Abstract
Purpose
Therapy resistance and associated liver disease make hepatocellular cancers (HCC) difficult to treat with traditional cytotoxic therapies, while newer targeted approaches offer only modest survival benefit. We focused on DNA-dependent protein kinase, DNA-PKcs, encoded by PRKDC and central to DNA damage repair by non-homologous end joining. Our aim was to explore its roles in hepatocarcinogenesis and as a novel therapeutic candidate.
Experimental Design
PRKDC was characterised in liver tissues from of 132 patients (normal liver (n=10), cirrhotic liver (n=13), dysplastic nodules (n=18), HCC (n=91)) using Affymetrix U133 Plus 2.0 and 500K Human Mapping SNP arrays (cohort 1). In addition, we studied a case series of 45 patients with HCC undergoing diagnostic biopsy (cohort 2). Histological grading, response to treatment and survival were correlated with DNA-PKcs quantified immunohistochemically. Parallel in vitro studies determined the impact of DNA-PK on DNA repair and response to cytotoxic therapy.
Results
Increased PRKDC expression in HCC was associated with amplification of its genetic locus in cohort 1. In cohort 2, elevated DNA-PKcs identified patients with treatment-resistant HCC, progressing at a median of 4.5 months compared to 16.9 months, while elevation of activated pDNA-PK independently predicted poorer survival. DNA-PKcs was high in HCC cell lines, where its inhibition with NU7441 potentiated irradiation and doxorubicin-induced cytoxicity, while the combination suppressed HCC growth in vitro and in vivo.
Conclusions
These data identify PRKDC/DNA-PKcs as a candidate driver of hepatocarcinogenesis, whose biopsy characterisation at diagnosis may impact stratification of current therapies, and whose specific future targeting may overcome resistance.
Keywords: HCC, DNA-PK, cirrhosis, targeted therapy, DNA Repair
Introduction
Hepatocellular carcinoma (HCC) is the second most common cause of cancer death (1). It arises on a background of chronic liver diseases such as viral hepatitis, alcoholic liver disease (ALD) and, increasingly, non-alcoholic liver disease (NAFLD) (2). Cirrhosis and advanced HCC stage at presentation in the majority of patients severely restricts both surgical and non-surgical therapeutic options. Curative treatments are limited to patients with early cancers who are fit enough for resection, liver transplantation or radiofrequency ablation (3). HCC resistance to conventional palliative cytotoxic agents is compounded by increased toxicity, attributed to hepatic metabolism and their reduced clearance in patients with impaired liver function. Sorafenib represents a major advance in the medical management of these patients (4) but the survival benefit is modest (4). The need to identify key drivers of hepatocarcinogenesis and chemoradioresistance, as well as biomarkers to target therapy effectively is of paramount importance.
The most widely used palliative treatment for fit patients with intermediate stage HCC (Barcelona Clinic for Liver Cancer (BCLC) stage ‘B’) is transarterial chemoembolization (TACE). Its overall efficacy, however, is questionable and patient selection is key (3). Tumour directed radiotherapy is promising (5, 6) but as yet has no proven benefit. Both radiotherapy and the cytotoxic drugs used in TACE cause DNA damage, to which the cell mounts a DNA damage response (DDR). DNA double strand breaks (DSB) are the most cytotoxic and are repaired by two major pathways, with non-homologous end-joining (NHEJ) being the most active in both replicating and non-replicating cells alike. Crucial to NHEJ is DNA-dependent protein kinase (DNA-PK), a heterodimeric enzyme consisting of Ku70, Ku80 and the catalytic subunit DNA-PKcs, encoded by protein kinase DNA activated catalytic polypeptide (PRKDC). DNA-PK phosphorylates a variety of cellular proteins, including autophosphorylation of DNA-PKcs at serine2056 (7). Up-regulated DNA repair activity is often evident in established cancers (8), potentially contributing to therapeutic resistance. DNA-PK inhibitors in pre-clinical development have been shown to sensitise human cancer cells and tumour xenografts to IR and topoisomerase II poisons (9). NU7441 is a potent and selective DNA-PK inhibitor (in vitro IC50 = 14 nM), demonstrating excellent sensitisation in breast and colon cancer cell lines (10). The association between PRKDC expression and DNA-PK levels or activity in HCC is scant, but some evidence for an increase is documented (11, 12). The aim of the current study was to determine the prognostic significance of DNA-PK expression and activity in human HCC and explore the therapeutic potential of DNA-PK inhibition in vitro and in vivo.
Methods
Patient cohorts
Cohort 1 included tissues from consented consecutive patients undergoing resection or liver transplantation at 3 university hospitals in the United States (Mount Sinai Hospital, New York, NY) and Europe (Hospital Clínic, Barcelona, Spain, and National Cancer Institute, Milan, Italy) as described previously (13, 14). Patients with extrahepatic spread were excluded. Specific analyses are as described in Figure 1. Cohort 2 was a case series of 45 patients (Table 1) undergoing pre-treatment diagnostic biopsy, either because there was no history/evidence of associated cirrhosis, or because of radiological diagnostic doubt, from a total of 632 patients managed in Newcastle between 2000 and 2010 (2). Those who did not consent to the use of their surplus tissues after diagnostic purposes were excluded. Patients were followed until 30/06/2013.
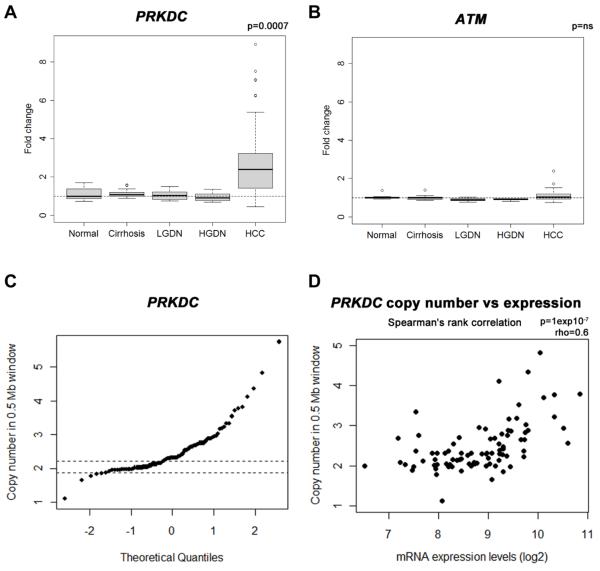
Figure 1. Increased PRKDC in HCC and amplification at the DNA locus.
PRKDC (a) and ATM (b) mRNA expression levels were analysed in 132 Human liver tissues using Affymetrix U133 Plus 2.0 arrays and expressed as fold change relative to normal liver. Tissues included normal liver (n=10), cirrhotic liver (n=13), low grade dysplastic nodules (LGDN; n=10), high grade dysplastic nodules (HGDN; n=8) and HCV-related HCC (n=91), PRKDC was significantly elevated in HCC; p=0.0007. Tumour PRKDC locus copy number was determined using the Affymetrix 500K Human Mapping Array (c). The maximum value of paired cirrhotic samples was used as a cut-off (mean DNA copy number in 0.5Mb around PRKDC gene locus, cut-off 2.25). Panel (d) shows the relationship between PRKDC locus copy number and mRNA levels (Spearman’s rank correlation ρ=0.6; p = 10−7).
Table 1. Clinical features of 45 patients undergoing diagnostic biopsy – cohort two.
| Variable | |
|---|---|
|
| |
| Age (mean ± S.E.) | 67.07 ± 1.7 |
|
| |
| Sex (male/female) | 39/6 |
|
| |
| BMI (mean ± S.E.) | 28.8 ± 1.2 |
|
| |
| Type 2 Diabetes Y/N | 24/21 |
|
| |
| Cirrhosis Y/N | 25/20 |
|
| |
| Etiology | |
| None | 18 |
| ALD | 9 |
| NAFLD | 7 |
| HCV | 3 |
| AIH | 2 |
| Haemochromatosis | 3 |
| Cryptogenic | 3 |
|
| |
| Edmondson Grade 1/2/3 | 13/23/9 |
|
| |
| Size (cm) (mean ± S.E.) | 6.3 ± 0.70 |
|
| |
| Number (mean ± S.E.) | 2.47 ± 0.47 |
|
| |
| PVT Y:N | 6/32 |
|
| |
| Extra-hepatic disease Y:N | 6/39 |
|
| |
| INR (mean ± S.E.) | 1.02 ± 0.02 |
|
| |
| Albumin (g/l) (mean ± S.E.) | 37.7 ± 0.89 |
|
| |
| Bilirubin (μmol/l) | 25.7 ± 10.50 |
|
| |
| Ascites Y:N | 5/40 |
|
| |
| Encephalopathy Y:N | 0/44 |
|
| |
| Childs-Pugh A/B/C | 39/5/1 |
|
| |
| BCLC A/B/C/D | 14/9/20/2 |
|
| |
| Median survival (months) | 24.60 |
Continuous data are presented as mean ± standard error. ALD – alcoholic liver disease; NAFLD – nonalcoholic fatty liver disease; HCV – hepatitis C virus; HBV – hepatitis B virus; AIH – autoimmune hepatitis; BMI – Body Mass Index; PVT – portal vein thrombosis; INR – international normalised ratio; BCLC – Barcelona Clinic for Liver Cancer
Immunohistochemistry
Using formalin fixed paraffin embedded (FFPE) tissues, HCC grading was by two pathologists (15). A detailed immunohistochemistry protocol is in supplementary methods. Briefly, antigen retrieval was with an Antigen Access Unit (A. Menarini diagnostics, Berkshire, UK). Antibodies: anti-DNA-PKcs (rabbit polycolonal, H-163; 1:500; Santa Cruz Biotechnology, Santa Cruz, CA), anti-phosphorylated Ser2056 DNA-PKcs (rabbit polyclonal, ab20407; 1:500; Abcam, Cambridge, UK), anti-ATM (rabbit polyclonal, MAT3-4G10/8; 1:800; Sigma, Poole, UK), anti-phosphorylated Ser1981 ATM (rabbit polyclonal, AF1655; 1:300; R&D Systems, Minneapolis, MN). Sections were analysed using Aperio® Image analysis. Hepatocyte nuclei were identified using a modified nuclear algorithm and staining quantified in pixels after background subtraction. The selection of normal versus tumour areas was by a pathologist, while the application of the quantification algorithm was by supervised researchers. Both pathologists and researchers were blinded until the study endpoint.
Cell lines and In vitro assays
HCC cell lines SNU-182, SNU-475, HepG2, Hep3B, Huh7 (ATCC, Manassas, Virginnia, USA) and PLC/PRF/5 (ECACC, Porton Down, UK) were maintained as per suppliers guidelines. All cell lines were authenticated (LGC Standards) and free of Mycoplasma contamination (MycoAlert Assay, Cambrex Bio Science, Nottingham, UK). Mean change in gene expression (±SEM) was using Human DNA Repair PCR Profiler Arrays (SA Biosciences, Qiagen, West Sussex, UK), expressed as ΔΔCt relative to HPRT1. Western blotting was as described previously (16). Image acquisition/densitometry was using a G-box chemi-luminescent image analyser (Syngene, Cambridge, UK). γH2AX and RAD51 foci detection was as previously described (17). Cell survival was assessed by colony formation and automated counting, normalised to untreated control (±SEM) (16). ShRNA mediated knock down of DNA-PKcs and subsequent analysis of DSBR activity using the traffic light reporter system (18, 19) are detailed in supplementary methods.
Xenograft Model
Female nude mice (CD1 nu/nu, Charles River, Wilmington, MA) were maintained as previously described (16). Huh7 cells (1 × 107 in 50 μL culture medium) were implanted subcutaneously. NU7441 (10 mg/kg i.p.) and/or doxorubicin (2 mg/kg i.p) were administered to tumour-bearing mice daily for 5 days.
Statistics
Data was analysed using SPSS statistics (version 19.0). 2-way ANOVA was used for clonogenic and immunofluorescence assays, paired t-tests to compare tumour versus normal tissue and log rank test (Mantel Cox) and Cox proportional hazards regression for survival analyses.
Chemicals
Chemicals were from Sigma (Poole, UK) unless stated otherwise. Antibodies: anti-RAD51 antibody (rabbit polyclonal, sc-8349), from Santa Cruz Biotechnology, Santa Cruz, CA); Ku70 (monoclonal, ab3114), Ku80 (monoclonal, ab3107), from Abcam, Cambridge, UK; actin (mouse, monoclonal, Ab-1; Calbiochem, Merck Biosciences, Nottingham, UK); anti-γH2AX (monoclonal, 05-636; Millipore, Durham, UK).
Results
Amplification of PRKDC in HCC in association with increased mRNA levels.
Expression of genes involved in the DDR, was evaluated in a cohort of 132 samples (13, 14) of normal, chronically diseased and tumour liver tissues (Figure 1a). PRKDC was up-regulated 2.4 fold in HCC relative to non-cancerous liver (p=0.0007) while the mRNA level of ATM - Ataxia Telangiectasia Mutated kinase, central to the DDR involving both homologous recombination repair (HRR) and NHEJ, was unchanged (Figure 1b). The PRKDC gene locus showed copy number gains in 55% of HCCs (56/101 samples compared to 83 paired cirrhotic HCV positive samples) (Figure 1c). PRKDC copy number correlated significantly with gene expression. (Spearman’s rho = 0.6, p=1×10−7, Figure 1d). There was no correlation between PRKDC mRNA levels and patient outcome. In a small number of supplementary cases from the Newcastle HPB Research Tissue bank, tumour specific PRKDC locus amplification determined by Multiplex Ligation-dependent Probe Amplification (MPLA®) was associated with DNA-PKcs protein over-expression, shown in Supplementary Figure 1.
Increased HCC nuclear DNA-PKcs and treatment resistance
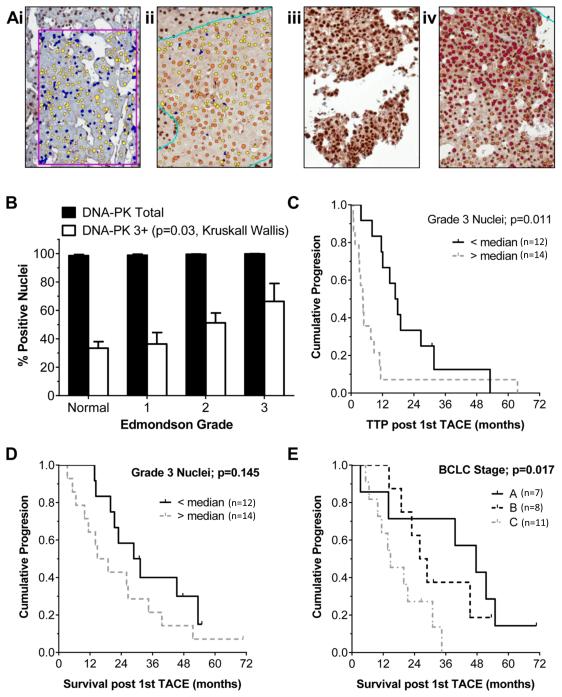
Nuclear DNA-PKcs protein levels assessed by IHC in paired tumour and non-tumour liver from an independent cohort of 45 patients (Table 1) were scored as negative or grades one to three based on the positive pixel count (Figure 2a). Most hepatocyte and HCC nuclei were positive, but the percentage of grade three nuclei was higher in tumour tissues (normal hepatocytes 33±5%, versus 50±5% of HCC nuclei; p=0.001) and increased stepwise with the histological grade (Figure 2b). The HCC DNA-PKcs level, or percentage of grade three nuclei, was not associated with overall survival (data not shown). In a subset analysis of patients receiving palliative doxorubicin in the form of TACE as their first line therapy (n=26; Supplementary Table 1), the time to radiological progression (EASL guidelines 2001 (20)) after the first treatment was significantly shorter in those with high DNA-PKcs (>48% HCC nuclei DNA-PKcs grade three) compared to those with lower DNA-PKcs (median 4.5 months versus 16.9 months, p=0.011, Kaplan Meier; Figure 2c). The high DNA-PKcs association was independent of tumour size, which was the only other factor also predictive of time to radiological progression in this selected subset of patients by univariate analysis (p=0.034) (multivariate Cox-regression: DNA-PKcs grade 3+ Hazards Ratio (HR) 2.5, confidence intervals (CI) 1.0-6.0, p=0.041; tumour size HR 1.12; CI 0.97-1.23; p=0.12). The BCLC stage (20), combining HCC features, liver function and patient performance, rather than any single factor, was predictive of survival in this treated group (Figure 2d+e). There were no differences in ATM levels between HCC and paired non-tumour liver tissues.
Figure 2. Increased DNA-PKcs protein levels in HCC and a shorter time to progression in patients receiving palliative TACE.
DNA-PKcs protein levels were determined immunohistochemically in 45 paired normal and HCC paraffin embedded tissues from cases described in Table 1. Hepatocyte nuclei were detected using an Aperio Imagescope nuclear algorithm. Level of nuclear expression was determined by pixel intensity, scored as un-elevated (0, blue), low (1, yellow), moderate (2, orange) or high (3, red). Application of the algorithm is shown in two paired normal and HCC tissues (a). DNA-PK was detected in the majority of hepatocyte nuclei and the % total nuclei positive (score 1-3) is shown, as % of nuclei scoring 3+ within each Edmondson tumour grade (normal n=45, grade 1 n=13, grade 2 n=23, grade 3 n=9). Grade 3+ nuclei increased significantly with tumour grade (b). Time to radiological progression (TTP) in patients receiving first line treatment with doxorubicin TACE (patients without extra-hepatic disease, no portal vein thrombosis, Childs-Pugh grade A, unsuitable for 1st line RFA) is shown in (c), where ‘High DNA-PKcs’ cases were those with > 48% HCC nuclei grade 3+ (n= 14; range 48-99%), compared to ‘Low DNA-PKcs’ cases, with <48% grade 3+ (n=12; range 0-43%). Median TTP was 4.5 versus 16.9 months (p=0.011 Kaplan Meier). Survival within this selected group was not significantly associated with any individual factor, including nuclear DNA-PK level (d), but was predicted by BCLC stage A-C, combining tumour, liver function and performance status (P=0.017 Kaplan Meier) (e).
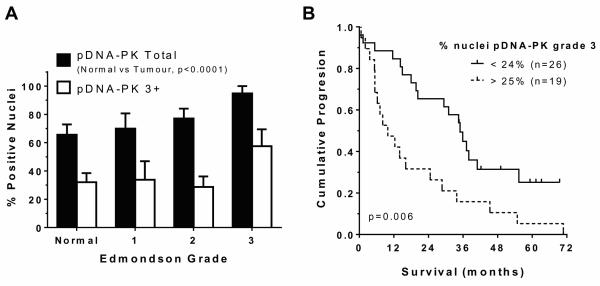
Elevated HCC DNA-PKcs activity was independently associated with poor survival
DNA-PK activity, assessed by IHC detection of pDNA-PKcss2056, was less prevalent and did not correlate significantly with levels of the native protein (data not shown). However, pDNA-PKcss2056 was elevated in tumour versus non-tumour liver (71±7% positive nuclei versus 29±12% respectively, p=0.003) and increased with histological tumour grade (p=0.011) (Figure 3a). The median survival of patients with low tumour pDNA-PKcss2056 (<25% 3+ nuclei; n=26) was 35 months compared to 9.9 months in those with >25% 3+ nuclei (n=19; p=0.007, Kaplan Meier)(Figure 3b). HCC pDNA-PKcss2056 was independently associated with survival (p=0.006; HR= 2.91; 95% CI 1.37-6.17 Supplementary Table 2). ATM activity (autophosphorylation at serine1981) (21) was unchanged in HCC compared to non-tumour liver and was not associated with tumour grade or survival.
Figure 3. Increased tumour pDNA-PK and poorer survival.
Immunohistochemical pDNA-PKcsserine2056 levels were determined by pixel intensity, scored as described in Figure 2. Compared to total DNA-PK, fewer nuclei scored positive for pDNA-PKcsserine2056 overall. The % mean±SEM of positive total nuclei (scores 1-3 combined) and nuclei scoring 3+ within each Edmondson tumour stage are shown in (a). Panel (b) shows median survival of patients grouped as high (>25% 3+ nuclei, n=19) was 9.9 months, compared to low (<25% 3+ nuclei, n=26) tumour pDNA-PK (35 months; p=0.007, Kaplan Meier, p=0.006 by multivariate Cox Regression, including other tumour factors that were significant by univariate analysis: tumour size, tumour number, presence of extra-hepatic disease, presence of portal vein thrombosis).
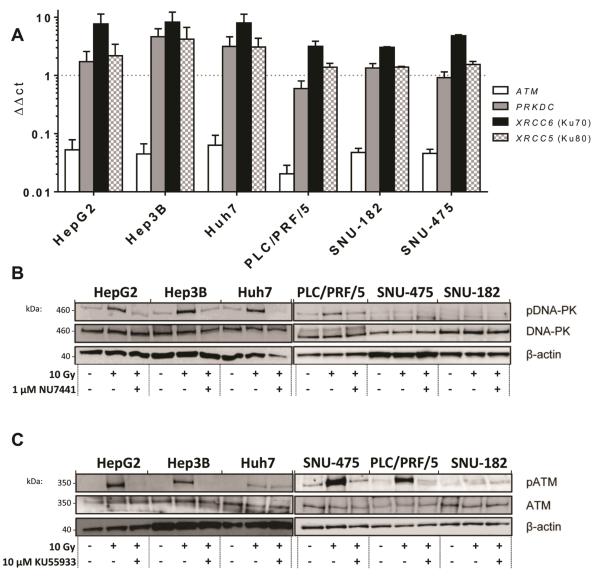
DNA-PK expression and activity in a panel of HCC cell lines
Measurement of DDR gene mRNA levels in a panel of 6 HCC cell lines revealed high PRKDC, XRCC6 (Ku70) and XRCC5 (Ku80), with Hep3B and PLC/PRF/5 having the highest and lowest expression respectively (Figure 4a). Other genes involved in NHEJ (XRCC4, LIG4 and XRCC6BP1) (Supplementary Figure 2a) and ATM (Figure 4a) were not as highly expressed. The expression of genes involved in homologous recombination repair (HRR) was modest and there was no indication that nucleotide excision repair (NER), base excision repair (BER) or mis-match repair (MMR) gene expression was altered across the panel (Supplementary Figure 2b-d). Western blot analysis of DNA-PKcs, Ku70, Ku80 (Figures 4b and 4c), XRCC4, Ligase 4 and ATM (Supplementary Figure 3) revealed abundant DNA-PKcs in all cell lines. Function is critical to the DDR and survival and IR (10 Gy) induced variable activation of DNA-PK (pDNA-PKser2056) and ATM (pATMser1981) (Figures 4b and 4c). Irradiation did not affect the levels of Ku70, Ku80, XRCC4 or Ligase 4. To establish a relationship between DNA-PK expression and NHEJ activity, the level of DNA-PKcs was suppressed (shRNA) causing a corresponding reduction in NHEJ repair, assessed using the traffic light reporter system (18), in HuH7 cells (Supplementary Figure 4 and supplementary methods).
Figure 4. Expression of DNA-PK and related genes in HCC cell lines and inhibition of DNA-PK and ATM activity by specific inhibitors.
DDR gene expression shown as ΔΔCt was determined by quantitative real-time PCR super-arrays relative to HPRT. High levels of DNA-PKcs and partners XRCC5 and XRCC6 were detected (a). Quantification of protein levels for DNA-PK (b) and ATM (c) was by western blotting relative to β-actin and activity was determined using phospho-specific antibodies. DNA-PK or ATM activation following 10 Gy irradiation was inhibited in each of the cell lines in the presence of 1 μM DNA-PK inhibitor NU7441 (b) or 10 μM ATM inhibitor KU55933 (c) respectively. Basal control activity (C) is also shown for comparison. NU7441 (gifted from Celine Cano, Newcastle University), KU55933 (from Marc Hummersone, KuDOS, Cambridge, UK) and doxorubicin were dissolved in dimethyl sulfoxide.
Effect of DNA-PK and ATM inhibitors on the repair of DNA DSBs
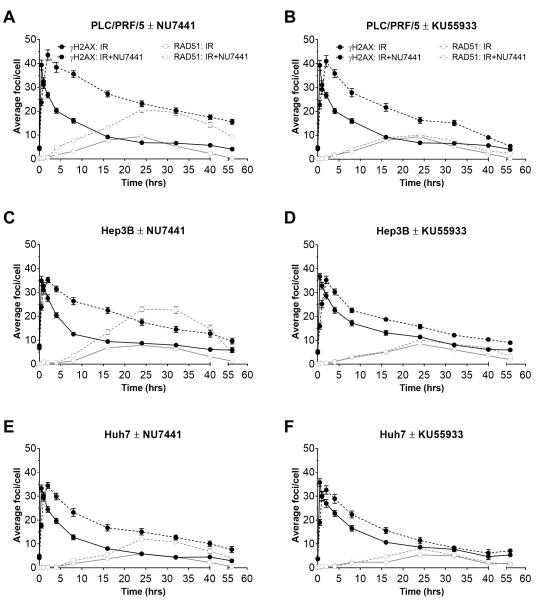
The effect of DNA-PK and ATM inhibitors, NU7441 and KU55933, on DSB repair was investigated in the cell lines that showed a substantial IR induction of ATM and/or DNA-PK activity. DNA DSB induction and repair by HRR was visualised by γH2AX and RAD51 foci respectively (Figure 5). The IR-induced rapid and substantial (5 to 10-fold) increase then gradual decline in γH2AX foci was followed by an increase in RAD51 foci, peaking at 24h, thereafter gradually declining. NU7441 slightly delayed the time to reach peak γH2AX and hindered resolution of the foci. This was particularly notable in PLC/PRF/5 cells where the foci were still 5× baseline at 24h (Figure 5a. Interestingly, inhibition of NHEJ by NU7441 caused a very substantial increase in RAD51 foci (Figures 5a, 5c and 5e), a marker of HRR, for which ATM is presumed to be key. Contrary to expectation, the ATM inhibitor KU55933 did not suppress RAD51 foci (Figures 5b, 5d and 5f).
Figure 5. Irradiation induced DSB repair, retarded by DNA-PK or ATM inhibition.
PLC/PR5, Hep3B or HuH7 cells were exposed to 2 Gy IR treatment in the absence (solid lines) or presence (broken lines) of 1 μM NU7441 (a+c+e) or 10 μM KU55933 (b+d+f) for 1 h prior to, during and after IR. DSB visualised by γH2AX foci (black symbols and lines), and RAD51 foci (grey symbols and lines) as a measure of HRR, were detected immunohistochemically over a 56 h time course and quantified using ImageJ software. Data are mean ± SEM of a minimum of 3 independent experiments.
Doxorubicin sensitivity, radiosensitivity and the effect of NU7441 and KU55933
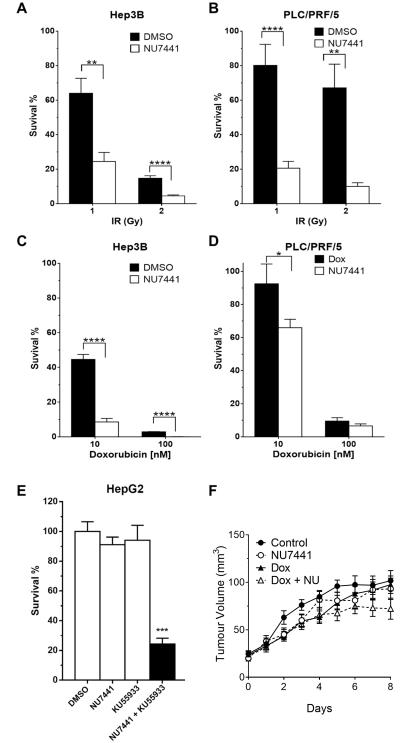
Measurement of the survival of HCC cells exposed to IR and doxorubicin (Supplementary Figure 5) revealed that HepG2 cells, with high levels of DNA-PK and ATM, were highly radio and chemo-resistant. SNU-182 cells were the most chemo-sensitive, consistent with their low DNA-PK and ATM activity. SNU-182 cells grew slowly and the more modest induction of DNA-PK autophosphorylation post irradiation may reflect different temporal kinetics of its activation. Otherwise, ATM and DNA-PK expression and activity were not critical determinants of sensitivity to IR or doxorubicin. NU7441 significantly potentiated IR-induced cytotoxicity 3 to 40-fold, and doxorubicin cytotoxicity 2 to 50-fold (Figure 6a-d, Supplementary Figure 6). Inhibition of ATM by KU55933 caused an up to 44-fold radio-sensitisation and 10-fold chemo-sensitisation (Supplementary Figure 7).
Figure 6. NU7441 potentiates chemotherapy and radiation induced cytotoxicity and delays xenograft tumour growth.
HCC cells were exposed to IR 1 or 2 Gy (a+b) or doxorubicin 10 or 100 nM (c+d) and incubated for 24h prior to reseeding in drug free medium and incubation for 14-21 days for colony formation. Colony forming assays with (white bars) or without (black bars) 1 μM NU7441 for 1h prior to IR exposure and for 24h after, or DMSO (black bars) versus 1 μM NU7441 (black bars) for 1 h prior to doxorubicin and for 24h after demonstrate NU7441 potentiation, with reduced cell survival. In the absence of IR or doxorubicin the combination of NU7441 (1 μM) and KU55933 (10 μM) inhibit cell survival (e). Data are plotted as mean±SEM for a minumum of 3 independent experiments, *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001. Mice with measurable Huh7 xenografts were treated (i.p.) with saline (control, n=9), 10 mg/kg NU7441 (n=9), 2 mg/kg doxorubicin (n=9) or both 10 mg/kg NU7441 and 2 mg/kg doxorubicin (n=9) daily for five days (f). Increasing tumour volume was delayed in treatment groups compared to control animals, with the trend in the combination treatment group in keeping with modest potentiation.
While NU7441 and KU55933 alone had only a marginal effect on survival, the two together profoundly inhibited survival, particularly in HepG2 and Huh7 cells (Figure 6e; Supplementary Figure 8). The combination of these inhibitors also contributed additional chemo- and radio-potentiation compared to either alone (Supplementary Figure 9).
The rank order of sensitivity to doxorubicin compared to IR in the cell lines was markedly different, as was sensitisation by NU7441 and KU55933. This was not due to effects on doxorubicin efflux as nuclear accumulation was similar in all cells and was not increased by either NU7441 or KU55933. In contrast, verapamil, an established MDR inhibitor, increased the nuclear accumulation of doxorubicin in all cell lines (Supplementary Figure 10).
In vivo studies
Huh7 cells, which reliably formed subcutaneous xenografts and had high DNA-PK levels and activity, were used to explore chemosensitisation by NU7441 in vivo. The poor solubility of KU55933 precluded in vivo evaluation of ATM inhibition. Neither doxorubicin nor NU7441, alone or in combination, caused any significant weight loss in the mice (Supplementary Figure11a). Scheduled killing based on tumour burden was 7 days post treatment allocation in the control group versus 12 in the NU7441 + doxorubicin group (Kaplan Meier, p=0.043)(Supplementary Figure11b). In mice treated with NU7441+doxorubicin, the increasing tumour volume trend was modestly reduced compared to control over the study period (Figure 6f).
Discussion
Here we show increased PRKDC mRNA expression in HCC in association with amplification of the PRKDC locus, supporting copy number gain as a potential mechanism. Having demonstrated tumour specific overexpression of DNA-PKcs, the protein encoded by PRKDC, in association with MPLA defined PRKDC locus amplification in resected HCC cases, we demonstrated increased HCC DNA-PKcs in a second cohort of patients undergoing pre-treatment diagnostic biopsies. Importantly, increased DNA-PKcs expression was associated with a shorter time to progression in patients receiving cytotoxic TACE therapy. These data support a role for DNA-PK in resistance of HCC to cytotoxic therapy. In all patients in the second cohort, DNA-PK activation (pDNA-PKcss2056) in the diagnostic pre-treatment biopsy was independently associated with poorer patient survival. This suggests activation of DNA-PKcs following gene amplification contributes to tumour progression. Although higher levels of genomic stress associated with inflammation and reactive oxygen species are well recognised in patients with chronic liver disease, and may predispose to carcinogenesis, recent evidence confirms even higher levels of oxidative DNA damage (22)(23) in HCC tissues. Thus endogenous activation of DNA-PK may reflect high levels of oxidative stress-induced tumour DNA damage in a subgroup of patients with a particularly poor prognosis. DNA-PK stabilisation of c-Myc (11), or promotion of genomic instability through competition with high-fidelity HRR (24), are candidate contributory mechanisms. Taken together, these data identify DNA-PK amplification and elevated expression, in the presence of either endogenous or exogenous activation, as a candidate driver of hepatocarcinogenesis or therapy resistance, respectively.
Similarly, HCC cell lines also had high levels of PRKDC mRNA and DNA-PKcs, with lower expression of other NHEJ genes and ATM, supporting a specific role for DNA-PK, rather than a general one for DDR genes, in hepatocarcinogenesis. Variation in the protein levels between the cell lines un-related to the mRNA expression was in keeping with translation and protein stability being dysregulated in cancer (25). Neither DNA-PK nor ATM expression/activity predicted the rate of DSB repair or chemo- or radio-sensitivity in the HCC cell lines, reflecting the complexity and multifactorial nature of the DDR. Nevertheless, suppression of DNA-PKcs levels in HuH7 cells reduced repair of I-SceI induced DSBs by NHEJ. Furthermore, inhibition of DNA-PK activity retarded the resolution of DSBs, sensitising HCC cells to the effects of doxorubicin and IR.
Both DNA-PK suppression or inhibition conferred a shift toward HRR, consistent with the hypothesis that NHEJ and HRR compete for DNA breaks (26). In contrast to previous reports in other cells (27, 28), KU55933 did not supress IR-induced RAD51 focus formation, suggesting that ATM kinase activity is not essential for HRR in HCC. Our subsequent demonstration that the combination of NU7441 and KU55933 had a substantial impact on HCC cell survival in the absence of a DNA damaging agent is indicative of synthetic lethality, as neither alone was significantly cytotoxic. These observations support recent studies indicating that ATM deficiency confers sensitivity to DNA-PK inhibition (29). Furthermore, the ability of NU7441 and KU55933 to chemo- and/or radio-sensitise in all the cell lines was encouraging for their potential as anticancer therapeutics. While extending these studies to the in vivo setting was hampered by the rapid growth of the tumour xenografts, the combination of NU7441 with doxorubicin did suppress the rate of xenograft growth.
These data are novel and suggest that (a) the combination of inhibitors of DNA repair pathway inhibitors may induce HCC cell death; and (b) the potentiation of tumour damage by cytotoxic agents may facilitate their use at doses much less toxic to non-tumour liver, warranting further exploration. Of more immediate clinical relevance, is the evidence that HCC DNA-PKcs expression by IHC in pre-treatment diagnostic biopsy material predicts responsiveness to TACE. TACE treatment has been controversial, although in fit (BCLC B) patients there is overall survival benefit (3). Here we report a biomarker that may identify patients with more aggressive tumours, potentially with a lesser response to TACE. Because confirmatory HCC biopsy is not routinely performed, our present observational study is hampered by a small cohort size of radiologically atypical patients, as well as heterogeneity in terms of HCC stage. However, in line with the management of other non-resectable cancers, where tissue-based molecular testing guides decision making (30), we suggest that the time is approaching where biopsy characterisation of HCC will be needed for better patient stratification.
Supplementary Material
Translational Relevance.
Hepatocellular carcinoma (HCC) has few treatment options and is the second most common cause of cancer death globally. This study demonstrates that an increase in DNA-PKcs, a key enzyme in DNA double strand break repair, drives this deadly cancer. Increased DNA-PKcs copy number and expression was associated with the malignant process and predicted resistance to hepatic trans-arterial chemo-embolisation (TACE) therapy. We found that increased DNA-PK activity was an independent indicator of poor survival and in pre-clinical studies, inhibition of DNA-PKcs profoundly radio- and chemosensitised HCC.
Our study identifies DNA-PKcs as a candidate biomarker that potentially could be used to stratify patients for TACE therapy and identifies a novel candidate for future therapeutic inhibition to reverse radio- and chemoresistance in HCC. Targeting a ‘driving’ process in hepatocarciongenesis may ultimately offer significantly improved survival benefit.
Acknowledgements
Liam Cornell was supported by the patient support group, LIVErNORTH. Helen Reeves and the creation of the Newcastle University Gastroenterology Research Tissue Bank were supported by the European Community’s Seventh Framework Programme (FP7/2001-2013) under grant agreement HEALTH-F2-2009-241762 for the project FLIP. Joanne Munk, Despina Televantou, Huw Thomas, Jennifer Jackson, David Newell were supported by programme grants from CR UK and Newcastle Experimental Cancer Medicine Center. We thank Professor John Lunec and Dr Debbie Hicks for help with MLPA analyses.
Author Contributions
Liam Cornell:
Involved in all aspects of conception and design, acquisition of majority of laboratory data, data analysis and interpretation. Major contribution to manuscript preparation and revision, approval of final version and agreement to be accountable for all aspects of the work.
Joanne Munck
Involved in aspects of conception and design, acquisition of laboratory data, data analysis and interpretation. Contribution to manuscript preparation and revision, approval of final version and agreement to be accountable for all aspects of the work.
Huw Thomas:
Involved in conception and design of the study with major contribution to design and acquisition of in-vivo studies/data. Agrees to be accountable for all aspects of the work.
Helen L Reeves:
Involved in all aspects of conception and design, acquisition of both clinical and laboratory data, data analysis and interpretation. Major contribution to manuscript preparation and revision, approval of final version and agreement to be accountable for all aspects of the work.
Derek M Manas:
Involved in conception of project, including grant application for funding. Significant contribution to patient identification and recruitment for samples collection and clinical data acquisition. Involved in data interpretation and critical revision of intellectual content. Agrees to be accountable for all aspects of the work.
John Rose:
Involved in conception of the project with a major contribution to clinical imaging acquisition and reporting. Involved in data interpretation and critical revision of intellectual content. Agrees to be accountable for all aspects of the work
Alastair Burt:
Involved in conception of the project with a major contribution to histology reporting, tumour staging and identification of tumour versus non-tumour liver identification for immunohistochemical data acquisition. Involved in data interpretation and critical revision of intellectual content. Agrees to be accountable for all aspects of the work
Despina Televantou:
Involved in design of the studies, with a major contribution to data acquisition from patient tissues – selection of tumour and normal liver areas for algorithm application, validation of algorithm for semi-quantitative reporting of level of expression of target proteins. . Involved in data interpretation and critical revision of intellectual content. Agrees to be accountable for all aspects of the work.
Jennifer Jackson:
Major contribution to tissue processing, immunohistochemistry optimisation and data acquisition.
Laura Ogle:
Involved in design of studies, acquisition of laboratory data, data analysis and interpretation. Contribution to manuscript preparation and revision, approval of final version and agreement to be accountable for all aspects of the work.
Catherine Willoughby:
Involved in design of studies, acquisition of data, data analysis and interpretation. Contribution to manuscript preparation and revision, approval of final version and agreement to be accountable for all aspects of the work.
Clara Alsinet:
Involved in design and acquisition of both clinical and laboratory data, data analysis and interpretation. Major contribution to manuscript preparation and revision, approval of final version and agreement to be accountable for all aspects of the work.
Augusto Villanueva:
Involved in design and acquisition of both clinical and laboratory data, data analysis and interpretation. Major contribution to manuscript preparation and revision, approval of final version and agreement to be accountable for all aspects of the work.
David Newell:
Involved in conception of the project, data analysis and interpretation. Contribution to manuscript preparation, revision and approval of final version. Agrees to be accountable for all aspects of the work.
Geoffrey Shapiro:
Involved in design of studies, acquisition of laboratory data, data analysis and interpretation. Contribution to manuscript preparation and revision, approval of final version and agreement to be accountable for all aspects of the work.
Nicola Curtin:
Involved in all aspects of conception and design, acquisition of data, data analysis and interpretation. Major contribution to manuscript preparation and revision, approval of final version and agreement to be accountable for all aspects of the work.
Footnotes
Conflicts of Interest
Nicola Curtin is co-inventor, but not owner or any financial interest, on the patent that includes NU7441. All other authors have no conflict of interest.
References
- 1.World Health Organization . International Agency for Research on Cancer, World cancer report. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dyson J, Jaques B, Chattopadyhay D, Lochan R, Graham J, Das D, et al. Hepatocellular cancer: The impact of obesity, type 2 diabetes and a multidisciplinary team. J Hepatol. 2014;60:110–7. doi: 10.1016/j.jhep.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 3.EASL-EORTC clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol. 2012;56:908–43. doi: 10.1016/j.jhep.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 4.Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378–90. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 5.Salem R, Lewandowski RJ, Mulcahy MF, Riaz A, Ryu RK, Ibrahim S, et al. Radioembolization for hepatocellular carcinoma using Yttrium-90 microspheres: a comprehensive report of long-term outcomes. Gastroenterology. 2010;138:52–64. doi: 10.1053/j.gastro.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 6.Bujold A, Dawson LA. Stereotactic radiation therapy and selective internal radiation therapy for hepatocellular carcinoma. Cancer Radiother. 2011;15:54–63. doi: 10.1016/j.canrad.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 7.Chen BP, Chan DW, Kobayashi J, Burma S, Asaithamby A, Morotomi-Yano K, et al. Cell cycle dependence of DNA-dependent protein kinase phosphorylation in response to DNA double strand breaks. J Biol Chem. 2005;280:14709–15. doi: 10.1074/jbc.M408827200. [DOI] [PubMed] [Google Scholar]
- 8.Curtin NJ. DNA repair dysregulation from cancer driver to therapeutic target. Nat Rev Cancer. 2012;12:801–17. doi: 10.1038/nrc3399. [DOI] [PubMed] [Google Scholar]
- 9.Davidson D, Amrein L, Panasci L, Aloyz R. Small Molecules, Inhibitors of DNA-PK, Targeting DNA Repair, and Beyond. Frontiers in pharmacology. 2013;4:5. doi: 10.3389/fphar.2013.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao Y, Thomas HD, Batey MA, Cowell IG, Richardson CJ, Griffin RJ, et al. Preclinical evaluation of a potent novel DNA-dependent protein kinase inhibitor NU7441. Cancer Res. 2006;66:5354–62. doi: 10.1158/0008-5472.CAN-05-4275. [DOI] [PubMed] [Google Scholar]
- 11.An J, Yang DY, Xu QZ, Zhang SM, Huo YY, Shang ZF, et al. DNA-dependent protein kinase catalytic subunit modulates the stability of c-Myc oncoprotein. Molecular cancer. 2008;7:32. doi: 10.1186/1476-4598-7-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu ZJ, Sui JG, Ding YQ, Cao ZS, Zhou PK, Wu DC. Expression of DNA-PK in hepato- and cholangio-neoplasms and its significance. Zhonghua Gan Zang Bing Za Zhi. 2004;12:652–5. [PubMed] [Google Scholar]
- 13.Wurmbach E, Chen YB, Khitrov G, Zhang W, Roayaie S, Schwartz M, et al. Genome-wide molecular profiles of HCV-induced dysplasia and hepatocellular carcinoma. Hepatology. 2007;45:938–47. doi: 10.1002/hep.21622. [DOI] [PubMed] [Google Scholar]
- 14.Chiang DY, Villanueva A, Hoshida Y, Peix J, Newell P, Minguez B, et al. Focal gains of VEGFA and molecular classification of hepatocellular carcinoma. Cancer Res. 2008;68:6779–88. doi: 10.1158/0008-5472.CAN-08-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edmondson HA, Steiner PE. Primary carcinoma of the liver: a study of 100 cases among 48,900 necropsies. Cancer. 1954;7:462–503. doi: 10.1002/1097-0142(195405)7:3<462::aid-cncr2820070308>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 16.Munck JM, Batey MA, Zhao Y, Jenkins H, Richardson CJ, Cano C, et al. Chemosensitization of cancer cells by KU-0060648, a dual inhibitor of DNA-PK and PI-3K. Mol Cancer Ther. 2012;11:1789–98. doi: 10.1158/1535-7163.MCT-11-0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tavecchio M, Munck JM, Cano C, Newell DR, Curtin NJ. Further characterisation of the cellular activity of the DNA-PK inhibitor, NU7441, reveals potential cross-talk with homologous recombination. Cancer Chemother Pharmacol. 2012;69:155–64. doi: 10.1007/s00280-011-1662-4. [DOI] [PubMed] [Google Scholar]
- 18.Certo MT, Ryu BY, Annis JE, Garibov M, Jarjour J, Rawlings DJ, et al. Tracking genome engineering outcome at individual DNA breakpoints. Nature methods. 2011;8:671–6. doi: 10.1038/nmeth.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–98. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 20.Bruix J, Sherman M, Llovet JM, Beaugrand M, Lencioni R, Burroughs AK, et al. Clinical management of hepatocellular carcinoma. Conclusions of the Barcelona-2000 EASL conference. European Association for the Study of the Liver. J Hepatol. 2001;35:421–30. doi: 10.1016/s0168-8278(01)00130-1. [DOI] [PubMed] [Google Scholar]
- 21.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 22.Romilda C, Marika P, Alessandro S, Enrico L, Marina B, Andromachi K, et al. Oxidative DNA damage correlates with cell immortalization and mir-92 expression in hepatocellular carcinoma. BMC cancer. 2012;12:177. doi: 10.1186/1471-2407-12-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsuda Y, Wakai T, Kubota M, Osawa M, Takamura M, Yamagiwa S, et al. DNA damage sensor gamma -H2AX is increased in preneoplastic lesions of hepatocellular carcinoma. TheScientificWorldJournal. 2013;2013:597095. doi: 10.1155/2013/597095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bunting SF, Nussenzweig A. End-joining, translocations and cancer. Nat Rev Cancer. 2013;13:443–54. doi: 10.1038/nrc3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Audic Y, Hartley RS. Post-transcriptional regulation in cancer. Biology of the cell/under the auspices of the European Cell Biology Organization. 2004;96:479–98. doi: 10.1016/j.biolcel.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 26.Hartlerode AJ, Scully R. Mechanisms of double-strand break repair in somatic mammalian cells. Biochem J. 2009;423:157–68. doi: 10.1042/BJ20090942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shrivastav M, Miller CA, De Haro LP, Durant ST, Chen BP, Chen DJ, et al. DNA-PKcs and ATM co-regulate DNA double-strand break repair. DNA Repair (Amst) 2009;8:920–9. doi: 10.1016/j.dnarep.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kass EM, Helgadottir HR, Chen CC, Barbera M, Wang R, Westermark UK, et al. Double-strand break repair by homologous recombination in primary mouse somatic cells requires BRCA1 but not the ATM kinase. Proc Natl Acad Sci U S A. 2013;110:5564–9. doi: 10.1073/pnas.1216824110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Riabinska A, Daheim M, Herter-Sprie GS, Winkler J, Fritz C, Hallek M, et al. Therapeutic targeting of a robust non-oncogene addiction to PRKDC in ATM-defective tumors. Science translational medicine. 2013;5:189ra78. doi: 10.1126/scitranslmed.3005814. [DOI] [PubMed] [Google Scholar]
- 30.Burstein HJ, Griggs JJ, Prestrud AA, Temin S. American society of clinical oncology clinical practice guideline update on adjuvant endocrine therapy for women with hormone receptor-positive breast cancer. Journal of oncology practice/American Society of Clinical Oncology. 2010;6:243–6. doi: 10.1200/JOP.000082. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.