Abstract
We have determined the DNA sequence of Klebsiella pneumoniae multidrug resistance plasmid pKPI-6, which is a self-transmissible IncN-type plasmid. pKPI-6 harboring blaIMP-6 and blaCTX-M-2 confers a stealth-type carbapenem resistance phenotype on members of the family Enterobacteriaceae that is not detectable with imipenem. pKPI-6 is already epidemic in Japan, favoring the dissemination of IMP-6 and CTX-M-2 in members of the family Enterobacteriaceae.
TEXT
Although still uncommon in Japan, carbapenem-resistant members of the family Enterobacteriaceae (CRE) have recently emerged and rapidly disseminated in the world (1). Stealth-type phenotypes, because of inability of automated systems to detect them, sometimes gave misleading results that caused a failure to stop the spread of CRE, e.g., Klebsiella pneumoniae carbapenemase-producing bacteria in Europe and the United States (2).
In 2011, five isolates of extended-spectrum β-lactamase (ESBL)-positive K. pneumoniae showed a paradoxical resistance phenotype, being highly resistant to almost all of the β-lactam antibiotics except imipenem (3). We therefore designated these isolates ISMRK (imipenem-susceptible but meropenem-resistant Klebsiella) (3). ISMRK bacteria carry the metallo-β-lactamase (MBL) gene blaIMP-6 in a cassette of a class 1 integron, In722, and the ESBL gene blaCTX-M-2; both are on a ca. 47-kb self-transmissible plasmid designated pKPI-6. The combination of IMP-6 (3, 4) and CTX-M-2 gives ISMRK isolates a stealth phenotype not detectable with imipenem. We also alarmingly reported possible false diagnosis of ISMRK by clinical laboratories because not only imipenem but meropenem resistance of ISMRK cannot be screened for with the Vitek and RAISUS rapid automated susceptibility test systems (5). This is a more serious threat, leading to inappropriate antimicrobial therapy and a failure to stop the spread of ISMRK.
ISMRK strain a26 (3), isolated from a urine sample at the Hiroshima University Hospital, was used as the source of the pKPI-6 plasmid sequenced.
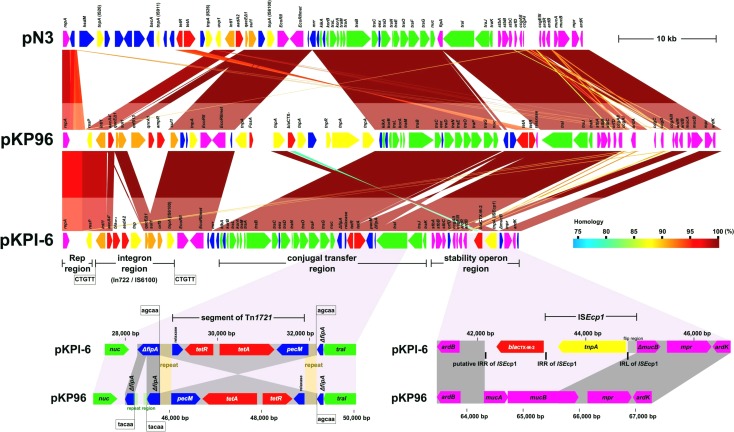
Whole-genome sequencing of pKPI-6 showed that the plasmid has a circularly closed 47,236-bp DNA sequence with an average G+C content of 50.99%, which is lower than that of the K. pneumoniae chromosome, which is about 57.7%, the highest G+C content of any species in the family Enterobacteriaceae (6). The numbering of pKPI-6 commences at the first nucleotide of the ATG start codon of repA (see Table S1 in the supplemental material). Annotation of the finished sequence data shows that pKPI-6 contains 53 putative genes, 4 of which encode proteins with no homology to known genes. Among the other 49 genes with known functions, the following 5 confer resistance to antibiotics: aacA4′ (a derivative of aacA4) (7), aadA2, tetR-tetA, blaIMP-6, and blaCTX-M-2. pKPI-6 possessed a typical incompatibility group N (IncN) plasmid backbone (Fig. 1; see Table S1 in the supplemental material). IncN plasmids are important vehicles of clinically relevant resistance genes, including MBL and ESBL genes (8). There are three insertions of acquired extra DNAs containing resistance genes, an integron region including blaIMP-6, a segment of Tn1721 including tetA and tetR, and an ISEcp1 region including blaCTX-M-2 (Fig. 1). Sequence analysis showed that the backbone gene organization of pKPI-6 is highly homologous to that of multidrug resistance-encoding IncN plasmids pKP96 (9) (accession no. EU195449; coverage, 100%; identity, 99%) and pN3 (accession no. FR850039; coverage, 97%; identity, 99%) (Fig. 1). Multilocus sequence typing (MLST) (8) shows that pKPI-6, pKP96, and pN3 belong to sequence type 5 (ST5). The class 1 integron of pKPI-6, designated In722 (10) (Fig. 2), and IS6100 are located at the same position on pKP96, suggesting that the class 1 integron was inserted by IS6100, creating a duplication of a 5-bp direct repeat (CTGTT) (Fig. 1). The fourth gene cassette of In722 contains a gene encoding a potential transposase related to the IS91 family of transposases (11). This transposase was identified as ISKpn22 by ISfinder (http://www-is.biotoul.fr//).
FIG 1.
Structural comparison of pKPI-6, pKP96, and pN3. The nucleotide sequence of pKPI-6 plasmid DNA purified from a transconjugant was determined by the random shotgun sequencing method as described previously (15). Collected sequences were assembled by using Phrap software (version 1.080730) (16). Gaps were closed by direct sequencing of PCR products amplified with oligonucleotide primers designed to anneal each end to the neighboring contigs. Sequence reads were processed by using MetaGeneAnnotator (17), the InSilico molecular cloning software package, genomics edition (InSilico Biology Inc., Yokohama, Japan), the program BLASTP (18), and GenomeMatcher software (19). Color shadings between plasmids indicate homologous regions. Sequences of the terminal direct repeats of the acquired region are shown. ORFs are represented by pentagons, and annotated genes are colored on the basis of predicted gene function as follows: antimicrobial resistance genes, red; conjugation genes, green; transposons, yellow; integrons, orange; plasmid maintenance genes, violet; hypothetical genes, blue. Two regions, segments of Tn1721 and blaCTX-M-2-ISEcp1, are visually extended at the bottom to show the difference between pKPI-6 and pKP96. Boxed nucleotide sequences represent direct duplications. Comparative analysis and generation of the image shown were performed with GenomeMatcher software (19). IRL, left inverted repeat; IRR, right inverted repeat.
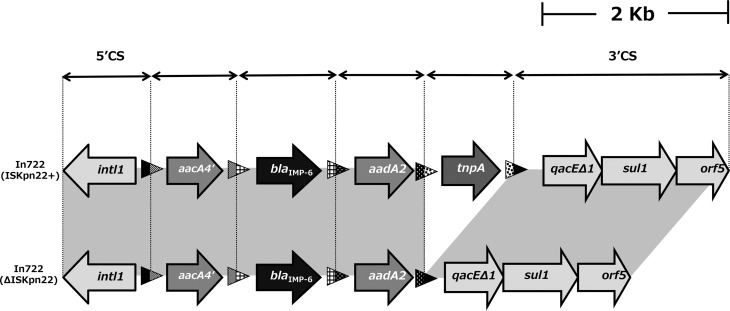
FIG 2.
Schematic representation of the genetic content of the blaIMP-6 integron (In722) lacking ISKpn22. The attI1 and attC elements are shown as triangles. 5′CS and 3′CS represent the 5′ and 3′ conserved segments, respectively.
The gene array of the conjugal transfer system of pKPI-6 and pKP96 was conserved, except for the orientation of a segment of Tn1721 including the tetA and tetR genes (GenBank accession no. X61367) that was inserted between nuc and traI (Fig. 1). Comparison of the pN3 DNA sequence to those of pKPI-6 and pKP96 shows that the 3′ segment of Tn1721 juxtaposed with two 245-bp short stretches of DNA in the opposite orientation is inserted into fipA by homologous recombination at TTGCT, resulting in direct repeat AGCAA at both ends (Fig. 1, lower left). Inversion of the segment between pKPI-6 and pKP96 may be due to the inverted repeat of the 245-bp short stretches. Another resistance gene, blaCTX-M-2, was mobilized with an ISEcp1 mobile element that appears to interrupt the mucA and mucB genes in the backbone structure of the IncN plasmid. There were 14-bp inverted repeats of ISEcp1 present at both ends where a 5′-flanking short stretch containing the left inverted repeat showed inversion in ISEcp1 (gray in Fig. 1).
A retrospective search for members of the family Enterobacteriaceae showing the ISMR phenotype in the Kinki region (the geographic center of Japan) identified 13 isolates of K. pneumoniae, 1 of K. oxytoca, and 6 of Escherichia coli; all 20 were positive for blaIMP-6 and blaCTX-M-2 (see Table S3 in the supplemental material). These strains were recognized primarily as ESBL producers but not as MBL producers because of their susceptibility to imipenem delineated by the automated susceptibility testing system.
Genomic profiling by pulsed-field gel electrophoresis performed as described elsewhere (12) and subsequent clustering analysis of K. pneumoniae carrying blaIMP-6 and blaCTX-M-2 identified a cluster with ≥80% similarity having eight isolates in the Kinki region and the representative K. pneumoniae isolate in Hiroshima, a26 (3). The MICs of a variety of antimicrobial agents clearly suggest that they have similar susceptibility profiles (shaded in Table S3 in the supplemental material). In addition, MLST (13) indicated that these isolates all belong to ST37.
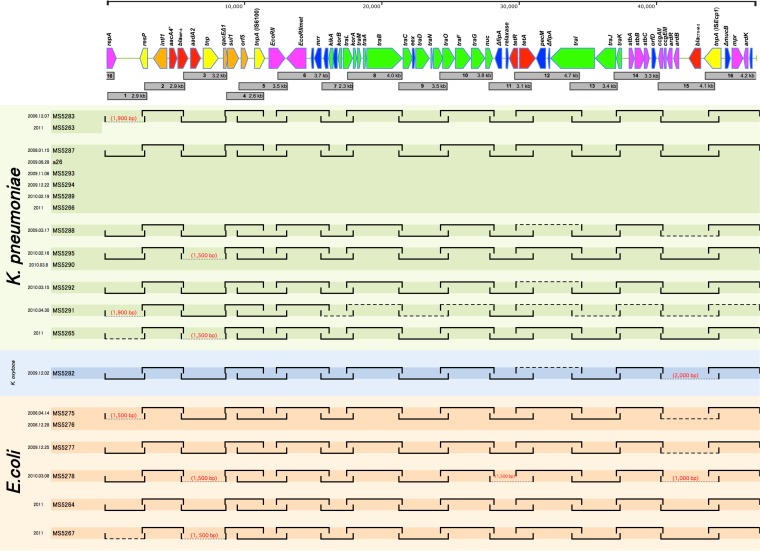
We examined the genome organization of the blaIMP-6- and blaCTX-M-2-bearing plasmids from the 20 isolates of K. pneumoniae, K. oxytoca, and E. coli by the PCR scanning method (12). We generated 16 pairs of primers whose PCR products cover the entire 47.2-kb plasmid (Fig. 3; see Table S2 in the supplemental material). All of the primer pairs yielded PCR products of the expected sizes for seven isolates, including K. pneumoniae a26. In the other plasmids, some differences in fragment size were noted (one or two segments in each plasmid). Size variations were observed in limited regions, mainly regions 1, 3, and 15 shown in Fig. 3. The size of amplified region 1 was remarkably diverse, ranging from 1.5 to ∼2.9 kb. This heterogeneity is due to the size variation in direct repeats called iteron sequences common in the replicative region of Gram-negative plasmids (14). Region 3 displayed two patterns, 3.2 and 1.5 kb. Sequencing analysis of the 1.5-kb DNA fragment showed that not only ISKpn22 but the entire gene cassette containing ISKpn22 was absent from the integron structure compared with the DNA sequence of In722; and this integron is designated In722ΔISKpn22 (Fig. 2).
FIG 3.
Summary of the PCR scanning analysis of IMP-6-positive plasmids from 13 K. pneumoniae, 1 K. oxytoca, and 6 E. coli clinical strains from the Kinki region. The plasmid structures in clinical isolates were examined by PCR scanning with 16 sets of primers (numbered 1 to 16; see Table S2 in the supplemental material) designed to cover all of plasmid pKPI-6. PCR amplification for the scan of pKPI-6 was performed with Quick Taq HS DyeMix (Toyobo, Tokyo, Japan) with 30 cycles of denaturing at 96°C for 20 s, annealing at 50°C for 30 s, and polymerization at 68°C for 5 min. By comparing the length of each amplified fragment to pKPI-6, the regional heterogeneity was determined. The gene arrangement of pKPI-6 is presented at the top. PCR products covering the entire plasmid sequence are depicted as numbered gray squares. The numbers represent the primer sets used for PCR scanning (see Table S2 in the supplemental material). Solid lines represent the regions amplified. Where the length of the amplified fragment is different from that of pKPI-6, the estimated size of the fragment is shown in parentheses. If no PCR product was obtained when a prolonged extension time was used, no estimated size is shown.
The In722 and In722ΔISKpn22 integrons both have a 6-bp insertion (CAGCAG) in the 5′ conserved segment that is not found in any other available class 1 integron sequence. This suggests that the two integrons are closely related and ISKpn22 is embedded within an integron gene cassette that can be excised through Intl-mediated recombination, which probably occurred in these strains, resulting in a shortened region 3. In722ΔISKpn22 was found in three isolates of K. pneumoniae and two isolates of E. coli from the Kinki region (Fig. 3). In Hiroshima, we reported five ISMRK isolates from four different institutions, all of which carried In722 but not In722ΔISKpn22. Some of the K. pneumoniae (two) and E. coli (three) isolates lack PCR products in region 15, which contains blaCTX-M-2. PCR amplification with a 5′ primer designed for the blaCTX-M-2 open reading frame (ORF) and a 3′ primer designed for tnpA yielded products of the expected size, suggesting that blaCTX-M-2 is present next to tnpA in these plasmids.
The identification of pKPI-6 in clinical isolates of K. oxytoca and E. coli from the Kinki region strongly suggests that pKPI-6 has spread across species in at least two regions, but the unique phenotype suggests that it may have already spread further without being recognized in Japan.
Nucleotide sequence accession number.
An annotated sequence of pKPI-6 has been submitted to the GenBank nucleotide sequence database under accession number AB616660.
Supplementary Material
ACKNOWLEDGMENT
We thank Jim Nelson for editorial assistance.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.04759-14.
REFERENCES
- 1.Gupta N, Limbago BM, Patel JB, Kallen AJ. 2011. Carbapenem-resistant Enterobacteriaceae: epidemiology and prevention. Clin Infect Dis 53:60–67. doi: 10.1093/cid/cir202. [DOI] [PubMed] [Google Scholar]
- 2.McKenna M. 2013. Antibiotic resistance: the last resort. Nature 499:394–396. doi: 10.1038/499394a. [DOI] [PubMed] [Google Scholar]
- 3.Shigemoto N, Kuwahara R, Kayama S, Shimizu W, Onodera M, Yokozaki M, Hisatsune J, Kato F, Ohge H, Sugai M. 2012. Emergence in Japan of an imipenem-susceptible, meropenem-resistant Klebsiella pneumoniae carrying blaIMP-6. Diagn Microbiol Infect Dis 72:109–112. doi: 10.1016/j.diagmicrobio.2011.09.019. [DOI] [PubMed] [Google Scholar]
- 4.Yano H, Kuga A, Okamoto R, Kitasato H, Kobayashi T, Inoue M. 2001. Plasmid-encoded metallo-β-lactamase (IMP-6) conferring resistance to carbapenems, especially meropenem. Antimicrob Agents Chemother 45:1343–1348. doi: 10.1128/AAC.45.5.1343-1348.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harino T, Kayama S, Kuwahara R, Kashiyama S, Shigemoto N, Onodera M, Yokozaki M, Ohge H, Sugai M. 2013. Meropenem resistance in imipenem-susceptible meropenem-resistant Klebsiella pneumoniae isolates not detected by rapid automated testing systems. J Clin Microbiol 51:2735–2738. doi: 10.1128/JCM.02649-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu KM, Li LH, Yan JJ, Tsao N, Liao TL, Tsai HC, Fung CP, Chen HJ, Liu YM, Wang JT, Fang CT, Chang SC, Shu HY, Liu TT, Chen YT, Shiau YR, Lauderdale TL, Su IJ, Kirby R, Tsai SF. 2009. Genome sequencing and comparative analysis of Klebsiella pneumoniae NTUH-K2044, a strain causing liver abscess and meningitis. J Bacteriol 191:4492–4501. doi: 10.1128/JB.00315-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lambert T, Ploy MC, Courvalin P. 1994. A spontaneous point mutation in the aac(6′)-Ib′ gene results in altered substrate specificity of aminoglycoside 6′-N-acetyltransferase of a Pseudomonas fluorescens strain. FEMS Microbiol Lett 115:297–304. [DOI] [PubMed] [Google Scholar]
- 8.García-Fernández A, Villa L, Moodley A, Hasman H, Miriagou V, Guardabassi L, Carattoli A. 2011. Multilocus sequence typing of IncN plasmids. J Antimicrob Chemother 66:1987–1991. doi: 10.1093/jac/dkr225. [DOI] [PubMed] [Google Scholar]
- 9.Shen P, Jiang Y, Zhou Z, Zhang J, Yu Y, Li L. 2008. Complete nucleotide sequence of pKP96, a 67,850 bp multiresistance plasmid encoding qnrA1, aac(6′)-Ib-cr and blaCTX-M-24 from Klebsiella pneumoniae. J Antimicrob Chemother 62:1252–1256. doi: 10.1093/jac/dkn397. [DOI] [PubMed] [Google Scholar]
- 10.Moura A, Soares M, Pereira C, Leitao N, Henriques I, Correia A. 2009. INTEGRALL: a database and search engine for integrons, integrases and gene cassettes. Bioinformatics 25:1096–1098. doi: 10.1093/bioinformatics/btp105. [DOI] [PubMed] [Google Scholar]
- 11.Mendiola MV, Jubete Y, de la Cruz F. 1992. DNA sequence of IS91 and identification of the transposase gene. J Bacteriol 174:1345–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kouda S, Ohara M, Onodera M, Fujiue Y, Sasaki M, Kohara T, Kashiyama S, Hayashida S, Harino T, Tsuji T, Itaha H, Gotoh N, Matsubara A, Usui T, Sugai M. 2009. Increased prevalence and clonal dissemination of multidrug-resistant Pseudomonas aeruginosa with the blaIMP-1 gene cassette in Hiroshima. J Antimicrob Chemother 64:46–51. doi: 10.1093/jac/dkp142. [DOI] [PubMed] [Google Scholar]
- 13.Diancourt L, Passet V, Verhoef J, Grimont PAD, Brisse S. 2005. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol 43:4178–4182. doi: 10.1128/JCM.43.8.4178-4182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chattoraj DK. 2000. Control of plasmid DNA replication by iterons: no longer paradoxical. Mol Microbiol 37:467–476. doi: 10.1046/j.1365-2958.2000.01986.x. [DOI] [PubMed] [Google Scholar]
- 15.Yamaguchi T, Hayashi T, Takami H, Nakasone K, Ohnishi M, Nakayama K, Yamada S, Komatsuzawa H, Sugai M. 2000. Phage conversion of exfoliative toxin A production in Staphylococcus aureus. Mol Microbiol 38:694–705. doi: 10.1046/j.1365-2958.2000.02169.x. [DOI] [PubMed] [Google Scholar]
- 16.Ewing B, Hillier LD, Wendl MC, Green P. 1998. Base-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res 8:175–185. [DOI] [PubMed] [Google Scholar]
- 17.Noguchi H, Taniguchi T, Itoh T. 2008. MetaGeneAnnotator: detecting species-specific patterns of ribosomal binding site for precise gene prediction in anonymous prokaryotic and phage genomes. DNA Res 15:387–396. doi: 10.1093/dnares/dsn027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ohtsubo Y, Ikeda-Ohtsubo W, Nagata Y, Tsuda M. 2008. GenomeMatcher: a graphical user interface for DNA sequence comparison. BMC Bioinformatics 9:376. doi: 10.1186/1471-2105-9-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.