immunoblot analyses (Figures 2a and 2b). We initially showed that STAT3 is required for extrinsic resistance by infecting parental K562S cells with a puromycin-selectable vector harboring shSTAT3 or scrambled control (shSCR), followed by culture in HS-5 CM or RM +/− imatinib. As expected, shSTAT3 reduced colony formation and increased apoptosis of K562S cells following culture in HS-5 CM but not RM, thereby abolishing the protective effects of BM-derived factors (Supplementary Figures 4a and 4b). Consistent with STAT3 activation in intrinsic TKI resistance (Figure 1), shSTAT3 reduced the clonogenicity of K562R and AR230R cells by 50-60% in the presence of 1.0 or 2.5 μM imatinib, with no effect in TKI-sensitive parental controls (Figures 2a and 2b). We then functionally inhibited STAT3 using dominant-negative mutants (dnSTAT3)17, 18, both a C-terminal truncated mutant17 and a STAT3-Y705F mutant18, yielding the same results. Similar to shSTAT3, dnSTAT3 reduced colony formation of K562R and AR230R cells by ~30-60%, without significant effects on parental TKI-sensitive controls, and this was correlated with reduction of pSTAT3Y705 but not pSTAT5Y694 (Figures 2c and 2d). We consistently observed a reduction of colony formation upon imatinib withdrawal from K562R and AR230R cells. This is reminiscent of previous observations in imatinib-resistant Ba/F3 cells, where increased cell death was observed following drug withdrawal19.
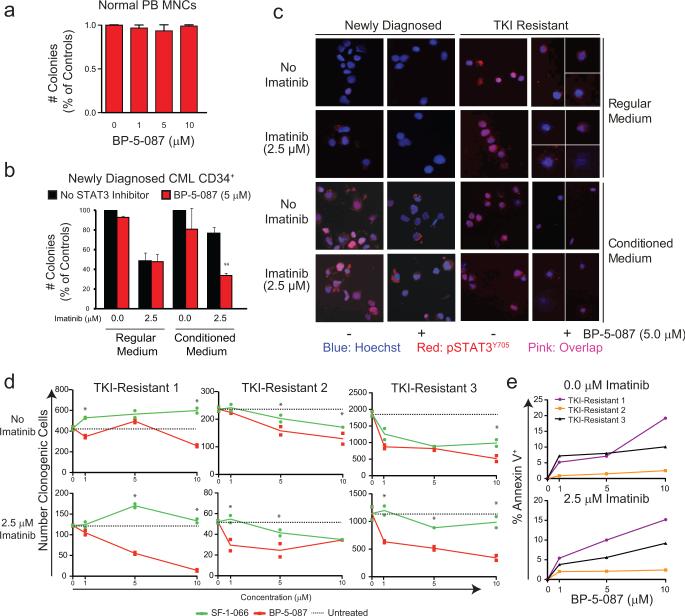
Figure 2. Inhibition of STAT3 reduces colony formation by TKI-resistant CML cells.
(a and b) TKI-resistant CML cell lines were retrovirally transduced with shRNA targeting STAT3 (shSTAT3) or scrambled control (shSCR), and cultured in semisolid medium +/− imatinib (1.0-2.5 μM). STAT3 and not STAT5 knockdown was confirmed by immunoblot analyses (a and b, right). shSTAT3 reduced colony formation of K562R (a, left, n=4) and AR230R (b, left, n=4) cells in the presence of imatinib, with no effect on parental TKI-sensitive controls. (c and d) TKI-resistant CML cell lines were transduced with dominant-negative STAT3 mutants (dnSTAT3) or empty vector (EV) and cultured in semisolid medium +/− imatinib (1.0 μM). Inhibition of pSTAT3Y705 was confirmed by immunoblot analyses (c and d, right). dnSTAT3 reduced colony formation of K562R (c, left, n=4) and AR230R (d, left, n=3) cells with no effect on parental TKI-sensitive controls (n=2). (e and f) K562R (e, n=4) and AR230R (f, n=4) cells were incubated in methylcellulose semisolid medium with SF-1-066 (1-10 μM) +/− imatinib (1.0 μM). SF-1-066 reduced colony formation of only TKI-resistant and not TKI-sensitive cells. (g) Mononuclear cells (MNCs) from peripheral blood of normal donors (n=2) or CMLCD34+ cells from newly diagnosed patients (n=4) were treated ex vivo with SF-1-066 (10 μM) +/− imatinib (2.5 μM) in RM or HS-5 CM for 96 hr followed by colony forming assays. All data are represented as percent of controls. Error bars represent SEM. *p<0.05; **p<0.01; ***p<0.001.
To pharmacologically target STAT3 in TKI resistance, we used S3I-201.1066 (SF-1-066), a salicylic acid-based STAT3 inhibitor20. SF-1-066 was previously shown to interact with the STAT3 SH2 domain, to reduce phosphorylation at Y705, and to reduce DNA-binding in human breast and pancreatic cancer cells21, 22. We reasoned that this inhibitor, while not sufficiently potent for clinical applications, would facilitate direct pharmacologic targeting of STAT3 in primary BCR-ABL1 kinase-independent resistance. SF-1-066 (10 μM) in combination with imatinib (1.0 μM) reduced colony formation of TKI-resistant K562R and AR230R cells by ~60-70%, with no significant effects on parental TKI-sensitive controls (Figures 2e and 2f). These data confirm the selectivity of SF-1-066 for STAT3 over STAT5, since STAT5 inhibition is expected to kill TKI-sensitive CML cells23, 24. Next we tested the effect of SF-1-066 in CMLCD34+ cells from newly diagnosed patients. Cells were cultured for 96 hr in HS-5 CM or RM, without additional cytokines, +/− SF-1-066 (10 μM) and/or imatinib (2.5 μM). In HS-5 CM, SF-1-066 in combination with imatinib reduced colony formation of CMLCD34+ cells by 42.5% compared to imatinib alone, thereby abrogating its protective effects (Figure 2g). However, SF-1-066 resulted in slight inhibition of cells cultured in RM (Figure 2g), and also impaired colony formation by mononuclear cells (MNCs) from healthy individuals (Figure 2g). These results provide proof of principle for synthetic lethality by combined inhibition of STAT3 and BCRABL1 in primary kinase-independent TKI resistance. However, the high dose of SF-1-066 (10 μM) required to achieve an effect, along with inhibition of normal MNCs, prompted us to identify more potent and selective STAT3 inhibitors.
Design, synthesis, and biochemical validation of STAT3 inhibitors
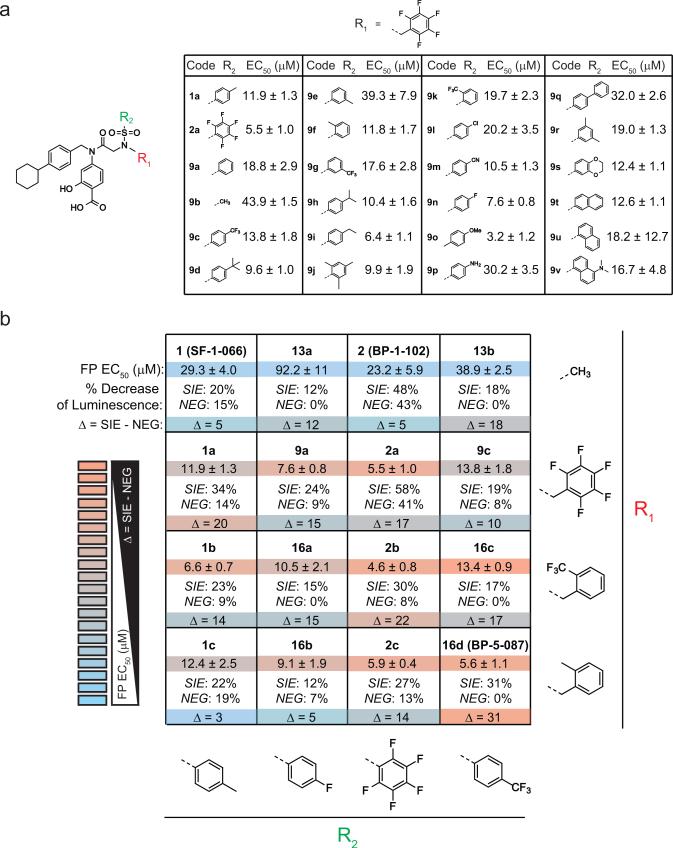
To identify more potent and selective STAT3 inhibitors, we employed an iterative SAR study to interrogate the effects of functional group alterations on the activity of SF-1-066 (Supplementary Figure 5a). STAT3 SH2-domain binding was assessed using fluorescence polarization (FP) assays, and cellular activity was evaluated in a luciferase reporter assay designed to quantify endogenous STAT3 transcriptional activity in TKI-resistant AR230R cells. AR230R cells were lentivirally transduced with a reporter construct harboring either two STAT3-inducible elements (AR230R-SIE) or a negative control reporter with two mutated STAT3 binding sequences (AR230R-NEG) (Supplementary Figure 5b). A difference in luminescence intensity in AR230RSIE cells indicates a change in potency compared to SF-1-066, whereas a difference in AR230R NEG cells indicates a change in selectivity.
We synthesized 74 putative STAT3 inhibitors and screened them in FP and/or luciferase reporter assays. We initially examined 10 STAT3 inhibitors based on SF-1-066 (1) and the more recent derivative, BP-1-102 (2)25, 26, in our luciferase assay at 10 μM. These compounds possessed additional functionality on the sulfonamide nitrogen position, designated as R1 (Supplementary Figure 5c)27. This initial screen revealed that substituting the sulfonamide sulfur position (designated as R2) with a pentafluorobenzene group (such as 2a) resulted in increased potency in AR230R-SIE cells (Supplementary Figure 5d), but also demonstrated activity in the AR230R-NEG cells, indicating a lack of selectivity. By contrast, the R1=pentafluorobenzyl substituent (such as 1a) increased inhibitor potency in AR230R-SIE cells without sacrificing selectivity in AR230R-NEG cells (Supplementary Figure 5d). To capitalize on this advance, we synthesized a library of 24 analogues incorporating the R1=pentafluorobenzyl group, imparting structural diversity at the R2 position (Figure 3a). Using FP for the initial screen, we selected molecules with 4-fluorobenzene and 4-trifluoromethylbenzene in the R2 position for further modification. A focused library of compounds containing the best R2 substituents was also synthesized to incorporate other promising R1 substituents. Figure 3b summarizes the FP EC50 values and luciferase data for select compounds categorized by functional group. Compounds with enhanced potency and selectivity were identified based on a substantial difference (Δ) in luminescence intensity for the AR230R-SIE cells and little difference in the AR230R-NEG cells (Figures 3b). Based on potency and selectivity, BP-5-087 (16d) was selected for testing in the context of BCR-ABL1 kinase-independent resistance.
Figure 3. A STAT3 compound library screen identifies compounds with greater potency and selectivity than SF-1-066.
(a) A library of 24 putative STAT3 inhibitors was synthesized to incorporate the R1=pentafluorobenzyl group, imparting structural diversity at the R2 position. Values represent the EC50 of each molecule in FP assays. (b) Subsequent STAT3 inhibitor libraries were compared by both FP and luciferase reporter assays. For the luciferase assay, TKI-resistant AR230R cells were transduced with a luciferase reporter harboring sequential STAT3-inducible elements (AR230R-SIE) or a mutated control sequence (AR230R-NEG) (see also Supplementary Figure 4b). For each compound, the table represents EC50 values as assessed by FP (top, n=3) and the percent inhibition that each compound achieved in AR230R-SIE versus AR230R-NEG cells at 5 μM in the presence of 1.0 μM imatinib (bottom, n=3).
BP-5-087 interacts with the SH2 domain of STAT3
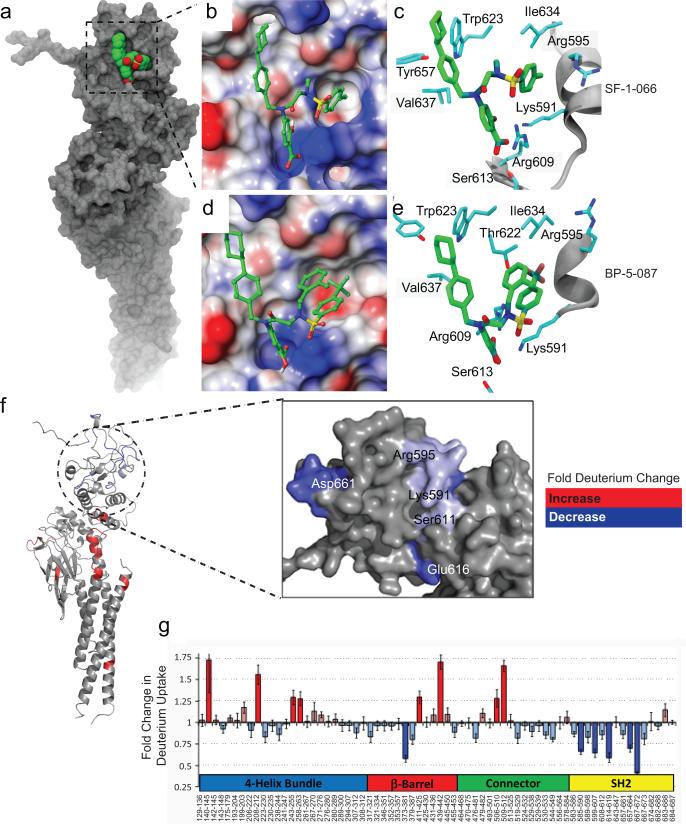
To confirm binding to the STAT3 SH2 domain, we performed high resolution computational docking simulations. Modeling was first performed using the Glide Extra Precision (GlideXP) algorithm, and the estimated docking scores for BP-5-087 and SF-1-066 were −4.9 kcal/mol and −3.8 kcal/mol, respectively. A more negative docking score for BP-5-087 reflects a higher propensity for ligand binding. In the second stage of simulation, Glide induced-fit docking was used to consider the inherent flexibility of the STAT3 SH2 domain, which we observed as thermal fluctuations in X-ray crystallography experiments (Supplementary Figures 6a and 6b). This significantly lowered the docking scores for BP-5-087 (−9.6 kcal/mol) and SF-1-066 (−7.6 kcal/mol), with the corresponding binding poses displayed in Figures 4a-e. As expected, the salicylic acid moiety common to BP-5-087 and SF-1-066 occupied the site in which the charged phosphotyrosine (pY) of a second STAT3 monomer normally binds, engaging in hydrogen bonding with R609 and S613. Both compounds displayed similar interactions with the hydrophobic site including W623, V637, and Y657, as well as hydrogen bonding with the amine group of K591. Although BP-5-087 and SF-1-066 both interact with T622, I634, and R595, the degree of induced fit observed in the two cases was remarkably distinct. Upon BP-5-087 binding, the side chain of R595 reorients, creating a more evident hydrophobic pocket. In addition, molecular modeling suggests that the R1=2-methylbenzyl group present in BP-5-087 may provide a stabilizing intramolecular aromatic interaction with the R2=4-trifluoromethylbenzene group, which may enhance inhibitor rigidity.
Figure 4. Computational modeling of STAT3 binding by BP-5-087.
(a) The entire STAT3 protein is represented in grey with the SH2 domain in green and red within the boxed region. (b and d) The protein surface of the STAT3 SH2 domain bound to either SF-1-066 (b) or BP-5-087 (d) is represented depending on the electrostatic potential with color-coding ranging from red (negative charge) to blue (positive charge). (c and e) The amino acid residues of the STAT3 SH2 domain predicted to interact with SF-1-0-66 (c) or BP-5-087 (e) are also shown. Importantly, BP-5-087 reorients various residues within the binding pocket, which may optimize inhibitor complementarity. (f) Site-specific change in % deuterium uptake observed by TRESI-MS/HDX following BP-5-087 binding to STAT3 in a 7:1 molar ratio color-coded onto the STAT3 X-ray crystal structure (PDB ID:1BG1; left). The enlarged region depicts the SH2 domain on the surface of the predicted BP-5-087 binding site (right). (g) Relative changes in deuterium uptake observed by TRESI-MS/HDX following BP-5-087 binding to STAT3 and grouped by domain. Sequence coverage was 71%. Changes considered significant (>25%) are highlighted by darker colors. The most pronounced decreases in deuterium uptake were observed in peptic peptides that line the BP-5-087 salicylic acid-binding and trifluoromethylbenzene-binding sub-pockets of the SH2 domain. However, the HDX experiment sequence coverage did not extend to peptides lining the BP-5-087 cyclohexylbenzyl-binding sub-pocket. Data represent the average of three independent replicates. Error bars represent SEM.
To further map the STAT3 amino acid residues important for compound action, binding of BP-5-087 to the STAT3 SH2 domain was experimentally confirmed by analyzing changes in site-specific deuterium uptake in STAT3 upon BP-5-087 binding. Specifically, we employed time-resolved electrospray ionization mass spectrometry (TRESI-MS) and hydrogen-deuterium exchange (HDX)28 to characterize the structural transitions that occur to STAT3 upon BP-5-087 binding. In three independent replicates, these data confirmed that the binding epitope for BP-5-087 is indeed located within the STAT3 SH2 domain (Figures 4f and 4g). Fold change in deuterium uptake was analyzed for 68 peptic peptides of STAT3, generating a 71% sequencing coverage, and mapped onto the X-ray crystal structure of STAT3 (Figure 4f). Significant decreases in deuterium uptake clustered almost exclusively to the STAT3 SH2 domain, indicating exclusion of solvent molecules or the formation of new backbone hydrogen bonds in this region (Figure 4g). A number of significant decreases in deuterium uptake were observed in SH2 domain regions proximal to the predicted BP-5-087 sub-pockets, which may result from allosteric changes in STAT3 induced by drug binding. A model for the proposed mechanism of action for BP-5-087 is presented in Supplementary Figure 6c.
BP-5-087 targets TKI-resistant CMLCD34+ stem and progenitor cells
Similar to the effects of shSTAT3 or dnSTAT3 on apoptosis of AR230R cells (Supplementary Figure 7a), BP-5-087 (1.0 μM) increased apoptosis of AR230R cells by 13.7% compared to SF-1-066, which had no significant effect even at 10 μM (Supplementary Figure 7b). Importantly, BP-5-087 had no effect on parental AR230S cells (Supplementary Figure 7a) or CD34+ progenitor cells from healthy individuals (Figure 5a; Supplementary Figure 7c), demonstrating a substantial improvement over the parent compound, SF-1-066.
Figure 5. BP-5-087 impairs colony formation of TKI-resistant CML progenitor cells.
(a) MNCs from healthy individuals (n=5) were plated in cytokine-supplemented methylcellulose semisolid medium with the indicated concentrations of BP-5-087. (b) CMLCD34+ cells from newly diagnosed patients (n=3) were treated ex vivo with BP-5-087 (5 μM) and/or imatinib (2.5 μM) for 96 hr followed by colony forming assays. Error bars represent SEM. **p<0.01. (c) Aliquots of CML CD34+ cells from newly diagnosed and TKI-resistant patients were harvested after 24 hr of treatment for immunofluorescence with a pSTAT3Y705 antibody. BP-5-087 reduced and excluded pSTAT3Y705 from the nucleus in primary cells with intrinsic or extrinsic TKI resistance, which correlated with a reduction in colony forming ability. For TKI-resistant samples treated with BP-5-087, it was difficult to obtain fields with more than two cells. Therefore, ≥2 fields are shown with white dividing lines. One representative experiment is shown. Blue: Hoechst; Red: pSTAT3Y705; Pink: Overlap. (d and e) CMLCD34+ cells from TKI-resistant (n=3) patients were treated ex vivo with BP-5-087 (1-10 μM) and/or imatinib (2.5 μM) for 96 hr followed by colony forming (d) and apoptosis (e) assays. *p<0.05. Error bars represent SEM.
We first assessed the effects of BP-5-087 in primary CMLCD34+ cells from newly diagnosed patients cultured in HS-5 CM. BP-5-087 had little effect on treatment-naïve patient cells cultured in RM, indicating no off-target effects on STAT5. However, BP-5-087 in combination with imatinib reduced colony formation of cells grown in HS-5 CM by 56%, thereby abrogating its protective effects (Figure 5b). We next analyzed the effects of BP-5-087 and imatinib on STAT3 phosphorylation and subcellular localization in CMLCD34+ progenitors by immunofluorescence. As expected, CMLCD34+ cells from newly diagnosed patients showed high levels of nuclear pSTAT3Y705 when cultured in HS-5 CM, but not in RM (Figure 5c). In contrast, CMLCD34+ cells from TKI-resistant patients demonstrated high levels of pSTAT3Y705 in the absence of BM-derived factors. In both cases, BP-5-087 reduced the overall levels of nuclear pSTAT3Y705, with the remaining signal located within the cytoplasm (Figure 5c).
To assess the effects of BP-5-087 in intrinsic TKI resistance, CMLCD34+ cells from TKI-resistant patients were cultured in BP-5-087 or SF-1-066 (1-10 μM) +/− imatinib (2.5 μM), and analyzed for colony formation following drug exposure. BP-5-087 in combination with imatinib reduced colony formation and increased apoptosis of TKI-resistant CMLCD34+ progenitors as low as 1.0 μM, a marked improvement in potency compared to SF-1-066 (Figures 5d and 5e). These data show that BP-5-087 exhibits increased potency over SF-1-066, without compromising selectivity, and without toxic effects to normal controls.
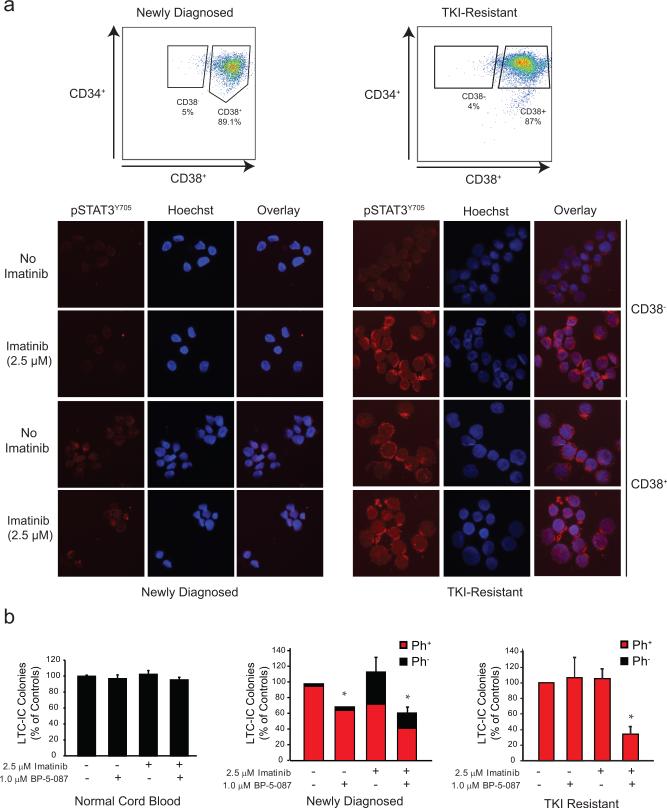
Ex vivo studies have shown that CML LSCs are not ‘addicted’ to BCR-ABL1 kinase activity and survive despite BCR-ABL1 inhibition7, 8. To assess STAT3 activation in the relevant stem and progenitor cell populations, we used CD38 to distinguish between primitive (CD34+38−) and mature (CD34+38+) CML progenitor cells (Figure 6a). FACS-sorted cells from newly diagnosed or TKI-resistant CML patients were treated with imatinib (2.5 μM) for 4 hr followed by immunofluorescence for pSTAT3Y705. No significant pSTAT3Y705 was detected in untreated CD34+38− cells from newly diagnosed or TKI-resistant patients. However, in CD34+38-cells from TKI-resistant patients, imatinib markedly induced nuclear and cytoplasmic pSTAT3Y705, whereas levels remained low in samples from newly diagnosed patients (Figure 6a). To determine whether BP-5-087 targets this primitive cell population, we performed long-term culture-initiating cell (LTC-IC) assays on CMLCD34+ cells from newly diagnosed and TKI-resistant patients. Following ex vivo exposure to BP-5-087 (1.0 μM) +/− imatinib (2.5 μM), cells were cultured on irradiated M210B4 stroma for 6 weeks and plated in colony forming assays as described15, 29. BP-5-087 had no effect on LTC-IC survival of normal cord blood CD34+ cells (Figure 6b, left). In samples from newly diagnosed CML patients, BP-5-087 reduced the number of LTC-IC colonies alone and in combination with imatinib to 69.9% and 61.6% of untreated controls, respectively (Figure 6b, middle). In samples from TKI-resistant CML patients, neither BP-5-087 nor imatinib alone had any effect on LTC-IC survival, whereas dual treatment reduced LTC-IC colonies to 34.2% of controls (Figure 6b, right). All TKI-resistant LTC-ICs were positive for BCR-ABL1, consistent with the low number of normal LTC-ICs that characterizes advanced CML. Altogether, these data suggest that LSCs from CML patients with kinase-independent resistance activate STAT3 upon challenge with imatinib, and that BP-5-087 may be a novel therapeutic approach for eradicating this TKI-resistant stem cell population (Figure 7).
Figure 6. BP-5-087 reduces survival of CML LSCs.
(a) CMLCD34+ cells from newly diagnosed (n=3) or TKI-resistant (n=4) CML patients were sorted by FACS for primitive (CD34+38−) and mature (CD34+38+) cells followed by immunofluorescence with a pSTAT3Y705 antibody. In CD34+38− cells, pSTAT3Y705 was only detectable in samples from TKI-resistant patients following exposure to imatinib. One representative experiment is shown. Blue: Hoechst; Red: pSTAT3Y705; Pink: Overlap. (b) CD34+ cells from normal cord blood (n=3, left), newly diagnosed (n=2, middle), or TKI-resistant (n=3, left) CML patients were treated ex vivo with BP-5-087 (1 μM) +/− imatinib (2.5 μM) in RM for 96 hr followed by plating in LTC-IC assays. Following 6 weeks of culture, remaining cells were plated in colony forming assays. Combined treatment with BP-5-087 and imatinib reduced LTC-IC survival in samples from newly diagnosed and TKI-resistant patients, but not normal cord blood. Bars represent percent of untreated controls. Ph+ colonies are represented in red; Ph- colonies are represented in black. Error bars represent SEM. *p<0.05.
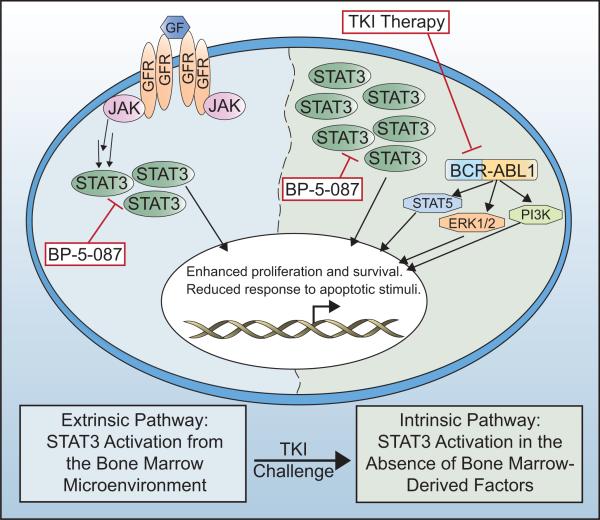
Figure 7. Model of the molecular network regulating kinase-independent TKI resistance +/− TKIs and the STAT3 inhibitor, BP-5-087.
In the absence of TKIs, BCR-ABL1 kinase activates canonical downstream signaling pathways, including pSTAT3S727, STAT5, ERK1/2, and PI3K, whereas pSTAT3Y705 activation occurs through interaction with the BM microenvironment. Upon long-term challenge with multiple TKIs, overt resistance develops when malignant cells establish intrinsic mechanisms to further activate STAT3 without a requirement for BM-derived factors. BP-5-087 is predicted to block STAT3 activation in both scenarios of TKI resistance.
DISCUSSION
BCR-ABL1 kinase-independent TKI resistance is associated with constitutive activation of various signaling pathways, including SRC family kinases30-33, STAT534, PI3K/AKT35, and Wnt/β-catenin36-38, but no uniform picture has emerged6. Furhermore, STAT3 activation by BM-derived factors confers TKI resistance to CML progenitor cells10, 39. Here, we demonstrate that STAT3 activation is a key feature of primary CML stem and progenitor cells with kinase-independent resistance. Using genetic, functional, and pharmacologic inhibition, we demonstrate that STAT3 inhibition in combination with BCR-ABL1 reduces survival of TKI-resistant CML stem and progenitor cells, highlighting a critical role for STAT3. Previous reports have implicated STAT5 in TKI resistance34, 40. However, in our patient specimens, pSTAT5Y694 remained under the control of BCR-ABL1 kinase activity (Figure 1). Importantly, pSTAT3Y705 was the only signaling node activated in both the presence and absence of BM-derived factors (Figure 1; Supplementary Figures 3 and 8). These data are consistent with a model whereby STAT3 is initially activated in CML stem cells through interaction with the BM microenvironment. However, upon long-term TKI challenge, cell-autonomous resistance develops when malignant cells establish intrinsic mechanisms to further activate STAT3 without a requirement for BM-derived factors (Figure 7).
STAT3 activation is implicated in malignant transformation and drug resistance in a variety of cancers41. In some cases, inactivation of negative STAT3 regulators has been demonstrated42, 43. In others, STAT3 is activated by autocrine production of IL-644 or through acquired activating mutations45, 46. SRC family kinases are known to activate STAT347, and have also been linked to imatinib resistance in CML cell lines and patient samples30-32, 48, 49. In both K562R and AR230R cells, treatment with dasatinib resulted in partial reduction of pSTAT3Y705, suggesting partial but not full dependence on SRC family kinases (Supplementary Figure 2). Since multiple mechanisms are known to activate STAT3, directly targeting STAT3 rather than upstream pathways is an attractive therapeutic approach50. Unlike classical enzyme active sites, the STAT3 transcription factor lacks a defined binding pocket, and relies on non-contiguous interactions across large surface areas for affinity with binding partners. The STAT3 SH2 domain is primarily hydrophobic, with a hydrophilic sub-pocket that binds to phosphotyrosine peptide sequences, most notably the one presented by its partner in the STAT3:STAT3 dimer. Precise placement of a small-molecule inhibitor within the STAT3 SH2 domain should therefore block SH2-dependent dimer formation, a step subsequent to phosphorylation by kinases such as JAK or SRC51. Incorporating drug-like characteristics into SH2 domain binders is challenging; however, development of a potent STAT3 inhibitor will have therapeutic value for treatment of many different diseases, including TKI-resistant CML. We developed a number of lead compounds to optimize STAT3 inhibitor potency and selectivity. Our high throughput screening system allowed us to evaluate STAT3 binding affinity in biochemical FP assays and in a cellular context with luciferase reporter assays (Supplementary Figure 5a). Beginning with the parent compound, SF-1-06620, we used SAR-based drug design and compound library screening to identify BP-5-087 as a potent and selective salicylic acid-based STAT3 inhibitor with activity against TKI-resistant CML. Using a computational induced-fit docking approach, the enhanced potency of BP-5-087 was traced to reorientation of the R595 side chain within the binding site (Figure 4), resulting in optimized inhibitor affinity. Importantly, TRESI-MS/HDX experiments precisely mapped binding of BP-5-087 to the STAT3 SH2 domain.
BP-5-087 exerts effects on TKI-resistant CML stem and progenitor cells at 1.0 μM, representing a 10-fold or greater improvement in potency compared to SF-1-066, and a marked improvement to other recently published STAT3 inhibitors26,52,53, 54,55,56,57. The combination of BP-5-087 and imatinib was required to reduce survival of CML progenitors and LTC-ICs from patients with kinase-independent resistance, suggesting that a situation of synthetic lethality is required to target these cells. The term synthetic lethality, while traditionally a genetics term, is more recently being used to describe combinatorial anticancer therapeutics6. In this particular case, the combined inhibition of both BCR-ABL1 and STAT3 is required to kill CML stem and progenitor cells with kinase-independent TKI resistance, while inhibition of only BCR-ABL1 or only STAT3 has very limited effects, consistent with a synthetically lethal situation.
In summary, our data unveil a novel mechanism of kinase-independent TKI resistance in primary CML stem and progenitor cells, and suggest that the STAT3 inhibitor, BP-5-087, intercepts survival signals that are intrinsic and extrinsic to the CML LSC. BP-5-087 may therefore have utility for the treatment of TKI-resistant CML and other diseases characterized by STAT3 activation.
Supplementary Material
ACKNOWLEDGEMENTS
The authors gratefully acknowledge Johanna Estrada, Kevin Gantz, Hannah Redwine, Hillary Finch, and Anthony Iovino for technical assistance, and Kimberly Snow and Candice Ott for clerical assistance. We thank Dr. Rob C. Laister and the Minden group for providing full-length purified STAT3 protein and a STAT3 expression construct. We also thank Dr. Il-Hoan Oh, Catholic University of Korea, for providing a dominant-negative STAT3 construct.
GRANT SUPPORT
M.W.D. was supported by grants from the National Institutes of Health (NIH), including HL082978-01, CA046939-23, and R01CA178397, was a Scholar in Clinical Research of the Leukemia & Lymphoma Society (LLS) (7036-01), and is funded by LLS grant SCOR7005-11. A.M.E. was supported by a NIH T32 training grant (CA093247), followed by a LLS Career Development Award (5090-12), and is currently funded through a Scholar Award from the American Society of Hematology. A.M.E. also acknowledges support from the NIH Loan Repayment Program. This research was supported in part by the LLS Screen-to-Lead Program awarded to M.W.D., T.O., and P.T.G. (SLP-8002-14). T.O. is supported by NIH grant R01CA178397. R.B. acknowledges a petascale computing Research Award at the Extreme Science and Engineering Discovery Environment (XSEDE) supercomputers (TG-CHE120086). XSEDE is supported by National Science Foundation grant OCI-1053575. R.B. acknowledges startup funds from the Department of Medicinal Chemistry, and technical support and computing allocations at the Center for High Performance Computing, The University of Utah. R.M. was supported by grant SFBF47 from the Austrian Science Fund (FWF). P.T.G. and B.D.G.P are supported by the National Sciences and Engineering Research Council. P.T.G. is also supported by the Canadian Breast Cancer Research Foundation. D.J.W. is supported by a Discovery Grant (257588) and by an Ontario Ministry of Research and Innovation Early Researcher Award. We acknowledge support of funds in conjunction with grant P30 CA042014 awarded to the Huntsman Cancer Institute, and 5P30CA042014-24 awarded to The University of Utah Flow Cytometry Facility.
Footnotes
Conflict of Interest Statement: M.W.D. is a consultant for BMS, Novartis, ARIAD, Pfizer and Incyte. His laboratory receives research funding from BMS and Novartis.
Supplementary information is available at Leukemia's website.
REFERENCES
- 1.Druker BJ, Guilhot F, O'Brien SG, Gathmann I, Kantarjian H, Gattermann N, et al. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N Engl J Med. 2006 Dec 7;355(23):2408–2417. doi: 10.1056/NEJMoa062867. [DOI] [PubMed] [Google Scholar]
- 2.de Lavallade H, Apperley JF, Khorashad JS, Milojkovic D, Reid AG, Bua M, et al. Imatinib for newly diagnosed patients with chronic myeloid leukemia: incidence of sustained responses in an intention-to-treat analysis. J Clin Oncol. 2008 Jul 10;26(20):3358–3363. doi: 10.1200/JCO.2007.15.8154. [DOI] [PubMed] [Google Scholar]
- 3.Gorre ME, Mohammed M, Ellwood K, Hsu N, Paquette R, Rao PN, et al. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001 Aug 3;293(5531):876–880. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- 4.Shah NP, Nicoll JM, Nagar B, Gorre ME, Paquette RL, Kuriyan J, et al. Multiple BCR-ABL kinase domain mutations confer polyclonal resistance to the tyrosine kinase inhibitor imatinib (STI571) in chronic phase and blast crisis chronic myeloid leukemia. Cancer Cell. 2002 Aug;2(2):117–125. doi: 10.1016/s1535-6108(02)00096-x. [DOI] [PubMed] [Google Scholar]
- 5.O'Hare T, Shakespeare WC, Zhu X, Eide CA, Rivera VM, Wang F, et al. AP24534, a pan-BCR-ABL inhibitor for chronic myeloid leukemia, potently inhibits the T315I mutant and overcomes mutation-based resistance. Cancer Cell. 2009 Nov 6;16(5):401–412. doi: 10.1016/j.ccr.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O'Hare T, Zabriskie MS, Eiring AM, Deininger MW. Pushing the limits of targeted therapy in chronic myeloid leukaemia. Nat Rev Cancer. 2012 Aug;12(8):513–526. doi: 10.1038/nrc3317. [DOI] [PubMed] [Google Scholar]
- 7.Corbin AS, Agarwal A, Loriaux M, Cortes J, Deininger MW, Druker BJ. Human chronic myeloid leukemia stem cells are insensitive to imatinib despite inhibition of BCR-ABL activity. J Clin Invest. 2011 Jan 4;121(1):396–409. doi: 10.1172/JCI35721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hamilton A, Helgason GV, Schemionek M, Zhang B, Myssina S, Allan EK, et al. Chronic myeloid leukemia stem cells are not dependent on Bcr-Abl kinase activity for their survival. Blood. 2012 Feb 9;119(6):1501–1510. doi: 10.1182/blood-2010-12-326843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bewry NN, Nair RR, Emmons MF, Boulware D, Pinilla-Ibarz J, Hazlehurst LA. Stat3 contributes to resistance toward BCR-ABL inhibitors in a bone marrow microenvironment model of drug resistance. Mol Cancer Ther. 2008 Oct;7(10):3169–3175. doi: 10.1158/1535-7163.MCT-08-0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Traer E, MacKenzie R, Snead J, Agarwal A, Eiring AM, O'Hare T, et al. Blockade of JAK2-mediated extrinsic survival signals restores sensitivity of CML cells to ABL inhibitors. Leukemia. 2012 May;26(5):1140–1143. doi: 10.1038/leu.2011.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schust J, Berg T. A high-throughput fluorescence polarization assay for signal transducer and activator of transcription 3. Anal Biochem. 2004 Jul 1;330(1):114–118. doi: 10.1016/j.ab.2004.03.024. [DOI] [PubMed] [Google Scholar]
- 12.Farid R, Day T, Friesner RA, Pearlstein RA. New insights about HERG blockade obtained from protein modeling, potential energy mapping, and docking studies. Bioorg Med Chem. 2006 May 1;14(9):3160–3173. doi: 10.1016/j.bmc.2005.12.032. [DOI] [PubMed] [Google Scholar]
- 13.Rob T, Liuni P, Gill PK, Zhu S, Balachandran N, Berti PJ, et al. Measuring dynamics in weakly structured regions of proteins using microfluidics-enabled subsecond H/D exchange mass spectrometry. Anal Chem. 2012 Apr 17;84(8):3771–3779. doi: 10.1021/ac300365u. [DOI] [PubMed] [Google Scholar]
- 14.Copland M, Pellicano F, Richmond L, Allan EK, Hamilton A, Lee FY, et al. BMS-214662 potently induces apoptosis of chronic myeloid leukemia stem and progenitor cells and synergizes with tyrosine kinase inhibitors. Blood. 2008 Mar 1;111(5):2843–2853. doi: 10.1182/blood-2007-09-112573. [DOI] [PubMed] [Google Scholar]
- 15.Hogge DE, Lansdorp PM, Reid D, Gerhard B, Eaves CJ. Enhanced detection, maintenance, and differentiation of primitive human hematopoietic cells in cultures containing murine fibroblasts engineered to produce human steel factor, interleukin-3, and granulocyte colony-stimulating factor. Blood. 1996 Nov 15;88(10):3765–3773. [PubMed] [Google Scholar]
- 16.Kaeda J, Chase A, Goldman JM. Cytogenetic and molecular monitoring of residual disease in chronic myeloid leukaemia. Acta Haematol. 2002;107(2):64–75. doi: 10.1159/000046635. [DOI] [PubMed] [Google Scholar]
- 17.Oh IH, Eaves CJ. Overexpression of a dominant negative form of STAT3 selectively impairs hematopoietic stem cell activity. Oncogene. 2002 Jul 18;21(31):4778–4787. doi: 10.1038/sj.onc.1205592. [DOI] [PubMed] [Google Scholar]
- 18.Corvinus FM, Orth C, Moriggl R, Tsareva SA, Wagner S, Pfitzner EB, et al. Persistent STAT3 activation in colon cancer is associated with enhanced cell proliferation and tumor growth. Neoplasia. 2005 Jun;7(6):545–555. doi: 10.1593/neo.04571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dengler MA, Staiger AM, Gutekunst M, Hofmann U, Doszczak M, Scheurich P, et al. Oncogenic stress induced by acute hyper-activation of Bcr-Abl leads to cell death upon induction of excessive aerobic glycolysis. PloS One. 2011;6(9):e25139. doi: 10.1371/journal.pone.0025139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fletcher S, Singh J, Zhang X, Yue P, Page BD, Sharmeen S, et al. Disruption of transcriptionally active Stat3 dimers with non-phosphorylated, salicylic acid-based small molecules: potent in vitro and tumor cell activities. Chembiochem. 2009 Aug 17;10(12):1959–1964. doi: 10.1002/cbic.200900172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang X, Yue P, Fletcher S, Zhao W, Gunning PT, Turkson J. A novel small-molecule disrupts Stat3 SH2 domain-phosphotyrosine interactions and Stat3-dependent tumor processes. Biochem Pharmacol. 2010 May 15;79(10):1398–1409. doi: 10.1016/j.bcp.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fletcher S, Page BD, Zhang X, Yue P, Li ZH, Sharmeen S, et al. Antagonism of the Stat3-Stat3 protein dimer with salicylic acid based small molecules. ChemMedChem. 2011 Aug 1;6(8):1459–1470. doi: 10.1002/cmdc.201100194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hantschel O, Warsch W, Eckelhart E, Kaupe I, Grebien F, Wagner KU, et al. BCR-ABL uncouples canonical JAK2-STAT5 signaling in chronic myeloid leukemia. Nat Chem Biol. 2012 Mar;8(3):285–293. doi: 10.1038/nchembio.775. [DOI] [PubMed] [Google Scholar]
- 24.Schafranek L, Nievergall E, Powell JA, Hiwase DK, Leclercq T, Hughes TP, et al. Sustained inhibition of STAT5, but not JAK2, is essential for TKI-induced cell death in chronic myeloid leukemia. Leukemia. 2014 May 12; doi: 10.1038/leu.2014.156. [DOI] [PubMed] [Google Scholar]
- 25.Page BD, Fletcher S, Yue P, Li Z, Zhang X, Sharmeen S, et al. Identification of a non-phosphorylated, cell permeable, small molecule ligand for the Stat3 SH2 domain. Bioorg Med Chem Lett. 2011 Sep 15;21(18):5605–5609. doi: 10.1016/j.bmcl.2011.06.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang X, Yue P, Page BD, Li T, Zhao W, Namanja AT, et al. Orally bioavailable small-molecule inhibitor of transcription factor Stat3 regresses human breast and lung cancer xenografts. Proc Natl Acad Sci U S A. 2012 Jun 12;109(24):9623–9628. doi: 10.1073/pnas.1121606109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Page BDG, Croucher DC, Li ZH, Haftchenary S, Jimenez-Zepeda VH, Atkinson J, et al. Inhibiting Aberrant Signal Transducer and Activator of Transcription Protein Activation with Tetrapodal, Small Molecule Src Homology 2 Domain Binders: Promising Agents against Multiple Myeloma. J Med Chem. 2013;56(18):7190–7200. doi: 10.1021/jm3017255. [DOI] [PubMed] [Google Scholar]
- 28.Resetca D, Wilson DJ. Characterizing rapid, activity-linked conformational transitions in proteins via sub-second hydrogen deuterium exchange mass spectrometry. FEBS J. 2013 Nov;280(22):5616–5625. doi: 10.1111/febs.12332. [DOI] [PubMed] [Google Scholar]
- 29.Petzer AL, Eaves CJ, Lansdorp PM, Ponchio L, Barnett MJ, Eaves AC. Characterization of primitive subpopulations of normal and leukemic cells present in the blood of patients with newly diagnosed as well as established chronic myeloid leukemia. Blood. 1996 Sep 15;88(6):2162–2171. [PubMed] [Google Scholar]
- 30.Donato NJ, Wu JY, Stapley J, Gallick G, Lin H, Arlinghaus R, et al. BCR-ABL independence and LYN kinase overexpression in chronic myelogenous leukemia cells selected for resistance to STI571. Blood. 2003 Jan 15;101(2):690–698. doi: 10.1182/blood.V101.2.690. [DOI] [PubMed] [Google Scholar]
- 31.Wu J, Meng F, Kong LY, Peng Z, Ying Y, Bornmann WG, et al. Association between imatinib-resistant BCR-ABL mutation-negative leukemia and persistent activation of LYN kinase. J Natl Cancer Inst. 2008 Jul 2;100(13):926–939. doi: 10.1093/jnci/djn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pene-Dumitrescu T, Smithgall TE. Expression of a Src family kinase in chronic myelogenous leukemia cells induces resistance to imatinib in a kinase-dependent manner. J Biol Chemistry. 2010 Jul 9;285(28):21446–21457. doi: 10.1074/jbc.M109.090043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hayette S, Chabane K, Michallet M, Michallat E, Cony-Makhoul P, Salesse S, et al. Longitudinal studies of SRC family kinases in imatinib- and dasatinib-resistant chronic myelogenous leukemia patients. Leuk Res. 2011 Jan;35(1):38–43. doi: 10.1016/j.leukres.2010.06.030. [DOI] [PubMed] [Google Scholar]
- 34.Casetti L, Martin-Lanneree S, Najjar I, Plo I, Auge S, Roy L, et al. Differential Contributions of STAT5A and STAT5B to Stress Protection and Tyrosine Kinase Inhibitor Resistance of Chronic Myeloid Leukemia Stem/Progenitor Cells. Cancer Res. 2013 Apr 1;73(7):2052–2058. doi: 10.1158/0008-5472.CAN-12-3955. [DOI] [PubMed] [Google Scholar]
- 35.Quentmeier H, Eberth S, Romani J, Zaborski M, Drexler HG. BCR-ABL1-independent PI3Kinase activation causing imatinib-resistance. J Hematol Oncol. 2011;4:6. doi: 10.1186/1756-8722-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neviani P, Harb JG, Oaks JJ, Santhanam R, Walker CJ, Ellis JJ, et al. PP2A-activating drugs selectively eradicate TKI-resistant chronic myeloid leukemic stem cells. J Clin Invest. 2013 Oct 1;123(10):4144–4157. doi: 10.1172/JCI68951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang B, Li M, McDonald T, Holyoake TL, Moon RT, Campana D, et al. Microenvironmental protection of CML stem and progenitor cells from tyrosine kinase inhibitors through N-cadherin and Wnt-beta-catenin signaling. Blood. 2013 Mar 7;121(10):1824–1838. doi: 10.1182/blood-2012-02-412890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McWeeney SK, Pemberton LC, Loriaux MM, Vartanian K, Willis SG, Yochum G, et al. A gene expression signature of CD34+ cells to predict major cytogenetic response in chronic-phase chronic myeloid leukemia patients treated with imatinib. Blood. 2010 Jan 14;115(2):315–325. doi: 10.1182/blood-2009-03-210732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nair RR, Tolentino JH, Hazlehurst LA. Role of STAT3 in Transformation and Drug Resistance in CML. Front Oncol. 2012;2:30. doi: 10.3389/fonc.2012.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Warsch W, Kollmann K, Eckelhart E, Fajmann S, Cerny-Reiterer S, Holbl A, et al. High STAT5 levels mediate imatinib resistance and indicate disease progression in chronic myeloid leukemia. Blood. 2011 Mar 24;117(12):3409–3420. doi: 10.1182/blood-2009-10-248211. [DOI] [PubMed] [Google Scholar]
- 41.Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, et al. Stat3 as an oncogene. Cell. 1999 Aug 6;98(3):295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 42.Nallar SC, Kalakonda S, Lindner DJ, Lorenz RR, Lamarre E, Weihua X, et al. Tumor-derived mutations in the gene associated with retinoid interferon-induced mortality (GRIM-19) disrupt its anti-signal transducer and activator of transcription 3 (STAT3) activity and promote oncogenesis. J Biol Chem. 2013 Mar 15;288(11):7930–7941. doi: 10.1074/jbc.M112.440610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rossa C, Jr., Sommer G, Spolidorio LC, Rosenzweig SA, Watson DK, Kirkwood KL. Loss of expression and function of SOCS3 is an early event in HNSCC: altered subcellular localization as a possible mechanism involved in proliferation, migration and invasion. PloS One. 2012;7(9):e45197. doi: 10.1371/journal.pone.0045197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hartman ZC, Yang XY, Glass O, Lei G, Osada T, Dave SS, et al. HER2 overexpression elicits a proinflammatory IL-6 autocrine signaling loop that is critical for tumorigenesis. Cancer Res. 2011 Jul 1;71(13):4380–4391. doi: 10.1158/0008-5472.CAN-11-0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koskela HL, Eldfors S, Ellonen P, van Adrichem AJ, Kuusanmaki H, Andersson EI, et al. Somatic STAT3 mutations in large granular lymphocytic leukemia. N Engl J Med. 2012 May 17;366(20):1905–1913. doi: 10.1056/NEJMoa1114885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Couronne L, Scourzic L, Pilati C, Della Valle V, Duffourd Y, Solary E, et al. STAT3 mutations identified in human hematological neoplasms induce myeloid malignancies in a mouse bone marrow transplantation model. Haematologica. 2013 Jul 19; doi: 10.3324/haematol.2013.085068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cao X, Tay A, Guy GR, Tan YH. Activation and association of Stat3 with Src in v-Src-transformed cell lines. Mol Cell Biol. 1996 Apr;16(4):1595–1603. doi: 10.1128/mcb.16.4.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu Y, Swerdlow S, Duffy TM, Weinmann R, Lee FY, Li S. Targeting multiple kinase pathways in leukemic progenitors and stem cells is essential for improved treatment of Ph+ leukemia in mice. Proc Natl Acad Sci U S A. 2006 Nov 7;103(45):16870–16875. doi: 10.1073/pnas.0606509103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Samanta AK, Chakraborty SN, Wang Y, Kantarjian H, Sun X, Hood J, et al. Jak2 inhibition deactivates Lyn kinase through the SET-PP2A-SHP1 pathway, causing apoptosis in drug-resistant cells from chronic myelogenous leukemia patients. Oncogene. 2009 Apr 9;28(14):1669–1681. doi: 10.1038/onc.2009.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Darnell JE., Jr Transcription factors as targets for cancer therapy. Nat Rev Cancer. 2002 Oct;2(10):740–749. doi: 10.1038/nrc906. [DOI] [PubMed] [Google Scholar]
- 51.Levy DE, Darnell JE., Jr Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002 Sep;3(9):651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 52.Song H, Wang R, Wang S, Lin J. A low-molecular-weight compound discovered through virtual database screening inhibits Stat3 function in breast cancer cells. Proc Natl Acad Sci U S A. 2005 Mar 29;102(13):4700–4705. doi: 10.1073/pnas.0409894102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schust J, Sperl B, Hollis A, Mayer TU, Berg T. Stattic: a small-molecule inhibitor of STAT3 activation and dimerization. Chem Biol. 2006 Nov;13(11):1235–1242. doi: 10.1016/j.chembiol.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 54.Pan Y, Zhou F, Zhang R, Claret FX. Stat3 inhibitor Stattic exhibits potent antitumor activity and induces chemo- and radio-sensitivity in nasopharyngeal carcinoma. PloS One. 2013;8(1):e54565. doi: 10.1371/journal.pone.0054565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fuh B, Sobo M, Cen L, Josiah D, Hutzen B, Cisek K, et al. LLL-3 inhibits STAT3 activity, suppresses glioblastoma cell growth and prolongs survival in a mouse glioblastoma model. Br J Cancer. 2009 Jan 13;100(1):106–112. doi: 10.1038/sj.bjc.6604793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang X, Sun Y, Pireddu R, Yang H, Urlam MK, Lawrence HR, et al. A novel inhibitor of STAT3 homodimerization selectively suppresses STAT3 activity and malignant transformation. Cancer Res. 2013 Mar 15;73(6):1922–1933. doi: 10.1158/0008-5472.CAN-12-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dave B, Landis MD, Tweardy DJ, Chang JC, Dobrolecki LE, Wu MF, et al. Selective small molecule Stat3 inhibitor reduces breast cancer tumor-initiating cells and improves recurrence free survival in a human-xenograft model. PloS One. 2012;7(8):e30207. doi: 10.1371/journal.pone.0030207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.