Summary
Plasmids harbor genes coding for specific functions including virulence factors and antibiotic resistance that permit bacteria to survive the hostile environment found in the host and resist treatment. Together with other genetic elements such as integrons and transposons, and using a variety of mechanisms, plasmids participate in the dissemination of these traits resulting in the virtual elimination of barriers among different kinds of bacteria. In this article we review the current information about physiology and role in virulence and antibiotic resistance of plasmids from the gram-negative opportunistic pathogen Klebsiella pneumoniae. This bacterium has acquired multidrug resistance and is the causative agent of serious communityand hospital-acquired infections. It is also included in the recently defined ESKAPE group of bacteria that cause most of US hospital infections.
Keywords: eskape, gram-negative, xer recombination, Klebsiella
The study of plasmids and their biology has had a decisive impact in the advance of molecular genetics contributing numerous fundamental discoveries beyond the field of plasmid biology (1). Interestingly, the study of plasmids was already well under way before the structure of DNA was known with the experiments that led to the discovery of conjugation and recombination in bacteria using as system the plasmid F, known at that time as “F factor” (2, 3). The continuation of these studies showed that bacterial plasmids are responsible for many of the particular properties of bacteria of medical, industrial, or agricultural interest. Their fundamental role in shaping the characteristics of the host bacteria and their ability to propagate led some authors to propose the somewhat controversial idea that they should be considered as independent organisms (4). The role of plasmids in antibiotic resistance was first recognized in Japan when strains that were susceptible or multiresistant were isolated from the same patient during a single epidemic of dysentery. This fact suggested that susceptible strains were becoming multiresistant, not through successive mutational steps, but rather by acquisition of the necessary genetic determinants in a single step. Watanabe and Fukasawa reported that this process was due to transfer of a plasmid (at that time called resistance transfer factor, RTF, or R-factor) that harbored the resistance genes (5, 6). Later it became clear that plasmids were carriers of not only antibiotic resistance genes but also genes or groups of genes that specify properties that are essential or contribute to the virulence of the host bacteria (7-20). Studies during the following few decades permitted to learn in some detail numerous biological characteristics of plasmids, as well as the high diversity of existing plasmids and their association with other genetic mobile elements.
There is currently an epidemic of antibiotic resistant bacterial infections, which has been identified as one of the greatest threats to human health by the World Health Organization (http://www.who.int/drugresistance/activities/wha66_side_event/en/index.html) (21-24). Within this epidemic, a group of pathogens has been individualized and collectively named ESKAPE (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter). These pathogens are the causative agents of the majority of hospital infections because they “escape” the antibiotic treatment by becoming resistant or persistent to antibiotic treatment (21, 25-27). Plasmids play a central role in the dissemination and acquisition of the resistant determinants in these bacteria. In this article we describe aspects of plasmids of K. pneumoniae, a representative of problematic gram-negative opportunistic pathogens belonging to the ESKAPE group (28-30), with emphasis on their role in virulence and resistance to antibiotics.
K. pneumoniae is the causative agent of serious community- and hospitalacquired infections including but not limited to urinary tract infections, pneumonia, septicemias, meningitis, and soft tissue infections (31-38). K. pneumoniae has also been identified as a causative agent of other less common, yet serious, infections such as liver abscess and invasive syndrome (39, 40), septic arthritis (41), or generalized pustulosis (42), and as the triggering factor in the initiation and development of ankylosing spondylitis and Crohn's disease (43-45). K. pneumoniae strains have accumulated plasmids that carry virulence and resistance genes that keep increasing its ability to resist the main antibiotics used for treatment such as cephalosporins, carbapenems, penicillins, aminoglycosides or fluoroquinolones (26, 37, 38, 46, 47).
K. pneumoniae strains usually harbor more than one plasmid, including small high copy number and low copy number plasmids that are usually large. Of all completed genomes so far, the multiresistant strain HS11286, isolated from human sputum, harbors the most plasmids with sizes 1.31, 3.35, 3.75, 105.97, 111.19, and 122.80 kbp, respectively (48). More than 70 K. pneumoniae plasmids have been completely sequenced and some have been further analyzed with particular attention to the presence of virulence and resistance genes as well as mobile elements.
Small plasmids
A large group of the plasmids found in K. pneumoniae isolates are small, with sizes spanning between less than 2-kbp and 25-kbp (Table 1). Most of these plasmids, which share homology at the replication regions (Fig. 1b), replicate through the ColE1-type mechanism (49-53), are non-self transmissible, and not always encode resistance genes as it is the case of plasmids pKPN2 (54) and pKlebBk17/ 80 (55) that encode a restriction-modification system and a bacteriocin, respectively (Table 1). The backbones of the plasmids within this group have a common general organization consisting of the ColE1-type replication region that includes the genes coding for RNA I and RNA II and in some of them the negative regulator rom (or rop), a transfer region consisting of oriT or this locus accompanied by the genes coding for the remaining relaxosome components (56), and a Xer sitespecific recombination site (Fig.1a). Most probably these plasmids are not confined to K. pneumoniae but they are able to replicate and be stably maintained in other Enterobacteriaceae or gram-negatives. One of the most thoroughly studied ColE1- type plasmids from K. pneumoniae is pJHCMW1 (Table 1), isolated from a neonate with meningitis (38, 57). This plasmid is 11,354-bp long, of which 7,992-bp are Tn1331, a multiresistance transposon that includes aac(6’)-Ib, ant(3”)-Ia, blaOXA-9, and blaTEM-1 (58-61). The backbone of pJHCMW1 is composed of a ColE1-type replication region lacking rom, a functional oriT that mediates mobilization when the remaining components of the relaxosome and the transferosome are supplied in trans by a helper plasmid (74), and a Xer site-specific recombination site named mwr, which has been studied in some detail (see below) (75-78). The aac(6’)-Ib, ant(3”)-Ia, and blaOXA-9 genes are organized in a region resembling the variable region of integrons (59, 60, 62-64). Tn1331, first found in pJHCMW1 (58), or its variations have been found in plasmids hosted by other gram negatives such as Enterobacter, Salmonella, Serratia, and Pseudomonas (37, 58, 63-73). pJHCMW1 was recently used as model of ColE1-type plasmids in microscopy studies to determine the mobility of the molecules inside the cells as well as their location and the implications in partition at the moment of cell division (79). Plasmid molecules were highly mobile but were mainly found located at the poles of the cell because they tend to be excluded from the nucleoid occupied space. In fact, in experiments where the nucleoid-free space was increased by using a dnaN159(ts) mutant at the non permissive temperature or by treatment with cephalexin the plasmid molecules occupied all the nucleoid-free space (79). These results confirmed that the pJHCMW1 plasmid molecules freely move and are not specifically targeted to the pole but rather they tend to occupy the nucleoid-free space. The molecules were no forming clusters and occasionally were able to move between poles (79). This tendency to preferentially locate at the nucleoid-free poles ensures that at cell division both daughter cells host the plasmid.
Table 1.
Completely sequenced ColE1-type plasmids of K. pneumoniae
| Plasmid1 | Size (bp) | Relevant genotype | Accession number | Reference |
|---|---|---|---|---|
| pKPN2 | 4,196 | kpn2kIR, kpn2kIM | AF300473 | (54) |
| pIP843 | 7,086 | bla CTX-M-17 | AY033516 | (98) |
| pH205 | 8,197 | bla CMY-36 | EU331426 | (154) |
| pKlebB-k17/80 | 5,258 | kba, kbi | AF156893 | (55) |
| pJHCMWl | 11,354 | aac(6′)-Ib, ant(3″)-Ia, bla OXA-9 , bla TEM | AF479774 | (61) |
| pS15 | 24,296 | aac(6')-Ib, blaKPC-2, cloacin, immunity protein | FJ223606 | (69) |
| pColEST258 | 13,636 | aac(6')-Ib, cloacin, immunity protein | JN247853 | (82) |
Plasmids that have been subjected to characterization studies beyond just sequencing
kpn2kIR, kpn2kIM, Type II restriction-modification system; kba, kbi, Klebicin B (a bacteriocin) and immunity.
Fig. 1.
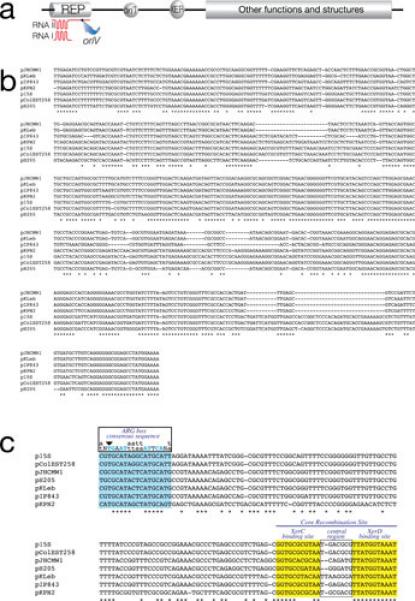
Small plasmids. a, General genetic organization of small ColE1-type plasmids from K. pneumoniae and other Enterobacteriaceae. b, Alignment of the nucleotide sequences of the replication regions of K. pneumoniae ColE1-type plasmids using CLUSTAL W (143). c, Alignment of the nucleotide sequences of Xer site-specific recombination sites of K. pneumoniae ColE1-type plasmids using CLUSTAL W. The ARG box, XerC and XerD binding sites are shown in color and the central regions are boxed. Blue capital letters indicate the most important conserved nucleotides in the ARG box. Downward pointing arrowhead shows the conserved T nucleotide that its found substituted by a C in several Xer site-specific recombination sites (76, 81, 88).
A BLAST analysis of the backbone region of pJHCMW1 shows that the Salmonella Typhimurium plasmid pFPTB1, which includes the blaTEM-135 and tetR/tetA genes in Tn3- and Tn1721-related transposons, is the most closely related with identity coverage including the replication region (99% identity) and the Xer site-specific recombination site fpr, but leaving a gap between these two regions (nucleotides 1770 and 3081) where the oriT and a deficient Xer site-specific recombination site are located in pJHCMW1 (Fig. 2) (80, 81).
Fig. 2.
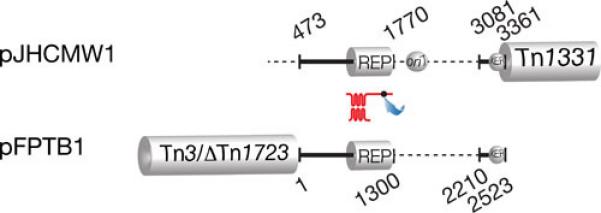
Comparison of pFPBT1 and pJHCMW1. The black lines, which represent regions of homology (coordinates 463-3361 in pJHCMW1), are drawn at scale. The Tn3-like transposons, Tn1331 and Tn3/DeltaTn1723, as well as the dots indicating oriT and the Xer target sites are shown at the correct locations but are not drawn to scale. The replication regions (REP) share 97% homology. The numbers indicate the coordinates in the GenBank database (pJHCMW1, accession number AF479774; pFPBT1, accession number AJ634602). The location of the similar but not identical Xer site-specific recombination sites (81) is indicated.
Fig. 1b shows the alignment of the replication regions of the seven plasmids listed in Table 1. Plasmid p15S completely contains pColEST258 and therefore they share a common block of DNA including the replication region (82). These two plasmids include the region of Tn1331 that encompasses the genes tnpA, tnpR, and aac(6’)-Ib (82). All other replication regions are related but not identical. Since small variations in the nucleotide sequence of the replication regions are sufficient to result in compatible plasmids, only experimental assays will determine whether these plasmids are incompatible. All seven plasmids include a Xer site-specific recombination site consisting of the core recombination site and the accessory sequences where the architectural proteins, usually ArgR and PepA, bind to help formation of the synaptic complex (Fig. 1c). The function of this site, at least in some plasmids, is to maximize stabilization by mediating resolution of multimers, a process that leads to a reduction in the effective number of molecules and results in segregational loss of the plasmid (83-87). Other plasmid-related functions of these sites may include exchange of DNA regions among plasmids (54, 88) as well as mediating the insertion of integrative mobile elements into chromosomes and plasmids (83, 89, 90).
The XerD binding sites, usually the most conserved fraction of Xer recombination target sites (91, 92), are identical in all seven plasmids listed in Table 1 (Fig. 1c). XerC binding sites are more divergent and the central regions are considerably different in sequence and size. While six sites include all 8 nucleotides that are highly conserved in all ARG boxes (Fig. 1c), pJHCMW1 and pH205 include a C instead of a T in one of those positions (arrowhead in Fig. 1c). This substitution has been shown to impair the efficiency of a site to act as target for Xer site-specific recombination (76). In particular, the pJHCMW1 Xer site-specific recombination site, called mwr, has been studied in more detail and shows some interesting characteristics. Resolution of dimers harboring this site is inefficient when the E. coli host cells are cultured in standard L broth (osmolality 209 (mmol/kg) (75,76).). However, the efficiency of resolution is inversely proportional to the osmolarity of the medium and all molecules appear as monomers when cells are cultured in L broth without NaCl added (osmolality 87 mmol/kg) (78). than ideal interaction of ArgR with the mwr ARG-box at higher osmolarity may, at least in part, be responsible for deficient formation of the synaptic complex; a problem that may be compensated when the cells grow in medium with lower osmolarity (Fig. 3). This compensation seems to occur through an increase in negative supercoiling density (Fig. 3) (78). In vitro recombination experiments suggest that the increase in efficiency of resolution occurs at the level of formation of Holliday junction, rather than at the level of Holliday junction resolution (Fig. 3) (78). It is of interest that numerous Xer site-specific recombination sites with identical deficiency in the ARG box have been detected and in some cases experiments demonstrated that they mediate dimer resolution at low efficiency (81). This fact, taken together with other findings like the presence of Xer site-specific recombination sites flanking blaOXA genes in Acinetobacter plasmids (90, 93-96) or the presence of different DNA fragments flanked by a Xer recombination site and an oriT in otherwise identical plasmids (54, 88) leads to the idea that not all Xer recombination sites stabilize plasmids by multimer resolution but may play other, or additional, roles related to plasmid evolution.
Fig. 3.
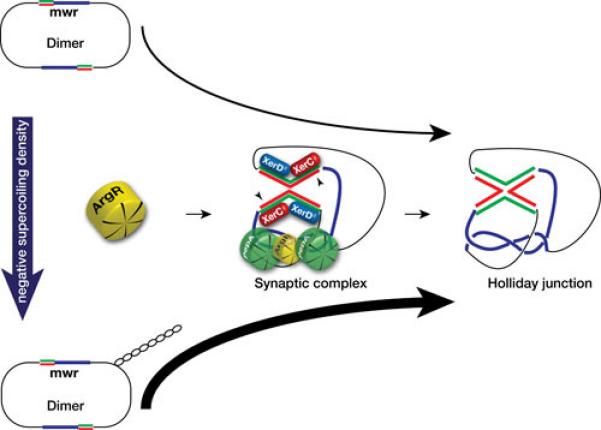
Effect of changes in osmolarity of the culture medium on Xer site-specific recombination at mwr. Schematic representation of the possible chain of events that lead to a higher efficiency of Xer site-specific recombination at the K. pneumoniae plasmid pJHCMW1 site mwr. A decrease in the NaCl concentration in the growth medium (L broth containing 0.5% NaCl added to no NaCl added) is correlated with an increase in supercoiling density, which facilitates interaction of ArgR with the substandard mwr ARG box leading to a more efficient formation of a productive synaptic complex and Holliday junction (78). Molecular models of the interwrapped synaptic complex are available at the following references (146-148). The two strands are shown only in the core recombination site (red and green lines), blue lines represent the accessory sequences.
Small plasmids may be less than the ideal vehicle for dissemination of resistance genes because a) some of the genes are located outside gene cassettes or mobile elements reducing the versatility shown by these elements to promote dissemination at the molecular level, and b) the plasmids may lack an oriT or possess one but lack all other conjugation functions including the proteins that form the specific relaxosome making dissemination at the cellular level by conjugation less efficient (97). Taken together, these factors must reduce the ability of some genes to be mobilized between molecules and cells. A recent report describes a way to reduce this constrain by cointegration of the small plasmid with another plasmid that can provide the machinery for conjugation. The K. pneumoniae plasmid pIP843, which harbors the extended spectrum β–lactamase gene blaCTX-M-17 (98), in spite of including an oriT locus was not transferred in mating experiments between the original K. pneumoniae isolate and a recipient E. coli (98). This result suggested that the presence in the same cells of helper plasmids that provide all necessary components, specific and nonspecific, to mobilize plasmids like pIP843 might not be the most usual situation. However, dissemination of blaCTX-M-17 is dramatically enhanced by its presence in a large conjugative plasmid, pE66An, isolated from an E. coli clinical strain (99). Interestingly, pE66An has the structure of a cointegrate formed between an original ~73-kbp plasmid and pIP843 (Fig. 4). The point of cointegration is the pIP843 RNA II gene. Therefore, this event resulted in inactivation of the ColE1-type replicon (Fig. 4), which must have been important for generating a stable large plasmid.
Fig. 4.
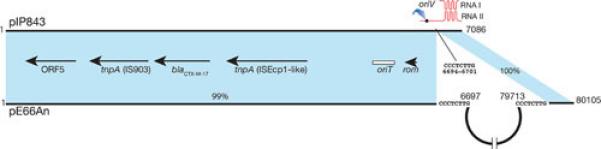
Genetic maps comparison of the K. pneumoniae pIP843 and the E. coli pE66An. The shadowed areas show regions of homology (6681/6701 identities and 6 gaps in the region with 99% homology). The ColE1-type replication region is schematically shown on top of the pIP843 map. The semicircle in pE66An represents the region encompassing nucleotides 6697 – 79713.
Large plasmids
Other plasmids belonging to diverse incompatibility groups, usually larger than those discussed in the previous section, are found in K. pneumoniae. While it was well known that antibiotic resistance as well as virulence genes are housed in several of these plasmids, the interest in studying them has recently increased with the realization they host genes responsible for resistance to last resort antibiotics such as blaKPC, blaNDM-1, and blaOXA (34, 46, 100). The virulence phenotype of a K. pneumoniae strain was first associated with the presence of a plasmid when it was determined that the 180-kbp plasmid, pKP100, harbors the genes coding for the aerobactin iron uptake system and the mucoid phenotype (101-104). Iron uptake systems are well-known virulence factors of numerous bacterial pathogens (12, 105-108). Loss of pKP100 resulted in concomitant loss of virulence, and the transfer of a mobilizable derivative of pKP100 resulted in reacquisition of the virulent phenotype (103). In another instance, a 185-kbp plasmid was isolated by conjugation using as donor a K. pneumoniae isolate that contains three plasmids. This plasmid includes several genes conferring resistance to β-lactams, kanamycin, neomycin, streptomycin, sulfonamides, and tetracyclines, the genes coding for the aerobactin system, and a gene encoding a 29-Kd protein responsible for the ability of this strain to adhere to intestinal cells (109). The association of the mucoid phenotype and production/utilization of aerobactin, and their relation to virulence has been observed in several but not all studies (101, 110, 111). A 219-kbp virulence plasmid, pLVPK, that carries the genes coding for the aerobactin system and is most probably related to those mentioned above was isolated from a highly virulent clinical isolate of K2 serotype and completely sequenced (112). In fact, it has been suggested that numerous K. pneumoniae blood isolates harbor a large virulence plasmid of about 200 kbp that includes the aerobactin system, the ability to express the mucoid phenotype, and resistance to antimicrobials (112). In addition to aerobactin, other iron uptake systems may be included in these plasmids. In the case of pLVPK, the plasmid also includes two more iron-transport systems: the iroBCDN cluster that mediates iron uptake through a catecholate siderophore (113), and an homolog of fecIRA, which encodes a Fur-dependent regulatory system for iron uptake (114). However, the FecR encoded by this plasmid is truncated at the C-proximal end and the FecA is active in transport but it is induction inactive (115). Interestingly, incomplete FecIRA systems are commonly found in plasmids in K. pneumoniae and are chromosomally mediated in Enterobacter (115). BLAST analyses against K. pneumoniae sequences using the pLVPK regions encompassing the iutA and iucDCBA, the iroBCDN, and the fecIRA genes showed 100% homology to the plasmids pK2044 (116) and pKCTC2242 (117) in all three regions and 99% homology to the plasmids pKN-LS6 (accession number JX442974.1), pKPN-IT (82), and pKPN_CZ (118) in the fecIRA region. A comparison of pLVKP, pK2044, and pKCTC2242 using the MAUVE aligner version in which different colors represent local collinear blocks (LCB) with the location of key genes or clusters is shown in Fig. 5. While pVLKP and pK2044 are the most related including all LCBs in the same location, in pKCTC2242 the LCB that in the other two plasmids includes a ter cluster similar to that found in E. coli O157:H7 (119) is truncated, losing the cluster. Furthermore the LCBs in pKCTC2242 are rearranged with respect to pVLKP and pK2044 (Fig. 5). All three plasmids also include an rmpA2 homolog and rmpA, the genes involved in the mucoid phenotype (41, 102, 104, 112). In addition, three physically linked gene clusters coding for resistance to lead, copper, and silver homologs to other known clusters found in Ralstonia metallidurans (120), E. coli (121), and Salmonella Typhimurium (122), respectively, were found in pLVPK as well as pK2044 and pKCTC2242 (Fig. 5). Several other K. pneumoniae plasmids, pKPX-1 (123), pUUH239.2 (124), pKN-LS6 (accession number JX442974), pBK32179 (66), pKPN-IT (82), and pKPN-CZ (118), include highly related but not identical fragments.
Fig. 5.
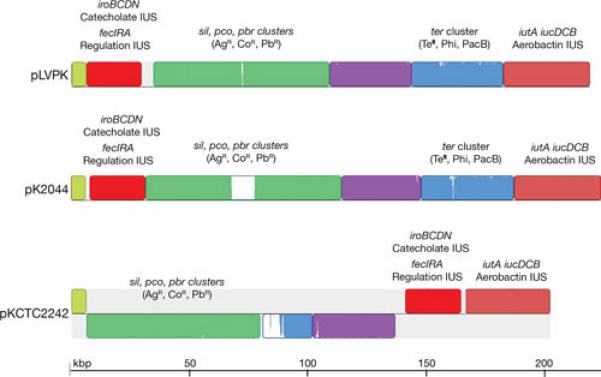
Multiple alignment of pLVPK, pK2044 and pKCTC2242. The nucleotide sequences of pLVPK (accession number AY378100.1)(112), pK2044 (accession number AP006726.1)(116), and pKCTC2242 (accession number CP002911.1)(117) were compared using the MAUVE aligner version 2.3.1 (149). Different colors represent local LCBs. Inside each block there is a similarity profile of the sequence, the height corresponds to the average level of conservation. Completely white areas are not aligned and most probably contain sequences specific to the particular molecule. In pKCTC2242 the LCBs drawn below the black line are inverted with respect to their homologs in pLVPK and pK2044. Some genes or clusters present in these blocks are identified by name. The terZ gene has been reported as “truncated” (112). The truncation is a consequence of an extra T in the sequence that could also be a sequencing error. The terBCDE genes are sufficient for the tellurite resistance phenotype (TeR). The ter cluster is also responsible for the phage inhibition (Phi) and colicin resistance (PacB) phenotypes (150). Copper (pco), silver (sil), lead (pbr), and tellurite (ter) resistance related genes; IUS, iron uptake system.
Replication of pLVPK most probably occurs through the iteron mechanism as a repA homolog located between two sets of iterons was found. The region shows higher than 90% homology to the K. pneumoniae plasmids, pKCTC2242 (117), pK2044 (116), and pNDM-MAR (72) and the RepA amino acid sequence show similarity to numerous proteins from plasmids isolated from enterobacteria (112). Another putative replication protein was found in pLVPK with homology to several replicator proteins in the database. In addition homologs to sopA and sopB strongly suggested the presence of an F plasmid-like partitioning system.
An interesting case is that of pClpk, a 150kbp self transferable plasmid isolated from the K. pneumoniae C132-98, a strain that caused critical infections over a twoyear period in a hospital and that is characterized for an unusual thermotolerance (125). K. pneumoniae C132-98 harbors at least eight plasmids, 4 of which with a size of 100 kbp or larger. The plasmid pClpk encodes ClpK, an ATPase responsible for the increased heat resistance that characterizes K. pneumoniae C132-98 (125). The clpK gene was then found in other K. pneumoniae strains and in at least another plasmid, the 220 kbp IncFIIk pUUH239.2, which also harbors several resistance genes including blaCTX-M-15 and aac(6’)-Ib-cr (124, 126).
Although cases such as that of the small plasmid pJHCMW1 (38, 57, 58, 61) are well known, numerous plasmids including multiple resistance genes are large and self transmissible. This conjugation machinery requires genes coding for the relaxosome, the type IV coupling protein, and the type IV secretion system (127). As illustrated in previous paragraphs the resistance genes may be included in various genetic elements and coexist with other genetic elements and genes coding for resistance to metals or virulence factors.
A well studied multiresistance K. pneumoniae plasmid is pMET1, a plasmid isolated form a clinical strain responsible for a high mortality hospital outbreak (37, 128). Its replication was extensively studied by cloning and deletion assays that permitted to identify a 1655-bp replication region that includes repA, which codes for a protein belonging to the RepA IncFII superfamily, and an AT-rich sequence likely to include ori (128). BLASTn analysis of this region showed extensive homology only with the Yersinia pestis plasmid pCRY (129), the Cronobacter turicensis plasmid pCTU2 (130) and the C. sakazakii plasmid pESA2 (accession number CP000784). In addition, a pMET1 segment including the type IV secretion system, which is related to those found in the ICEKp1 and the HPIECOR31 (131), and the mobB and mobC genes shared high similarity with the cryptic Yersinia pestis plasmid pCRY. The homology shared between the cryptic pCRY and the multidrug resistance pMET1 suggests that the latter plasmid can be replicated and stably maintained in Yersinia species, representing a high public health and biodefense threat due to transfer of multiple resistance genes to pathogenic Yersinia strains. Absent in pCRY are the pMET1 partition genes parF and parG. The resistance genes in pMET1 reside in Tn1331.2, an 11,042-bp transposon highly related to Tn1331, the transposon found in pJHCMW1 (see above). Tn1331.2 has a perfect duplication of the 3,047-bp DNA region that includes aac(6’)-Ib, aadA1, and blaOXA-9 (37, 128).
The resistance to antibiotics encoded by some of the large K. pneumoniae plasmids plays a central role in enhancing the morbidity and mortality of K. pneumoniae due to the advent of the carbapenemases, in particular KPC (K. pneumoniae carbapenemase) and NDM-1 (New Delhi metallo-β-lactamase-1), creating a significant public health threat (66, 132, 133). Both blaKPC and blaNDM-1 were first detected in K. pneumoniae but they are present in other gram-negatives. The emergence of carbapenemases has virtually eliminated the possibility of using β-lactams to treat multidrug resistance gram-negative infections (134). Furthermore, the presence of genes coding for carbapenemases is usually accompanied by others that code for resistance to other β-lactams, aminoglycosides, and fluoroquinolones, which leaves few treatment options (34, 100, 135). In particular the common localization of carbapenemase-coding genes in plasmids has renewed the interest in their study and numerous large plasmids coding for carbapenemases (KPC, VIM, NDM, IMP, and OXA types) and CTX-M type extended spectrum β-lactamases have been fully sequenced (132). Since there is consensus that currently we are witnessing a convergence of two plasmid-driven epidemics, one of blaKPC and the other of blaNDM carrying K. pneumoniae (100, 134, 136-138), in the following paragraphs we describe some representative characteristics about plasmids including blaNDM and blaKPC genes.
The carbapenemase NDM-1 is a broad-spectrum metallo-β-lactamase that mediates inactivation of nearly all β-lactams with the exception of aztreonam. It was first identified in a K. pneumoniae isolate from a urinary tract infection (139) and has rapidly disseminated among Enterobacteriaceae and Acinetobacter spp (140). Plasmids carrying blaNDM-1 belonging to different incompatibility groups have already been isolated. Fig. 6 shows a comparative diagram of sequenced K. pneumoniae plasmids that harbor this gene. Although plasmids carrying the gene can be largely unrelated belonging to different incompatibility groups (Fig. 6a), the immediate environment of blaNDM-1 is highly related in all plasmids (Fig. 6b). The blaKPC gene is found as part of the 10-kbp transposon Tn4401, which is present in numerous multiresistance plasmids belonging to different incompatibility groups. It may be worth mentioning that Tn4401 has not yet been found in any chromosome. The large number of different conjugative plasmids allowed the rapid and successful spreading of blaKPC to other gram-negatives. Of special interest is the recent report of the IncI plasmid pBK15692 in which Tn4401 has inserted within the tnpA gene of Tn1331 (Fig. 7) creating a transposon with the capability to confer resistance to virtually all β-lactams and aminoglycosides. The dissemination of pBK15692 will increase the already complicated landscape of gram-negative infections.
Fig. 6.
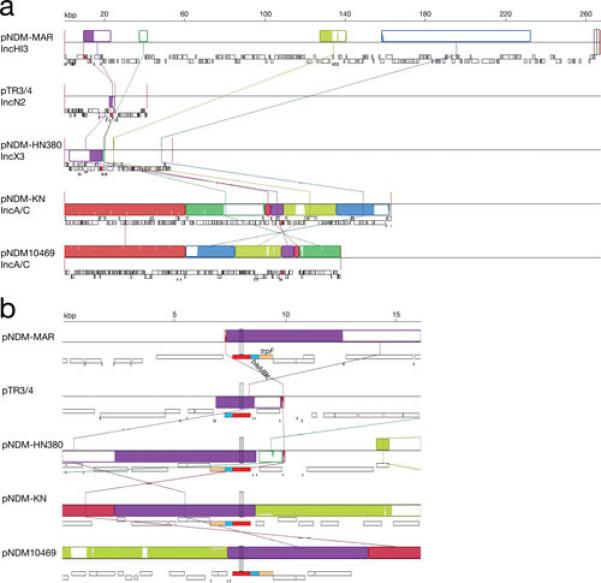
Multiple alignment of pNDM-MAR, pTR3/4, pNDM-HN380, pNDM-KN, and pNDM10469. The nucleotide sequences of pNDM-MAR (accession number JN420336) (72), pTR3/4 (accession number JQ349086) (151), pNDM-HN380 (accession number JX104760) (152), pNDM-KN (accession number JN157804) (153), and pNDM10469 (accession number JN861072) were compared using the MAUVE aligner version 2.3.1 (149). The blaNDM-1 gene is represented in red, genes bleMBL and trpF are represented in light blue and light brown, respectively. Plasmids pTR3 and pTR4, originally thought to be similar but not identical were later proved to be identical and renamed pTR3/4 (151). a. The comparison of the complete nucleotide sequence is shown with LCBs represented in blocks of different colors. b. Zoom in the region including the blaNDM-1 gene.
Fig. 7.
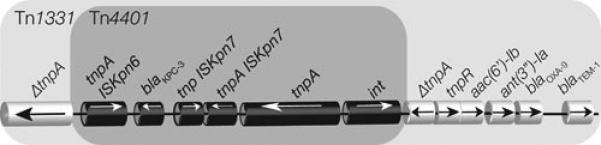
Genetic map of the Tn1331: :Tn4401 region in the K. pneumoniae plasmid pBK692.
Concluding remarks
Most interactions between humans and bacteria do not result in disease and many of them are beneficial for one or both interacting partners. When the interaction results in disease, the relation between humans and pathogenic bacteria has been one resembling an arms race through evolution. As the human body was developing and evolving numerous strategies to defend against bacterial infections, bacteria were doing the same to counter those defenses that limit or prevent the establishment of invading bacteria. In addition an artificial defense that humans have counted for a few decades is the utilization of antibiotics. The evading strategies that permit bacteria to colonize or cause damage to the host are known as virulence factors. The presence or absence of a virulence factor can determine whether a bacterium behaves as a pathogen or not in normal conditions. Numerous virulence factors as well as antibiotic resistance genes are usually part of plasmids or genetic elements located in plasmids that have the capability to disseminate at the molecular level such as integrons or transposons. This intense activity permits the generation of new variations of plasmid molecules that can accumulate genetic determinants for several virulence factors and resistance. This dissemination in combination with the tremendous tendency that plasmids have to disseminate at the cellular level results in the virtual elimination of barriers among different kinds of bacteria permitting these genes to reach nearly all bacteria (141, 142). Plasmids play an essential role in this fluid situation in which genes behave as a pool being able to reach any bacteria transforming them from friend into foe or from susceptible into resistant. This chapter illustrates the rich variety of plasmids that can harbor numerous virulence factors and resistance genes in K. pneumoniae. However, careful examination of the existing knowledge shows that we are still barely scratching the surface. There is a wealth of information that still needs to be acquired about these plasmids. Their study using the most modern technologies will enhance the possibilities to design new strategies to deal with emerging infectious diseases that represent a serious threat to human health.
Acknowledgements
The authors’ work cited in this review article was funded by Public Health Service grant 2R15AI047115 (to MET) from the National Institutes of Health and and PIP2011/2013 N° 11420100100152 (to MSR).
References
- 1.Cohen SN. Bacterial plasmids: their extraordinary contribution to molecular genetics. Gene. 1993;135:67–76. doi: 10.1016/0378-1119(93)90050-d. [DOI] [PubMed] [Google Scholar]
- 2.Lederberg J, Tatum EL. Gene recombination in Escherichia coli. Nature. 1946;158:558. doi: 10.1038/158558a0. [DOI] [PubMed] [Google Scholar]
- 3.Tatum EL, Lederberg J. Gene recombination in the bacterium Escherichia coli. J. Bacteriol. 1947;53:673–684. doi: 10.1128/jb.53.6.673-684.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Datta N. Plasmids as organisms. In: Helinski D, Cohen S, Clewell D, Jackson D, Hollaender A, editors. Plasmids in bacteria. Vol. 30. Plenum Press; New York, NY.: 1985. pp. 3–16. [Google Scholar]
- 5.Watanabe T, Fukasawa T. Episome-mediated transfer of drug resistance in Enterobacteriaceae. I. Transfer of resistance factors by conjugation. J. Bacteriol. 1961;81:669–678. doi: 10.1128/jb.81.5.669-678.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Watanabe T. Infective heredity of multiple drug resistance in bacteria. Bacteriol. Rev. 1963;27:87–115. doi: 10.1128/br.27.1.87-115.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hammerl JA, Freytag B, Lanka E, Appel B, Hertwig S. The pYV virulence plasmids of Yersinia pseudotuberculosis and Y. pestis contain a conserved DNA region responsible for the mobilization by the selftransmissible plasmid pYE854. Environ Microbiol Rep. 2012;4:433–438. doi: 10.1111/j.1758-2229.2012.00353.x. [DOI] [PubMed] [Google Scholar]
- 8.Stephens C, Murray W. Pathogen evolution: How good bacteria go bad. Curr. Biol. 2001;11:R53–56. doi: 10.1016/s0960-9822(01)00012-4. [DOI] [PubMed] [Google Scholar]
- 9.Elwell LP, Shipley PL. Plasmid-mediated factors associated with virulence of bacteria to animals. Annu. Rev. Microbiol. 1980;34:465–496. doi: 10.1146/annurev.mi.34.100180.002341. [DOI] [PubMed] [Google Scholar]
- 10.Guiney DG, Fang FC, Krause M, Libby S. Plasmid-mediated virulence genes in non-typhoid Salmonella serovars. FEMS Microbiol. Lett. 1994;124:1–9. doi: 10.1111/j.1574-6968.1994.tb07253.x. [DOI] [PubMed] [Google Scholar]
- 11.Johnson TJ, Nolan LK. Pathogenomics of the virulence plasmids of Escherichia coli. Microbiol. Mol. Biol. Rev. 2009;73:750–774. doi: 10.1128/MMBR.00015-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tolmasky ME, Crosa JH. Regulation of plasmid-mediated iron transport and virulence in Vibrio anguillarum. Biol. Met. 1991;4:33–35. doi: 10.1007/BF01135554. [DOI] [PubMed] [Google Scholar]
- 13.Actis LA, Tolmasky ME, Crosa JH. Vibriosis. In: Woo PT, Bruno DW, editors. Fish Diseases and Disorders, vol. 3, viral, bacterial and fungal infections. Cab International Publishing; Wallingford, United Kingdom: 2011. pp. 570–605. [Google Scholar]
- 14.Shannon JG, Hasenkrug AM, Dorward DW, Nair V, Carmody AB, Hinnebusch BJ. Yersinia pestis subverts the dermal neutrophil response in a mouse model of bubonic plague. MBio. 2013;4:e00170–00113. doi: 10.1128/mBio.00170-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsui H, Bacot CM, Garlington WA, Doyle TJ, Roberts S, Gulig PA. Virulence plasmid-borne spvB and spvC genes can replace the 90-kilobase plasmid in conferring virulence to Salmonella enterica serovar Typhimurium in subcutaneously inoculated mice. J. Bacteriol. 2001;183:4652–4658. doi: 10.1128/JB.183.15.4652-4658.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fabrega A, Vila J. Salmonella enterica serovar Typhimurium skills to succeed in the host: virulence and regulation. Clin. Microbiol. Rev. 2013;26:308–341. doi: 10.1128/CMR.00066-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Waters VL, Crosa JH. Colicin V virulence plasmids. Microbiol. Rev. 1991;55:437–450. doi: 10.1128/mr.55.3.437-450.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drancourt M. Plague in the genomic area. Clin. Microbiol. Infect. 2012;18:224–230. doi: 10.1111/j.1469-0691.2012.03774.x. [DOI] [PubMed] [Google Scholar]
- 19.Wajima T, Sabui S, Kano S, Ramamurthy T, Chatterjee NS, Hamabata T. Entire sequence of the colonization factor coli surface antigen 6-encoding plasmid pCss165 from an enterotoxigenic Escherichia coli clinical isolate. Plasmid. 2013 doi: 10.1016/j.plasmid.2013.07.006. in press. [DOI] [PubMed] [Google Scholar]
- 20.Crosa JH, Actis LA, Mitoma Y, Perez-Casal J, Tolmasky ME, Valvano M. Plasmid-mediated iron sequestering systems in pathogenic strains of Vibrio anguillarum and Escherichia coli. In: Helinski D, Cohen S, Clewell D, Jackson D, Hollaender A, editors. Plasmids in Bacteria. Plenum Press; New York, NY.: 1985. pp. 759–774. [DOI] [PubMed] [Google Scholar]
- 21.Infectious Diseases Society of A The 10 x ′20 Initiative: pursuing a global commitment to develop 10 new antibacterial drugs by 2020. Clin. Infect. Dis. 2010;50:1081–1083. doi: 10.1086/652237. [DOI] [PubMed] [Google Scholar]
- 22.Spellberg B, Guidos R, Gilbert D, Bradley J, Boucher HW, Scheld WM, Bartlett JG, Edwards J, Jr., Infectious Diseases Society of A The epidemic of antibiotic-resistant infections: a call to action for the medical community from the Infectious Diseases Society of America. Clin. Infect. Dis. 2008;46:155–164. doi: 10.1086/524891. [DOI] [PubMed] [Google Scholar]
- 23.Shlaes DM, Sahm D, Opiela C, Spellberg B. The FDA reboot of antibiotic development. Antimicrob. Agents Chemother. 2013;57:4605–4607. doi: 10.1128/AAC.01277-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spellberg B, Bartlett JG, Gilbert DN. The future of antibiotics and resistance. N. Engl. J. Med. 2013;368:299–302. doi: 10.1056/NEJMp1215093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boucher HW, Talbot GH, Benjamin DK, Jr., Bradley J, Guidos RJ, Jones RN, Murray BE, Bonomo RA, Gilbert D, Infectious Diseases Society of A. 10 x ′20 Progress--Development of New Drugs Active Against Gram- Negative Bacilli: An Update From the Infectious Diseases Society of America. Clin. Infect. Dis. 2013;56:1685–1694. doi: 10.1093/cid/cit152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 2009;48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 27.Rice LB. Progress and challenges in implementing the research on ESKAPE pathogens. Infect. Control Hosp. Epidemiol. 31 Suppl. 2010;1:S7–10. doi: 10.1086/655995. [DOI] [PubMed] [Google Scholar]
- 28.Kuehn BM. “Nightmare” bacteria on the rise in US hospitals, long-term care facilities. JAMA. 2013;309:1573–1574. doi: 10.1001/jama.2013.2922. [DOI] [PubMed] [Google Scholar]
- 29.Rice LB. The clinical consequences of antimicrobial resistance. Curr. Opin. Microbiol. 2009;12:476–481. doi: 10.1016/j.mib.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 30.Perez F, Endimiani A, Hujer KM, Bonomo RA. The continuing challenge of ESBLs. Curr. Opin. Pharmacol. 2007;7:459–469. doi: 10.1016/j.coph.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coque TM, Oliver A, Perez-Diaz JC, Baquero F, Canton R. Genes encoding TEM-4, SHV-2, and CTX-M-10 extended-spectrum beta-lactamases are carried by multiple Klebsiella pneumoniae clones in a single hospital (Madrid, 1989 to 2000). Antimicrob. Agents Chemother. 2002;46:500–510. doi: 10.1128/AAC.46.2.500-510.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Daza R, Gutierrez J, Piedrola G. Antibiotic susceptibility of bacterial strains isolated from patients with community-acquired urinary tract infections. Int. J. Antimicrob. Agents. 2001;18:211–215. doi: 10.1016/s0924-8579(01)00389-2. [DOI] [PubMed] [Google Scholar]
- 33.Liam CK, Lim KH, Wong CM. Community-acquired pneumonia in patients requiring hospitalization. Respirology. 2001;6:259–264. doi: 10.1046/j.1440-1843.2001.00336.x. [DOI] [PubMed] [Google Scholar]
- 34.Nordmann P, Cuzon G, Naas T. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect. Dis. 2009;9:228–236. doi: 10.1016/S1473-3099(09)70054-4. [DOI] [PubMed] [Google Scholar]
- 35.Poulou A, Voulgari E, Vrioni G, Koumaki V, Xidopoulos G, Chatzipantazi V, Markou F, Tsakris A. Outbreak Caused by an Ertapenem-Resistant, CTX-M-15-Producing Klebsiella pneumoniae ST101 Clone Carrying an OmpK36 Porin Variant. J. Clin. Microbiol. 2013 doi: 10.1128/JCM.01244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramirez MS, Xie G, Marshall SH, Hujer KM, Chain PS, Bonomo RA, Tolmasky ME. Multidrug-resistant (MDR) Klebsiella pneumoniae clinical isolates: a zone of high heterogeneity (HHZ) as a tool for epidemiological studies. Clin. Microbiol. Infect. 2012;18:E254–258. doi: 10.1111/j.1469-0691.2012.03886.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tolmasky ME, Chamorro RM, Crosa JH, Marini PM. Transposonmediated amikacin resistance in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 1988;32:1416–1420. doi: 10.1128/aac.32.9.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Woloj M, Tolmasky ME, Roberts MC, Crosa JH. Plasmid-encoded amikacin resistance in multiresistant strains of Klebsiella pneumoniae isolated from neonates with meningitis. Antimicrob. Agents Chemother. 1986;29:315–319. doi: 10.1128/aac.29.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hsueh PR, Wu JJ, Teng LJ, Chen YC, Yang PC, Ho SW, Luh KT. Primary liver abscess caused by one clone of Klebsiella pneumoniae with two colonial morphotypes and resistotypes. Emerg. Infect. Dis. 2002;8:100–102. doi: 10.3201/eid0801.010167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Siu LK, Yeh KM, Lin JC, Fung CP, Chang FY. Klebsiella pneumoniae liver abscess: a new invasive syndrome. Lancet Infect. Dis. 2012;12:881–887. doi: 10.1016/S1473-3099(12)70205-0. [DOI] [PubMed] [Google Scholar]
- 41.Suzuki K, Nakamura A, Enokiya T, Iwashita Y, Tomatsu E, Muraki Y, Kaneko T, Okuda M, Katayama N, Imai H. Septic arthritis subsequent to urosepsis caused by hypermucoviscous Klebsiella pneumoniae. Intern. Med. 2013;52:1641–1645. doi: 10.2169/internalmedicine.52.0175. [DOI] [PubMed] [Google Scholar]
- 42.Huang HY, Wu YH, Kuo CF. Klebsiella pneumoniae sepsis with unusual cutaneous presentation of generalized pustulosis. Clin. Exp. Dermatol. 2013;38:626–629. doi: 10.1111/ced.12092. [DOI] [PubMed] [Google Scholar]
- 43.Rashid T, Wilson C, Ebringer A. The Link between Ankylosing Spondylitis, Crohn's Disease, Klebsiella, and Starch Consumption. Clin. Dev. Immunol. 2013;2013:872632. doi: 10.1155/2013/872632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ebringer A, Rashid T, Tiwana H, Wilson C. A possible link between Crohn's disease and ankylosing spondylitis via Klebsiella infections. Clin. Rheumatol. 2007;26:289–297. doi: 10.1007/s10067-006-0391-2. [DOI] [PubMed] [Google Scholar]
- 45.Rashid T, Ebringer A. Ankylosing spondylitis is linked to Klebsiella-- the evidence. Clin. Rheumatol. 2007;26:858–864. doi: 10.1007/s10067-006-0488-7. [DOI] [PubMed] [Google Scholar]
- 46.Patel G, Bonomo RA. “Stormy waters ahead”: global emergence of carbapenemases. Front Microbiol. 2013;4:48. doi: 10.3389/fmicb.2013.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pendleton JN, Gorman SP, Gilmore BF. Clinical relevance of the ESKAPE pathogens. Expert Rev. Anti Infect. Ther. 2013;11:297–308. doi: 10.1586/eri.13.12. [DOI] [PubMed] [Google Scholar]
- 48.Liu P, Li P, Jiang X, Bi D, Xie Y, Tai C, Deng Z, Rajakumar K, Ou HY. Complete genome sequence of Klebsiella pneumoniae subsp. pneumoniae HS11286, a multidrug-resistant strain isolated from human sputum. J. Bacteriol. 2012;194:1841–1842. doi: 10.1128/JB.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tolmasky ME, Actis LA, Crosa JH. Plasmid DNA replication. In: Flickinger M, editor. Encyclopedia of Industrial Biotechnology: Bioprocess, Bioseparation, and Cell Technology. Vol. 6. John Wiley and Sons, Inc.; New York, NY.: 2010. pp. 3931–3953. [Google Scholar]
- 50.Actis LA, Tolmasky ME, Crosa JH. Bacterial plasmids: replication of extrachromosomal genetic elements encoding resistance to antimicrobial compounds. Front. Biosci. 1999;4:D43–62. doi: 10.2741/actis. [DOI] [PubMed] [Google Scholar]
- 51.Polisky B. ColE1 replication control circuitry: sense from antisense. Cell. 1988;55:929–932. doi: 10.1016/0092-8674(88)90235-8. [DOI] [PubMed] [Google Scholar]
- 52.Allen JM, Simcha DM, Ericson NG, Alexander DL, Marquette JT, Van Biber BP, Troll CJ, Karchin R, Bielas JH, Loeb LA, Camps M. Roles of DNA polymerase I in leading and lagging-strand replication defined by a highresolution mutation footprint of ColE1 plasmid replication. Nucleic Acids Res. 2011;39:7020–7033. doi: 10.1093/nar/gkr157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eguchi Y, Tomizawa J. Complexes formed by complementary RNA stem-loops. Their formations, structures and interaction with ColE1 Rom protein. J. Mol. Biol. 1991;220:831–842. doi: 10.1016/0022-2836(91)90356-b. [DOI] [PubMed] [Google Scholar]
- 54.Zakharova MV, Beletskaya IV, Denjmukhametov MM, Yurkova TV, Semenova LM, Shlyapnikov MG, Solonin AS. Characterization of pECL18 and pKPN2: a proposed pathway for the evolution of two plasmids that carry identical genes for a Type II restriction-modification system. Mol Genet Genomics. 2002;267:171–178. doi: 10.1007/s00438-002-0644-y. [DOI] [PubMed] [Google Scholar]
- 55.Riley MA, Pinou T, Wertz JE, Tan Y, Valletta CM. Molecular characterization of the klebicin B plasmid of Klebsiella pneumoniae. Plasmid. 2001;45:209–221. doi: 10.1006/plas.2001.1519. [DOI] [PubMed] [Google Scholar]
- 56.Smillie C, Garcillan-Barcia MP, Francia MV, Rocha EP, de la Cruz F. Mobility of plasmids. Microbiol. Mol. Biol. Rev. 2010;74:434–452. doi: 10.1128/MMBR.00020-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tolmasky ME, Roberts M, Woloj M, Crosa JH. Molecular cloning of amikacin resistance determinants from a Klebsiella pneumoniae plasmid. Antimicrob. Agents Chemother. 1986;30:315–320. doi: 10.1128/aac.30.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tolmasky ME, Crosa JH. Tn1331, a novel multiresistance transposon encoding resistance to amikacin and ampicillin in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 1987;31:1955–1960. doi: 10.1128/aac.31.12.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tolmasky ME. Sequencing and expression of aadA, bla, and tnpR from the multiresistance transposon Tn1331. Plasmid. 1990;24:218–226. doi: 10.1016/0147-619x(90)90005-w. [DOI] [PubMed] [Google Scholar]
- 60.Tolmasky ME, Crosa JH. Genetic organization of antibiotic resistance genes (aac(6')-Ib, aadA, and oxa9) in the multiresistance transposon Tn1331. Plasmid. 1993;29:31–40. doi: 10.1006/plas.1993.1004. [DOI] [PubMed] [Google Scholar]
- 61.Sarno R, McGillivary G, Sherratt DJ, Actis LA, Tolmasky ME. Complete nucleotide sequence of Klebsiella pneumoniae multiresistance plasmid pJHCMW1. Antimicrob. Agents Chemother. 2002;46:3422–3427. doi: 10.1128/AAC.46.11.3422-3427.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ramirez MS, Parenteau TR, Centron D, Tolmasky ME. Functional characterization of Tn1331 gene cassettes. J. Antimicrob. Chemother. 2008;62:669–673. doi: 10.1093/jac/dkn279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ramirez MS, Tolmasky ME. Aminoglycoside modifying enzymes. Drug Resist Updat. 2010;13:151–171. doi: 10.1016/j.drup.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ramirez MS, Nikolaidis N, Tolmasky ME. Rise and dissemination of aminoglycoside resistance: the aac(6′)-Ib paradigm. Front Microbiol. 2013;4:121. doi: 10.3389/fmicb.2013.00121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Alavi MR, Antonic V, Ravizee A, Weina PJ, Izadjoo M, Stojadinovic A. An Enterobacter plasmid as a new genetic background for the transposon Tn1331. Infect Drug Resist. 2011;4:209–213. doi: 10.2147/IDR.S25408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen L, Chavda KD, Al Laham N, Melano RG, Jacobs MR, Bonomo RA, Kreiswirth BN. Complete nucleotide sequence of a blaKPC-harboring IncI2 plasmid and its dissemination in New Jersey and New York hospitals: a hidden threat. Antimicrob. Agents Chemother. 2013 doi: 10.1128/AAC.01397-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Garcia DC, Woloj M, Kaufman S, Sordelli DO, Pineiro S. Sequences related to Tn1331 associated with multiple antimicrobial resistance in different Salmonella serovars. Int. J. Antimicrob. Agents. 1995;5:199–202. doi: 10.1016/0924-8579(95)00005-s. [DOI] [PubMed] [Google Scholar]
- 68.Garcia DC, Catalano M, Pineiro S, Woloj M, Kaufman S, Sordelli DO. The emergence of resistance to amikacin in Serratia marcescens isolates from patients with nosocomial infection. Int. J. Antimicrob. Agents. 1996;7:203–210. doi: 10.1016/s0924-8579(96)00322-6. [DOI] [PubMed] [Google Scholar]
- 69.Gootz TD, Lescoe MK, Dib-Hajj F, Dougherty BA, He W, Della-Latta P, Huard RC. Genetic organization of transposase regions surrounding blaKPC carbapenemase genes on plasmids from Klebsiella strains isolated in a New York City hospital. Antimicrob. Agents Chemother. 2009;53:1998–2004. doi: 10.1128/AAC.01355-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Poirel L, Cabanne L, Collet L, Nordmann P. Class II transposon-borne structure harboring metallo-beta-lactamase gene blaVIM-2 in Pseudomonas putida. Antimicrob. Agents Chemother. 2006;50:2889–2891. doi: 10.1128/AAC.00398-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rice LB, Carias LL, Hutton RA, Rudin SD, Endimiani A, Bonomo RA. The KQ element, a complex genetic region conferring transferable resistance to carbapenems, aminoglycosides, and fluoroquinolones in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2008;52:3427–3429. doi: 10.1128/AAC.00493-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Villa L, Poirel L, Nordmann P, Carta C, Carattoli A. Complete sequencing of an IncH plasmid carrying the blaNDM-1, blaCTX-M-15 and qnrB1 genes. J. Antimicrob. Chemother. 2012;67:1645–1650. doi: 10.1093/jac/dks114. [DOI] [PubMed] [Google Scholar]
- 73.Warburg G, Hidalgo-Grass C, Partridge SR, Tolmasky ME, Temper V, Moses AE, Block C, Strahilevitz J. A carbapenem-resistant Klebsiella pneumoniae epidemic clone in Jerusalem: sequence type 512 carrying a plasmid encoding aac(6′)-Ib. J. Antimicrob. Chemother. 2012;67:898–901. doi: 10.1093/jac/dkr552. [DOI] [PubMed] [Google Scholar]
- 74.Dery KJ, Chavideh R, Waters V, Chamorro R, Tolmasky LS, Tolmasky ME. Characterization of the replication and mobilization regions of the multiresistance Klebsiella pneumoniae plasmid pJHCMW1. Plasmid. 1997;38:97–105. doi: 10.1006/plas.1997.1303. [DOI] [PubMed] [Google Scholar]
- 75.Tolmasky ME, Colloms S, Blakely G, Sherratt DJ. Stability by multimer resolution of pJHCMW1 is due to the Tn1331 resolvase and not to the Escherichia coli Xer system. Microbiology. 2000;146:581–589. doi: 10.1099/00221287-146-3-581. [DOI] [PubMed] [Google Scholar]
- 76.Pham H, Dery KJ, Sherratt DJ, Tolmasky ME. Osmoregulation of dimer resolution at the plasmid pJHCMW1 mwr locus by Escherichia coli XerCD recombination. J. Bacteriol. 2002;184:1607–1616. doi: 10.1128/JB.184.6.1607-1616.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bui D, Ramiscal J, Trigueros S, Newmark JS, Do A, Sherratt DJ, Tolmasky ME. Differences in resolution of mwr-containing plasmid dimers mediated by the Klebsiella pneumoniae and Escherichia coli XerC recombinases: potential implications in dissemination of antibiotic resistance genes. J. Bacteriol. 2006;188:2812–2820. doi: 10.1128/JB.188.8.2812-2820.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Trigueros S, Tran T, Sorto N, Newmark J, Colloms SD, Sherratt DJ, Tolmasky ME. mwr Xer site-specific recombination is hypersensitive to DNA supercoiling. Nucleic Acids Res. 2009;37:3580–3587. doi: 10.1093/nar/gkp208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Reyes-Lamothe R, Tran T, Meas D, Lee L, Li AM, Sherratt DJ, Tolmasky ME. High-copy bacterial plasmids diffuse in the nucleoid-free space, replicate stochastically and are randomly partitioned at cell division. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt918. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pasquali F, Kehrenberg C, Manfreda G, Schwarz S. Physical linkage of Tn3 and part of Tn1721 in a tetracycline and ampicillin resistance plasmid from Salmonella Typhimurium. J. Antimicrob. Chemother. 2005;55:562–565. doi: 10.1093/jac/dkh553. [DOI] [PubMed] [Google Scholar]
- 81.Tran T, Sherratt DJ, Tolmasky ME. fpr, a deficient Xer recombination site from a Salmonella plasmid, fails to confer stability by dimer resolution: comparative studies with the pJHCMW1 mwr site. J. Bacteriol. 2010;192:883–887. doi: 10.1128/JB.01082-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Garcia-Fernandez A, Villa L, Carta C, Venditti C, Giordano A, Venditti M, Mancini C, Carattoli A. Klebsiella pneumoniae ST258 producing KPC-3 identified in italy carries novel plasmids and OmpK36/OmpK35 porin variants. Antimicrob. Agents Chemother. 2012;56:2143–2145. doi: 10.1128/AAC.05308-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Das B, Martinez E, Midonet C, Barre FX. Integrative mobile elements exploiting Xer recombination. Trends Microbiol. 2013;21:23–30. doi: 10.1016/j.tim.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 84.Summers DK, Sherratt DJ. Multimerization of high copy number plasmids causes instability: CoIE1 encodes a determinant essential for plasmid monomerization and stability. Cell. 1984;36:1097–1103. doi: 10.1016/0092-8674(84)90060-6. [DOI] [PubMed] [Google Scholar]
- 85.Colloms SD, Sykora P, Szatmari G, Sherratt DJ. Recombination at ColE1 cer requires the Escherichia coli xerC gene product, a member of the lambda integrase family of site-specific recombinases. J. Bacteriol. 1990;172:6973–6980. doi: 10.1128/jb.172.12.6973-6980.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cornet F, Mortier I, Patte J, Louarn JM. Plasmid pSC101 harbors a recombination site, psi, which is able to resolve plasmid multimers and to substitute for the analogous chromosomal Escherichia coli site dif. J. Bacteriol. 1994;176:3188–3195. doi: 10.1128/jb.176.11.3188-3195.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Summers D. Timing, self-control and a sense of direction are the secrets of multicopy plasmid stability. Mol. Microbiol. 1998;29:1137–1145. doi: 10.1046/j.1365-2958.1998.01012.x. [DOI] [PubMed] [Google Scholar]
- 88.Tran T, Andres P, Petroni A, Soler-Bistue A, Albornoz E, Zorreguieta A, Reyes-Lamothe R, Sherratt DJ, Corso A, Tolmasky ME. Small plasmids harboring qnrB19: a model for plasmid evolution mediated by sitespecific recombination at oriT and Xer sites. Antimicrob. Agents Chemother. 2012;56:1821–1827. doi: 10.1128/AAC.06036-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Val ME, Bouvier M, Campos J, Sherratt D, Cornet F, Mazel D, Barre FX. The single-stranded genome of phage CTX is the form used for integration into the genome of Vibrio cholerae. Mol. Cell. 2005;19:559–566. doi: 10.1016/j.molcel.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 90.Grosso F, Quinteira S, Poirel L, Novais A, Peixe L. Role of common blaOXA-24/OXA-40-carrying platforms and plasmids in the spread of OXA-24/OXA-40 among Acinetobacter species clinical isolates. Antimicrob. Agents Chemother. 2012;56:3969–3972. doi: 10.1128/AAC.06255-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Blakely GW, Sherratt DJ. Interactions of the site-specific recombinases XerC and XerD with the recombination site dif. Nucleic Acids Res. 1994;22:5613–5620. doi: 10.1093/nar/22.25.5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hayes F, Sherratt DJ. Recombinase binding specificity at the chromosome dimer resolution site dif of Escherichia coli. J. Mol. Biol. 1997;266:525–537. doi: 10.1006/jmbi.1996.0828. [DOI] [PubMed] [Google Scholar]
- 93.D'Andrea MM, Giani T, D'Arezzo S, Capone A, Petrosillo N, Visca P, Luzzaro F, Rossolini GM. Characterization of pABVA01, a plasmid encoding the OXA-24 carbapenemase from Italian isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 2009;53:3528–3533. doi: 10.1128/AAC.00178-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Merino M, Acosta J, Poza M, Sanz F, Beceiro A, Chaves F, Bou G. OXA-24 carbapenemase gene flanked by XerC/XerD-like recombination sites in different plasmids from different Acinetobacter species isolated during a nosocomial outbreak. Antimicrob. Agents Chemother. 2010;54:2724–2727. doi: 10.1128/AAC.01674-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Povilonis J, Seputiene V, Krasauskas R, Juskaite R, Miskinyte M, Suziedelis K, Suziedeliene E. Spread of carbapenem-resistant Acinetobacter baumannii carrying a plasmid with two genes encoding OXA-72 carbapenemase in Lithuanian hospitals. J. Antimicrob. Chemother. 2013;68:1000–1006. doi: 10.1093/jac/dks499. [DOI] [PubMed] [Google Scholar]
- 96.Montealegre MC, Maya JJ, Correa A, Espinal P, Mojica MF, Ruiz SJ, Rosso F, Vila J, Quinn JP, Villegas MV. First identification of OXA-72 carbapenemase from Acinetobacter pittii in Colombia. Antimicrob. Agents Chemother. 2012;56:3996–3998. doi: 10.1128/AAC.05628-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Francia MV, Varsaki A, Garcillan-Barcia MP, Latorre A, Drainas C, de la Cruz F. A classification scheme for mobilization regions of bacterial plasmids. FEMS Microbiol. Rev. 2004;28:79–100. doi: 10.1016/j.femsre.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 98.Cao V, Lambert T, Courvalin P. ColE1-like plasmid pIP843 of Klebsiella pneumoniae encoding extended-spectrum beta-lactamase CTX-M-17. Antimicrob. Agents Chemother. 2002;46:1212–1217. doi: 10.1128/AAC.46.5.1212-1217.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Le TM, Baker S, Le TP, Le TP, Cao TT, Tran TT, Nguyen VM, Campbell JI, Lam MY, Nguyen TH, Nguyen VV, Farrar J, Schultsz C. High prevalence of plasmid-mediated quinolone resistance determinants in commensal members of the Enterobacteriaceae in Ho Chi Minh City, Vietnam. J. Med. Microbiol. 2009;58:1585–1592. doi: 10.1099/jmm.0.010033-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nordmann P, Naas T, Poirel L. Global spread of carbapenemaseproducing Enterobacteriaceae. Emerg. Infect. Dis. 2011;17:1791–1798. doi: 10.3201/eid1710.110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nassif X, Sansonetti PJ. Correlation of the virulence of Klebsiella pneumoniae K1 and K2 with the presence of a plasmid encoding aerobactin. Infect. Immun. 1986;54:603–608. doi: 10.1128/iai.54.3.603-608.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nassif X, Honore N, Vasselon T, Cole ST, Sansonetti PJ. Positive control of colanic acid synthesis in Escherichia coli by rmpA and rmpB, two virulence-plasmid genes of Klebsiella pneumoniae. Mol. Microbiol. 1989;3:1349–1359. doi: 10.1111/j.1365-2958.1989.tb00116.x. [DOI] [PubMed] [Google Scholar]
- 103.Nassif X, Fournier JM, Arondel J, Sansonetti PJ. Mucoid phenotype of Klebsiella pneumoniae is a plasmid-encoded virulence factor. Infect. Immun. 1989;57:546–552. doi: 10.1128/iai.57.2.546-552.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wacharotayankun R, Arakawa Y, Ohta M, Tanaka K, Akashi T, Mori M, Kato N. Enhancement of extracapsular polysaccharide synthesis in Klebsiella pneumoniae by RmpA2, which shows homology to NtrC and FixJ. Infect. Immun. 1993;61:3164–3174. doi: 10.1128/iai.61.8.3164-3174.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Perry RD, Fetherston JD. Yersiniabactin iron uptake: mechanisms and role in Yersinia pestis pathogenesis. Microbes Infect. 2011;13:808–817. doi: 10.1016/j.micinf.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zimbler DL, Penwell WF, Gaddy JA, Menke SM, Tomaras AP, Connerly PL, Actis LA. Iron acquisition functions expressed by the human pathogen Acinetobacter baumannii. Biometals. 2009;22:23–32. doi: 10.1007/s10534-008-9202-3. [DOI] [PubMed] [Google Scholar]
- 107.Crosa JH. Signal transduction and transcriptional and posttranscriptional control of iron-regulated genes in bacteria. Microbiol. Mol. Biol. Rev. 1997;61:319–336. doi: 10.1128/mmbr.61.3.319-336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chen Q, Wertheimer AM, Tolmasky ME, Crosa JH. The AngR protein and the siderophore anguibactin positively regulate the expression of irontransport genes in Vibrio anguillarum. Mol. Microbiol. 1996;22:127–134. doi: 10.1111/j.1365-2958.1996.tb02662.x. [DOI] [PubMed] [Google Scholar]
- 109.Darfeuille-Michaud A, Jallat C, Aubel D, Sirot D, Rich C, Sirot J, Joly B. R-plasmid-encoded adhesive factor in Klebsiella pneumoniae strains responsible for human nosocomial infections. Infect. Immun. 1992;60:44–55. doi: 10.1128/iai.60.1.44-55.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yu VL, Hansen DS, Ko WC, Sagnimeni A, Klugman KP, von Gottberg A, Goossens H, Wagener MM, Benedi VJ, International Klebseilla Study G Virulence characteristics of Klebsiella and clinical manifestations of K. pneumoniae bloodstream infections. Emerg. Infect. Dis. 2007;13:986–993. doi: 10.3201/eid1307.070187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Vernet V, Madoulet C, Chippaux C, Philippon A. Incidence of two virulence factors (aerobactin and mucoid phenotype) among 190 clinical isolates of Klebsiella pneumoniae producing extended-spectrum betalactamase. FEMS Microbiol. Lett. 1992;75:1–5. doi: 10.1016/0378-1097(92)90447-v. [DOI] [PubMed] [Google Scholar]
- 112.Chen YT, Chang HY, Lai YC, Pan CC, Tsai SF, Peng HL. Sequencing and analysis of the large virulence plasmid pLVPK of Klebsiella pneumoniae CG43. Gene. 2004;337:189–198. doi: 10.1016/j.gene.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 113.Sorsa LJ, Dufke S, Heesemann J, Schubert S. Characterization of an iroBCDEN gene cluster on a transmissible plasmid of uropathogenic Escherichia coli: evidence for horizontal transfer of a chromosomal virulence factor. Infect. Immun. 2003;71:3285–3293. doi: 10.1128/IAI.71.6.3285-3293.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Braun V, Mahren S, Ogierman M. Regulation of the FecI-type ECF sigma factor by transmembrane signalling. Curr. Opin. Microbiol. 2003;6:173–180. doi: 10.1016/s1369-5274(03)00022-5. [DOI] [PubMed] [Google Scholar]
- 115.Mahren S, Schnell H, Braun V. Occurrence and regulation of the ferric citrate transport system in Escherichia coli B, Klebsiella pneumoniae, Enterobacter aerogenes, and Photorhabdus luminescens. Arch. Microbiol. 2005;184:175–186. doi: 10.1007/s00203-005-0035-y. [DOI] [PubMed] [Google Scholar]
- 116.Wu KM, Li LH, Yan JJ, Tsao N, Liao TL, Tsai HC, Fung CP, Chen HJ, Liu YM, Wang JT, Fang CT, Chang SC, Shu HY, Liu TT, Chen YT, Shiau YR, Lauderdale TL, Su IJ, Kirby R, Tsai SF. Genome sequencing and comparative analysis of Klebsiella pneumoniae NTUH-K2044, a strain causing liver abscess and meningitis. J. Bacteriol. 2009;191:4492–4501. doi: 10.1128/JB.00315-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shin SH, Kim S, Kim JY, Lee S, Um Y, Oh MK, Kim YR, Lee J, Yang KS. Complete genome sequence of the 2,3-butanediol-producing Klebsiella pneumoniae strain KCTC 2242. J. Bacteriol. 2012;194:2736–2737. doi: 10.1128/JB.00027-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Dolejska M, Villa L, Dobiasova H, Fortini D, Feudi C, Carattoli A. Plasmid content of a clinically relevant Klebsiella pneumoniae clone from the Czech Republic producing CTX-M-15 and QnrB1. Antimicrob. Agents Chemother. 2013;57:1073–1076. doi: 10.1128/AAC.01886-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Taylor DE, Rooker M, Keelan M, Ng LK, Martin I, Perna NT, Burland NT, Blattner FR. Genomic variability of O islands encoding tellurite resistance in enterohemorrhagic Escherichia coli O157:H7 isolates. J. Bacteriol. 2002;184:4690–4698. doi: 10.1128/JB.184.17.4690-4698.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Borremans B, Hobman JL, Provoost A, Brown NL, van Der Lelie D. Cloning and functional analysis of the pbr lead resistance determinant of Ralstonia metallidurans CH34. J. Bacteriol. 2001;183:5651–5658. doi: 10.1128/JB.183.19.5651-5658.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Brown NL, Barrett SR, Camakaris J, Lee BT, Rouch DA. Molecular genetics and transport analysis of the copper-resistance determinant (pco) from Escherichia coli plasmid pRJ1004. Mol. Microbiol. 1995;17:1153–1166. doi: 10.1111/j.1365-2958.1995.mmi_17061153.x. [DOI] [PubMed] [Google Scholar]
- 122.Gupta A, Matsui K, Lo JF, Silver S. Molecular basis for resistance to silver cations in Salmonella. Nat. Med. 1999;5:183–188. doi: 10.1038/5545. [DOI] [PubMed] [Google Scholar]
- 123.Huang TW, Chen TL, Chen YT, Lauderdale TL, Liao TL, Lee YT, Chen CP, Liu YM, Lin AC, Chang YH, Wu KM, Kirby R, Lai JF, Tan MC, Siu LK, Chang CM, Fung CP, Tsai SF. Copy Number Change of the NDM-1 sequence in a multidrug-resistant Klebsiella pneumoniae clinical isolate. PLoS One. 2013;8:e62774. doi: 10.1371/journal.pone.0062774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sandegren L, Linkevicius M, Lytsy B, Melhus A, Andersson DI. Transfer of an Escherichia coli ST131 multiresistance cassette has created a Klebsiella pneumoniae-specific plasmid associated with a major nosocomial outbreak. J. Antimicrob. Chemother. 2012;67:74–83. doi: 10.1093/jac/dkr405. [DOI] [PubMed] [Google Scholar]
- 125.Bojer MS, Struve C, Ingmer H, Hansen DS, Krogfelt KA. Heat resistance mediated by a new plasmid encoded Clp ATPase, ClpK, as a possible novel mechanism for nosocomial persistence of Klebsiella pneumoniae. PLoS One. 2010;5:e15467. doi: 10.1371/journal.pone.0015467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bojer MS, Hammerum AM, Jorgensen SL, Hansen F, Olsen SS, Krogfelt KA, Struve C. Concurrent emergence of multidrug resistance and heat resistance by CTX-M-15-encoding conjugative plasmids in Klebsiella pneumoniae. APMIS. 2012;120:699–705. doi: 10.1111/j.1600-0463.2012.02885.x. [DOI] [PubMed] [Google Scholar]
- 127.Guglielmini J, de la Cruz F, Rocha EP. Evolution of conjugation and type IV secretion systems. Mol. Biol. Evol. 2013;30:315–331. doi: 10.1093/molbev/mss221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Soler Bistue AJ, Birshan D, Tomaras AP, Dandekar M, Tran T, Newmark J, Bui D, Gupta N, Hernandez K, Sarno R, Zorreguieta A, Actis LA, Tolmasky ME. Klebsiella pneumoniae multiresistance plasmid pMET1: similarity with the Yersinia pestis plasmid pCRY and integrative conjugative elements. PLoS One. 2008;3:e1800. doi: 10.1371/journal.pone.0001800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Song Y, Tong Z, Wang J, Wang L, Guo Z, Han Y, Zhang J, Pei D, Zhou D, Qin H, Pang X, Han Y, Zhai J, Li M, Cui B, Qi Z, Jin L, Dai R, Chen F, Li S, Ye C, Du Z, Lin W, Wang J, Yu J, Yang H, Wang J, Huang P, Yang R. Complete genome sequence of Yersinia pestis strain 91001, an isolate avirulent to humans. DNA Res. 2004;11:179–197. doi: 10.1093/dnares/11.3.179. [DOI] [PubMed] [Google Scholar]
- 130.Stephan R, Lehner A, Tischler P, Rattei T. Complete genome sequence of Cronobacter turicensis LMG 23827, a food-borne pathogen causing deaths in neonates. J. Bacteriol. 2011;193:309–310. doi: 10.1128/JB.01162-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Schubert S, Dufke S, Sorsa J, Heesemann J. A novel integrative and conjugative element (ICE) of Escherichia coli: the putative progenitor of the Yersinia high-pathogenicity island. Mol. Microbiol. 2004;51:837–848. doi: 10.1046/j.1365-2958.2003.03870.x. [DOI] [PubMed] [Google Scholar]
- 132.Carattoli A. Plasmids and the spread of resistance. Int. J. Med. Microbiol. 2013;303:298–304. doi: 10.1016/j.ijmm.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 133.Schultsz C, Geerlings S. Plasmid-mediated resistance in Enterobacteriaceae: changing landscape and implications for therapy. Drugs. 2012;72:1–16. doi: 10.2165/11597960-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 134.Munoz-Price LS, Poirel L, Bonomo RA, Schwaber MJ, Daikos GL, Cormican M, Cornaglia G, Garau J, Gniadkowski M, Hayden MK, Kumarasamy K, Livermore DM, Maya JJ, Nordmann P, Patel JB, Paterson DL, Pitout J, Villegas MV, Wang H, Woodford N, Quinn JP. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect. Dis. 2013;13:785–796. doi: 10.1016/S1473-3099(13)70190-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Nordmann P, Poirel L, Walsh TR, Livermore DM. The emerging NDM carbapenemases. Trends Microbiol. 2011;19:588–595. doi: 10.1016/j.tim.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 136.Giakkoupi P, Papagiannitsis CC, Miriagou V, Pappa O, Polemis M, Tryfinopoulou K, Tzouvelekis LS, Vatopoulos AC. An update of the evolving epidemic of blaKPC-2-carrying Klebsiella pneumoniae in Greece (2009-10). J. Antimicrob. Chemother. 2011;66:1510–1513. doi: 10.1093/jac/dkr166. [DOI] [PubMed] [Google Scholar]
- 137.Johnson AP, Woodford N. Global spread of antibiotic resistance: the example of New Delhi metallo-beta-lactamase (NDM)-mediated carbapenem resistance. J. Med. Microbiol. 2013;62:499–513. doi: 10.1099/jmm.0.052555-0. [DOI] [PubMed] [Google Scholar]
- 138.Snitkin ES, Zelazny AM, Thomas PJ, Stock F, Group NCSP, Henderson DK, Palmore TN, Segre JA. Tracking a hospital outbreak of carbapenemresistant Klebsiella pneumoniae with whole-genome sequencing. Sci. Transl. Med. 2012;4:148ra116. doi: 10.1126/scitranslmed.3004129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 2009;53:5046–5054. doi: 10.1128/AAC.00774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Nordmann P, Poirel L, Toleman MA, Walsh TR. Does broadspectrum beta-lactam resistance due to NDM-1 herald the end of the antibiotic era for treatment of infections caused by Gram-negative bacteria? J. Antimicrob. Chemother. 2011;66:689–692. doi: 10.1093/jac/dkq520. [DOI] [PubMed] [Google Scholar]
- 141.Tolmasky ME. Overview of dissemination mechanisms of genes coding for resistance to antibiotics. In: Bonomo RA, Tolmasky ME, editors. Enzyme-mediated resistance to antibiotics: mechanisms, dissemination, and prospects for inhibition. ASM Press; Washington, DC.: 2007. pp. 267–270. [Google Scholar]
- 142.Levy S. The antibiotic paradox. How the misuse of antibiotics destroys their curative powers. 2nd ed. Perseus Publishing; Cambridge, MA.: 2002. [Google Scholar]
- 143.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Reyes-Lamothe R, Possoz C, Danilova O, Sherratt DJ. Independent positioning and action of Escherichia coli replisomes in live cells. Cell. 2008;133:90–102. doi: 10.1016/j.cell.2008.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sutton MD. The Escherichia coli dnaN159 mutant displays altered DNA polymerase usage and chronic SOS induction. J. Bacteriol. 2004;186:6738–6748. doi: 10.1128/JB.186.20.6738-6748.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Colloms SD. The topology of plasmid-monomerizing Xer site-specific recombination. Biochem. Soc. Trans. 2013;41:589–594. doi: 10.1042/BST20120340. [DOI] [PubMed] [Google Scholar]
- 147.Minh PN, Devroede N, Massant J, Maes D, Charlier D. Insights into the architecture and stoichiometry of Escherichia coli PepA*DNA complexes involved in transcriptional control and site-specific DNA recombination by atomic force microscopy. Nucleic Acids Res. 2009;37:1463–1476. doi: 10.1093/nar/gkn1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Reijns M, Lu Y, Leach S, Colloms SD. Mutagenesis of PepA suggests a new model for the Xer/cer synaptic complex. Mol. Microbiol. 2005;57:927–941. doi: 10.1111/j.1365-2958.2005.04716.x. [DOI] [PubMed] [Google Scholar]
- 149.Darling AE, Mau B, Perna NT. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One. 2010;5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Whelan KF, Colleran E, Taylor DE. Phage inhibition, colicin resistance, and tellurite resistance are encoded by a single cluster of genes on the IncHI2 plasmid R478. J. Bacteriol. 1995;177:5016–5027. doi: 10.1128/jb.177.17.5016-5027.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Chen YT, Lin AC, Siu LK, Koh TH. Sequence of closely related plasmids encoding bla(NDM-1) in two unrelated Klebsiella pneumoniae isolates in Singapore. PLoS One. 2012;7:e48737. doi: 10.1371/journal.pone.0048737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ho P, Li Z, Lo W, Cheung Y, Lin C, Sham P, Cheng V, Ng T, Que T, Chow K. Identification and characterization of a novel incompatibility group X3 plasmid carrying blaNDM-1 in Enterobacteriaceae isolates with epidemiological links to multiple geographical areas in China. Emerg Microbes Infect. 2012;1:e39. doi: 10.1038/emi.2012.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Carattoli A, Villa L, Poirel L, Bonnin RA, Nordmann P. Evolution of IncA/C blaCMY-(2)-carrying plasmids by acquisition of the blaNDM-(1) carbapenemase gene. Antimicrob. Agents Chemother. 2012;56:783–786. doi: 10.1128/AAC.05116-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zioga A, Whichard JM, Kotsakis SD, Tzouvelekis LS, Tzelepi E, Miriagou V. CMY-31 and CMY-36 cephalosporinases encoded by ColE1-like plasmids. Antimicrob. Agents Chemother. 2009;53:1256–1259. doi: 10.1128/AAC.01284-08. [DOI] [PMC free article] [PubMed] [Google Scholar]


