Abstract
MukB, a divergent structural maintenance of chromosomes (SMC) protein, is important for chromosomal segregation and condensation in γ-proteobacteria. MukB and canonical SMC proteins share a characteristic five-domain structure. Globular N- and C-terminal domains interact to form an ABC-like ATPase or “head” domain, which is connected to a smaller dimerization or “hinge” domain by a long, antiparallel coiled coil. In addition to mediating dimerization, this hinge region has been implicated in both conformational flexibility and dynamic protein-DNA interactions. We report here the first crystallographic model of the MukB hinge domain. This model also contains approximately 20% of the coiled coil domain, including an unusual coiled coil deviation. These results will facilitate studies to clarify the roles of both the hinge and coiled coil domains in MukB function.
Keywords: MukB, Structural Maintenance of Chromosomes (SMC), hinge, coiled coil, condensin
Chromosomes must be faithfully replicated and segregated during cell proliferation. A growing body of evidence has shown that structural maintenance of chromosomes (SMC) proteins and their non-SMC accessory proteins play a crucial role in this process 1; 2. In eukaryotes, SMC proteins function as heterodimers and can be categorized into three groups: cohesin (SMC1/SMC3), condensin (SMC2/SMC4) and a DNA repair complex (SMC5/SMC6) 1; 2. In contrast, most prokaryotes contain a single SMC protein that functions as a homodimer 3; 4.
In γ-proteobacteria, including Escherichia coli (E. coli), MukB and its accessory proteins take the place of the canonical SMC protein complex 5. In spite of its limited sequence similarity with other bacterial SMC proteins, MukB performs similar cellular functions. Both E. coli mukB− strains and smc− strains from Bacillus subtilis and Caulobacter crescentus show temperature sensitive colony formation and an increase in the number of anucleate cells at the permissive temperature, suggesting a deficiency in chromosome segregation 6–12. As is the case with bacterial SMC proteins, two non-SMC accessory proteins, MukE and MukF, are required for full MukB function 13–15. Finally, a convincing array of experiments have demonstrated that MukB can condense DNA both in vitro and in vivo16–21. Nonetheless, the mechanism of this MukB activity remains largely unknown22.
In addition to its functional similarity to SMC proteins, MukB shares the five-domain structure found in all SMC proteins 6; 23–30. The globular N- and C-terminal domains combine to form an ATP Binding Cassette (ABC) ATPase domain; this “head” region is connected to a smaller globular dimerization domain, also called the “hinge”, by an unusual, 50 nm long, antiparallel coiled coil domain 6; 23–30. Although this structure has been established clearly by electron microscopy (EM) and biochemical experiments28; 29; 31, high-resolution structural data are available only for the ATPase “head” domain of MukB32; 33.
The ATPase domain of MukB shares both sequence homology and structural features with other SMC proteins and ABC transporters 3; 34. In contrast, the hinge domain of MukB has no sequence homology with any protein of known structure, making it difficult to accurately predict its structure and function35. The available crystal structures of the hinge domains from Thermotoga maritima SMC (TmSMC) and a distantly related SMC-like protein, Pyrococcus furiosus Rad50 (PfRad50), show completely different structures. The hinge domain of TmSMC is a large globular domain of 169 residues 28. In contrast, the hinge domain of Rad50 is a simple “zinc-hook” made of only 14 residues 30. Though the hinge domain of MukB has a comparable size to that of other bacterial SMC protein as judged by EM25, their primary sequences are not obviously related. Thus, in spite of the functional similarity between MukB and prokaryotic SMC proteins, the structural relationship between their hinge domains has not been clear.
We have previously obtained a chemical crosslink between two residues (S570-N859) on the hinge-proximal end of the MukB coiled-coil domain31. This information allowed us to design a well-behaved, soluble fragment of MukB containing the entire hinge domain and roughly matched segments of the coiled coil. This truncation fragment successfully led to the first crystallographic model for the hinge domain of MukB (3.1 Å resolution), including not only the globular hinge domain, but also a stretch of ~ 8 heptads of the adjacent coiled coil domain.
Overall Structure of MukB-D
On the basis of our earlier studies, we designed a stable MukB construct (566–863) using the crosslinked residue pair S570-N859C as a guidepost31. This construct, denoted MukB-D, was expected to include the entire hinge domain, along with a portion of the MukB coiled coil domain31. Equilibrium sedimentation studies confirm that MukB-D forms a dimer in solution31. In addition, the circular dichroism (CD) spectrum of MukB-D shows the minima at 208 and 222 nm that are characteristic of an α-helical conformation, suggesting the presence of coiled coil (Fig S1). MukB-D also undergoes a cooperative thermal unfolding transition with a midpoint Tm of approximately 52 °C and appears to be fully folded at room temperature (Fig S1). Taken together, these data suggest that our construct is autonomously folded into the proper conformation in solution.
MukB-D was crystallized using PEG 20K as the precipitant. It forms tetragonal crystals with the P4322 spacegroup (a = b = 56 Å, c = 343 Å). In the crystal structure, the dimer is related by a crystallographic C2 axis. The region 572–854 is visible in the refined structure and accounts for 95% of the construct (566–863), which suggests our domain indeed contains two coiled-coil strands of roughly equal length.
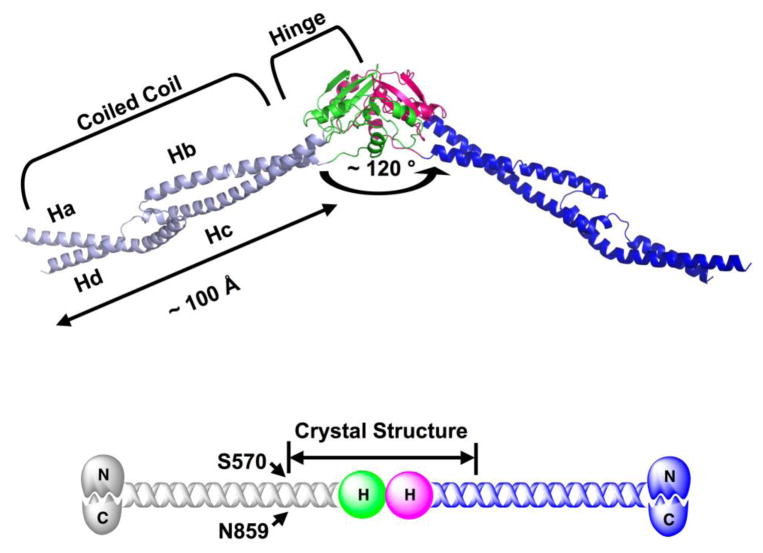
Overall, the MukB-D dimer adopts an open V-shape (~ 120) with the globular hinge domain in the center (Fig 1). EM studies have demonstrated that the MukB dimer adopts a broad range of angles from 0° to 180° 25, suggesting that there is a flexible “hinge” within or close to the hinge domain. The observed, fixed V-shape of the MukB dimer could be the result of crystal packing interactions or it could represent a stable conformation in the absence of ATPase head domain. Interestingly, the crystal structure of the TmSMC dimer in a different space group shows an almost identical angle between the coiled coil segments of each monomer28. This angle may serve a role for the higher order architecture of MukB multimers, which are occasionally observed with EM36.
Fig 1.
Overall structure of MukB-D (566–863). Each MukB-D monomer contains the complete hinge domain (666–779) and two coiled coil strands (572–665 and 780–854). The angle of the V-shaped dimer is ~ 120°. The length of coiled coil domain in each monomer is ~ 100 Å. The hinge domains of two monomers are colored green and magenta respectively; the coiled coil domains for the same monomers are colored light blue and blue respectively. All figures involving crystal structures were prepared with PyMol (DeLano Scientific LLC). Experimental details for protein crystallization and structure determination are included in the supplementary material.
The Hinge Domain of MukB
The central domain of each monomer consists of three α-helices (H1–H3) and five β-strands (B1–B5) (Fig S2A) and folds into a compact globular shape (Fig 1). This globular domain is significantly smaller than the TmSMC hinge region (114 and 169 residues, respectively). Nonetheless, it is solely responsible for mediating MukB dimerization (Fig 1). This result is consistent with previous EM and biochemical studies of MukB suggesting that the coiled coil is intramolecular25; 31; 36. Moreover, the same arrangement was observed in the crystal structures of the hinge domains from TmSMC and PfRad50 28; 30. Thus, MukB shares this structural arrangement with other SMC and SMC-like proteins.
Although the MukB hinge region has no detectable sequence homology with proteins of known structure, a structure-based search of the protein database 37 reveals that it shares significant structural features with the TmSMC hinge domain (Fig 2A). This result is somewhat unexpected given the low sequence identity between the two domains (Fig S3). Indeed, a phylogenetic analysis of SMC proteins suggests that MukB is less closely related to canonical SMC proteins than is Rad50, and has even led to the proposal that the overall structural similarity between MukB and SMC proteins may be the result of convergent evolution 3.
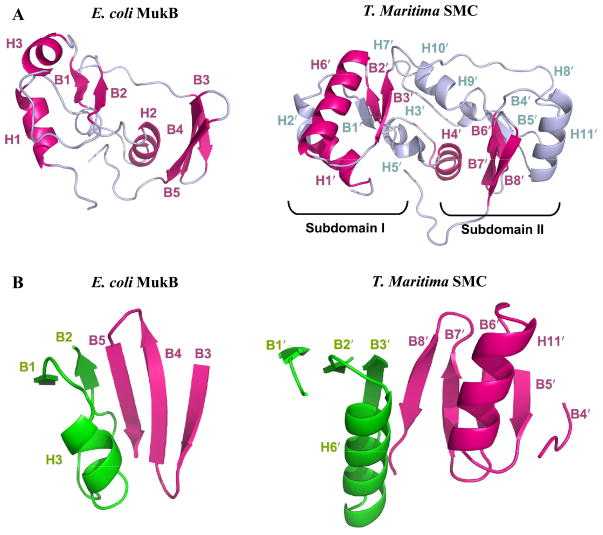
Fig 2.
Secondary structural features of E. coli MukB and TmSMC hinge domain. (A) Highlighted view of the shared features (magenta) between MukB and TmSMC. (B) Comparison of the major dimerization interfaces in MukB and TmSMC. Two monomers from the same dimer are labeled as green and magenta respectively.
Both the MukB and TmSMC hinge domains contain similarly positioned N-terminal α-helices (H1 and H1′, respectively; Fig 2A). This region is connected to similarly positioned α-helices (H2 and H4′), followed by a common β–α–β motif (B1-H3-B2 and B2′-H6′-B3′). Thus, the N-terminal portion of MukB hinge domain has a similar structure to that of TmSMC hinge subdomain I 28, even though MukB is missing several recognizable elements of secondary structure relative to TmSMC. In contrast, the remainder of the MukB hinge domain structure has little similarity with TmSMC hinge subdomain II, with the exception of a three-stranded antiparallel β-sheet that correlates to three of the five strands in TmSMC subdomain II (B3–B5 and B6′–B8′). In total, about two-thirds of the hinge domain of MukB consists of structural elements shared with the much larger TmSMC hinge domain, even though their sequences are only 13% identical (Fig. S3).
Strikingly, the inter-monomer contacts are very similar in MukB and TmSMC (Fig 2B). In MukB, the two-stranded β-sheet (B1, B2) of one MukB monomer combines with the three-stranded β-sheet (B3–B5) of the second monomer to form a five-stranded β-sheet (Fig 2B). Similarly, in TmSMC, the three-stranded β sheet from subdomain I of one monomer combines with the five-stranded β sheet from subdomain II of the other monomer to form an eight-stranded β-sheet 28. In both cases, the strands that form the contact interface with the opposite monomer (B2 and B5; B3′ and B8′) associate in an antiparallel orientation. The TmSMC dimer interface also contains two helices (H6′ and H11′) that pack against one another along one face of the β-sheet28 (Fig 2B). However, this element is absent from the MukB dimer interface, as MukB lacks a helix corresponding to H11′ of TmSMC.
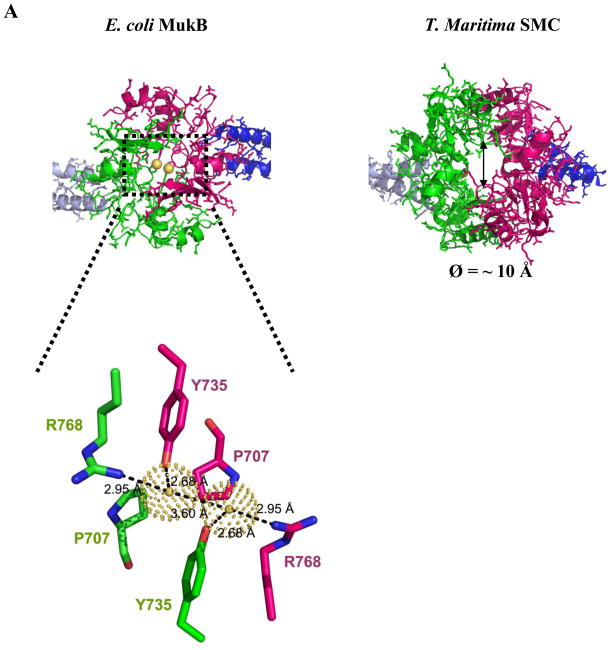
The hinge domain of TmSMC forms a unique “donut-like” shape28 with a hole in the middle (Ø = ~ 10 Å) (Fig 3A). This structural arrangement provides two independent dimerization interfaces in the TmSMC dimer, resulting in the burial of 4250 Å2 of surface area. Interestingly, MukB lacks this “donut hole”. Instead, two interactions appear to bridge the gap between the symmetrical five-stranded β-sheets (Fig 3A). First, the Pro 707 residues from each monomer (H2-B1 loop) are in position to pack against one another, effectively plugging the “hole.” Second, a water-mediated H-bond network is formed by Y735 (B2) and R768 (B5) from each monomer. All three residues are absolutely conserved among MukB proteins from different organisms (Fig S2A), suggesting that these interactions are important for dimer formation. This continuous interface results in the burial of approximately 2930 Å2 of total surface area upon dimer formation.
Fig 3.
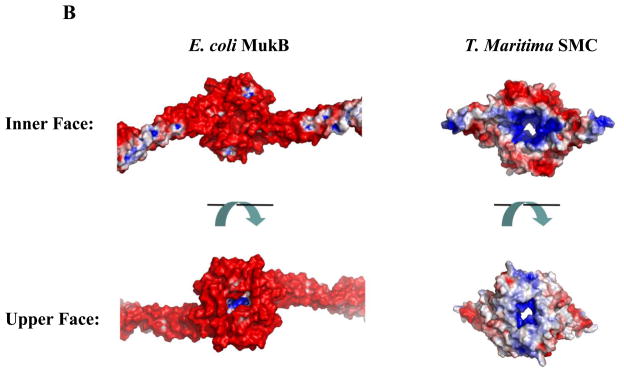
Comparison of E. coli MukB and TmSMC hinge domains. Color coding is the same as in Fig 1. (A) The MukB hinge domain has an overall shape different from the “donut” shaped TmSMC hinge domain. View is from the upper face of Fig 1. Two water molecules in MukB dimer interface are shown as yellow spheres (inset). (B) Surface potential analysis of MukB and TmSMC, generated by APBS46. Views are from the inner (bottom) and upper faces of Fig 1. Positively-charged regions are colored blue, while negatively-charged regions are colored red.
The hinge domains of MukB and TmSMC also have completely distinct surface potential features. There are two basic patches on the inner surface of the TmSMC hinge domain (Fig 3B), which are conserved across different organisms 38. It has been suggested that these basic patches are crucial for dynamic protein-DNA interactions in Bacillus subtilis SMC (BsSMC) 38. In contrast, the accessible inner face of the MukB hinge domain is predominantly negatively charged (Fig 3B). The upper face of the MukB hinge domain is also much more negatively charged than the corresponding face of TmSMC (Fig 3B). Thus, although the MukB hinge domain incorporates structural features of the canonical SMC hinge fold, it is clearly a highly divergent domain with respect to size, primary amino acid sequence, and surface potential.
Coiled Coil Domain of MukB-D
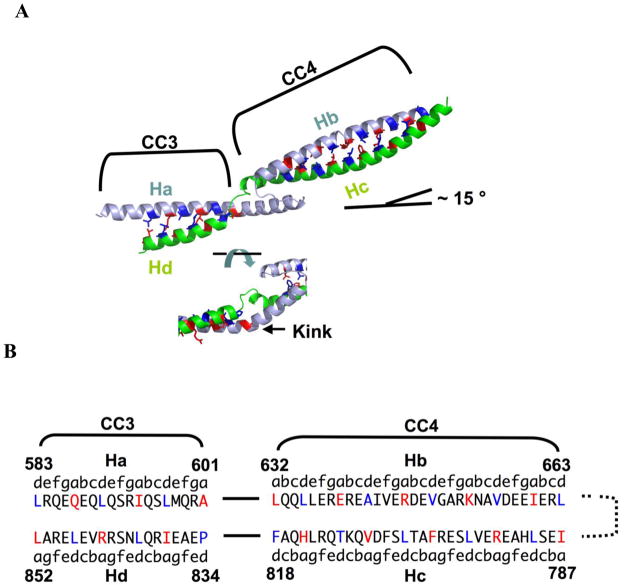
It has proven challenging to crystallize long coiled coils. In our structure, each monomer includes a ~ 100 Å long antiparallel coiled coil domain, accounting for ~ 20 % of the MukB coiled coil (Fig 1). This region is discontinuous (Fig 4A), with two coiled coil segments that are connected by an unusual coiled coil deviation. On the basis of coiled coil prediction algorithms and crosslinking data, we have previously denoted these two regions as coiled coil region 3 (CC3; Ha and Hd) and coiled coil region 4 (CC4; Hb and Hc)31.
Fig 4.
Register analysis of coiled coil domain in E. coli MukB. (A) Close-up view of the coiled coil segment highlighting inter-strand packing (top) and the coiled coil discontinuity (bottom). The N- and C-terminal helical strands within the same monomer are colored light blue and green respectively. The coiled coil regions were assigned by the computer algorithm SOCKET 41 (packing cut-off = 7.4 Å). The two regions of the coiled coil are denoted CC3 (583–601 and 834–852) and CC4 (632–663 and 787–818). The side chains of residues at a (red) and d (dark blue) positions are shown as sticks. (B) Sequence of coiled coil strands highlighting residues involved in inter-strand packing interactions.
Like their parallel counterparts, the sequences of antiparallel coiled coil are characterized by a heptad repeat (abcdefg)n of amino acid residues, in which hydrophobic residues at the a and d positions mediate inter-strand interactions through knobs-into-holes packing 39; 40. An analysis of the MukB coiled coil regions with the structure-based algorithm SOCKET 41 recognizes both segments as coiled coil (Fig 4B) but suggests that the strands of these segments associate more loosely than is the case for canonical coiled coils. Indeed, at a default packing cutoff distance of 7.0 Å, only four of the eight a-d′ packing layers in CC4 were recognized as knobs-into-holes packing; none of the layers in CC3 were recognized. At a more liberal cutoff distance of 7.4 Å, seven of ten layers in CC4 and 3 of 6 in CC3 were recognized as knobs-into-holes packing.
Our construct was likely to contain only a portion of CC331, and indeed, the residues predicted to interact by crosslinking (S570-N859C) are not visible in our model. However, as L583 (d) is packed against L852 (a), we can predict that N859 (a) packs against A576 (d), which is within the length of the crosslinker31 from S570. Although the loose packing in CC3 may derive from the fact that it is a fragment of a longer coiled coil, CC4 is intact in our model. Thus, the non-canonical association of its constituent strands (Hb and Hc) may be of functional significance, allowing for the exposure of interior residues or for conformational changes that could be important for protein-protein or protein-nucleic acid interactions.
CC3 and CC4 are not divided by a typical coiled coil deviation such as a stutter, stammer, or skip42. Instead, the N-terminal helix in CC3 (Ha) is extended by 17 residues relative to the C-terminal helix (Hd). A 7-resiude loop then connects this extended helix to the N-terminal helix of CC4 (Hb) (Fig 4A). On the C-terminal strand, a 5-residue loop connects CC4 (Hc) to CC3 (Hd). This α-helical extension (602–618) contains an absolutely conserved Pro residue (602), which introduces a 40 ° kink and allows the helix to partially pack against CC4 (Fig 4A, Fig S4). As a result of this interruption, the coiled coil axis changes direction by approximately 15°. Although the functional role of this unusual interruption awaits further investigation, the high degree of conservation among MukB proteins in this region suggests that it is significant.
Role of the Hinge Domain in MukB Function
In addition to mediating dimerization, three roles for the hinge domain of MukB and other SMC proteins have been proposed. First, as the name “hinge” suggests, this domain is thought to play a critical role in allowing the dramatic range of angles observed between the two coiled coils by EM25. To provide such flexibility, the globular hinge domain could undergo a dramatic change in conformation. However, the hinge regions of both MukB and TmSMC adopt similar fixed conformations with a ~120° angle between coiled coils. Thus, if a conformational change takes place, the alternative structure has not yet been structurally characterized.
Alternatively, the region connecting the coiled coil to the hinge domain could be flexible in solution. Indeed, in our structural model, both strands of the coiled coil are separated from the globular hinge domain by short loops that could mediate a change in conformation without disrupting the packing of either the dimerization interface or of the coiled coil. Finally, it is also worth noting that the unusual disruption in the middle of the coiled coil could provide such flexibility (Fig 4A). Because this disruption is only ~ 65 Å away from the globular hinge domain, it might be difficult to distinguish the two by EM. Thus, the disruption could also serve as a “hinge” for the opening and closing motions observed with SMC proteins.
SMC and MukB proteins adopt a variety of conformations not only between the two coiled coil domains, but also within each coiled coil itself 6; 25; 27; 28; 36; 43. These disruptions are associated with increased flexibility in the Rad50 coiled coil and it has been proposed that several conserved disruptions in the SMC coiled coil are responsible for flexibility31; 43; 44. Our structure identifies one of these disruptions for the first time. Using disulfide crosslinking experiments, we have localized four additional short regions in the MukB coiled coil in which the N-terminal strand has extra ~ 15 residues relative to the C-terminal strand (Weitzel, C., Waldman, V., Graham, T., Berger, A., and Oakley, M., unpublished results), suggesting that this coiled coil deviation may also be responsible for flexibility in other regions of the coiled coil.
Second, Hirano and co-workers have shown that BsSMC lacking the ATPase domain retains the ability to bind both single- and double-stranded DNA, while BsSMC lacking the hinge region binds DNA poorly23. In addition, the hinge domain is required for ssDNA-stimulated ATPase activity23; 38. These observations have led to the proposal that the hinge region is crucial for transient protein-DNA interactions38. Conserved basic residues on the inner surface of the hinge domain of BsSMC are required for these activities38. In contrast, the equivalent surface of the MukB hinge domain is overwhelmingly negatively charged (Fig 3B). Moreover, gel-shift analysis suggests that the DNA-binding affinity of MukB-D for DNA is dramatically lower than that of intact MukB (Fig S5), even though the inner face of the coiled coil domain is positively charged (Fig S6). Thus, the hinge region of MukB is not likely to be a major contributor to protein-DNA interactions.
Although DNA binding triggers ATP hydrolysis in BsSMC, it remains unclear how these two domains at opposite ends of a long coiled coil can communicate with each other. Interestingly, the recent crystal structure of MukB ATPase domain shows that its top surface is predominantly positively charged. Mutagenesis studies suggested that this region mediates interactions with DNA33. It is also possible the acidic surface of the MukB hinge domain is involved in a direct physical interaction with the ATPase domain. Such a direct interaction has been observed with yeast Schizosaccharomyces pombe SMC proteins by atomic force microscopy (AFM)24.
Finally, Nasmyth and co-workers have suggested that cohesin binds to DNA by trapping it within a ring-shaped complex of SMC1, SMC3 and their accessory proteins2, and that opening of the hinge domain is essential for loading this complex on to DNA45. These investigators proposed that the “donut-like” SMC hinge domain opens up by dissociating its two independent dimerization interfaces in a stepwise fashion45. Our results strongly suggest that, although MukB bears structural similarities with TmSMC, its dimerization interface is continuous. Thus, it is unlikely that MukB opens in the stepwise manner that has been proposed for SMC proteins45.
With availability of this crystal structure, more sophisticated studies on the MukB coiled coil and hinge domain will enable us to understand how MukB interacts with DNA and performs its functions both in vitro and in vivo.
Supplementary Material
Acknowledgments
The authors thank Dr. Hua Yuan, Dr. Joel Ybe and Dr. Faming Zhang for their technical suggestions and assistances. The authors also acknowledge Dr. Jay Nix for assistance with collecting the initial round of datasets and Vince Waldman for assistance with DNA binding studies. This work was supported by the National Institutes of Health (GM57571 to M.G.O and 5P01GM051487 to J.M.B.) and the Packard Foundation (J.M.B.).
Abbreviations
- SMC
Structural Maintenance of Chromosomes
- ABC
ATP Binding Cassette
- CC3
coiled coil region 3
- CC4
coiled coil region 4
- TmSMC
Thermotoga maritima SMC
- BsSMC
Bacillus subtilis SMC
- PfRad50
Pyrococcus furiosus Rad50
- LB
Luria Bertani
- IPTG
isopropyl-β-D-thiogalactopyranoside
- DTT
dithiothreitol
- Ni-NTA
nickel-nitrilotriacetic acid
- Tris
2-Amino-2-hydroxymethyl-propane-1,3-diol
- PBS
Phosphate-Buffered Saline
- MOPS
3-(N-Morpholino)propanesulfonic acid
- PEG
Polyethylene glycol
- OD600
Optical density at 600 nm
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- PCR
Polymerase Chain Reaction
- EM
electron microscopy
- E. coli
Escherichia coli
Footnotes
Accession Number:
Coordinates and structure factors have been deposited in the Protein Data Bank under accession number 3IBP.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hirano T. At the heart of the chromosome: SMC proteins in action. Nat Rev Mol Cell Biol. 2006;7:311–22. doi: 10.1038/nrm1909. [DOI] [PubMed] [Google Scholar]
- 2.Nasmyth K, Haering CH. The structure and function of SMC and kleisin complexes. Annu Rev Biochem. 2005;74:595–648. doi: 10.1146/annurev.biochem.74.082803.133219. [DOI] [PubMed] [Google Scholar]
- 3.Cobbe N, Heck MMS. The evolution of SMC proteins: phylogenetic analysis and structural implications. Mol Biol Evol. 2004;21:332–347. doi: 10.1093/molbev/msh023. [DOI] [PubMed] [Google Scholar]
- 4.Hirano T. SMC-mediated chromosome mechanics: a conserved scheme from bacteria to vertebrates? Genes Dev. 1999;13:11–19. doi: 10.1101/gad.13.1.11. [DOI] [PubMed] [Google Scholar]
- 5.Soppa J. Prokaryotic structural maintenance of chromosomes (SMC) proteins: distribution, phylogeny, and comparison with MukBs and additional prokaryotic and eukaryotic coiled-coil proteins. Gene. 2001;278:253–264. doi: 10.1016/s0378-1119(01)00733-8. [DOI] [PubMed] [Google Scholar]
- 6.Niki H, Imamura R, Kitaoka M, Yamanaka K, Ogura T, Hiraga S. E. coli MukB protein involved in chromosome partition forms a homodimer with a rod-and-hinge structure having DNA binding and ATP/GTP binding activities. EMBO J. 1992;11:5101–9. doi: 10.1002/j.1460-2075.1992.tb05617.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Niki H, Jaffe A, Imamura R, Ogura T, Hiraga S. The new gene mukB codes for a 177 kd protein with coiled-coil domains involved in chromosome partitioning of E. coli. EMBO J. 1991;10:183–93. doi: 10.1002/j.1460-2075.1991.tb07935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weitao T, Dasgupta S, Nordstrom K. Role of the mukB gene in chromosome and plasmid partition in Escherichia coli. Mol Microbiol. 2000;38:392–400. doi: 10.1046/j.1365-2958.2000.02138.x. [DOI] [PubMed] [Google Scholar]
- 9.Britton RA, Lin DC-H, Grossman AD. Characterization of a prokaryotic SMC protein involved in chromosome partitioning. Genes Dev. 1998;12:1254–1259. doi: 10.1101/gad.12.9.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graumann PL, Losick R, Strunnikov AV. Subcellular localization of Bacillus subtilis SMC, a protein involved in chromosome condensation and segregation. J Bacteriol. 1998;180:5749–5755. doi: 10.1128/jb.180.21.5749-5755.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moriya S, Tsujikawa E, Hassan AKM, Asai K, Kodama T, Ogasawara N. A Bacillus subtilis gene-encoding protein homologous to eukaryotic SMC motor protein is necessary for chromosome partition. Mol Microbiol. 1998;29:179–187. doi: 10.1046/j.1365-2958.1998.00919.x. [DOI] [PubMed] [Google Scholar]
- 12.Jensen RB, Shapiro L. The Caulobacter crescentus smc gene is required for cell cycle progression and chromosome segregation. Proc Natl Acad Sci USA. 1999;96:10661–10666. doi: 10.1073/pnas.96.19.10661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamanaka K, Ogura T, Niki H, Hiraga S. Identification of two new genes, mukE and mukF, involved in chromosome partitioning in Escherichia coli. Mol Gen Genet. 1996;250:241–51. doi: 10.1007/BF02174381. [DOI] [PubMed] [Google Scholar]
- 14.Yamazoe M, Onogi T, Sunako Y, Niki H, Yamanaka K, Ichimura T, Hiraga S. Complex formation of MukB, MukE and MukF proteins involved in chromosome partitioning in Escherichia coli. EMBO J. 1999;18:5873–5884. doi: 10.1093/emboj/18.21.5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fennell-Fezzie R, Gradia SD, Akey D, Berger JM. The MukF subunit of Escherichia coli condensin: architecture and functional relationship to kleisins. EMBO J. 2005;24:1921–30. doi: 10.1038/sj.emboj.7600680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu KH, Liu E, Dean K, Gingras M, DeGraff W, Trun NJ. Overproduction of three genes leads to camphor resistance and chromosome condensation in Escherichia coli. Genetics. 1996;143:1521–1532. doi: 10.1093/genetics/143.4.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Q, Mordukhova EA, Edwards AL, Rybenkov VV. Chromosome condensation in the absence of the non-SMC subunits of MukBEF. J Bacteriol. 2006;188:4431–41. doi: 10.1128/JB.00313-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen N, Zinchenko AA, Yoshikawa Y, Araki S, Adachi S, Yamazoe M, Hiraga S, Yoshikawa K. ATP-induced shrinkage of DNA with MukB protein and the MukBEF complex of Escherichia coli. J Bacteriol. 2008;190:3731–7. doi: 10.1128/JB.01863-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cui Y, Petrushenko ZM, Rybenkov VV. MukB acts as a macromolecular clamp in DNA condensation. Nat Struct Mol Biol. 2008;15:411–8. doi: 10.1038/nsmb.1410. [DOI] [PubMed] [Google Scholar]
- 20.Petrushenko ZM, Lai CH, Rai R, Rybenkov VV. DNA reshaping by MukB. Right-handed knotting, left-handed supercoiling. J Biol Chem. 2006;281:4606–15. doi: 10.1074/jbc.M504754200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petrushenko ZM, Lai CH, Rybenkov VV. Antagonistic interactions of kleisins and DNA with bacterial Condensin MukB. J Biol Chem. 2006;281:34208–17. doi: 10.1074/jbc.M606723200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rybenkov VV. Towards the architecture of the chromosomal architects. Nat Struct Mol Biol. 2009;16:104–5. doi: 10.1038/nsmb0209-104. [DOI] [PubMed] [Google Scholar]
- 23.Hirano M, Hirano T. Hinge-mediated dimerization of SMC protein is essential for its dynamic interaction with DNA. EMBO J. 2002;21:5733–5744. doi: 10.1093/emboj/cdf575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoshimura SH, Hizume K, Murakami A, Sutani T, Takeyasu K, Yanagida M. Condensin Architecture and Interaction with DNA Regulatory Non-SMC Subunits Bind to the Head of SMC Heterodimer. Curr Biol. 2002;12:508–513. doi: 10.1016/s0960-9822(02)00719-4. [DOI] [PubMed] [Google Scholar]
- 25.Melby TE, Ciampaglio CN, Briscoe G, Erickson HP. The symmetrical structure of structural maintenance of chromosomes SMC and MukB proteins: long, antiparallel coiled coils, folded at a flexible hinge. J Cell Biol. 1998;142:1595–1604. doi: 10.1083/jcb.142.6.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirano T, Mitchison TJ. A heterodimeric coiled-coil protein required for mitotic chromosome condensation in vitro. Cell. 1994;79:449–58. doi: 10.1016/0092-8674(94)90254-2. [DOI] [PubMed] [Google Scholar]
- 27.Anderson DE, Losada A, Erickson HP, Hirano T. Condensin and cohesin display different arm conformations with characteristic hinge angles. J Cell Biol. 2002;156:419–424. doi: 10.1083/jcb.200111002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haering CH, Lowe J, Hochwagen A, Nasmyth K. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol Cell. 2002;9:773–788. doi: 10.1016/s1097-2765(02)00515-4. [DOI] [PubMed] [Google Scholar]
- 29.Hirano M, Anderson DE, Erickson HP, Hirano T. Bimodal activation of SMC ATPase by intra- and inter-molecular interactions. EMBO J. 2001;20:3238–3250. doi: 10.1093/emboj/20.12.3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hopfner KP, Craig L, Moncalian G, Zinkel RA, Usui T, Owen BAL, Karcher A, Henderson B, Bodmer JL, McMurray CT, Carney JP, Petrini JHJ, Tainer JA. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature. 2002;418:562–566. doi: 10.1038/nature00922. [DOI] [PubMed] [Google Scholar]
- 31.Li Y, Weitzel CS, Arnold RJ, Oakley MG. Identification of Interacting Regions within the Coiled Coil of the Escherichia Coli Structural Maintenance of Chromosomes Protein MukB. J Mol Biol. 2009;391:57–73. doi: 10.1016/j.jmb.2009.05.070. [DOI] [PubMed] [Google Scholar]
- 32.Van den Ent F, Lockhart A, Kendrick-Jones J, Lowe J. Crystal structure of the N-terminal domain of MukB: a protein involved in chromosome partitioning. Structure. 1999;7:1181–1187. doi: 10.1016/s0969-2126(00)80052-0. [DOI] [PubMed] [Google Scholar]
- 33.Woo JS, Lim JH, Shin HC, Suh MK, Ku B, Lee KH, Joo K, Robinson H, Lee J, Park SY, Ha NC, Oh BH. Structural studies of a bacterial condensin complex reveal ATP-dependent disruption of intersubunit interactions. Cell. 2009;136:85–96. doi: 10.1016/j.cell.2008.10.050. [DOI] [PubMed] [Google Scholar]
- 34.Lowe J, Cordell SC, van den Ent F. Crystal Structure of the SMC Head Domain: An ABC ATPase with 900 Residues Antiparallel Coiled-coil Inserted. J Mol Biol. 2001;306:25–35. doi: 10.1006/jmbi.2000.4379. [DOI] [PubMed] [Google Scholar]
- 35.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 36.Matoba K, Yamazoe M, Mayanagi K, Morikawa K, Hiraga S. Comparison of MukB homodimer versus MukBEF complex molecular architectures by electron microscopy reveals a higher-order multimerization. Biochem Biophys Res Commun. 2005;333:694–702. doi: 10.1016/j.bbrc.2005.05.163. [DOI] [PubMed] [Google Scholar]
- 37.Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–1. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hirano M, Hirano T. Opening closed arms: long-distance activation of SMC ATPase by hinge-DNA interactions. Mol Cell. 2006;21:175–86. doi: 10.1016/j.molcel.2005.11.026. [DOI] [PubMed] [Google Scholar]
- 39.Parry DA, Fraser RD, Squire JM. Fifty years of coiled-coils and alpha-helical bundles: a close relationship between sequence and structure. J Struct Biol. 2008;163:258–69. doi: 10.1016/j.jsb.2008.01.016. [DOI] [PubMed] [Google Scholar]
- 40.Oakley MG, Hollenbeck JJ. The design of antiparallel coiled coils. Curr Opin Struct Biol. 2001;11:450–457. doi: 10.1016/s0959-440x(00)00232-3. [DOI] [PubMed] [Google Scholar]
- 41.Walshaw J, Woolfson DN. Socket: a program for identifying and analysing coiled-coil motifs within protein structures. J Mol Biol. 2001;307:1427–50. doi: 10.1006/jmbi.2001.4545. [DOI] [PubMed] [Google Scholar]
- 42.Brown JH, Cohen C, Parry DAD. Heptad Breaks in alpha-Helical Coiled Coils: Stutters and Stammers. Proteins. 1996;26:134–145. doi: 10.1002/(SICI)1097-0134(199610)26:2<134::AID-PROT3>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 43.van Noort J, van Der Heijden T, de Jager M, Wyman C, Kanaar R, Dekker C. The coiled-coil of the human Rad50 DNA repair protein contains specific segments of increased flexibility. Proc Natl Acad Sci USA. 2003;100:7581–6. doi: 10.1073/pnas.1330706100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beasley M, Xu H, Warren W, McKay M. Conserved disruptions in the predicted coiled-coil domains of eukaryotic SMC complexes: implications for structure and function. Genome Res. 2002;12:1201–1209. doi: 10.1101/gr107302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gruber S, Arumugam P, Katou Y, Kuglitsch D, Helmhart W, Shirahige K, Nasmyth K. Evidence that loading of cohesin onto chromosomes involves opening of its SMC hinge. Cell. 2006;127:523–37. doi: 10.1016/j.cell.2006.08.048. [DOI] [PubMed] [Google Scholar]
- 46.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98:10037–41. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.