Abstract
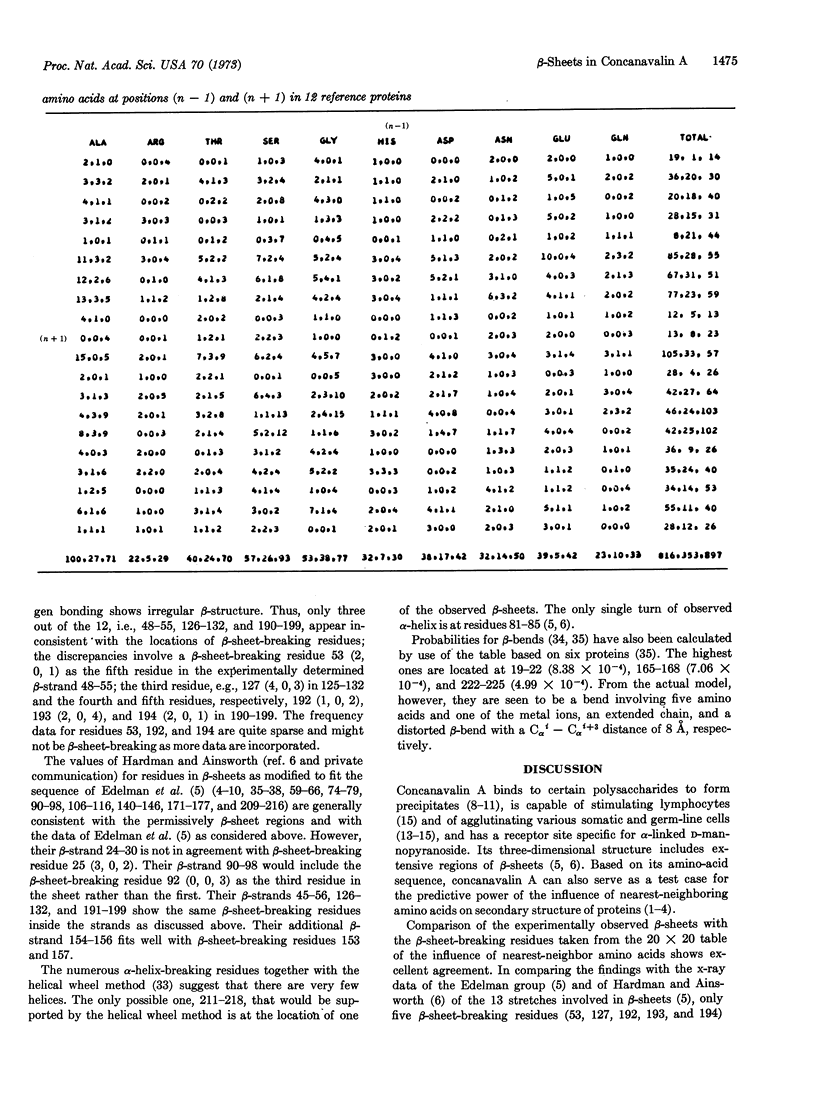
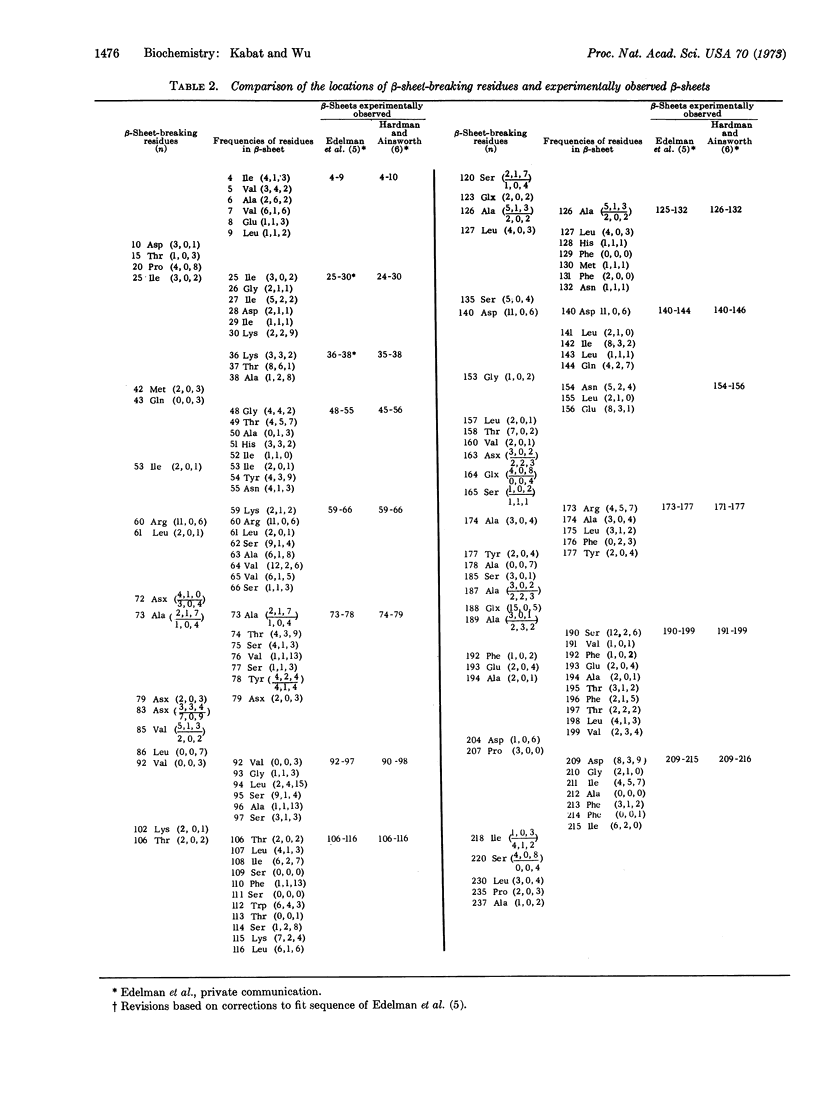
A 20 × 20 table of tripeptides has been compiled that may be used to locate β-sheet breaking and α-helix breaking residues in proteins. It is based on the definition of an α-helical and a β-sheet domain on the (ϕ, Ψ) map based on the occurrences of α-helices and β-sheets in 12 known proteins whose sequence and three-dimensional structure have been determined. Each entry in the 20 × 20 table lists three numbers, the frequency of occurrences of the middle amino acid (n) in relation to its nearest neighbors (n - 1) and (n + 1) in the α-helical domain, the β-sheet domain and outside these regions. The regions between two β-sheet-breaking residues would be permissively β-sheet regions. The sequence of concanavalin A has been examined in this manner and of the 13 β-strands defined by x-ray crystallography, 10 were in agreement with the permissively β-sheet regions and, in the remaining three, β-sheet-breaking residues were the third in one, and the third, fourth, and fifth residues in another, and the sixth residue in the third from the beginning of the β-strands. The findings provide strong support for the role of nearest-neighboring amino acids in determining secondary structure of proteins.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alden R. A., Wright C. S., Kraut J. A hydrogen-bond network at the active site of subtilisin BPN'. Philos Trans R Soc Lond B Biol Sci. 1970 Feb 12;257(813):119–124. doi: 10.1098/rstb.1970.0014. [DOI] [PubMed] [Google Scholar]
- Arnone A., Bier C. J., Cotton F. A., Day V. W., Hazen E. E., Jr, Richardson D. C., Yonath A., Richardson J. S. A high resolution structure of an inhibitor complex of the extracellular nuclease of Staphylococcus aureus. I. Experimental procedures and chain tracing. J Biol Chem. 1971 Apr 10;246(7):2302–2316. [PubMed] [Google Scholar]
- Birktoft J. J., Blow D. M. Structure of crystalline -chymotrypsin. V. The atomic structure of tosyl- -chymotrypsin at 2 A resolution. J Mol Biol. 1972 Jul 21;68(2):187–240. doi: 10.1016/0022-2836(72)90210-0. [DOI] [PubMed] [Google Scholar]
- Birktoft J. J., Matthews B. W., Blow D. M. Atomic co-ordinates for tosyl-alpha-chymotrypsin. Biochem Biophys Res Commun. 1969 Jul 7;36(1):131–137. doi: 10.1016/0006-291x(69)90659-7. [DOI] [PubMed] [Google Scholar]
- Blake C. C., Mair G. A., North A. C., Phillips D. C., Sarma V. R. On the conformation of the hen egg-white lysozyme molecule. Proc R Soc Lond B Biol Sci. 1967 Apr 18;167(1009):365–377. doi: 10.1098/rspb.1967.0034. [DOI] [PubMed] [Google Scholar]
- Blundell T. L., Cutfield J. F., Cutfield S. M., Dodson E. J., Dodson G. G., Hodgkin D. C., Mercola D. A., Vijayan M. Atomic positions in rhombohedral 2-zinc insulin crystals. Nature. 1971 Jun 25;231(5304):506–511. doi: 10.1038/231506a0. [DOI] [PubMed] [Google Scholar]
- Blundell T. L., Dodson G. G., Dodson E., Hodgkin D. C., Vijayan M. X-ray analysis and the structure of insulin. Recent Prog Horm Res. 1971;27:1–40. doi: 10.1016/b978-0-12-571127-2.50025-0. [DOI] [PubMed] [Google Scholar]
- Bunting J. R., Athey T. W., Cathou R. E. Backbone folding of immunoglobulin light and heavy chains: a comparison of predicted -bend positions. Biochim Biophys Acta. 1972 Nov 28;285(1):60–71. doi: 10.1016/0005-2795(72)90180-8. [DOI] [PubMed] [Google Scholar]
- Burger M. M., Noonan K. D. Restoration of normal growth by covering of agglutinin sites on tumour cell surface. Nature. 1970 Nov 7;228(5271):512–515. doi: 10.1038/228512a0. [DOI] [PubMed] [Google Scholar]
- Dickerson R. E., Takano T., Eisenberg D., Kallai O. B., Samson L., Cooper A., Margoliash E. Ferricytochrome c. I. General features of the horse and bonito proteins at 2.8 A resolution. J Biol Chem. 1971 Mar 10;246(5):1511–1535. [PubMed] [Google Scholar]
- Edelman G. M., Cunningham B. A., Reeke G. N., Jr, Becker J. W., Waxdal M. J., Wang J. L. The covalent and three-dimensional structure of concanavalin A. Proc Natl Acad Sci U S A. 1972 Sep;69(9):2580–2584. doi: 10.1073/pnas.69.9.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelman G. M., Millette C. F. Molecular probes of spermatozoan structures. Proc Natl Acad Sci U S A. 1971 Oct;68(10):2436–2440. doi: 10.1073/pnas.68.10.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GOLDSTEIN I. J., HOLLERMAN C. E., SMITH E. E. PROTEIN-CARBOHYDRATE INTERACTION. II. INHIBITION STUDIES ON THE INTERACTION OF CONCANAVALIN A WITH POLYSACCHARIDES. Biochemistry. 1965 May;4:876–883. doi: 10.1021/bi00881a013. [DOI] [PubMed] [Google Scholar]
- Goldstein I. J., So L. L. Protein-carbonhydrate interaction. 3. Agar gel-diffusion studies on the interaction of Concanavalin A, a lectin isolated from jack bean, with polysaccharides. Arch Biochem Biophys. 1965 Aug;111(2):407–414. doi: 10.1016/0003-9861(65)90203-1. [DOI] [PubMed] [Google Scholar]
- Hardman K. D., Ainsworth C. F. Structure of concanavalin A at 2.4-A resolution. Biochemistry. 1972 Dec 19;11(26):4910–4919. doi: 10.1021/bi00776a006. [DOI] [PubMed] [Google Scholar]
- Hendrickson W. A., Love W. E. Structure of lamprey haemoglobin. Nat New Biol. 1971 Aug;232(33):197–203. doi: 10.1038/newbio232197a0. [DOI] [PubMed] [Google Scholar]
- Inbar M., Sachs L. Interaction of the carbohydrate-binding protein concanavalin A with normal and transformed cells. Proc Natl Acad Sci U S A. 1969 Aug;63(4):1418–1425. doi: 10.1073/pnas.63.4.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabat E. A., Wu T. T. Construction of a three-dimensional model of the polypeptide backbone of the variable region of kappa immunoglobulin light chains. Proc Natl Acad Sci U S A. 1972 Apr;69(4):960–964. doi: 10.1073/pnas.69.4.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabat E. A., Wu T. T. The influence of nearest-neighboring amino acid residues on aspects of secondary structure of proteins. Attempts to locate -helices and -sheets. Biopolymers. 1973 Apr;12(4):751–774. doi: 10.1002/bip.1973.360120406. [DOI] [PubMed] [Google Scholar]
- Lewis P. N., Momany F. A., Scheraga H. A. Folding of polypeptide chains in proteins: a proposed mechanism for folding. Proc Natl Acad Sci U S A. 1971 Sep;68(9):2293–2297. doi: 10.1073/pnas.68.9.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipscomb W. N., Reeke G. N., Jr, Hartsuck J. A., Quiocho F. A., Bethge P. H. The structure of carboxypeptidase A. 8. Atomic interpretation at 0.2 nm resolution, a new study of the complex of glycyl-L-tyrosine with CPA, and mechanistic deductions. Philos Trans R Soc Lond B Biol Sci. 1970 Feb 12;257(813):177–214. doi: 10.1098/rstb.1970.0020. [DOI] [PubMed] [Google Scholar]
- Lloyd K. O., Kabat E. A., Beychok S. Immunochemical studies on blood groups. 43. The interaction of blood group substances from various sources with a plant lectin, concanavalin A. J Immunol. 1969 Jun;102(6):1354–1362. [PubMed] [Google Scholar]
- Mathews F. S., Levine M., Argos P. The structure of calf liver cytochrome b 5 at 2.8 A resolution. Nat New Biol. 1971 Sep 1;233(35):15–16. doi: 10.1038/newbio233015a0. [DOI] [PubMed] [Google Scholar]
- Mathews F. S., Levine M., Argos P. Three-dimensional Fourier synthesis of calf liver cytochrome b 5 at 2-8 A resolution. J Mol Biol. 1972 Mar 14;64(2):449–464. doi: 10.1016/0022-2836(72)90510-4. [DOI] [PubMed] [Google Scholar]
- Powell A. E., Leon M. A. Reversible interaction of human lymphocytes with the mitogen concanavalin A. Exp Cell Res. 1970 Oct;62(2):315–325. doi: 10.1016/0014-4827(70)90560-4. [DOI] [PubMed] [Google Scholar]
- Schiffer M., Edmundson A. B. Use of helical wheels to represent the structures of proteins and to identify segments with helical potential. Biophys J. 1967 Mar;7(2):121–135. doi: 10.1016/S0006-3495(67)86579-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumner J. B., Howell S. F. Identification of Hemagglutinin of Jack Bean with Concanavalin A. J Bacteriol. 1936 Aug;32(2):227–237. doi: 10.1128/jb.32.2.227-237.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright C. S., Alden R. A., Kraut J. Structure of subtilisin BPN' at 2.5 angström resolution. Nature. 1969 Jan 18;221(5177):235–242. doi: 10.1038/221235a0. [DOI] [PubMed] [Google Scholar]
- Wu T. T., Kabat E. A. An attempt to locate the non-helical and permissively helical sequences of proteins: application to the variable regions of immunoglobulin light and heavy chains. Proc Natl Acad Sci U S A. 1971 Jul;68(7):1501–1506. doi: 10.1073/pnas.68.7.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyckoff H. W., Tsernoglou D., Hanson A. W., Knox J. R., Lee B., Richards F. M. The three-dimensional structure of ribonuclease-S. Interpretation of an electron density map at a nominal resolution of 2 A. J Biol Chem. 1970 Jan 25;245(2):305–328. [PubMed] [Google Scholar]


