Abstract
The linguistically distinctive Haida and Tlingit tribes of Southeast Alaska are known for their rich material culture, complex social organization, and elaborate ritual practices. However, much less is known about these tribes from a population genetic perspective. For this reason, we analyzed mtDNA and Y-chromosome variation in Haida and Tlingit populations to elucidate several key issues pertaining to the history of this region. These included the genetic relationships of Haida and Tlingit to other indigenous groups in Alaska and Canada; the relationship between linguistic and genetic data for populations assigned to the Na-Dene linguistic family, specifically, the inclusion of Haida with Athapaskan, Eyak, and Tlingit in the language family; the possible influence of matrilineal clan structure on patterns of genetic variation in Haida and Tlingit populations; and the impact of European entry into the region on the genetic diversity of these indigenous communities. Our analysis indicates that, while sharing a “northern” genetic profile, the Haida and the Tlingit are genetically distinctive from each other. In addition, Tlingit groups themselves differ across their geographic range, in part due to interactions of Tlingit tribes with Athapaskan and Eyak groups to the north. The data also reveal a strong influence of maternal clan identity on mtDNA variation in these groups, as well as the significant influence of non-native males on Y-chromosome diversity. These results yield new details about the histories of the Haida and Tlingit tribes in this region.
Keywords: haplogroup, haplotype, lineage, SNP, STR, genealogy, founder, moiety
Genographic Consortium members include: Syama Adhikarla (Madurai Kamaraj University, Madurai, Tamil Nadu, India), Christina J. Adler (University of Adelaide, South Australia, Australia), Elena Balanovskaya (Research Centre for Medical Genetics, Russian Academy of Medical Sciences, Moscow, Russia), Oleg Balanovsky (Research Centre for Medical Genetics, Russian Academy of Medical Sciences, Moscow, Russia), Jaume Bertranpetit (Universitat Pompeu Fabra, Barcelona, Spain), Andrew C. Clarke (University of Otago, Dunedin, New Zealand), David Comas (Universitat Pompeu Fabra, Barcelona, Spain), Alan Cooper (University of Adelaide, South Australia, Australia), Clio S. I. Der Sarkissian (University of Adelaide, South Australia, Australia), ArunKumar GaneshPrasad (Madurai Kamaraj University, Madurai, Tamil Nadu, India), Angela Hobbs (National Health Laboratory Service, Johannesburg, South Africa), Asif Javed (IBM, Yorktown Heights, New York, United States), Li Jin (Fudan University, Shanghai, China), Matthew E. Kaplan (University of Arizona, Tucson, Arizona, United States), Daniela R. Lacerda (Universidade Federal de Minas Gerais, Belo Horizonte, Minas Gerais, Brazil), Shilin Li (Fudan University, Shanghai, China), Begoña Martínez-Cruz (Universitat Pompeu Fabra, Barcelona, Spain), Elizabeth A. Matisoo-Smith (University of Otago, Dunedin, New Zealand), Marta Melé (Universitat Pompeu Fabra, Barcelona, Spain), Nirav C. Merchant (University of Arizona, Tucson, Arizona, United States), R. John Mitchell (La Trobe University, Melbourne, Victoria, Australia), Laxmi Parida (IBM, Yorktown Heights, New York, United States), Ramasamy Pitchappan (Madurai Kamaraj University, Madurai, Tamil Nadu, India), Daniel E. Platt (IBM, Yorktown Heights, NY, USA), Lluis Quintana-Murci (Institut Pasteur, Paris, France), Colin Renfrew (University of Cambridge, Cambridge, UK), Daniela R. Lacerda (Universidade Federal de Minas Gerais, Belo Horizonte, Minas Gerais, Brazil), Ajay K. Royyuru (IBM, Yorktown Heights, New York, United States), Fabrício R. Santos (Universidade Federal de Minas Gerais, Belo Horizonte, Minas Gerais, Brazil), Himla Soodyall (National Health Laboratory Service, Johannesburg, South Africa), David F. Soria Hernanz (National Geographic Society, Washington, DC, USA), Pandikumar Swamikrishnan (IBM, Somers, New York, United States), Chris Tyler-Smith (The Wellcome Trust Sanger Institute, Hinxton, UK), Arun Varatharajan Santhakumari (Madurai Kamaraj University, Madurai, Tamil Nadu, India), Pedro Paulo Vieira (Universidade Federal do Rio de Janeiro, Rio de Janeiro, Brazil), R. Spencer Wells (National Geographic Society, Washington, DC, USA), Pierre A. Zalloua (Lebanese American University, Chouran, Beirut, Lebanon), Janet S. Ziegle (Applied Biosystems, Foster City, California, United States).
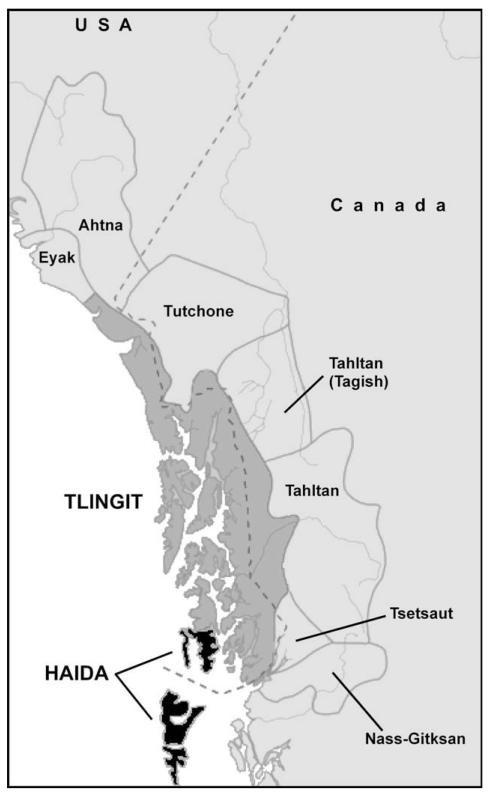
Southeast Alaska has long been known for its diversity of Native American communities. Prior to European contact, Eyak, Haida, Tlingit, and Tsimshian tribes lived in coastal and island portions of Southeast Alaska and northern British Columbia (Fig. 1). These groups are known for their distinctive totem poles, house carvings, masks and ceremonial garb, and material culture as well as their complex social organization and elaborate potlatches (Garfield and Wingert, 1966; Kaplan and Barsness, 1986; Fairbanks, 1989; Kan, 1989; Halpin and Seguin, 1990; Emmons, 1991; MacDonald, 1994; Miller, 2000; Reid, 2002), and are considered to be part of a “Northwest Coast Culture.” However, while this region has been well studied from ethnographic (Swanton, 1905, 1908; Krause, 1956 [1885]; de Laguna et al., 1972; Oberg, 1973; Olsen, 1976; Emmons, 1991) and linguistic perspectives (Sapir, 1915; Krause 1979; Krause and Golla, 1980, 1981; Leer et al., 2001), much less is known about these tribes from a population genetic perspective.
Fig. 1.
A map showing the historical territories of the Tlingit and Haida in relation to neighbor tribes of Canada and Alaska (based on a map presented in Goddard (1996) and modified at en.wikipedia.org/wiki/File:Tlingit-map.png).
The ancestors of the Haida and Tlingit populations probably did not establish themselves in Southeast Alaska until after the end of the last glacial maximum. The earliest Paleoarctic sites, including Chuck Lake and Thorne River, Groundhog Bay in Icy Strait (Chichagof Island) and Hidden Falls on Baranof Island near Sitka, date to 10,000–6,500 years before present (YBP) and are marked by microblade and cobble stone tools (Carlson 1990; Davis, 1990; Ackerman, 1992, 1996; Moss, 1998; Ames and Maschner, 1999). During this period, the Northwest Coast developed into a distinct culture area (Davis, 1990; Ackerman, 1992, 1996; Moss, 1998; Ames and Maschner, 1999). Over the next 2,000 years, the culture and social organization associated with Northwest Coast peoples began to emerge (Davis, 1990; Ackerman, 1992). Around 5,000 years ago, the coastal art and aesthetic styles associated with the Northwest Coast began to appear (Davis, 1990; Ackerman, 1992). By 3,000 years ago, the cultures of the Northwest Coast were largely the same as those observed at the time of contact (Davis, 1990; Ames and Maschner, 1999). The indigenous populations living in the region today are the direct cultural, and possibly biological, descendants of these prehistoric Northwest Coast groups (Davis, 1990; Ames and Maschner, 1999).
LINGUISTIC DIVERSITY IN SOUTHEAST ALASKA
Efforts to understand the history of the indigenous groups from this region have also involved the examination of linguistic evidence. Such research has revealed that these groups speak languages belonging to the Athapaskan-Eyak-Tlingit (AET) family. The Tlingit language, recognized as one of the oldest branches of the family, may have split from the Eyak-Athapaskan branch about 5,000 years ago (Krause, 1979; Greenberg et al., 1986; Krause and Golla, 1980) (Supporting Information Fig. S1). It is subdivided into Gulf Coast, Northern, Southern, and Inland dialects, which are largely mutually intelligible (Krause and Golla, 1980, 1981; de Laguna, 1990a; Leer et al., 2001). Sixteen component “tribes” (qwaan) comprise the three coastal subdialect regions. Each of these tribes is centered on a primary village (Krause and Golla, 1981; Goldschmidt and Haas, 1986; de Laguna, 1990a; Hope and Thornton, 2000). These are, north to south, the Gulf Coast region with Yakutat and Lituya Bay; the Northern region with Hoonah, Chilkat, Auk, Sitka, Hutsnuwu, Taku, and Sawdum; and the Southern region with Kake, Kuiu, Henya, Klawak, Stikine, Tongass, and Sanya (Supporting Information Fig. S2).
By contrast, Eyak is essentially an extinct language, historically spoken in the Copper River region of southcentral Alaska (Krause and Golla, 1981; de Laguna, 1990b). According to linguistic evidence, even current place names surrounding the Gulf of Alaska (including Yakutat) came from Eyak sources (de Laguna, 1972, 1990a; Krause and Golla, 1981; Emmons, 1991). This evidence, and Tlingit oral history, suggests that Eyak was once more widespread in Southeast Alaska than at the time of European contact (de Laguna, 1972, 1990a, b; Krause and Golla, 1981; Emmons, 1991).
Athapaskan is the youngest branch of the AET grouping within Na-Dene. Populations speaking Athapaskan languages now occupy large areas of northern North America, with most living in Alaska and Canada (Supporting Information Fig. S1). Proto-Athapaskan itself may have originated in the subarctic region of North America some 3,000 years ago (Hoijer, 1956; Krause and Golla, 1981). The current distribution of Athapaskan languages in North America likely reflects a series of migrations from this source area within the past 1,000 years (Greenberg et al., 1986; Ives, 2003, 2010; Matson and Magne, 2007).
Most researchers also support the “Na-Dene hypothesis,” which asserts a distant “genetic” relationship between AET and Haida languages (Sapir, 1915; Krause and Golla, 1981; Ruhlen, 1998). According to this model, Haida and AET languages resemble each other because of sharing a common ancestry (Greenberg et al., 1986; Ruhlen, 1994, 1998; Manaster Ramer, 1996; Enrico, 2003). However, some have questioned the inclusion of Haida in the Na-Dene linguistic family, viewing it as a linguistic isolate (Krause, 1979; Levine, 1979). The Haida language itself is divided into Northern and Southern dialects, with Northern Haida being split into Alaskan (Kaigani) Haida and Masset (North Graham Island) Haida dialects (Enrico, 2003, 2005).
Tribal and clan structure
All Northwest Coast groups have some form of an exogamous matrilineal clan system. In these systems, clan status is passed from mothers to their children. Based on this status, individuals have access to specific clan territories for hunting and fishing, and can use clan-specific crests for decorating clothing and dwellings (Olson, 1967; Emmons, 1991; Kaplan and Barsness, 1986; de Laguna 1990b). The clan system is comprised by two moieties or reciprocating descent groups. For the Tlingit and Eyak, clans are grouped into Raven (Yéil) or Eagle/Wolf (Ch’aak’/Gooch) moieties (Kaplan and Barsness, 1986; Emmons, 1991; de Laguna, 1990a, b). Traditionally, moiety membership influenced marriage practices, whereby persons belonging to Eagle clans would marry those from Raven clans, and vice versa.
Like the Tlingit and Eyak, the Haida are grouped into two moieties, although the clans belonging to the Haida Eagle moiety would belong to the Tlingit Raven moiety (Blackman, 1990). By contrast, the Tsimshian have four phratries instead of two moieties (Miller and Eastman, 1984; Halpin and Seguin, 1990; Miller, 2000). Thus, all Northwest Coast populations are organized into maternally linked clans, grouped into exogamous units, which influence their social and ritual practices, and probably also their genetic make-up.
Historical period in Southeast Alaska
European entry into the region created new patterns of cultural and economic interactions in Southeast Alaska. In the mid-1700s, Russians began exploring the region for exploitation of its natural resources (Frost, 2003). By the late 18th century, Russian, Spanish, English, and French explorers had made contact with Tlingit populations (Fedorova, 1973; Tikhmenev, 1978; de Laguna 1990a; Wilber, 1993; Hope and Thornton, 2000; Haycox, 2002; Black, 2004; Dauenhauer et al., 2008; Grinev, 2008). The American purchase of Alaska from Russia in 1867 led to further settlement and exploitation of the region (Haycox, 2002; Borneman, 2004). The Klondike Gold Rush of 1898 opened Alaska to further natural resource exploitation, including fishing, timbering, mining and oil drilling, which continues today, along with the influx of many people of non-native descent to find work in these industries since that time (Berton, 1959; Borneman, 2004).
Research objectives
To better understand the complex prehistory of Southeast Alaska and the genetic relationships among Northwest Coast tribes, we undertook a genetic analysis of Haida and Tlingit populations. We analyzed mtDNA and Y-chromosome variation to elucidate the maternal and paternal genetic histories of these indigenous populations and compared our findings to archeological, ethnographic, and linguistic data for them. Our results shed light on the abovementioned aspects of Southeast Alaskan prehistory and the history of Na-Dene speaking populations in the region, as well as the influence of social organization on genetic diversity in Haida and Tlingit populations. They further reveal the genetic impact of European movement into the region over the past three centuries.
METHODS
Sample locations
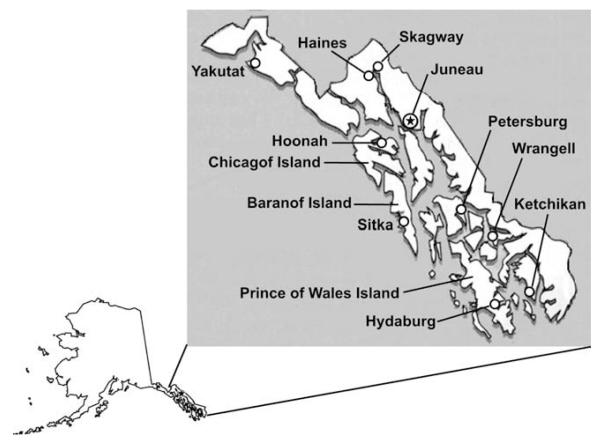
Between April 2009 and October 2011, we conducted field research in the Tlingit communities of Yakutat and Hoonah, the city of Juneau (Tlingit), and the Haida community of Hydaburg (Fig. 2). Based on ethnographic information, present day Tlingit communities may actually represent populations originating from 2 to 8 villages within each Tlingit tribe (qwaan) that were consolidated over the past 150 years (Goldschmidt and Haas, 1998). We also worked with several other Tlingit individuals from other communities around the region (Angoon, Sitka).
Fig. 2.
A map of southeast Alaska indicating the locations of study populations.
Yakutat
Eyak-speaking people from the Copper River area considered Yakutat to be part of their traditional homeland (de Laguna et al., 1964; de Laguna, 1972, 1990b; Emmons, 1991). Shortly before contact with Europeans, the Tlingit moved into this area and began absorbing Eyak groups. At that time, a number of Tlingit and mixed Tlingit-Eyak settlements were present in the region, although only Yakutat has survived to the present day. Based on the 2010 US census, there are 662 people living in the city and borough, with 35.8% of them (237) being native by ancestry.
Hoonah
Hoonah is located on the north shore of Chichagof Island, one of the larger islands of the Inner Passage. It is the principal village for the Huna Tlingits. Forced to leave their traditional home at Glacier Bay 200 years ago because of advancing glaciers, they relocated to Hoonah 20 miles to the south of this area (de Laguna 1990a; Emmons, 1991). Based on the 2010 US census, there are 760 people living in the city, with 52.5% of them (399) being native by ancestry.
Juneau
Juneau is located on the Gastineau Channel in Southeast Alaska, and faces the town of Douglas across the channel, with which it forms the current municipality of Juneau. It is also close to Auke Bay to the north and the Taku River to the south. Prior to European arrival in the late 18th century, Tlingit belonging to the Auke and Taku tribes fished in the Gastineau Channel (Emmons, 1991; Goldschmidt and Haas, 1998). Since becoming the capital of Alaska in 1906, Juneau has been a cultural center for Tlingit, Haida, and Tsimshian groups living in Alaska.
Hydaburg
The Haida first arrived on the Prince of Wales Island in the 1700s from Graham Island in British Columbia. By doing so, they pushed the Tongass Tlingit into the northern portion of the Prince of Wales Island. Hydaburg was formed in 1912 through the consolidation of the Kaigani Haida communities of Howkan (Dall Island), Klinkwan, and Sukkwan that were located along Cordova Bay on the western side of the Prince of Wales Island (Blackman, 1990). According to the 2010 US census, there are 376 people living in the city, with 77.1% of them (290) being native by ancestry.
Sample and data collection
During our work in Southeast Alaska, we obtained buccal samples from a total of 97 individuals following informed consent, of whom 64 were women and 33 were men. We also collected oral histories and genealogical data from Tlingit and Haida participants and community members, and examined archival materials, primary and secondary historical sources, and ethnological information from the region. This information provided a crucial genealogical and historical context with which to interpret the genetic data. All samples and genealogical data were obtained with approval from the University of Pennsylvania IRB, the Alaska Area IRB, and Alaska Native governments.
Tlingit and Haida samples were characterized for variation in the mitochondrial DNA (mtDNA) and the nonrecombining portion of the Y-chromosome (NRY) using the methods outlined below. All DNA test results were returned to the participants. For the purpose of this analysis, we reduced the mtDNA and NRY data sets to include only tribal members from each location. Accordingly, we excluded five of the participants because they were not Native Alaskans or tribal members. Based on genealogical information, we also excluded the mtDNAs of eight other individuals because they were maternally related to other participants, and three additional Y-chromosomes because they were paternally related to other participants. In addition, the data from individuals sampled in Hoonah and Juneau were combined, since individuals from each sampling location had clan and familial relationships. We were thus left with 84 tribal members (female and male) for the analysis of mtDNA variation, and 28 male tribal members for the Y-chromosome analysis.
Haida and Tlingit clan history
We obtained information about clan history of each community and its members to assess whether the mtDNA data corresponded with the matrilineal clan status of the participants. This information was also important because the different clans in the Haida and Tlingit clans often have distinct histories and geographic origins.
Yakutat Tlingit
There are several major clans in the Yakutat area. Among them, the Teikweidí (Brown Bear) and Galyax Kaagwaantaan (Beaver/Wolf) are subunits of the Eagle/Wolf moiety, while the L’unax.ádi (Silver Salmon) and Kwáashk’ikwáan (Humpback Salmon) are subunits of the Raven moiety (de Laguna, 1972, 1990a; Emmons, 1991; Goldschmidt and Haas, 1998).
Hoonah Tlingit
As seen in Yakutat, the Hoonah Tlingit have clans belonging to both the Eagle and Raven moieties. The Eagle moiety contains three clans, including Wooshkeetaan (Shark), Chookaneidi (Brown Bear), and Kaagwaantaan (Wolf), while the Raven moiety consists of two main clans, the T’akdeintaan (Kittywake) and L’uknax.a’di (Coho Salmon) (Swanton, 1908; Krause, 1956 [1885]; Goldschmidt and Haas, 1998; Dauenhauer et al., 2008).
Haida
Ethnographic information indicates that the Haida are divided into two moieties also called Raven and Eagle, with each having numerous lineages (Swanton, 1905; Blackman, 1990). Haida villages on the Prince of Wales Island historically contained representatives of several different lineages, and most contained members of both moieties. Based on oral histories, Swanton (1904, 1905) also speculated that the Eagle moiety might have come from a different tribe.
Mitochondrial DNA analysis
To elucidate the maternal genetic ancestry of the Native Alaskan individuals, we examined mtDNA variation in both male and female participants through direct sequencing of the control region (CR), and analysis of single nucleotide polymorphisms (SNPs) in the coding region of the mtDNA genome. The SNP analysis involved screening the samples for markers that identified the basal structure of the mtDNA phylogeny using TaqMan® assays (Applied Biosystems). All assays were read on an ABI 7900HT Fast Real-Time polymerase chain reaction (PCR) system. Once assigned to one of these basal lineages, the samples were screened for additional SNPs that define specific haplogroups and their subbranches using both custom TaqMan assays and polymerase chain reaction - restriction fragment length polymorphism (PCR-RFLP) analysis (Zhadanov et al., 2010; Gaieski et al., 2011). All haplogroup assignments were made using the nomenclature presented in PhyloTree.org (mtDNA Tree Build 12; 07-20-12; van Oven and Kayser, 2009).
In addition, for each sample, the entire mtDNA CR (1121 bp), encompassing hypervariable regions I and II (HVS1 and HVS2), was sequenced using previously published methods (Zhadanov et al., 2010; Gaieski et al., 2011). All sequences were read on an ABI 3130XL Gene Analyzer, and aligned and edited with the Sequencher 4.9 software tool (Gene Codes Corporation). The SNP assays and CR sequencing defined the maternal haplogroup and haplotypes, respectively, for each individual.
Y-chromosome analysis
We investigated the paternal genetic ancestry of Native Alaskans by screening the male samples for phylogenetically informative SNPs in the non-recombining portion of the Y-chromosome (NRY) that define major paternal haplogroups and their sub-branches. Biallelic markers defining each of the major NRY haplogroups were tested in all men, and classification into smaller, more refined haplogroups was accomplished using a hierarchical approach following published information (Y Chromosome Consortium, 2002; Karafet et al., 2008) and methods (Zhadanov et al., 2010; Gaieski et al., 2011). In addition, each sample was screened for seventeen short tandem repeats (STRs) using the multiplex AmpFlSTR Y-filer PCR Amplification Kit (ABI), and six additional fragment length polymorphisms and two additional Y-STRs (M17, M60, M91, M139, M175, M186, DYS388, and DYS426) through a separate multiplex reaction (Zhadanov et al., 2010; Gaieski et al., 2011). The combination of SNP and STR data defined paternal lineages in these individuals.
Comparative population data
For statistical and phylogenetic analyses, we compared Haida and Tlingit mtDNA and Y-chromosome data with those from other indigenous populations in the region (Supporting Information Fig. S3). These populations included Bella Coola (Ward et al., 1993), Greenland Inuit (Salliard et al., 2000), Alaskan Athapaskans (Shields et al., 1993), Apache and Navajo (Budowle et al., 2002), Yakima (Ward et al., 1993), Aleuts (Rubicz et al., 2003; Zlojutro et al., 2006; Zlojutro, 2008), Haida (Ward et al., 1993), and Nuu-chah-nulth (Ward et al., 1991). In addition, we compared our Y-chromosome STR data with other data sets (Bolnick et al., 2006; Redd et al., 2006; Zlojutro, 2008) to assess haplotype diversity in paternal lineages from indigenous populations of North America.
Statistical and phylogenetic analysis
Pairwise FST genetic distances between the Haida, Tlingit, and comparative populations were estimated with HVS1 sequence data using the Tamura-Nei model of sequence evolution (Tamura and Nei, 1993) in Arlequin v.3.11 software (http://cmpg.unibe.ch/software/arle-quin3/) (Excoffier et al., 2005). The inter-population FST genetic distances were then represented in two dimensions through multi-dimensional scaling (MDS) using SPSS 17.0 (SPSS 2009). In addition, haplotype diversity, nucleotide diversity, MPDs, Tajima’s D and Fu’s FS were calculated from mtDNA HVS1 sequences, using Arlequin 3.11.
We also conducted a hierarchical analysis of molecular variance (AMOVA) to evaluate the extent of population genetic structure in the data sets from Tlingit and Haida. For the Tlingit tribes, all indigenous mtDNA haplotypes were sorted by geographic location and clan membership. Any clan made up of a single individual was excluded from the analysis. All of the remaining clans were specific to a single moiety and geographic location, except for the L’uknax.ádi (Coho) clan of the Raven moiety, which was found in the Hoonah and Yakutat regions. This analysis thus involved data from 52 Tlingit (38 Hoonah [16 Eagle, 22 Raven]; 14 Yakutat [0 Eagle, 14 Raven]) individuals for whom we had both moiety and clan information. Similar comparisons using moiety membership were also examined as variables shaping the mtDNA diversity in the Tlingit and Haida, the latter being comprised of 16 Haida individuals (11 Eagle, 5 Hydaburg Raven) with indigenous haplotypes.
To compare our Y-STR data with other published data sets from Native American populations, we reduced the number of analyzed Y-STR loci to 10 of the 11 recommended by the Scientific Working Group on DNA Analysis Methods, namely, DYS19, DYS385a, DYS385b, DYS389I, DYS389II, DYS390, DYS391, DYS392, DYS393, and DYS439. In the published data sets, DYS385 was used in the diversity estimates, and the shorter repeat allele was consistently associated with DYS385a, although the assignment of the two-repeat alleles cannot be made accurately without further genotyping. Data sets were restricted to only indigenous Y-chromosomes. All participants who claimed Tlingit or Haida paternal ancestry were included in the statistical analysis, whereas those participants who claimed paternal ancestry tracing back to other populations were not. A haplogroup predictor (http://www.hprg.com/hapest5/) (Athey, 2007) was used to assign Y-STR haplotypes from Zlojutro (2008) into Y-chromosome haplogroups. After making the data sets comparable, we estimated haplotype diversity and MPDs from the Y-chromosome STR haplotypes using Arlequin 3.11.
Mitochondrial DNA CR sequences were used to construct a network of A2 haplotypes using NETWORK 4.6.0.0 (www.fluxus-engineering.com; Bandelt et al., 1995, 1999). The weighting scheme suggested by Bandelt et al. (2002) was used with slight modifications. According to this scheme, fast-evolving sites were given lower weights relative to other less mutable sites. Networks were created using both median joining and reduced median joining approaches with MP processing (Polzin and Daneschmand, 2003).
The time to most recent common ancestor (TMRCA) for each paternal haplogroup was estimated from Y-STR data using the calculation of rho statistics with Network 4.6.0.0, where the founder haplotype was inferred as in work by Sengupta et al. (2006). Networks were generated as described by Dulik et al. (2011). The evolutionary mutation rate was used to estimate coalescence dates with a generation time of 25 years.
RESULTS
mtDNA diversity in Southeast Alaska
Our analysis of mtDNA variation in Southeast Alaskan populations provided a number of insights into the maternal history of these groups. Haida and Tlingit populations had mtDNA profiles that resembled those of other northern populations from Alaska and Canada (Starikovskaya et al., 1998; Schurr et al., 1999; Rubicz et al., 2003, 2010; Schurr and Wallace, 2003; Zlojutro, et al., 2006; Crawford et al., 2010; Raff et al., 2011). The vast majority of individuals had mtDNAs belonging to Native American haplogroup A2, while most of the rest belonged to haplogroups C and D, which are also indigenous in origin (Supporting Information Tables S1 and S2; Fig. S4). None of these populations possessed haplogroup B2 or X2a mtDNAs, which appear in Amerindian populations to the south. Several individuals had mtDNAs belonging to West Eurasian (e.g., H, T, U) or East Eurasian (E1a1a) haplogroups. These persons either had Tlingit or Haida fathers and non-native mothers or had been adopted into the tribe.
A comparison of CR haplotypes in the Haida and Tlingit also revealed intriguing similarities between them (Supporting Information Table S2). All three groups shared two A2 haplotypes, the first being the founder HVS1 haplotype (#1) and the second having additional 16129 and 16311 mutations (#3). Two other A2 HVS1 haplotypes (#2 and #4) were observed in Hoonah and Hydaburg, although the Hydaburg individuals bearing these haplotypes had maternal Tlingit ancestry. These observations suggested that the founder A2 mtDNAs were originally present in both the Tlingit and the Haida, but that the derived HVS1 haplotypes (#2, #3, #4) arose in the Tlingit and were then later spread into the Haida.
A2 haplotypes possessing the 16129 mutation have previously been observed in North, Central, and South American populations (e.g., Ward et al., 1991, 1993; Torroni et al., 1993; Starikovskaya et al., 1998; Johnson and Lorenz, 2006; Tamm et al., 2007). If all of these haplotypes belong to the same maternal lineage, then this distribution suggests that it likely arose in founding Native American populations and was spread with initial settlement of the Americas. However, given the polymorphic nature of the 16129 site (Bandelt et al., 2002; RuizPesini et al., 2007; Achilli et al., 2008; van Oven and Kayser, 2009), the haplotypes bearing this mutation could appear similar because of the recurrence of this site in the HVS1, hence, would have only distant genetic relationships.
Interestingly, there were differences in the patterns of A2 variation between the two Tlingit populations and between the Tlingit and Haida populations. For example, HVS1 haplotype #7 only appeared in Yakutat Tlingit. This haplotype represents an Athapaskan-specific sublineage of A2 that has been found in Alaska and Canada, and other areas where Athapaskan populations expanded (e.g., US Southwest) (Shields et al., 1993; Torroni et al., 1993; Starikovskaya et al., 1998; Budowle et al., 2002; Rubicz et al., 2003; Schurr and Wallace, 2003). In addition, HVS1 haplotype #9, which is characterized by the 16355 mutation, was observed in only the Haida. It appears to have arisen in this tribe, as it occurs in no other population in the Northwest Coast region, aside from the neighboring Bella Coola, in which similar haplotypes has been observed (Ward et al., 1993).
Native Americans of different ethnic backgrounds also contributed several other A2 haplotypes to the Haida and Tlingit communities. For example, HVS1 haplotype #5 represented an Inuit individual from Hoonah, while HVS1 haplotype #8 belonged to an Inupiaq individual from Yakutat. These A2 mtDNAs are commonly seen in Yupik and Inupiaq populations (Starikovskaya et al., 1998; Schurr et al., 1999; Saillard et al., 2000; Helgaon et al., 2006; Gilbert et al., 2008). Likewise, the unique HVS1 haplotype #6 occurred in a Haida individual with Tsimshian maternal ancestry.
By contrast, haplogroup C and D haplotypes appeared mostly in the Yakutat Tlingit. These included a unique version of C1 (#11) in a Hispanic/Mexican individual who traced her maternal ancestry to Chihuahua, Mexico, and a C1b type (#10) in a Haida individual that differed from the founder C1 by two steps. In addition, a Tlingit individual from Yakutat had a D1 haplotype (#12), while another with Alaskan Athapaskan maternal ancestry had a D2 haplotype (#13) similar to those commonly seen in Chukchi, Siberian Yupik, and Aleut populations (Starikovskaya et al., 1998; Schurr et al., 1999; Rubicz et al., 2003; Zlojutro et al., 2006). However, D4h3 haplotypes were not observed in any of these populations, despite their presence in an ancient sample from the Northwest Coast and modern populations from southwestern North America (Kemp et al., 2007; Perego et al., 2009).
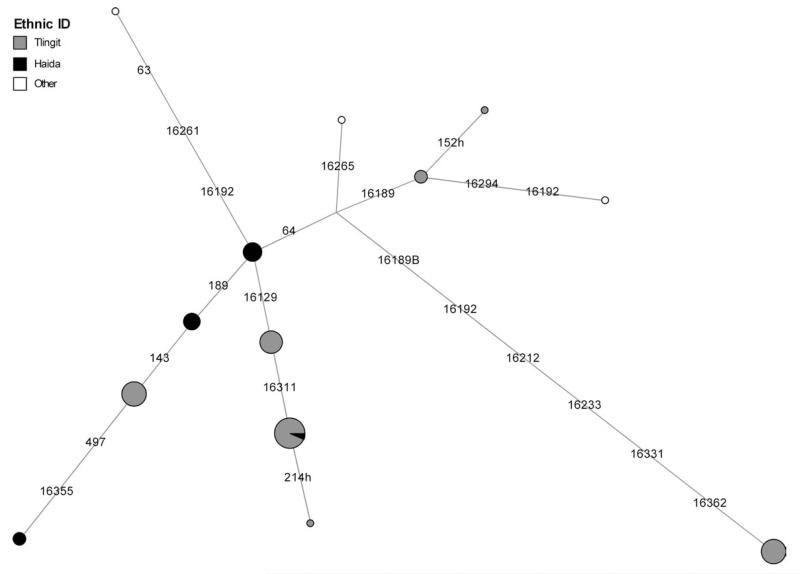
The network of A2 haplotypes confirmed these observations (Fig. 3; Supporting Information Fig. S5). The Haida and Tlingit shared several haplotypes, including #3, although Haida HVS1 haplotype #9 was clearly distinctive relative to others in Tlingit populations. Indeed, the whole mtDNA genome sequence for HVS1 haplotype #9 was quite distinctive relative to any other A2 haplotype that has been characterized in the same way (Supporting Information Table S3). Athapaskan HVS1 haplotype #7, Inupiaq HVS1 haplotype #5, and Tsimshian HVS1 haplotype #6 were also clearly delineated in the network (Supporting Information Fig. S5). Interestingly, the latter three haplotypes lacked the mutation at np 64, which occurs just before the start of the HVS2 region, and which was present in all of the A2 mtDNAs from the Haida and most of those from the Tlingit. The phylogenetic significance of this mutation is not entirely clear. It has been reported as being hypervariable in A2 mtDNAs throughout the Americas (Tamm et al., 2007; Achilli et al., 2008; Perego et al., 2009), although it might possibly delineate two sets of founding A2 haplotypes, one with and one without the mutation (Vilar et al., 2011).
Fig. 3.
A reduced-median network of HVS1 sequences from Tlingit and Haida populations. The nodes are shaded to indicate the Alaskan Native population(s) in which an HVS1 sequence appears, with the key appearing in the upper left corner of the figure.
The statistical analysis of Haida and Tlingit HVS1 sequence data also yielded new insights into the genetic diversity within the region. Comparisons of the Haida and Tlingit mtDNA data with those from published data sets showed that populations in Southeast Alaska were generally less diverse than populations from other regions (Supporting Information Table S4). The Nuu-chah-nulth, Bella Coola, and Yakima had greater haplotype diversities than the two Tlingit and two Haida populations. Aleuts and Greenlandic Inuit also had lower diversity estimates, although the Alaskan Athapaskans by Shields et al. (1993) had a higher value, probably because the samples were collected from various tribes in Alaska.
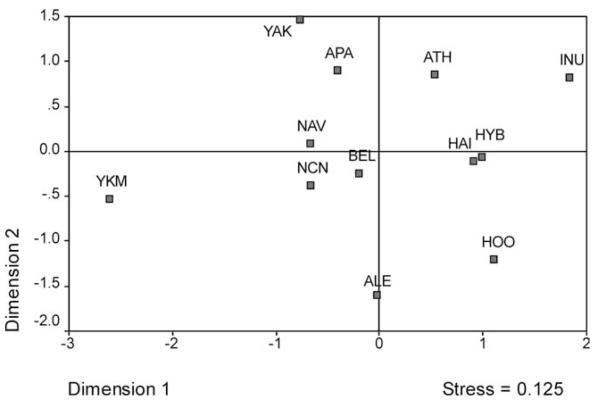
The MDS plot of these FST estimates further illustrated the genetic relationships among populations from the circumarctic region (Supporting Information Table S5; Fig. 4). Haida from Hydaburg and Queen Charlotte Island clustered tightly together and separately from Tlingit groups, and were genetically close to Bella Coola, a Northwest Coast Amerindian population. The Yakutat and Hoonah Tlingit were also genetically distant from each other, with Yakutat being closer to Alaskan Athapaskans, Apache and Navajo, in part due to sharing Athapaskan A2 haplotypes with these groups, and Hoonah showing some similarities to both NW Coast Amerindians and the Haida. The Greenland Inuit, Yakima, and Aleuts were all positioned at some distance from the general cluster of Northwest Coast populations. Overall, the FST values were generally high, indicating substantial population differentiation.
Fig. 4.
A MDS plot of FST estimates for Tlingit, Haida and comparative indigenous populations.
Influence of social organization on mtDNA diversity
When mtDNA data were sorted according to the descent groups of Tlingit and Haida individuals, we noted a strong correspondence between mitochondrial haplotype and maternal moiety affiliation (Supporting Information Table S6). In the Yakutat Tlingit, there were striking differences between the Eagle and Raven members, and between clans within each of these moieties. All individuals from the Kwáashk’ikwáan clan had A2 haplotype #7, those from the L’uknax.ádi clan had A2 haplotype #3, and an individual with Eyak maternal ancestry who belonged to the Galyax Kaagwaantaan clan (Raven moiety) had D1 haplotype #12. Within the Eagle moiety, individuals adopted into the Teikweidi clan included those having Koyukon Athapaskan (haplotype #13), Inupiaq (haplotype #8), and non-native (haplotype #16) maternal ancestry.
Similar to those for the Yakutat Tlingit, the Hoonah mtDNA data showed a clear distinction between the Eagle and Raven moiety members. Aside from those persons adopted into an Eagle clan, all but one of its members had only the A2 founder HVS1 haplotype #1. The remaining mtDNA haplotypes belonged to Raven clan members. Among these, L’uknax.ádi (Coho Salmon) and T’akdeintaan (Kittywake) members had A2 HVS1 haplotype #3, Kaach.ádi (Land Otter) members had HVS1 haplotype #2, and L’eeneidi (Dog Salmon) had HVS1 haplotype #4. Thus, judging from the clan and mtDNA data, the Eagle and Raven individuals shared virtually no specific mtDNA haplotypes.
In Hydaburg, the Eagle and Raven moieties were also genetically distinguishable from each other. Aside from the A2 founder HVS1 haplotype #1, they shared no other maternal lineages. In fact, based on full CR sequences, even the A2 founder haplotypes in Raven and Eagle individuals differed by the presence or absence of the 189 mutation in the HVS2. Thus, members of the Raven and Eagle moieties in the Kaigani Haida actually shared no specific haplotypes.
Comparison of the mtDNA sequence data at the moiety level for Haida and Tlingit populations hinted at deeper connections between them. In both the Hoonah and Yakutat, A2 HVS1 haplotype #3 occurred in Raven clan members, suggesting a common source for these mtDNAs. By contrast, these kinds of A2 haplotypes belonged to Haida Eagle moiety members. This was an intriguing finding in light of the hypothesized “foreign” origin of the Eagle moiety in the Haida (Swanton, 1905).
When assessing the distribution of mtDNA haplotypes in Haida and Tlingit relative to geography and moiety status using AMOVA, a more complex picture of maternal genetic variation emerged. The first AMOVA involved Hoonah and Yakutat Tlingit, with each clan being grouped into its respective moiety. The resulting Within Group variation was very low Table 1(a), reflecting low levels of diversity within each maternal clan. When the clans were grouped by moiety, the Between Population-Within Group component rose dramatically, indicating significant differences between at least two of the clans in one of the moieties. When the clans were grouped by geography, a significant portion of the variation was shifted into the Among Group component, although the differences were not significant (P-value >0.1).
TABLE 1. Results of Tlingit and Haida AMOVAs.
| Groups | % variation | P-value |
|---|---|---|
| (a) AMOVA of Hoonah and Yakutat Tlingit mtDNA diversity | ||
| Moiety | ||
| Among group | 0.17 | 0.519 |
| Between population within group | 97.60 | 0.000 |
| Within Group | 2.23 | 0.000 |
| Geography | ||
| Among group | 44.50 | 0.143 |
| Between population within group | 53.79 | 0.000 |
| Within Group | 1.71 | 0.000 |
| (b) AMOVA of Hoonah Tlingit mtDNA diversity by moiety | ||
| Among group | 71.68 | 0.101 |
| Between population within group | 22.29 | 0.000 |
| Within Group | 6.03 | 0.000 |
| (c) AMOVA of Tlingit and Haida mtDNA diversity Moiety | ||
| Among group | −9.39 | 0.900 |
| Between population within group | 55.30 | 0.000 |
| Within Group | 54.09 | 0.000 |
| Geography | ||
| Among group | 27.94 | 0.200 |
| Between population within group | 23.46 | 0.000 |
| Within Group | 48.60 | 0.000 |
| Ethnicity | ||
| Among group | −12.63 | 1.000 |
| Between population within group | 57.23 | 0.000 |
| Within Group | 55.41 | 0.000 |
The most likely candidate for the clan influencing the AMOVA results was the Humpback Salmon clan from Yakutat. As noted above, this clan belongs to the Raven moiety and has primarily the Athapaskan A2 haplotypes (#7) that is very different from the others present in the Hoonah and Hydaburg areas. These results were also consistent with the large FST value between the Hoonah and Yakutat Tlingit (Supporting Information Table S5).
To further assess the effects of the Yakutat samples on these results, data from only the Hoonah region Tlingit clans were subjected to AMOVA. They were grouped only by moiety. This AMOVA produced a very low Within Group, a moderate Between Population–Within Group, and also a very large Among Group value [Table 1(b)]. These results confirmed the genetic differences between the Eagle and Raven moieties in the Hoonah Tlingit, while also pointing to the skewing of the earlier Between Population–Within Group estimate by another clan.
A third AMOVA was conducted with both Haida and Tlingit samples, grouping them by geographic region and moiety. Both moiety and ethnicity produced considerable Between Population–Within Group estimates, while geography generated the highest Among Group value. Interesting, in all analyses, there were never significant Among Group differences at the 5% significance level [Table 1(c)]. These results could have been influenced by the small sample sizes of the populations as well as the relative lack of Eagle moiety members in Yakutat.
Y-chromosome diversity in Southeast Alaska
The analysis of NRY variation in these same populations using SNPs defined a variety of paternal haplotypes in male individuals (Supporting Information Tables S7 and S8; Fig. S4). Some of these haplotypes belonged to haplogroups C3*, C3b, Q1a*, and Q1a3a*, which have been observed in various Native American populations (e.g., Karafet et al., 2001; Lell et al., 2002; Bortolini et al., 2003; Zegura et al., 2004; Bolnick et al., 2006). Others belonged to haplogroups E1b1b1, I1, I2b, J1e and R1b1b2, which are common among European populations (Rosser et al., 2000; Karlsson et al., 2006), or O3a3*, which is common in Southeast and East Asia groups (Shi et al., 2005; Xue et al., 2006; Zhong et al., 2011). Haplogroups C and Q were found in participants claiming Tlingit or Haida paternal ancestry, with the exception of a single C3* lineage (Y-STR haplotype #1). All other haplogroups were found in men claiming a non-indigenous paternal ancestor.
Comparison of haplogroup C and Q haplotypes with those present in other Native American communities revealed a distinct pattern of male genetic diversity in Southeast Alaska. As noted above, we identified two branches of haplogroup Q in Southeast Alaskans, these being Q1a*, defined by the MEH2 SNP, and its derivative Q1a3a*, defined by the M346 and M3 SNPs. Q1a* is a paragroup made up of only a handful of individuals, with all other Native Americans falling into the more derived Q1a3* (M346) and Q1a3a* haplogroups (Bortolini et al., 2003; Zhadanov et al., 2010; Bisso-Machado et al., 2011; Gaieski et al., 2011). In Southeast Alaska, Q1a* appeared in only the Tlingit, while Q1a3a* was present in both the Haida and Tlingit.
The Q1a* and Q1a3a* haplotypes were quite diverse when considering their STR profiles. The differences between the two Q1a* haplotypes were remarkable, as they varied at 12 out of 19 STR loci, while the Q1a3a* haplotypes differed amongst themselves at 6–8 STR loci, on average. In fact, based on these data, every single Q haplotype in the Haida and Tlingit was found to be unique. This level of diversity suggested that these Q Y-chromosomes might represent paternal lineages that have persisted and diversified in the region for thousands of years. Consistent with this view, the TMRCA for the Q1a3a* Y-chromosomes present in the Tlingit and Haida was 10,120 years ± 2,200, using rho statistics and the evolutionary mutation rate, a date consistent with earlier estimates for the age of the M3 mutation in the Americas (Zegura et al., 2004).
In addition, we detected haplogroup C3* (M217) and C3b (P39) haplotypes in the Tlingit. These findings were consistent with other studies that have shown haplogroup C to be present in Athapaskan and Algonquian populations from North America (Bortolini et al., 2003; Zegura et al., 2004; Bolnick et al., 2006; Malhi et al., 2008). However, previous studies did not analyze a sufficient number of SNP markers to delineate the different kinds of C haplotypes that were present in their study populations.
Approximately half of the NRY haplotypes were non-indigenous in origin. Of these, haplogroups I1 and I2b occurred only in Hydaburg. This observation was consistent with a number of Haida families having male ancestors who, over the past several generations, came to the region from Scandinavia or northern Europe, where such haplogroups are common (Rosser et al., 2000; Karlsson et al., 2006). The other West Eurasian haplogroups appeared largely in individuals from Hoonah and Yakutat. Haplogroup R1b1b2 was especially common in these tribes, which was not surprising given that the male ancestors of these families traced their roots directly to Denmark, Germany, Ireland, Netherlands, Norway, Russia, Spain, or Sweden, where this paternal haplogroup is ubiquitous (Balaresque et al., 2010; Myres et al., 2010).
Interestingly, one Tlingit man from Yakutat possessed a Y-chromosome from haplogroup O3a3, which is typically seen in Southeast and East Asia. Although he self-identified as a Tlingit (having a Tlingit mother), he had a Filipino father. In fact, a number of Tlingit men and women had Filipino grandparents on the maternal or paternal sides of their families based on their genealogies, particularly in Yakutat. In such cases, women who had Filipino ancestry traced it through their paternal grandfathers, or men traced it through their paternal grandmothers. Thus, given the inheritance patterns of mtDNA and Y-chromosomes, the Filipino genetic lineages from these family members were not sampled.
To further explore Y-chromosome diversity in circumarctic populations, we compared our data with those from other native North American populations for which core Y-STR data were available. Our results revealed that Y-STR diversity in the Tlingit and Haida was relatively similar to that seen in other Native American populations (Table 2). The diversity value for the Hoonah Tlingit was 0.978, whereas that for the Yakutat Tlingit and Hydaburg Haida was 1.000 because of small sample sizes. Diversity estimates for Aleuts (Zlojutro, 2008) were recalculated without non-indigenous haplotypes. The resulting diversity estimate for Aleut haplogroup Q lineages was 0.924 instead of 0.990, well below the estimates seen in Southeast Alaska. The Tlingit and Haida values fell in the range of estimates for indigenous eastern North American populations (Bolnick et al., 2006) as well as those from Native Americans analyzed by Redd et al. (2006). It is likely that estimates from the latter are inflated, as recent admixture was not accounted for in this analysis.
TABLE 2. Y-STR haplotype diversity of indigenous Y-chromosomes in Haida, Tlingit and Aleut.
| Hoonah Tlingit | Yakutat Tlingit | Haida | Aleuts | |
|---|---|---|---|---|
| Sample number | 10 | 2 | 1 | 21 |
| Haplotype number | 9 | 2 | 1 | 12 |
| Haplotype diversity | 0.978 ± 0.054 | 1.000 ± 0.500 | 1.000 ± 0.000 | 0.924 ± 0.035 |
| Mean pairwise differences | 6.000 ± 3.125 | 9.000 ± 6.708 | 0.000 ± 0.000 | 4.771 ± 2.429 |
To conduct a comparable analysis with the Aleut data from Zlojutro (2008), we had to run the Aleut Y-STR haplotypes through a haplogroup predictor to identify the indigenous paternal lineages to which they belonged (Athey, 2007). Overall, this haplogroup predictor worked well, although a few of the Y-STR haplotypes seemed to be misclassified. Nevertheless, we identified most, if not all, indigenous Y-chromosomes in the Aleut villages, which represented only 15.3% of Aleut men (Zlojutro, 2008). The indigenous Y-chromosomes from Southeast Alaska made up approximately 50% of the total, which is similar to frequencies of indigenous Y-chromosomes in eastern North America (Bolnick et al., 2006).
DISCUSSION
Genetic variation in Southeast Alaska
Our analysis of mtDNA and Y-chromosome variation in Haida and Tlingit individuals provides new insights into the genetic history of indigenous populations from Southeast Alaska. Although the sample sizes for these populations are small, the resulting data, viewed in the context of ethnographic and genealogical evidence, appear to be generally representative of the communities in which we worked, thereby allowing us to make reasonably strong inferences about their population histories.
A primary objective of this study was to assess the genetic affinities of Haida and Tlingit populations with other native communities in North America. Overall, the Haida and Tlingit show a mostly “northern” genetic profile similar to that of circumarctic populations. This profile is marked by the high frequency of haplogroup A2 and the presence of haplogroup D2. Yet, specific A2 haplotypes differed. For example, A2b lineages that are common in Arctic populations were not found in Tlingit and Haida. The Southeast Alaskan groups also lack haplogroup B2 and X2a mtDNAs and exhibit very low frequencies of C1 and D1 haplotypes. These haplogroups are commonly seen in Amerindian populations such as the Nuu-chah-nulth, Bella Coola, Yakima, and other tribes located south of this region (Ward et al., 1991, 1993; Shields et al., 1993), and are also present in the Navajo and Apache due to their intermarriage with Puebloan and other Amerindian population since entering the American Southwest some 500–1,000 years ago (e.g., Budowle et al., 2002).
In addition, while observed in certain Amerindian populations living in mainly coastal areas in southwestern North America and western South America (Rickards et al., 1999; Schurr, 2004; Perego et al., 2009), D4h3 haplotypes are not present in either the Haida or Tlingit, despite its presence in the prehistoric On Your Knees Cave male from Prince of Wales Island (Kemp et al., 2007). This pattern of mtDNA diversity suggests that the haplogroups initially brought to the Northwest Coast have been replaced by those ancestral to the Haida, Tlingit, and other Northwest Coast tribes in the region.
Tlingit population history
Our genetic data provide new insights into the population history of Tlingit populations from Southeast Alaska. In some respects, the Hoonah and Yakutat tribes are genetically similar to each other, with both having two of the most common HVS1 haplotypes present in the region (#1 and #3). This finding suggests that these A2 mtDNAs could have been part of the original maternal gene pool for Tlingit populations, and is consistent with ethnographic evidence for the origins of certain Yakutat clans in Hoonah (de Laguna, 1972, 1990b). They both also have Q1a, Q1a3a*, and C3* Y-chromosome haplotypes, suggesting they could potentially have a common or similar paternal ancestry, although no specific STR haplotypes are shared between them.
At the same time, while being linguistically and culturally similar, the two Tlingit tribes are genetically distinctive from each other. The Yakutat Tlingit show stronger maternal genetic ties to Athapaskan groups from interior Alaska than do the Hoonah Tlingit, and is the only population with the Athapaskan-specific A2 haplotype (#7). A single Yakutat individual with Koyukon Athapaskan maternal ancestry also has a D2 haplotype (#13), which appear mostly in Alaskan Aleut, Athapaskan, and Inupiaq groups, as well as the Siberian Chukchi (Starikovskaya et al., 1998; Saillard et al., 2000; Rubicz et al., 2003; Zlojutro et al., 2006; Gilbert et al., 2008). In addition, as previously noted, a number of Yakutat participants have parents or grandparents who were born in the Alsek River or Dry Bay area, while others were noted to have Eyak maternal grandparents. Thus, the Yakutat haplotype distribution likely reflects extensive interactions with Athapaskan populations from interior of south-central Alaska, and possibly also the assimilation of smaller ethnic groups like the Eyaks (de Laguna, 1972, 1990b; Emmons, 1991).
The Hoonah Tlingit, on the other hand, exhibit mainly A2 founder haplotypes (#1) and those derived from it (#2 and #3), one of which is also seen in the Yakutat Tlingit (#3). They also lack the A2 sublineage that is observed in many other Athapaskan-speaking tribes. As a consequence, they show greater genetic similarities to the Haida and Northwest Coast Indian populations than to the Yakutat Tlingit. These affinities might possibly reflect the sharing of mtDNA haplotypes with the Haida and Tsimshian, with whom they have exchanged marital partners in the recent past, and which are mirrored by cultural influences on Tlingit populations from these neighboring tribes in the form of iconography and clan name identification (de Laguna, 1972, 1990a; Blackman, 1990; Emmons, 1991). This pattern also generally supports evidence suggesting that certain Tlingit clans originated in the area defined by the Nass River in British Columbia and Stikine River in Southeast Alaska (de Laguna, 1990b).
Such differences could also possibly reflect more ancient population subdivisions that resulted from cultural and geographic isolation. Previous linguistic studies noted several distinct dialect groups in Southeast Alaska, with the Yakutat belonging to the Gulf Coast group and Hoonah belonging to the Northern group (de Laguna, 1990a). This linguistic divergence would imply that Tlingit groups underwent regional genetic differentiation within Southeast Alaska. Under this scenario, the Tlingit emerged from a common Northwest Coast gene pool that had developed by 5,000 years ago, and then diverged from each other linguistically and genetically as they expanded throughout the region, while also being influenced by neighboring indigenous populations.
Haida population history
Our data indicate that the Haida are genetically distinctive from Tlingit populations. They have mostly mtDNA A2 founder haplotypes (#1), which, interestingly, are more diverse at the HVS2 level than those in the Tlingit, as well as others that were obtained through intermarriage with Tlingit groups (#2 and #4), based on genealogical information. They also possess a haplotype (#9) that is not seen in the Tlingit or other Northwest Coast populations, aside from the geographically adjacent Bella Coola (Ward et al., 1993), and also have a Q1a3a* haplotype distinct from those seen in the Tlingit. In addition, the Haida from Hydaburg are genetically closest to Haida from Queen Charlotte Island in Canada. This is not a surprising finding because the Haida only began expanding into the Prince of Wales Island during the past few centuries, displacing the Tongass Tlingit groups living there.
Furthermore, genetic distance estimates based on mtDNA HVS1 data show the Haida to have greater genetic affinities with Northwest Coast Amerindian than Athapaskan tribes. In this regard, Ward et al. (1993) believed that the similarities between the Haida and Bella Coola were due to a recent shared origin of the two populations, despite the fact that the Haida speak a Na-Dene language and the Bella Coola an Amerind one. By contrast, Lorenz and Smith (1997) asserted that the similarities between the Bella Coola and the Haida were not due a recent common origin of both populations, but rather reflected recent gene flow between them after the Bella Coola migrated into the area. Given the lack of haplogroup B2 and D1 mtDNAs in the Haida and the distribution of the A2 haplotypes described above, our data support the view that recent gene flow has produced the observed patterns of mtDNA diversity in the Haida and Bella Coola. Furthermore, based on the current genetic evidence, the Haida appear not to be closely related to adjacent Tlingit populations, with the pattern of genetic diversity in this population reflecting some degree of isolation from neighboring tribes.
Genetic and linguistic diversity in the Na-Dene linguistic family
Another goal of this study was to investigate the relationship between linguistic and genetic diversity for populations assigned to the Na-Dene linguistic family, specifically, the inclusion of Haida in the family with Athapaskan, Eyak, and Tlingit. As noted above, we observe differences in mtDNA composition between the Haida and the Tlingit. In addition to their distinct mtDNA profile, the Haida lack A2a haplotypes (#7) seen in many other Athapaskan tribes (Shields et al., 1993; Torroni et al., 1993; Starikovskaya et al., 1998; Budowle et al., 2002; Schurr and Wallace, 2003), although this is also true for the Hoonah Tlingit. The Q1a3a* Y-chromosome observed in the Haida (#20) is also distinctive compared to those in Tlingit groups, although it represents a single male lineage in this population. Together, these data suggest that the Haida do not share a recent common ancestry with the Tlingit.
Thus, we observe only modest genetic evidence for the composition of the Na-Dene stock as originally proposed by Sapir (1915), in which Haida and AET populations share a common genetic ancestry. However, this interpretation is not supported by previous analyses of Haida and AET languages. While linguistically influenced by Tlingit tribes as they expanded northward from their Canadian homeland, the Haida are viewed as speaking a Na-Dene language by a number of Native American linguists (Greenberg et al., 1986; Manaster Ramer, 1996; Ruhlen, 1998; Enrico, 2003). Thus, if the Haida share deeper linguistic ties with AET speaking populations, then they have since diverged from them genetically, albeit with some gene flow due to interactions with neighboring populations.
mtDNA diversity and clan status
As noted previously, ethnographic studies indicated that clan status in the Haida and Tlingit is based on matrilineal descent. Traditionally, a member of the Eagle Clan married someone from the Raven clan, and vice versa. However, the strictness of this practice has waned somewhat over the past century, partly due to intermarriage with non-native persons. For this reason, we were uncertain as to whether the mtDNA data would show any correspondence with the clan status of the participants.
In fact, there was a strong association between the maternal clan status and mtDNA haplotype background of individuals from Yakutat, Hoonah, and Hydaburg. In general, it was possible to distinguish members of the Tlingit Eagle and Raven moieties from each other based on the mtDNA haplotype composition of each social grouping. Members of the Raven moiety in Yakutat had either #2 and #3 haplotypes (L’uknax.ádi clan) or Athapaskan-specific A2 haplotypes (#7) (Kwáashk’ikwáan clan) because of previous interactions with Ahtna Athapaskan populations, with another Raven individual from the Gaanax.ádi clan having D1 haplotype (#12). Because all of the Eagle clan members involved in this study had been adopted into the Tekiweidi clan, they exhibited a mixture of different maternal haplotypes.
We observed the same pattern for Eagle and Raven moieties in the Hoonah Tlingit. The Eagle and Raven members shared no mtDNA haplotypes, except for #4. This was an intriguing finding because, while many of their clans apparently arose in the Hoonah region, the ancestors of a number of tribal members came from clans originating in other parts of the traditional Tlingit territory that encompassed the Northern and Southern subdialect regions (de Laguna, 1990a). Furthermore, both the Hoonah and Yakutat A2 haplotypes #3 occurred only in Raven clan members, suggesting a common source for these mtDNAs, and the genetic divergence of these two moieties some time ago. These results are consistent with information obtained through ethnographic studies of Tlingit tribes (de Laguna, 1972, 1990a; Emmons, 1991) and recent genealogical data, and indicate that matrilineal kinship and clan-based marriage practices continue to shape mtDNA variation in the Tlingit today.
Similarly, we observed genetic differences by moiety in the Kaigani Haida from Hydaburg. Accordingly, Raven and Eagle moiety members did not share any haplotypes when considering the entire CR sequences of the participants. However, the correspondence between moiety status and specific mtDNA haplotypes is not as strong as seen in the Tlingit populations. This pattern might reflect the mixing of lineages from formerly separated groups that resulted from the consolidation of smaller Haida communities in the early 20th century. In addition, whereas A2 haplotype #3 occurred in Tlingit Raven clan members, these kinds of mtDNAs appeared in Haida Eagle clan members. This observation is intriguing in light of the fact that, based on ethnographic information, clans belonging to the Haida Eagle moiety would be placed in the Tlingit Raven moiety (Blackman, 1990) and might even have originated in a different tribe (Swanton, 1905). Hence, judging from this evidence, these Tlingit and Haida subdivisions might possibly have deeper genetic connections between them than indicated here.
Historical admixture
The genetic data from the Haida and Tlingit confirm genealogical and historical evidence of intermarriage with non-indigenous persons. A number of Tlingit women have married men of non-native descent over the past several generations. These men, who have paternal Swedish, Danish, German, Scotch-Irish-English, and Filipino descent, came to Alaska to fish commercially, hunt whales, work in mines, timber forests, or serve at American military bases in Alaska. In the case of the O3a3* haplotype in the Tlingit, this finding is consistent with the economic history of Southeast Alaska where, during the early to mid-1900s, mostly male Filipinos were hired to work in Alaskan salmon canneries. These men subsequently established ties with members of such Alaska Native tribes as the Tlingit, Haida, Aleut, and Tsimshian, and often married into these communities (Cordova, 1983; Bacho, 1988; Bucholdt, 1996; Barton, 2011). In addition, some Haida and Tlingit men have married non-native women, most of whom having been formally adopted into their respective communities. Regardless of their non-native ancestry, these individuals are considered members of Haida and Tlingit tribes.
CONCLUSIONS
In summary, our genetic data reinforce perspectives on Haida and Tlingit history that come from archeological, ethnographic, and linguistic studies. The Tlingit are genetically different from the Haida, are themselves genetically subdivided, and have distinct genetic affinities with surrounding Athapaskan and Northwest Coast populations. The limited sharing of certain mtDNA haplotypes by Haida and Tlingit populations suggests only in recent times has there been population interactions leading to intermarriage, hence, gene flow, between them. In addition, we observed considerable diversity in the Q1a* and Q1a3a* paternal haplotypes in the Tlingit, implying their antiquity in the region and possibly greatly diversity in founding Y-chromosome haplogroups than previously appreciated. In this regard, our estimates of Q-M3 diversity were consistent with those obtained from other Native American populations. Additional details about the history of Haida and Tlingit peoples will emerge from comparative studies with other Alaskan Native populations, such as Yupik, Inupiaq, and Athapaskan groups from Alaska and Canada, and through whole mtDNA genome sequencing, expanded NRY SNP genotyping and analysis of autosomal loci in these populations.
Supplementary Material
ACKNOWLEDGMENTS
The authors express their gratitude for the participation by Tlingit and Haida individuals in this study, and the support of this study by the Huna Indian Association, the Huna Totem Corporation, the Huna Heritage Foundation, the Yakutat Tlingit Tribe, and the Hydaburg Cooperative Association. The authors also thank Kathy Miller and Kathryn Hurtley, past and present Executive Directors of the Huna Heritage Foundation, and Grace Villareal and Robert Starbard from the Huna Indian Association, for their facilitation of our research in Southeast Alaska. In addition, they thank Merritt Ruhlen for his constructive comments on the manuscript, and Janet Ziegle from Applied Biosystems for providing technical assistance with the DNA analysis. Finally, the authors thank two anonymous reviewers for their constructive feedback on this manuscript.
Grant sponsors: National Geographic Society, IBM, Waitt Family Foundation, University of Pennsylvania.
Footnotes
Additional Supporting Information may be found in the online version of this article.
LITERATURE CITED
- Achilli A, Perego UA, Bravi CM, Coble MD, Kong QP, Wood-ward SR, Salas A, Torroni A, Bandelt HJ. The phylogeny of the four pan-American MtDNA haplogroups: implications for evolutionary and disease studies. PLoS One. 2008;3:e1764. doi: 10.1371/journal.pone.0001764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackerman RE. Earliest stone industries on the North Pacific coast of North America. Arctic Anthropol. 1992;29:18–27. [Google Scholar]
- Ackerman RE. Ground Hog Bay 2. In: West FH, editor. American beginnings: prehistory and paleoecology of Beringia. University of Chicago Press; Chicago: 1996. pp. 424–430. [Google Scholar]
- Ames KM, Maschner HDG. Peoples of the northwest coast. Their archaeology and prehistory. Thames and Hudson; London: 1999. [Google Scholar]
- Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23:147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- Athey W. [Accessed on 23 February 2012];Haplogroup predictor. 2007 ( /www.hprg.com/hapest5/)
- Bacho P. Alaskeros: a documentary exhibit on pioneer Filipino cannery workers. IBU/ILWU Region 37; Seattle: 1988. [Google Scholar]
- Balaresque P, Bowden GR, Adams SM, Leung H-Y, King TE, Rosser ZH, Goodwin J, Moisan J-P, Richard C, Millward A, Demaine AG, Barbujani G, Previdere C, Wilson IJ, TylerSmith C, Jobling MA. A predominantly Neolithic origin for European paternal lineages. PLoS Biol. 2010;8:e1000285. doi: 10.1371/journal.pbio.1000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Sykes BC, Richards MB. Mitochondrial portraits of human populations. Genetics. 1995;141:743–753. doi: 10.1093/genetics/141.2.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandelt H-J, Quintana-Murci L, Salas A, Macaulay V. The fingerprint of phantom mutations in mitochondrial DNA data. Am J Hum Genet. 2002;71:1150–1160. doi: 10.1086/344397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton MP. [Accessed on 22 February 2012];Alaska. Encyclopedia of immigration. 2011 ( http://immigration-online.org/330-alaska.html)
- Berton P. The Klondike fever; the life and death of the last great gold rush. Knopf; New York: 1959. [Google Scholar]
- Bisso-Machado R, Jota MS, Ramallo V, Paixâo-Côrtes VR, Lacerda DR, Salzano FM, Bonatto SL, Santos FR, Bortolini MC. Distribution of Y-chromosome Q lineages in Native Americans. Am J Hum Biol. 2011;23:563–566. doi: 10.1002/ajhb.21173. [DOI] [PubMed] [Google Scholar]
- Black LT. Russians in America: 1732-1867. University of Alaska Press; Anchorage: 2004. [Google Scholar]
- Blackman MB. Haida: traditional culture. In: Suttles W, editor. Handbook of North American Indians. Vol. 7. Smithsonian Institution Press; Northwest Coast. Washington, DC: 1990. pp. 240–260. [Google Scholar]
- Bolnick DA, Bolnick DI, Smith DG. Asymmetric male and female genetic histories among Native Americans from eastern North America. Mol Biol Evol. 2006;23:2161–2174. doi: 10.1093/molbev/msl088. [DOI] [PubMed] [Google Scholar]
- Borneman WR. Alaska: saga of a bold land from Russian fur traders to the Gold Rush, extraordinary railroads, World War II, the oil boom, and the fight over ANWR. Harper-Collins; New York: 2004. [Google Scholar]
- Bortolini MC, Salzano FM, Thomas MG, Stuart S, Nasanen SP, Bau CH, Hutz MH, Layrisse Z, Petzl-Erler ML, Tsuneto LT, Hill K, Hurtado AM, Castro-de-Guerra D, Torres MM, Groot H, Michalski R, Nymadawa P, Bedoya G, Bradman N, Labuda D, Ruiz-Linares A. Y-chromosome evidence for differing ancient demographic histories in the Americas. Am J Hum Genet. 2003;73:524–539. doi: 10.1086/377588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch E, Calafell F, Rosser ZH, Nørby S, Lynnerup N, Hurles ME, Jobling MA. High level of male-biased Scandinavian admixture in Greenlandic Inuit shown by Y-chromosomal analysis. Hum Genet. 2003;112:353–363. doi: 10.1007/s00439-003-0913-9. [DOI] [PubMed] [Google Scholar]
- Buchholdt T. Filipinos in Alaska, 1788-1958. Aboriginal Press; Anchorage: 1996. [Google Scholar]
- Budowle B, Allard MW, Fisher CL, Isenberg AR, Monson KL, Stewart JE, Wilson MR, Miller KW. HVI and HVII mitochondrial data in Apaches and Navajos. Int J Legal Med. 2002;116:212–215. doi: 10.1007/s00414-001-0283-6. [DOI] [PubMed] [Google Scholar]
- Carlson RL. Cultural antecedents. In: Suttles W, editor. Handbook of North American Indians. Vol. 7. Smithsonian Institution Press; Northwest Coast. Washington, DC: 1990. pp. 60–69. [Google Scholar]
- Cordova F. Filipinos: forgotten Asian Americans: a pictorial essay 1763-circa 1963. Kendall/Hunt; Dubuque: 1983. [Google Scholar]
- Crawford MH, Rubicz RC, Zlojutro M. Origins of Aleuts and the genetic structure of populations of the archipelago: molecular and archaeological perspectives. Hum Biol. 2010;82:695–717. doi: 10.3378/027.082.0511. [DOI] [PubMed] [Google Scholar]
- Dauenhauer NM, Dauenhauer RL, Black LT, editors. Anooshi Lingit Aani Ka, Russians in Tlingit America: the Battles of Sitka, 1802 and 1804. University of Washington Press; Seattle: 2008. [Google Scholar]
- Davis SD. Prehistory of southeastern Alaska. In: Suttles W, editor. Handbook of North American Indians. Vol. 7. Smithsonian Institution Press; Northwest Coast. Washington, DC: 1990. pp. 197–202. [Google Scholar]
- de Laguna F. Under Mount Saint Elias: the history and culture of the Yakutat Tlingit. Smithsonian Contributions to Anthropology. Vol. 7. Smithsonian Institution Press; Washington, DC: 1972. [Google Scholar]
- de Laguna F. Tlingit. In: Suttles W, editor. Handbook of North American Indians. Vol. 7. Smithsonian Institution Press; Northwest Coast. Washington, DC: 1990a. pp. 203–239. [Google Scholar]
- de Laguna F. Eyak. In: Suttles W, editor. Handbook of North American Indians. Vol. 7. Smithsonian Institution Press; Northwest Coast. Washington, DC: 1990b. pp. 189–196. [Google Scholar]
- de Laguna F, Riddell FA, McGeein DF, Lane KS, Freed JA, Osborne C. Archeology of the Yakutat Bay Area, Alaska. U.S. Government Printing Office; Washington, DC: 1964. [Google Scholar]
- Dulik MC, Osipova LP, Schurr TG. Y-chromosome variation in Altaian Kazakhs reveals a common paternal gene pool for Kazakhs and the influence of Mongol expansions. PLoS One. 2011;6:e17548. doi: 10.1371/journal.pone.0017548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmons GE. In: The Tlingit Indians. Anthropological Papers of the American Museum of Natural History. De Laguna Frederica., editor. Vol. 70. American Museum of Natural History; New York: 1991. [Google Scholar]
- Enrico J. Haida syntax, (2 volumes) University of Nebraska Press; Lincoln: 2003. [Google Scholar]
- Enrico J. Haida dictionary: Skidegate, Masset, and Alaskan dialects, (2 volumes) Alaska Native Language Center; Fairbanks: 2005. [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol Bioinformat Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Fairbanks JA. Art of the Northern Tlingit. University of Washington Press; Seattle: 1989. [Google Scholar]
- Fedorova LG. In: The Russian population in Alaska and California: late 18th century—1867. Pierce RA, Donnelly MS, editors. Limestone Press; Kingston, Ontario: 1973. [Google Scholar]
- Frost O. Bering: the Russian discovery of America. Yale University Press; New Haven: 2003. [Google Scholar]
- Gaieski JB, Owings AC, Vilar MG, Dulik MC, Gaieski D, Gittelman RM, Dulik MC, Lindo J, Gau L, Schurr TG, The Genographic Consortium Genetic ancestry and indigenousheritage in a Native American descendant community in Bermuda. Am J Phys Anthropol. 2011;46:392–405. doi: 10.1002/ajpa.21588. [DOI] [PubMed] [Google Scholar]
- Garfield VE, Wingert PS. The Tsimshian Indians and their arts. University of Washington Press; Seattle: 1966. [Google Scholar]
- Gilbert MTP, Kivisild T, Bjarne G, Andersen PK, Metspalu E, Reidla M, Tamm E, Axelsson E, Götherström A, Campos PF, Rasmussen M, Metspalu M, Higham TFG, Schwenninger J-L, Nathan R, De Hoog C-J, Koch A, Møller LN, Andreasen C, Meldgaard M, Villems R, Bendixen C, Willerslev E. Paleo-Eskimo mtDNA genome reveals matrilineal discontinuity in Greenland. Science. 2008;320:1787–1789. doi: 10.1126/science.1159750. [DOI] [PubMed] [Google Scholar]
- Goddard I, Sturtevant WC, editors. Handbook of North American Indians, Volume 17: Languages. Smithsonian Institution; Washington, DC: 1996. The classification of the native languages of North America; pp. 290–323. [Google Scholar]
- Goddard I, editor. Native languages and language families of North America. University of Nebraska Press; Lincoln: 1999. [Google Scholar]
- Goldschmidt W, Haas T. Possessory rights of the native of Southeast Alaska. Reprinted as Haa aaní 5 Our land: Tlingit and Haida land rights and use. Sealaska Heritage Foundation; Juneau: 1998. [Google Scholar]
- Greenberg JH, Turner CG, II, Zegura SL. The settlement of the Americas: a comparison of linguistic, dental, and genetic evidence. Curr Anthropol. 1986;27:477–497. [Google Scholar]
- Grinev AV. The Tlingit Indians in Russian America, 1741-1867. University of Nebraska Press; Lincoln: 2008. [Google Scholar]
- Halpin MM, Seguin M. Tsimshian peoples: Southern Tsimshian, Coast Tsimshian, Nishga, and Gitksan. In: Suttles W, editor. Handbook of North American Indians, Volume 7: Northwest Coast. Smithsonian Institution Press; Washington, DC: 1990. pp. 267–284. [Google Scholar]
- Haycox S. Alaska: an American colony. University of Washington Press; Seattle: 2002. [Google Scholar]
- Helgason A, Pálsson G, Pedersen HS, Angulalik E, Gunnarsdóttir ED, Yngvadóttir B, Stefánsson K. Variation in Inuit populations of Greenland and Canada: migration history and population structure. Am J Phys Anthropol. 2006;130:123–134. doi: 10.1002/ajpa.20313. [DOI] [PubMed] [Google Scholar]
- Hoijer H. The chronology of the Athapaskan languages. Int J Anthropol Linguist. 1956;22:219–232. [Google Scholar]
- Hope A III, Thornton TF, editors. Will the time ever come? A Tlingit source book. : Alaska Native Knowledge Network; Fairbanks: 2000. [Google Scholar]
- Ives JW. Alberta, Athapaskans and Apachean Origins. In: Brink JW, Dormaar JF, editors. Archaeology in Alberta, a view from the new millennium. The Archaeological Society of Alberta; Medicine Hat: 2003. pp. 256–289. [Google Scholar]
- Ives JW. Dene-Yeneseian, migration and prehistory. In: Kari J and Potter B, editors. Special Issue, The Dene-Yeneseian connection. Anthropol Pap Univ Alaska. 2010;5:324–334. [Google Scholar]
- Johnson JR, Lorenz JG. Genetics, linguistics, and prehistoric migrations: an analysis of California Indian mitochondrial DNA lineages. J Cal Great Basin Anthropol. 2006;26:31–62. [Google Scholar]
- Kan S. Symbolic immortality: Tlingit potlatch of the nineteenth century. Smithsonian Institution Press; Washington, DC: 1989. [Google Scholar]
- Kaplan SA, Barsness KJ. Raven’s journey: the world of Alaska’s native people. University of Pennsylvania Museum Press; Philadelphia: 1986. [Google Scholar]
- Karafet T, Xu L, Du R, Wang W, Feng S, Wells RS, Redd AJ, Zegura SL, Hammer MF. Paternal population history of East Asia: sources, patterns, and microevolutionary processes. Am J Hum Genet. 2001;69:615–628. doi: 10.1086/323299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karafet TM, Mendez FL, Meilerman MB, Underhill PA, Zegura SL, Hammer MF. New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree. Genome Res. 2008;18:830–838. doi: 10.1101/gr.7172008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson AO, Wallerström T, Götherström A, Holmlund G. Y-chromosome diversity in Sweden—a long-time perspective. Eur J Hum Genet. 2006;14:963–970. doi: 10.1038/sj.ejhg.5201651. [DOI] [PubMed] [Google Scholar]
- Kemp BM, Mahli RS, McDonough J, Bolnick DA, Eshleman JA, Richards O, Martinez-Labarga C, Johnson JR, Lorenz J, Dixon EJ, Fifield TE, Heaton TH, Worl R, Smith DG. Genetic analysis of early Holocene skeletal remains from Alaska and its implications for the settlement of the Americas. Am J Phys Anthropol. 2007;132:605–621. doi: 10.1002/ajpa.20543. [DOI] [PubMed] [Google Scholar]
- Krause A. In: The Tlingit Indians. Results of a trip to the northwest coast of America and the Bering Straits. Gunther E, editor. University of Washington Press; Seattle: 1885. 1956. [Google Scholar]
- Krause M, Golla V. Alaska Native languages: past, present, and future. Alaska Native Language Center Research Papers 4. University of Alaska Press; Fairbanks: 1980. [Google Scholar]
- Krause M, Golla V. Northern Athapaskan languages. In: Helm J, editor. Handbook of North American Indians, Volume 6, Subarctic. Smithsonian Institution Press; Washington, DC: 1981. pp. 67–87. [Google Scholar]
- Krauss ME. Na-Dene and Eskimo-Aleut. In: Campbell L, Mithun M, editors. The languages of Native America. University of Texas Press; Austin: 1979. pp. 803–901. [Google Scholar]
- Leer J, Hitch D, Ritter J. Interior Tlingit noun dictionary: the dialects spoken by Tlingit elders of Carcross and Teslin, Yukon, and Atlin, British Columbia. Yukon Native Language Centre; Whitehorse: 2001. [Google Scholar]
- Lell JT, Sukernik RI, Starikovskaya YB, Jin L, Su B, Schurr TG, Underhill P, Wallace DC. The dual origins and Siberian affinities of Native American Y-chromosomes. Am J Hum Genet. 2002;70:192–206. doi: 10.1086/338457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine RD. Haida and Na-Dene: a new look at the evidence. Int J Am Linguist. 1979;45:157–170. [Google Scholar]
- MacDonald G. Haida monumental art. University of Washington Press; Seattle: 1994. [Google Scholar]
- Malhi RS, Gonzalez-Oliver A, Schroeder KB, Kemp BM, Greenberg JA, Dobrowski SZ, Smith DG, Resendez A, Karafet T, Hammer H, Zegura S, Brovko T. Distribution of Y chromosomes among native North Americans: a study of Athapaskan population history. Am J Phys Anthropol. 2008;137:412–424. doi: 10.1002/ajpa.20883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manaster Ramer A. Sapir’s classifications: Haida and the other Na Dene languages. Anthropol Linguist. 1996;38:179–215. [Google Scholar]
- Matson RG, Magne PR. Athapaskan migrations: the archaeology of Eagle Lake, British Columbia. University of Arizona Press; Tucson: 2007. [Google Scholar]
- Miller J. Tsimshian culture: a light through the ages. University of Nebraska Press; Lincoln: 2000. [Google Scholar]
- Miller J, Eastman C, editors. The Tsimshian and their neighbors of the north Pacific coast. : University of Washington Press; Seattle: 1984. [Google Scholar]
- Moss ML. Northern northwest coast regional overview. Arctic Anthropol. 1998;35:88–111. [Google Scholar]
- Myres NM, Rootsi S, Lin AA, Jarve M, King RJ, Kutuev I, Cabrera VM, Khusnutdinova EK, Pshenichnov A, Yunusbayev B, Balanovsky O, Balanovska E, Rudan P, Baldovic M, Herrera RJ, Chiaroni J, Di Cristofaro J, Villems R, Kivisild T, Underhill PA. A major Y-chromosome haplogroup R1b Holocene era founder effect in Central and Western Europe. Eur J Hum Genet. 2010;19:95–101. doi: 10.1038/ejhg.2010.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberg K. The social economy of the Tlingit Indians. Monograph 55, The American Ethnological Society. University of Washington Press; Seattle: 1973. [Google Scholar]
- Olsen RL. Social structure and social life of the Tlingit in Alaska. University of California; Berkeley: 1976. Anthropological Records 26. [Google Scholar]
- Perego UA, Achilli A, Angerhofer N, Accetturo N, Pala M, Olivieri A, Kashani BH, Ritchie KH, Scozzari R, Kong QP, et al. Distinctive Paleo-Indian migration routes from Beringia marked by two rare mtDNA haplogroups. Curr Biol. 2009;19:1–8. doi: 10.1016/j.cub.2008.11.058. [DOI] [PubMed] [Google Scholar]
- Polzin T, Daneschmand SV. On Steiner trees and minimum spanning trees in hypergraphs. Oper Res Lett. 2003;31:12–20. [Google Scholar]
- Raff JA, Bolnick DA, Tackney J, O’Rourke DH. Ancient DNA perspectives on American colonization and population history. Am J Phys Anthropol. 2011;146:503–514. doi: 10.1002/ajpa.21594. [DOI] [PubMed] [Google Scholar]
- Redd AJ, Chamberlain VF, Kearney VF, Stover D, Karafet T, Calderon K, Walsh B, Hammer MF. Genetic structure among 38 populations from the United States based on 11 U.S. core Y chromosome STRs. J Forensic Sci. 2006;51:580–585. doi: 10.1111/j.1556-4029.2006.00113.x. [DOI] [PubMed] [Google Scholar]
- Reid M. Myths and legends of the Haida Indians of the northwest coast. Bellerophon Books; Santa Barbara: 2002. [Google Scholar]
- Rickards O, Martinez-Labarga C, Lum JK, De Stefano GF, Cann RL. MtDNA history of the Cayapa Amerinds of Ecuador: detection of additional founding lineages for the Native American populations. Am J Hum Genet. 1999;65:519–530. doi: 10.1086/302513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosser ZH, Zerjal T, Hurles ME, Adojaan M, Alavantic D, Amorim A, Amos W, Armenteros M, Arroyo E, Barbujani G, Beckman G, Beckman L, Bertranpetit J, Bosch E, Bradley DG, Brede G, Cooper G, Corte-Real HB, de Knijff P, Decorte R, Dubrova YE, Evgrafov O, Gilissen A, Glisic S, Golge M, Hill EW, Jeziorowska A, Kalaydjieva L, Kayser M, Kivisild T, Kravchenko SA, Krumina A, Kucinskas V, Lavinha J, Livshits LA, Malaspina P, Maria S, McElreavey K, Meitinger TA, Mikelsaar AV, Mitchell RJ, Nafa K, Nicholson J, Norby S, Pandya A, Parik J, Patsalis PC, Pereira L, Peterlin B, Pielberg G, Prata MJ, Previdere C, Roewer L, Rootsi S, Rubinsztein DC, Saillard J, Santos FR, Stefanescu G, Sykes BC, Tolun A, Villems R, Tyler-Smith C, Jobling MA. Y-chromosomal diversity is clinal and influenced primarily by geography, rather than language. Am J Hum Genet. 2000;67:1526–1243. doi: 10.1086/316890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubicz R, Melton PE, Spitsyn V, Sun G, Deka R, Crawford MH. Genetic structure of native circumpolar populations based on autosomal, mitochondrial, and Y chromosome DNA markers. Am J Phys Anthropol. 2010;143:62–74. doi: 10.1002/ajpa.21290. [DOI] [PubMed] [Google Scholar]
- Rubicz R, Schurr TG, Babb P, Crawford MH. Mitochondrial DNA diversity in modern Aleuts, and their genetic relationship with other circumarctic populations. Hum Biol. 2003;75:809–835. doi: 10.1353/hub.2004.0009. [DOI] [PubMed] [Google Scholar]
- Ruhlen M. Na-Dene etymologies. In: Ruhlen M. On the Origin of Languages: Studies in Linguistic Taxonomy. Stanford University Press; Stanford: 1994. pp. 93–110. [Google Scholar]
- Ruhlen M. The origin of the Na-Dene. Proc Natl Acad Sci USA. 1998;95:13994–13996. doi: 10.1073/pnas.95.23.13994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Pesini E, Lott MT, Procaccio V, Poole JC, Brandon MC, Mishmar D, Yi C, Kreuziger J, Baldi P, Wallace DC. An enhanced MITOMAP with a global mtDNA mutational phylogeny. Nucleic Acids Res. 2007;35:D823–D828. doi: 10.1093/nar/gkl927. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saillard J, Forster P, Lynnerup N, Bandelt H-J, Nørby S. MtDNA variation among Greenland Eskimos: the edge of the Beringian expansion. Am J Hum Genet. 2000;67:718–726. doi: 10.1086/303038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapir E. The Na-Dene languages: a preliminary report. Am Anthropol. 1915;17:534–558. [Google Scholar]
- Schurr TG, Sukernik RI, Starikovskaya EB, Wallace DC. Mitochondrial DNA diversity in Koryaks and Itel’men: population replacement in the Okhotsk Sea-Bering Sea region during the Neolithic. Am J Phys Anthropol. 1999;108:1–40. doi: 10.1002/(SICI)1096-8644(199901)108:1<1::AID-AJPA1>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Schurr TG, Wallace DC. Genetic prehistory of Paleoasiatic-speaking peoples of northeastern Siberia and their links to Native American populations. In: Kendall L, Krupnik I, editors. Constructing cultures then and now: celebrating Franz Boas and the Jesup North Pacific Expedition. Smithsonian Institution Press; Baltimore: 2003. pp. 239–258. [Google Scholar]
- Schurr TG. The peopling of the New World: perspectives from molecular anthropology. Ann Rev Anthropol. 2004;33:551–583. [Google Scholar]
- Sengupta S, Zhivotovsky LA, King R, Mehdi SQ, Edmonds CA, Chow CE, Lin AA, Mitra M, Sil SK, Ramesh A, et al. Polarity and temporality of high-resolution Y-chromosome distributions in India identify both indigenous and exogenous expansions and reveal minor genetic influence of Central Asian pastoralists. Am J Hum Genet. 2006;78:202–221. doi: 10.1086/499411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Dong YL, Wen B, Xiao CJ, Underhill PA, Shen P, Chakraborty R, Jin L, Su B. Y-chromosome evidence of southern origin of the East Asian-specific haplogroup O3-M122. Am J Hum Genet. 2005;77:408–419. doi: 10.1086/444436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields G, Schmiechen A, Frazier B, Redd A, Voevoda MI, Reed JK, Ward RH. MtDNA sequences suggest a recent evolutionary divergence for Beringian and northern North American populations. Am J Hum Genet. 1993;53:549–562. [PMC free article] [PubMed] [Google Scholar]
- Starikovskaya YB, Sukernik RI, Schurr TG, Wallace DC. Mitochondrial DNA diversity in Chukchi and Siberian Eskimos: implications for the genetic prehistory of ancient Beringia. Am J Hum Genet. 1998;63:1473–1491. doi: 10.1086/302087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanton JR. Contributions to the ethnology of the Haida. Publications of the Jesup North Pacific Expedition 5(1); American Museum of Natural History Memoirs. G.E. Stechert; New York: 1905. [Google Scholar]
- Swanton JR. 26th Annual Report of the Bureau of American Ethnology. 1908. Social conditions, beliefs, and linguistic relationship of the Tlingit Indians; pp. 391–485. [Google Scholar]
- Tamm E, Kivisild T, Reidla M, Smith DG, Mulligan CJ, Bravi CM, Rickards O, Martinez-Labarga C, Khusnutdinova EK, Fedorova SA, Golubenko MV, Stepanov VA, Gubina MA, Zhadanov SI, Osipova LP, Damba L, Voevoda MI, Dipierri JE, Villems R, Malhi RS. Beringian standstill and spread of Native American founders. PLoS One. 2007;2:e829. doi: 10.1371/journal.pone.0000829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- Tikhmenev PA. In: A history of the Russian-American Company. Pierce RA, Donnelly MS, editors. University of Washington; Seattle: 1978. [Google Scholar]
- Torroni A, Schurr TG, Cabell MF, Brown MD, Neel JV, Larsen M, Smith DG, Vullo C, Wallace DC. Asian affinities and continental radiation of the four founding Native American mitochondrial DNAs. Am J Hum Genet. 1993;53:563–590. [PMC free article] [PubMed] [Google Scholar]
- van Oven M, Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum Mut. 2009;29:E386–E394. doi: 10.1002/humu.20921. [DOI] [PubMed] [Google Scholar]
- Vilar MG, Lum JK, Melendez C, Owings A, Gaieski JB, Schurr TG. Analysis of mtDNA haplogroup A2 frequency, distribution and diversity among populations from North and Central America and the Caribbean. Am J Phys Anthropol Suppl. 2011;144(Suppl 52):300. [Google Scholar]
- Ward RH, Frazier BL, Dew-Jager K, Pääbo S. Extensive mitochondrial diversity within a single Amerindian tribe. Proc Natl Acad Sci USA. 1991;88:8720–8724. doi: 10.1073/pnas.88.19.8720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward RH, Redd A, Valencia D, Frazier B, Pääbo S. Genetic and linguistic differentiation in the Americas. Proc Natl Acad Sci USA. 1993;90:10663–10667. doi: 10.1073/pnas.90.22.10663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilber G. The Sitka story: crown jewel of Baranof Island. Alaska Publications; Sitka: 1993. [Google Scholar]
- Xue Y, Zerjal T, Bao W, Zhu S, Shu Q, Xu J, Du R, Fu S, Li P, Hurles ME, Yang H, Tyler-Smith C. Male demography in East Asia: a north-south contrast in human population expansion times. Genetics. 2006;172:2431–2439. doi: 10.1534/genetics.105.054270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Y Chromosome Consortium A nomenclature system for the tree of human Y-chromosomal binary haplogroups. Genome Res. 2002;12:339–348. doi: 10.1101/gr.217602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegura SL, Karafet TM, Zhivotovsky LA, Hammer MF. High-resolution NSPs and microsatellite haplotypes point to a single, recent entry of Native American Y chromosomes into the Americas. Mol Biol Evol. 2004;21:164–175. doi: 10.1093/molbev/msh009. [DOI] [PubMed] [Google Scholar]
- Zhadanov SI, Dulik MC, Markley M, Jennings G, Gaieski JB, Elias G, Schurr TG, The Genographic Consortium Genetic heritage and native identity of the Seaconke Wampanoag tribe of Massachusetts. Am J Phys Anthropol. 2010;142:579–589. doi: 10.1002/ajpa.21281. [DOI] [PubMed] [Google Scholar]
- Zhong H, Shi H, Qi X, Duan Z, Tan P, Jin L, Su B, Ma RZ. Extended Y chromosome investigation suggests postglacial migrations of modern humans into East Asia via the northern route. Mol Biol Evol. 2011;28:717–727. doi: 10.1093/molbev/msq247. [DOI] [PubMed] [Google Scholar]
- Zlojutro M, Rubicz R, Devor EJ, Spitsyn VA, Makarov SV, Wilson K, Crawford MH. Genetic structure of the Aleuts and circumpolar populations based on mitochondrial DNA sequences: a synthesis. Am J Phys Anthropol. 2006;129:446–464. doi: 10.1002/ajpa.20287. [DOI] [PubMed] [Google Scholar]
- Zlojutro M, University of Kansas . Master’s Thesis. Lawrence: 2008. Mitochondrial DNA and Y-chromosome variation of eastern Aleut populations: implications for the genetic structure and peopling of the Aleutian Archipelago. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.