Abstract
Cytochrome c proteins, as enzymes to exchange electrons with substrates or as pure electron carriers to shuttle electrons, play vital roles in bacterial respiration and photosynthesis. In Shewanella oneidensis, a research model for the respiratory diversity, at least 42 c-type cytochromes are predicted to be encoded in the genome and are regarded to be the foundation of its highly branched electron transport pathways. However, only a small number of c-type cytochromes have been extensively studied. In this study, we identify soluble cytochrome c ScyA as an important factor influencing the nitrite resistance of a strain devoid of the bd oxidase by utilizing a newly developed transposon mutagenesis vector, which enables overexpression of the gene(s) downstream of the insertion site. We show that when in overabundance ScyA facilitates growth against nitrite inhibition by enhancing nitrite resistance of the cbb3 oxidase. Based on the data presented in this study, we suggest two possible mechanisms underlying the observed effect of ScyA: (1) ScyA increases electron flow to the cbb3 oxidase; (2) ScyA promotes nitrite resistance of the cbb3 oxidase, possibly by direct interaction.
Keywords: Cytochrome c, nitrite resistance, Shewanella, the cytochrome cbb3 oxidase
Introduction
Shewanella oneidensis, a facultative Gram-negative anaerobe, is renowned for its remarkable respiration versatility (Fredrickson et al. 2008). Linked to this unique characteristic is a high content of c-type cytochromes (Heidelberg et al. 2002; Meyer et al. 2004; Romine et al. 2008; Gao et al. 2010a), which are characterized by the covalent attachment of the prosthetic cofactor (the heme group) to the protein polypeptide at the cysteines within the signature heme c binding motif (HBM) CX2CH although exceptions exist (Kranz et al. 2009). These proteins, existing as membrane-bound or soluble in the periplasm, play vital roles in bacterial respiration as enzymes to exchange electrons with the bound substrates or as pure electron carriers to shuttle electrons.
Many of the functionally defined S. oneidensis c-type cytochromes are terminal reductases as their corresponding deletion mutants display distinguishable phenotypes, such as NrfA (nitrite reductase), NapB (small subunit of nitrate reductase), FccA (fumarate reductase), and DmsE (subunit of dimethyl sulfoxide [DMSO] reductase), to name a few (Tsapin et al. 2001; Gralnick et al. 2006; Gao et al. 2009). Furthermore, those involved in the respiration of insoluble electron acceptors (EAs), such as metal oxide, are relatively better understood because the subject has been under intensive investigation in the microorganism since its isolation (Fredrickson et al. 2008; Richardson et al. 2012; Richter et al. 2012). Nevertheless, a large fraction of the entire c-type cytochrome pool remains poorly defined since the loss of many of these c-type cytochromes individually does not result in distinct phenotypes (Bretschger et al. 2007; Gao et al. 2010a; Jin et al. 2013). Multiple mechanisms may underlie this phenomenon. First, a number of c-type cytochromes are not produced to a level of physiological relevance under the experimental conditions revealed in both transcriptomic and proteomic analyses, of which the cytochrome caa3 oxidase serves as a good example (Nissen et al. 2012; Jin et al. 2013; Zhou et al. 2013; Le Laz et al. 2014). Second, there exists the promiscuity of c-type cytochromes given that a large number of proteins sharing similar features coexist in the proteome, especially in the case of small periplasmic c-type cytochromes (Fonseca et al. 2013; Fu et al. 2014). Moreover, certain terminal reductases may be able to reduce multiple substrates, functionally overlapping substrate-specific reductases. For instance, in vitro analyses reveal that Otr, octaheme tetrathionate reductase (SO4144), is able to reduce nitrite and hydroxylamine and NrfA can carry out the reduction of sulfite to sulfide, although in both cases whether these substrates are the real physiological substrates is yet determined (Atkinson et al. 2007; Lukat et al. 2008). The situation is further complicated by the finding that c-type cytochromes can be functionally replaced by noncytochrome proteins (Cordova et al. 2011). In this case, an NrfD/PsrC-like protein (SirD), in association with an iron–sulfur redox partner (SirC), is in large part able to substitute CymA, a c-type cytochrome that couples to multiple terminal reductases and periplasmic electron transfer proteins (Marritt et al. 2012).
It is well known that nitrite is toxic to bacterial cells as it has been used as preservative in meat products for centuries. Although nitrite (≤2 mmol/L) can be used as sole EA to support growth of S. oneidensis, it imposes a greatly negative impact on viability and growth at higher concentrations (Gao et al. 2009; Dong et al. 2012; Fu et al. 2013; Zhang et al. 2013). The S. oneidensis genome encodes three terminal oxidases: a bd-type quinol oxidase encoding by the cydABX genes and two heme–copper oxidases (HCOs), a caa3-type and a cbb3-type encoded by the SO4606-9 and ccoNOQP genes, respectively (Heidelberg et al. 2002; Buschmann et al. 2010; Chen et al. 2014). According to their predicted affinity for molecular oxygen, the caa3-HCO and cbb3-HCO should operate under aerobic and microaerobic conditions, respectively. Interestingly, the cbb3-HCO is abundant in membranes under both aerobic and microaerobic conditions, whereas the caa3-HCO is not present (Zhou et al. 2013; Le Laz et al. 2014). The bd oxidase, like its Escherichia coli counterpart, plays a role in oxygen respiration under microaerobic but not aerobic conditions, and probably more importantly, confers nitrite and nitric oxide (NO) resistance to S. oneidensis (Borisov et al. 2011; Fu et al. 2013; Zhang et al. 2013; Chen et al. 2014).
Recently, we identified the cytochrome bc1 complex as a functional replacement of CymA in respiration of nitrate and nitrite by utilizing a transposon-based screening (Fu et al. 2014). Given that a large number of proteins remain functionally undefined, we reason that proteins capable of enhancing nitrite resistance, in absence or excess, may be present in S. oneidensis. In this study, we tested this hypothesis by screening the genome for genes encoding such proteins with a newly developed transposon vector, which contains a strong promoter embedded within the transposable segment. We found that overproduction of ScyA, a periplasmic cytochrome c, elevated nitrite resistance of a mutant lacking the bd oxidase. Subsequent analysis revealed that this effect was associated with the cbb3-HCO. We further showed that ScyA in excess did not affect the abundance of the cbb3-HCO. Instead, the protein when overproduced enhanced the resistance of the cbb3-HCO to nitrite either by increasing electron flow to the enzyme or by modulating biochemical properties of the enzyme via direct contact.
Experimental Procedures
Bacterial strains, plasmids, polymerase chain reaction primers, and culture conditions
Bacterial strains and plasmids used in this study are listed in Table1. Sequences of the primers used for generating polymerase chain reaction (PCR) products are available upon request. Chemicals were acquired from Sigma unless otherwise noted. For genetic manipulation purpose, E. coli and S. oneidensis strains were grown in Luria–Bertani (LB; Difco, Beijing, China) medium at 37°C and 30°C, respectively. When needed, growth medium was supplemented with chemicals at the following concentrations: 2,6-diaminopimelic acid, 0.3 mmol/L; ampicillin, 50 μg/mL; kanamycin, 50 μg/mL; and gentamycin, 15 μg/mL.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Escherichia coli strains | ||
| DH5α | Host strain for cloning | Lab stock |
| WM3064 | Donor strain for conjugation; ΔdapA | W. Metcalf, UIUC |
| Shewanella oneidensis strains | ||
| MR-1 | Wild-type | ATCC 700550 |
| HG0264 | ΔscyA derived from MR-1 | Jin et al. (2013) |
| HG0608-10 | Δpet derived from MR-1 | Fu et al. (2014) |
| HG0624 | Δcrp derived from MR-1 | Gao et al. (2010b) |
| HG2364-1 | Δcco derived from MR-1 | This study |
| HG3286-4 | Δcyd derived from MR-1 | This study |
| HG3980 | ΔnrfA derived from MR-1 | Gao et al. (2009) |
| HG3896 | ΔompS38 derived from MR-1 | This study |
| HG4591 | ΔcymA derived from MR-1 | Gao et al. (2009) |
| HGCCO-CYD | ΔccoΔcyd derived from MR-1 | This study |
| HGSCYA-CYD | ΔscyAΔcyd derived from MR-1 | This study |
| HGCYD-CYMA | ΔcydΔcymA derived from MR-1 | This study |
| HGCYD-PET | ΔcydΔpet derived from MR-1 | This study |
| HGCYD-NRFA | ΔcydΔnrfA derived from MR-1 | This study |
| Plasmids | ||
| pHGM01 | ApR, GmR, CMR, suicide vector | Jin et al. (2013) |
| pHG101 | Promoterless broad-host KmR vector | Wu et al. (2011) |
| pHG102 | pHG101 containing the S. oneidensis arcA promoter | Wu et al. (2011) |
| pFAC | Mariner-based transposon vector | Wong and Mekalanos (2000) |
| pHGC01 | Integrative vector for complementation | Fu et al. (2013) |
| pTP327 | Multicopy E. coli lacZ reporter vector | Gao et al. (2010b) |
| pHGEI01 | Integrative E. coli lacZ reporter vector | Fu et al. (2014) |
| pHGE-Ptac | Broad-host IPTG-inducible expression vector | Luo et al. (2013) |
| pHGT01 | Promoter-embedded Mariner-based transposon vector | This study |
| pHGE-ScyA-His6 | His-tagged ScyA expression vector | This study |
IPTG, isopropyl β-d-1-thiogalactopyranoside.
M1 defined medium containing 0.02% (w/v) of vitamin-free casamino acids and 15 mmol/L lactate as electron donor was used to evaluate impacts of mutations of interest on aerobic growth as described before (Gao et al. 2008). Growth of the deletion strains was determined by recording the optical density of cultures at 600 nm (OD600) with the wild type as the control. Fresh media were inoculated with exponential phase cultures to ∼0.01 of OD600 and shaken at 200 rpm 30°C. For nitrite sensitivity purpose, LB medium was used.
In-frame deletion mutagenesis and complementation
In-frame deletion strains were constructed according to the att-based Fusion PCR method described previously (Jin et al. 2013). In brief, two fragments flanking the targeted gene were amplified with primers containing attB and the gene-specific sequence, and then joined by a second round of PCR. The resulting fusion fragment was introduced into plasmid pHGM01 by site-specific recombination using the BP Clonase (Invitrogen, Shanghai, China) and maintained in E. coli WM3064. The resulting mutagenesis vector was then transferred from E. coli into S. oneidensis by conjugation. Integration of the mutagenesis construct into the chromosome was selected by gentamycin resistance and confirmed by PCR. Verified transconjugants were grown in LB broth in the absence of NaCl and plated on LB supplemented with 10% sucrose. Gentamycin-sensitive and sucrose-resistant colonies were screened by PCR for deletion of the targeted gene. The deletion mutations were then verified by sequencing.
Plasmids pHG101 and pHG102 were used in genetic complementation of mutants and overexpression of gene of interest (Wu et al. 2011). For complementation of genes adjacent to their promoters, a fragment containing the gene of interest and its native promoter was generated by PCR and cloned into pHG101. Otherwise, the gene of interest was amplified and inserted into MCS of pHG102 under the control of the S. oneidensis arcA promoter, which is constitutively active (Gao et al. 2010b). To single-copy complementation, pHGC01 was used as described previously (Fu et al. 2014). The resulting complementation vector was transferred into its relevant mutant strains via conjugation and its presence was confirmed by plasmid purification and restriction enzyme digestion.
Transposon mutagenesis
A random mutagenesis vector with an embedded robust promoter was developed in this study. We previously noticed that pFAC, a mariner-based random mutagenesis vector (Wong and Mekalanos 2000), was substantially lower in the transfer rate via conjugation from E. coli to S. oneidensis than pHGC01 (Fu et al. 2013). To increase the transfer efficiency of the new vector derived from pFAC, we amplified the R6K replicon and the mob gene with pHGC01 as the template and used the resulting fragment to replace the DNA sequence of pFAC beyond that for IS elements, gentamycin-resistant gene, and Himar1 transposase. Subsequently, we replaced Ptn, the promoter embedded in the transposable sequence of pFAC, with a promoter that is constitutively robust under most conditions so as to overexpress the gene after the insertion. An array of microarray data from our prior studies were examined and three genes from single gene operons with high signal intensities under all conditions were chosen for further validation (Gao et al. 2004, 2006, 2008, 2009; Jiang et al. 2014). These include SO2593, SO3896, and cymA (SO4591), encoding a hypothetical protein, an outer membrane porin (Maier and Myers 2004), and a well-studied tetraheme cytochrome c, respectively.
An analysis of sequences upstream of these three genes using Neural Network Promoter Prediction revealed that their predicted transcription starting sites of most confidence were within 350 bp relative to the translation initiation codon (Reese 2001). Consequently, for each gene a fragment covering 400 bp of sequence upstream of the coding region was amplified and cloned into a lacZ reporter system for the quantification of the promoter activity. As shown in Figure S2A, within multiple-copy pTP327 all of three promoters were much more robust than Ptn and ParcA, a constitutively active promoter for global regulator ArcA (Gao et al. 2010b). Among them, Pso3896 appeared to be particularly strong, approximately 400% over other two promoters. To further confirm this, we transferred these promoters to an integrative lacZ reporter system pHGEI01 such that their activities can be assessed in a single copy (Fu et al. 2014). Once transferred into S. oneidensis strains, pHGEI01 containing promoter of interest integrates into the chromosome and the antibiotic marker is then removed by an established approach (Fu et al. 2013). While these promoters in a single copy drove expression of β-galactosidase substantially less effectively compared to them in multiple copies, they were consistently more active than Ptn and ParcA (Fig. S2A). As Pso3896 is the strongest, we further examined the expression of the gene by analyzing the abundance of its product in the outer membrane. The insoluble membrane fractions from cells cultured in normal and high-osmolarity media were harvested and applied to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) for separation (Fig. S2B). SO3896, validated by the loss of the band in an SO3896 mutant, was apparently one of the major outer membrane proteins under test conditions, confirming that Pso3896 is robust and relatively constitutive. We, therefore, chose it for the subsequent construction. To drive transcription of genes immediately downstream of the insertion, Pso3896 was placed outwards within the transposable fragment using conventional cloning approaches, resulting in pHGT01 (Fig. S2C). By transferring pHGT01 into the Δcyd strain by conjugation, a random mutation library was constructed and nitrite resistance suppressor strains were selected with gentamycin and 3 mmol/L nitrite. According to the number of colonies on the control plates (Gm+Nitrite−), the transposon library was estimated to contain more than 100,000 individual insertion mutants. Colonies, obtained from Gm+Nitrite+ plates, were subjected to the mapping of the transposon insertion sites using the arbitrary PCR (Das et al. 2005).
SDS-PAGE analysis of outer membrane proteins
Envelope fractions were prepared according to an established method with modifications (Slauch and Silhavy 1989). Briefly, cultures of mid-log phase (∼0.4 of OD600) were normalized to the lowest OD to allow for comparison of outer membrane protein quantities across strain backgrounds. Normalized cultures were pelleted, washed once in 30 mmol/L Tris-HCl (pH 8.1), and pelleted again at 3800g. Cell pellets were then resuspended in 30 mmol/L Tris-HCl–20% sucrose buffer (500 μL), followed by the addition of 20 μL of 20-mg/mL lysozyme–0.1 mmol/L Ethylenediaminetetraacetic acid (EDTA) (pH 7.3), and incubated on ice for 30 min. Following lysozyme treatment, 1 mL of 3 mmol/L ETDA (pH 7.3) was added and the resulting extract was disrupted with a single 20-s pulse using a microtip sonicator (Haishu Instrument, Ningbo, China). A 1.5-mL fraction of the extract was then centrifuged at 16,000g for 60 min. Envelope fractions were collected as centrifuged precipitate and resuspended in 30 μL of Laemmli SDS sample buffer. After boiled for 5 min, 15-μL samples were subjected to SDS-PAGE (10% acrylamide, 6 mol/L urea, 1% SDS).
Expression and purification of S. oneidensis ScyA
An isopropyl β-d-1-thiogalactopyranoside (IPTG) inducible vector, pHGE-Ptac, was used for expressing recombinant ScyA with 6x His-tag fused to its C-terminus in S. oneidensis (Luo et al. 2013). Cells were grown to late-exponential phase (∼0.8 of OD600), induced with 0.5 mmol/L IPTG for 2 h, collected by centrifugation, resuspended in lysis buffer (50 mmol/L Tris/HCl, pH 7.5, 200 mmol/L NaCl, 1 mmol/L MgCl2, 10 mmol/L β-mercaptoethanol, 1 mmol/L phenylmethanesulfonylfluoride (PMSF), 5 mg/mL DNase I), and broken by passage twice through a French press (10,000 psi). The soluble recombinant ScyA protein was purified using a talon resin column (BD Biosciences, Beijing, China) according to the manufacturer's instructions. The protein concentration in the collected samples here, and all other samples mentioned in this study, was determined using a Bradford assay with bovine serum albumin (BSA) as a standard (Bio-Rad, Shanghai, China). Sample purity was checked by SDS-PAGE.
Site-directed mutagenesis
Plasmid pHG101-scyA-His6 was used as the template for site-directed mutagenesis with a QuikChange II XL site-directed mutagenesis kit (Stratagene, Santa Clara, CA, USA) as described previously (Sun et al. 2013).
Expression analyses
The activity of promoters of interest was assessed using both multiple-copy and single-copy integrative lacZ reporter systems as described previously (Gao et al. 2010b; Fu et al. 2014). Based on promoter prediction, transcription start sites of all high-confident promoters are located within −350 relative to the translational initiation codon. A fragment covering the sequence upstream of each operon tested from −400 to +1 was then amplified and cloned into the reporter vectors pTP327 and pHGEI01, verified by sequencing, and the correct plasmid was then transferred into S. oneidensis strains by conjugation. Cells grown to the mid-exponential phase under experimental settings were collected and β-galactosidase activity was performed with an assay kit as described previously (Wu et al. 2011).
Immunoblotting assay
Rabbit polyclonal antibodies against S. oneidensis ScyA were prepared in accordance with standard protocols by Genscript (Nanjing, China) and used for the immunoblotting analyses. Pellets of mid-log phase cells were washed once with phosphate-buffered saline (PBS), and resuspended in 2x Laemmli SDS sample buffer. After boiled for 5 min, samples were loaded onto 10% SDS-PAGE and either stained with Coomassie Brilliant Blue or electrophoretically transferred to polyvinylidene difluoride (PVDF) according to the manufacturer's instructions (Bio-Rad). Processing of the immunoblots was performed essentially as described previously (Dong et al. 2012). For estimation of relative abundance of proteins in the gel, band intensity was measured using ImageJ software (NIH).
Cytochrome oxidase activity assay
Visual analysis of the cbb3-HCO activity was done by staining colonies with the agents for the Nadi Assay. Nadi reactions were carried out by the addition of α-naphthol and N′,N′-dimethyl-p-phenylenediamine (DMPD) on LB agar plates (Marrs and Gest 1973). Colonies were timed for formation of the indophenol blue.
Solubilized membranes were prepared for quantitative analysis of the cytochrome oxidase activity as described previously (Chen et al. 2014). In brief, cell pellets were resuspended in 20 mmol/L Tris-HCl (pH 7.6) supplemented with DNase I and protease inhibitors and disrupted by French pressure. After debris and unbroken cells removing, the membranes were pelleted by ultracentrifugation for 1 h at 230,000g at 4°C and subsequently resuspended in 20 mmol/L Tris-HCl pH 7.6 with 5% glycerol to a protein concentration of 10 mg/mL. Solubilization was performed with n-dodecyl β-d-maltoside (DDM) to a final concentration of 1% (w/v) on a rotary tube mixer for 2 h at 4°C. The DDM-solubilized membranes were obtained by collecting the supernatant after ultracentrifuging for 1 h at 230,000g at 4°C. The cytochrome oxidase activity was assayed as a measure of oxygen consumption rates using an OxyGraph oxygen electrode (Hansatech, Norfolk, UK) using either ubiquinol-1 or N,N,N′,N′-tetramethyl-p-phenylenediamine dihydrochloride (TMPD) as electron donor according to the methods described previously (Mason et al. 2009; Chen et al. 2014; Le Laz et al. 2014; Xie et al. 2014). The IC50 values of the cytochrome bd and cbb3-HCO for nitrite were obtained from plots of rates against nitrite concentrations.
Heme Staining
Late-log phase cultures (∼0.8 of OD600) were pelleted by centrifugation at 5000g for 10 min. Heme staining of proteins from an aliquot of the pellet separated by SDS-PAGE (12.5%) was carried out with 3,3′,5,5′-tetramethylbenzidine (TMBZ) as described elsewhere (Thomas et al. 1976).
Nitrite sensitivity assay and concentration determination
Cells of S. oneidensis strains grown to an OD600 of ∼0.4 were adjusted to approximately 107 CFUs/mL, followed by 10-fold serial dilutions. Ten microliter of each dilution was spotted onto LB plates containing nitrite of varying concentrations. The plates were incubated at 30°C before being read. The assays were repeated at least three times with similar results. Concentrations of nitrite were determined by Ion Chromatography (IC) analysis (Gao et al. 2009).
Statistical analyses
Values are presented as means ± SD (standard deviation). Student's t-test was performed with statistical significance set at the 0.05 confidence level.
Results
Screening for genes relevant to nitrite resistance in S. oneidensis
Our prior studies demonstrated that loss of the cytochrome bd oxidase (Δcyd) results in a hypersensitive phenotype to nitrite (Fu et al. 2013; Zhang et al. 2013). As oxygen is the EA for growth under the experimental condition, this observation suggests that the cytochrome bd oxidase rather than other oxidases confers nitrite resistance to S. oneidensis, a notion supported by biochemical analyses of E. coli oxidases (Mason et al. 2009). S. oneidensis hosts a large number of electron transport proteins, some of which can enhance the activities of certain terminal reductases by offering additional electron transport pathways (Gao et al. 2009; Cordova et al. 2011; Fonseca et al. 2013; Fu et al. 2014). Moreover, although extensively studied, S. oneidensis remains poorly understood, having ∼40% of the proteome labeled as “functionally unknown” or “hypothetical”, a pool of candidates involved in nitrite resistance. Based on these, we reasoned that we could identify genes that play a role in nitrite resistance by either annulling or enhancing their expression.
Plasmid pFAC, a mariner-based transposon vector widely used for the construction of random insertion libraries in various bacteria, contains a promoter (Ptn) embedded in the transposable sequence (Wong and Mekalanos 2000; Fu et al. 2013). However, in the presence of 3 mmol/L nitrite, we failed to obtain any colonies from a transposon insertion library derived from the Δcyd strain. This is likely due to the fact that Ptn is intrinsically a rather weak promoter and thus unable to elevate transcription of gene of interest sufficiently high for conferring an evident increase in the resistance to nitrite (Fu et al. 2013). We therefore developed a new vector, pHGT01, by replacing i) Ptn with the SO3896 promoter that is constitutively robust so that substantial overexpression of genes after the insertion can be achieved and ii) the sequence for replication and conjugation with the R6K replicon and the mob gene which is more efficient, as detailed in Experimental procedures. The SO3896 gene encodes a mature porin polypeptide of 37.7 kDa (39.9 kDa before the signal peptide cleavage), and we therefore name the porin as OmpS38 (outer membrane porin of Shewanella, 38K).
With pHGT01 we made attempts to identify genes involved in the nitrite resistance of S. oneidensis when expressed at substantially elevated levels. The vector was transferred into the Δcyd strain by conjugation to construct a random mutation library. A total of ∼100,000 individual insertion mutants, estimated by colony-forming units (CFUs) on the nitrite-free control plates, were screened and 12 colonies were obtained on the plates supplemented with 3 mmol/L nitrite. Insertion sites of these 12 mutants were exclusively mapped to the region upstream of the scyA gene, which is flanked by genes encoding the cytochrome c maturation (Ccm) system (Jin et al. 2013) (Fig.1A). All colonies were orange-colored and displayed aerobic growth comparable to that of both the wild-type and Δcyd strains (data not shown), ruling out a possibility that the Ccm system is damaged (Jin et al. 2013). A nitrite susceptibility assay confirmed that the nitrite resistance of these suppressors of the Δcyd strain increased significantly compared to their parental strain (Fig.1B). However, they were still substantially more sensitive than the wild type, supporting the notion that the bd oxidase dictates the resistance (Fu et al. 2013). Given that all of the insertion sites are located within the intergenic rather than coding sequences, it is conceivable that enhanced expression of the scyA gene resulting from transposon insertion accounts for the elevated resistance of the Δcyd strain to nitrite. To rule out the possibility that the increased resistance results from disruption of the scyA gene, we examined the nitrite susceptibility of strains lacking the scyA gene in either the wild-type or cyd− background. Abilities of both ΔscyA and ΔcydΔscyA strains to respire on a variety of EAs, including oxygen, were similar to that of the wild type (data not shown). With respect to nitrite resistance, the ΔscyA and ΔcydΔscyA strains displayed a capacity indistinguishable from those of the wild-type and Δcyd strains, respectively (Fig.1B), indicating that the loss of ScyA does not impair the nitrite resistance of S. oneidensis.
Figure 1.
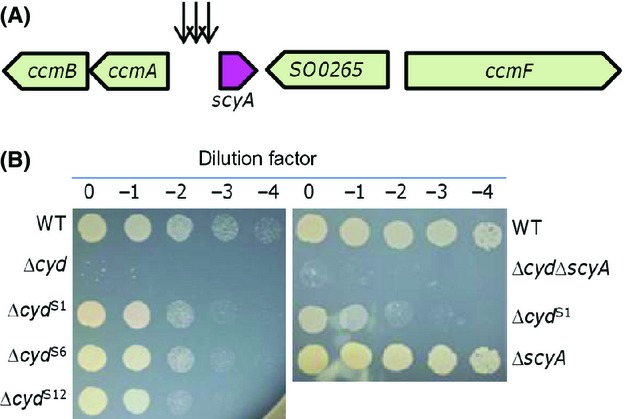
Transposon screens and terminal phenotypes. (A) Schematics indicating the approximate locations of the transposon insertions. Arrows represent transposon insertion points. (B) Nitrite sensitivity of representative Δcyd suppressors isolated in the screen. Densities of mid-log phase cultures were normalized, 10-fold serial dilutions were prepared, and 5 μL of each dilution was spotted onto LB containing 3 mmo/L nitrite. Experiments were repeated at least three times and similar results were obtained. LB, Luria–Bertani.
Overexpression of the scyA gene increases nitrite resistance
ScyA is a small monoheme cytochrome c, which has been suggested to be one of the most abundant periplasmic proteins found in cells grown under various conditions (Meyer et al. 2004; Rosenbaum et al. 2012). Up to date, the only verified role for ScyA has been to function as a mediator of electron transport between CymA and CcpA, a periplasmic diheme c-type cytochrome peroxidase (Schütz et al. 2011; Fonseca et al. 2013).
To provide direct evidence that overproduction of ScyA confers nitrite resistance to the Δcyd strain, we cloned the scyA gene with its native promoter into pHG101 and introduced the resulting plasmid pHG101-PscyA-scyA into the wild-type and Δcyd strains. Reportedly, this multiple-copy plasmid enables overproduction by approximately 5- to 15-fold (Wu et al. 2011; Dong et al. 2012; Fu et al. 2013; Sun et al. 2014). Although the expression of the scyA gene within pHG101 was sufficient to significantly elevate the nitrite resistance of the Δcyd strain, it failed to elicit any noticeable augment in the nitrite resistance in the wild type (Fig.2A). To validate the overproduction of ScyA, western blotting was performed with antibodies against ScyA. As shown in Figure2B, amounts of ScyA from pHG101-PscyA-scyA in both WT and Δcyd strains were similar, approximately 12-fold relative to those produced by these strains carrying the empty vector. To test whether the heme c of ScyA is essential to this effect, we replaced the 35th Cys residue of the heme-binding site CXXCH (CTVCH in ScyA) with a Ser, resulting in CTVSH. ScyAC35S failed to enhance nitrite resistance of the Δcyd strain, confirming that the observed effect depends on the heme c cofactor (Fig.2A).
Figure 2.
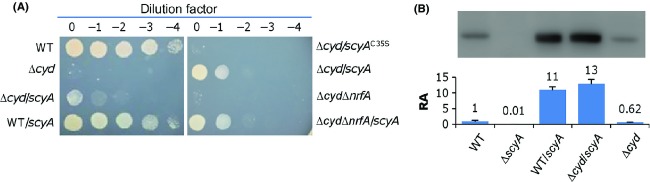
ScyA in excess increases nitrite resistance of the Δcyd strain. All strains without overexpressing ScyA carried empty vectors. The assays were repeated at least three times and similar results were obtained. (A) Nitrite sensitivity of various Shewanella oneidensis strains as described in (B). ScyAC35S is the a ScyA mutant whose the heme-binding site CTVCH was mutated to CTVSH. (B) Western blotting analysis for ScyA. To determine the extent of ScyA overproduction in the cultures from (A), extracts were prepared from cells grown to an OD600 of ∼0.8 under aerobic conditions. Immunoblot analysis was performed with antibodies against ScyA. Relative abundance (RA) of ScyA in these strains was estimated using software Image J. The values are the mean ± SD (error bars) (n = 4).
In S. oneidensis, two other soluble c-type cytochromes are also among the most abundant, FccA and CctA (SO2727, small tetraheme cytochrome c, also known as STC) (Meyer et al. 2004; Rosenbaum et al. 2012). Both proteins are capable of mediating electron transfer from CymA to outer membrane terminal reductase MtrA (Fonseca et al. 2013). To test whether the overproduction of these two c-type cytochromes has a similar effect as ScyA, we repeated the experiments with their coding genes on the same overexpression vectors. However, nitrite resistance of the Δcyd strain carrying the overexpression vectors remained the same (data not shown). These data, collectively, conclude that increased nitrite resistance of the Δcyd strain is specifically due to the overproduction of ScyA.
Under aerobic conditions, S. oneidensis cells are able to convert toxic nitrite to nonharmful ammonium using periplasmic nitrite reductase NrfA (Dong et al. 2012; Zhang et al. 2013). Therefore, it is possible that the increased nitrite resistance of the Δcyd strain resulting from overproduction of ScyA may be due to the fast removal of nitrite. To test this possibility, we constructed a ΔcydΔnrfA double mutant and compared to the Δcyd strain for the resistance to nitrite when the scyA gene was overexpressed. As shown in Figure2A, under the circumstance of overproduction of ScyA, the ΔcydΔnrfA and Δcyd strains displayed similar levels of the resistance to nitrite, indicating that the increased nitrite resistance of the Δcyd strain conferred by overproduced ScyA is unrelated to nitrite reduction.
ScyA is functionally linked to the cbb3-HCO but unlikely serves as the direct electron donor for the oxidase
In S. oneidensis, the cbb3-HCO is the predominant system to support growth under aerobic and microaerobic conditions and the bd oxidase primarily functions to facilitate survival of cells through a wide variety of stresses, nitrite in particular (Fu et al. 2013; Zhou et al. 2013; Le Laz et al. 2014). Given that the cbb3-HCO is the only oxidase present in the Δcyd strain to support growth and ScyA has been proposed to be an electron donor to terminal oxidases (Meyer et al. 2004), we therefore hypothesized that ScyA in excess may enhance the activity of the oxidase, allowing growth in the presence of nitrite. The activity of the cbb3-HCO was visualized using the Nadi plate assay, which specifically detects cytochrome c oxidase-dependent respiration (Marrs and Gest 1973). We have previously shown that the cbb3-HCO is the only enzyme reacting with the Nadi reagents in S. oneidensis (Zhou et al. 2013). Consistently, the wild-type and Δcyd colonies generated a blue ring in less than 1 min (Fig.3). As a negative control, a Δcco strain, which lacks the cbb3-HCO, could not generate a faint blue coloration in an incubation of 20 min. The cbb3-HCO activity in both the wild-type and Δcyd strains with overproduction of ScyA increased only modestly. This appears to be surprising given its significant effect on the resistance of the Δcyd strain to nitrite. Nevertheless, the results suggest that ScyA is functionally relevant to but unlikely serves as an exclusive electron donor for the cbb3-HCO.
Figure 3.
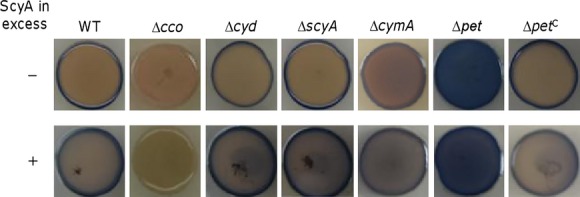
Effect of overproduction of ScyA on activities of the cytochrome c oxidase in test strains by the Nadi assay. The method is based on the rapid formation of indophenol blue from colorless a-naphthol catalyzed by cytochrome c oxidase, using N′,N′-dimethyl-p-phenylenediamine monohydrochloride as an exogenous electron donor. Nadi-positive and -negative strains were photographed 1 and 30 min after the reaction, respectively. The wild-type and Δcco strains serve as positive and negative controls. Mutants showing a distinct phenotype have been successfully complemented previously. Strains without overproducing ScyA contain the empty vector. ΔpetC represents the mutant carrying a copy of pet which is integrated for complementation.
In S. oneidensis, both the cytochrome bc1 complex, encoded by the pet operon comprising the petABC genes, and CymA can mediate electron transport from the quinol (Q) pool to the cbb3-HCO (Fu et al. 2014). The bc1 complex is predicted to be the dominant electron donor for the cbb3-HCO because loss of the bc1 complex and the oxidase results in similar growth defect, whereas the impacts of loss of CymA on aerobic growth are rather modest (Zhou et al. 2013; Fu et al. 2014). Indeed, the Nadi assay revealed that the strain lacking CymA was comparable from the wild type, confirming that the electron transfer capacity of CymA to the cbb3-HCO is negligible when the bc1 complex is present (Fig.3). Similarly, the wild-type and ΔcymA strains were indistinguishable under the conditions when ScyA was overproduced. In contrast, the loss of the bc1 complex drastically increased the activity of the cbb3-HCO, with the whole colony turning blue in 1 min. Although all of these mutants have been confirmed by successful complementation in our previous studies, the striking phenotype prompted us to further validate the mutation. A copy of the pet operon was cloned into an integrative vector (Fu et al. 2013), which allows a single-copy complementation without antibiotic interference. The cbb3-HCO hyperactive phenotype of the Δpet strain was fully corrected when this copy of the operon was expressed in trans (Fig.3), indicating that the phenotype observed from the Δpet strain was due to the loss of the bc1 complex. The effect of overproduced ScyA in the Δpet strain on the activity of the cbb3-HCO was not observed, because of the hyperactivity of the oxidase. Although further investigations are needed, we speculate that the cbb3-HCO in the strain missing the bc1 complex is idle, ready to react to the electron donor provided in the Nadi reagents (a model is provided in the discussion).
Increased production of the cbb3 oxidase has a modest effect on nitrite resistance of the Δcyd strain
Our results presented thus far suggest that the cbb3-HCO plays a critical role in nitrite resistance of the Δcyd strain in the presence of overabundant ScyA. In an attempt to offer direct evidence, we tested whether or not the cco operon in multiple copies from pHG101-Pcco-cco is able to elevate the nitrite resistance of the Δcyd strain. Overproduction of the cbb3-HCO from pHG101-Pcco-cco was evidenced by substantially increased cytochrome c oxidase activity using the Nadi assay (Fig.4A). In the wild-type, Δcco, and Δcyd strains with pHG101-Pcco-cco, the cbb3-HCO activities were significantly higher than those in the same strains without (Fig.4A). In the case of nitrite resistance, however, no apparent increase was observed from the Δcyd strain with the overproduced cbb3-HCO in the presence of 3 mmol/L nitrite (Fig.4B). We reasoned that this was likely caused by that nitrite used in the experiment overwhelmed the overproduction of the oxidase. Therefore, we retested the effect of the cbb3-HCO in overabundance on nitrite resistance of the Δcyd strain with nitrite of varying concentrations. We found that nitrite at approximately 1 mmol/L but not at higher concentrations was able to elicit a difference in the resistance between cells overexpressing the oxidase or not (Fig.4B), suggesting that the amount of the cbb3-HCO is a factor in determination of the nitrite resistance of the Δcyd strain.
Figure 4.
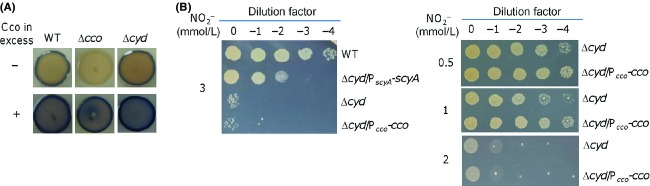
Cco in excess modestly increases nitrite resistance. The assays were repeated at least three times and similar results were obtained. (A) Effect of overproduction of the cbb3-HCO on its activity by the Nadi assay. Strains without overproducing Cco contain the empty vector. (B) Effect of overproduction of the cbb3-HCO on the nitrite resistance of the Δcyd strain in the presence of nitrite of various concentrations. HCO, heme–copper oxidases.
ScyA in excess does not affect the production of the cbb3-HCO
The overabundant cbb3-HCO in the experiment described above is insufficient to confer nitrite resistance to levels comparable to those resulting from ScyA in excess, explaining why we did not obtain any suppressors of the Δcyd strain in proximity of the cco operon during the transposon screening. Nonetheless, whether or not the increased cbb3-HCO activity in the presence of excess ScyA results from overabundance of the enzyme merits further investigation. To this end, we assessed the expression of the cco operon using the integrative lacZ reporter in strains overproducing ScyA. The promoter of the cco operon, which was defined before (Zhou et al. 2013), was cloned into pHGEI01 and the resulting vector was introduced into both the wild-type and Δcyd strains. A strain (Δcrp) lacking the crp gene, whose product mediates the expression of the cco and cyd operons in S. oneidensis (Fu et al. 2013; Zhou et al. 2013), was included in the analysis as a control. After integration and removal of the antibiotic marker, pHG101-PscyA-scyA was introduced. As shown in Figure5A, the activities of Pcco were comparable in the mid-log phase cells of all test strains except for the Δcrp strain. The compromised Pcco activity resulting from the loss of Crp was consistent with the finding in our previous report (Zhou et al. 2013). To confirm, we extracted proteins from these samples, separated them by SDS-PAGE, and stained for covalently bound heme with 3,3′,5,5′-tetramethylbenzidine (TMBZ). Similarly, the amounts of CcoP, a c-type cytochrome subunit of the cbb3-HCO, were comparable in all samples except for the Δcrp strain (Fig.5B). These data conclude that the abundance of ScyA has little effect on the production of the cbb3-HCO.
Figure 5.
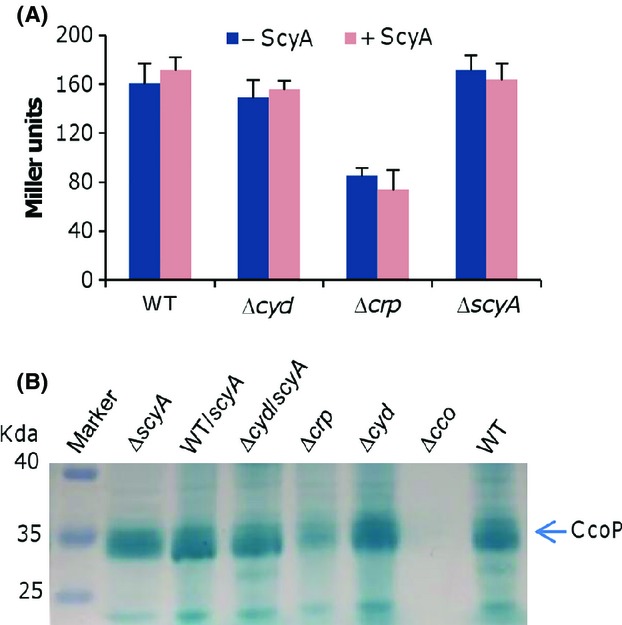
ScyA in excess affects the activity but not amount of the cbb3-HCO. (A) The activities of the cco promoter under indicated conditions determined by the integrative lacZ reporter. Error bars represent the SD of at least three independent experiments. (B) Production levels of the cbb3-HCO revealed by heme staining of CcoP. Proteins (10 μg per lane) extracted from indicated strains were separated on SDS-PAGE and analyzed by heme staining. The assays were repeated at least three times and similar results were obtained. HCO, heme–copper oxidases; SDS-PAGE, sodiumdodecyl sulphate polyacrylamide gel electrophoresis.
Effect of overproduced ScyA depends on a complete electron transport chain to cbb3-HCO
Although ScyA unlikely serves as the predominant electron donor for the cbb3-HCO, it is functionally linked to the oxidase. To further confirm this, we intended to address whether the observed effect of overproduced ScyA requires functional cbb3-HCO. Unfortunately, the effect of overproduced ScyA on the activity of the cbb3-HCO in the Δpet strain could not be examined using the Nadi assay because of the hyperactivity of the oxidase (Fig.3A). We then utilized an alternative method to test this. Attempts were made to generate double mutants lacking the bd oxidase and either the bc1 complex or CymA. While a ΔcydΔcymA strain was obtained smoothly, we failed to delete the cyd and pet operons under aerobic conditions. Given that the cbb3-HCO and the bd oxidase are a combination of synthetic lethal, these observations suggest that CymA alone is insufficient to ensure the function of the cbb3-HCO (Zhou et al. 2013). By performing mutagenesis under anaerobic conditions, we successfully obtained a strain (ΔcydΔpet) lacking both the bc1 complex and the bd oxidase, which could hardly grow aerobically (Fig.6A). Expression of either system in trans in the double mutant enabled growth under aerobic conditions, confirming that the inability to grow with oxygen is due to intended mutations. In line with the proposed role of the bc1 complex (Dibrova et al. 2013), these results conclude that the cbb3-HCO is, largely but not exclusively, dependent on the bc1 complex for electrons from the quinol pool. The residual activity of the cbb3-HCO in the absence of the bc1 complex is probably from CymA, which has been recently shown to overlap with the bc1 complex to some extent in respiration of nitrate and nitrite (Fu et al. 2014). ScyA in excess failed to rescue the aerobic growth of the ΔcydΔpet strain, a scenario also observed from the ΔcydΔcco strain. To rule out a possibility that loss of the bc1 complex prevents the production of the cbb3-HCO, cellular levels of the oxidase were examined. As shown in Figure6B, amounts of the cbb3-HCO in strains containing the bc1 complex or not were comparable. These data, collectively, indicate that ScyA in overabundance is unable to exert its effect unless the electron transfer to the cbb3-HCO is in place.
Figure 6.
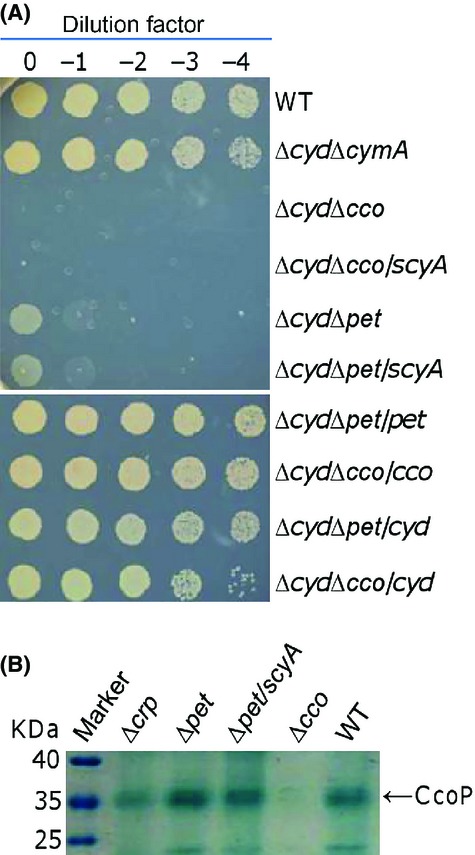
Function of ScyA in excess relies on a complete electron transfer to the cbb3-HCO. The assays were repeated at least three times and similar results were obtained. (A) Effect of overproduction of ScyA on the nitrite resistance of the ΔcydΔcco and ΔcydΔpet strain in the presence of 3 mmol/L nitrite. (B) Effect of overproduction of ScyA on production levels of the cbb3-HCO revealed by heme staining of CcoP. Proteins (10 μg per lane) extracted from indicated strains were separated on SDS-PAGE and analyzed by heme staining. HCO, heme–copper oxidases; SDS-PAGE, sodiumdodecyl sulphate polyacrylamide gel electrophoresis.
ScyA in excess alleviates inhibition of nitrite on the cbb3-HCO
As the overabundance of the cbb3-HCO is not sufficient to confer the Δcyd strain nitrite resistance comparable to that resulting from ScyA in excess, we hypothesized that overproduced ScyA may increase resistance of the cbb3-HCO to nitrite. To investigate this possibility, we assessed the effects of ScyA in overabundance on the half-maximal inhibitory concentration of the cbb3-HCO and the bd oxidase for nitrite (IC50).
Membrane preparations of S. oneidensis strains expressing only one of the terminal oxidases were used to measure oxygen reduction with ubiquinol-1 for the bd oxidase or with a combination of ascorbate and TMPD for the cbb3-HCO (Mason et al. 2009; Chen et al. 2014; Xie et al. 2014). At ∼70 μmol/L O2, the IC50 value of the S. oneidensis bd oxidase was approximately 0.18 μmol/L, whereas the IC50 value for the cbb3-HCO was about 0.05 μmol/L (Fig.7A), consistent with the observation that the Δcyd strain is more sensitive to nitrite than the Δcco strain. No difference in the IC50 values between the wild-type and the Δcco strain was observed, further validating that the bd oxidase is the enzyme that dictates nitrite resistance. Unfortunately, membrane preparations of S. oneidensis strains overexpressing ScyA did not show any effect on the IC50 values of the Δcyd or Δcco strain (data not shown). We suspected that this may be due to the loss of ScyA during the preparation for membranes. To circumvent this problem, we added a His6-tag after the C-terminus of ScyA and expressed the fusion protein in S. oneidensis for purification (Fig. S2). When in excess the His6-tag ScyA was able to elevate the nitrite resistance of the Δcyd strain to levels obtained from ScyA (Fig. S1), indicating that the tagged protein is biologically active. With purified His6-ScyA, the IC50 values of membrane preparations from the Δcyd and Δcco strains were measured (Fig.7B). While the addition of His6-ScyA had no effect of the IC50 values for the bd oxidase (Δcco), it increased those for the cbb3-HCO (Δcyd) proportionally with 4 μg/mL or less and further improvement was not observed with more proteins. Notably, the IC50 values for the bd oxidase exceeded those for the cbb3-HCO at all protein concentrations. These data, all together, indicate that ScyA improves nitrite resistance of S. oneidensis by reducing the sensitivity of the cbb3-HCO to nitrite.
Figure 7.
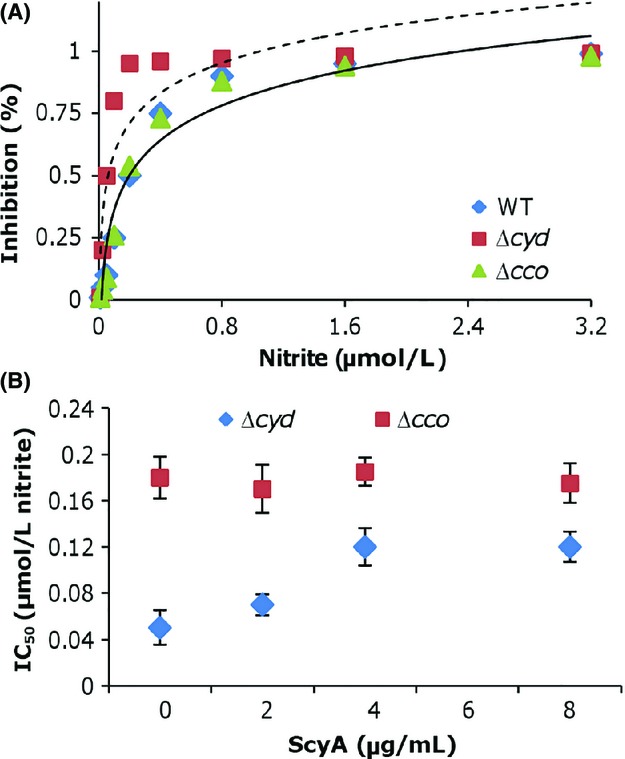
ScyA in excess improves the nitrite resistance of the cbb3-HCO. Error bars represent the SD of at least three independent experiments. A. Nitrite sensitivity of the cytochrome bd and cbb3-HCO. Respiration rates of membranes were measured in the presence of nitrite of various concentrations. The dash line was derived from the Δcyd strain and the solid lines were derived from WT and Δcco strains, which were undistinguishable. B. Effect of overproduction of ScyA on nitrite IC50 values of the cytochrome bd and cbb3-HCO. The half-maximal inhibitory concentrations for nitrite were measured as described in Experimental procedures. HCO, heme–copper oxidases; WT, wild type; IC, ion chromatography.
Discussion
The purpose of this study was to identify proteins other than the bd oxidase that facilitate aerobic growth of S. oneidensis in the presence of nitrite. We have shown previously that the bd oxidase is the primary force conferring this microorganism resistance to nitrite (Fu et al. 2013), a scenario similar to that the E. coli counterpart does on NO resistance (Mason et al. 2009). S. oneidensis is able to reduce nitrite to ammonia under aerobic condition, leading to detoxification (Dong et al. 2012; Zhang et al. 2013; Fu et al. 2014). However, there is a caveat: cells have to be in the stationary phase, even with the forced production of periplasmic nitrite reductase NrfA (Zhang et al. 2013). Thus, mutations that enable cells to respire on nitrite before the stationary phase are likely to improve growth with nitrite. It is also possible that the loss of some porins and/or transporters by which nitrite enters the periplasm would make a difference by slowing nitrite influx (Song and Niederweis 2012). Moreover, there may be an alternative oxidase, similar to that found in Vibrio fischeri which is highly resistant to NO (Dunn et al. 2010), encoded by a gene hidden in the genome as up to 40% genes are for hypothetical proteins (Heidelberg et al. 2002).
Given that multiple means for enhancing nitrite resistance possibly exist, we anticipated that we would identify a few. To our surprise, only ScyA, a monoheme cytochrome c5 that is suggested to be one of the most abundant periplasmic proteins found in cells under anaerobic conditions (Tsapin et al. 2001; Meyer et al. 2004; Fonseca et al. 2013), was caught. Such a finding is enabled by a new mariner-based transposon vector, developed in this study, which carries a highly active promoter for the ompS38 gene within the transposable segment. In S. oneidensis, OmpS38 is evidently one of the most abundance porins although its expression is barely affected by the EnvZ-OmpR two-component regulatory system (Yuan et al. 2011). Interestingly, production of the protein is upregulated under anaerobic conditions (Maier and Myers 2004). These features, altogether, render the vector suitability for screening for cryptic or quiescent operons in addition to knockout of active ones.
Despite intensive studies of S. oneidensis c-type cytochromes as the foundation for its respiratory versatility, we are only just beginning to uncover the molecular mechanisms underlying their physiological roles. This is particularly true for soluble c-type cytochromes as they are featured by a large number, up to 28 (Meyer et al. 2004), and diffusibility in the periplasm, the latter providing opportunities to interact with many similarly soluble and/or membrane-bound proteins. Two such proteins which are also among the most abundant, FccA and CctA serve as good examples. FccA is a soluble tetraheme flavocytochrome c fumarate reductase carrying heme cores that are distinct from those observed in well-studied tetraheme cytochrome c3 of Desulfovibrio (Leys et al. 1999; Taylor et al. 1999). In addition to catalyzing fumarate reduction, FccA mediates electron transfer from CymA to MtrA of the metal reduction complex MtrABC (Schuetz et al. 2009; Fonseca et al. 2013). Moreover, FccA, by binding to CymA to form a redox complex, regulates the direction of catalysis and electron transfer of the latter (McMillan et al. 2013). Similar to FccA, CctA is also able to interact with both CymA and MtrA, playing an important role in reduction of metal oxide particles (Gordon et al. 2000; Leys et al. 2002; Fonseca et al. 2013).
ScyA, like FccA and CctA, is also able to interact with CymA and proposed to be the only periplasmic electron donor for CcpA, a diheme c-type cytochrome peroxidase (Schütz et al. 2011; Fonseca et al. 2013). Here, we showed that ScyA in excess enhances the activity of the cbb3-HCO. Multiple insertions were mapped into the promoter region of the scyA gene, indicating that the random mutant pool is sufficiently large to completely cover all genes and intergenic sequences in between. Single catch implies that ScyA is likely the only one which in excess is able to improve nitrite resistance of the cbb3-HCO to the observed extent.
It is generally accepted that cytochrome c oxidases do not directly oxidize the quinol pool but via soluble cytochrome c electron shuttles, which are functionally essential to the enzymes (Ekici et al. 2012). Although proposed to be an electron donor to cytochrome c oxidases (Meyer et al. 2004), ScyA is unlikely to be predominant one based on the drastic difference in the Nadi staining between strains missing the bc1 complex and ScyA. Given that S. oneidensis is equipped with many cytochromes of similar redox potentials and the electron transport processes mediated by c-type cytochrome are suggested to be unspecific (Firer-Sherwood et al. 2008; Coursolle and Gralnick 2012; Fonseca et al. 2013), we would expect that more than one cytochrome c are able to serve as the electron donor for the cbb3-HCO. Moreover, the bc1 complex and CymA overlap each other in mediating electron transfer from the quinone pool to terminal reductases (HCOs actually are oxygen reductases) to some extent and are exchangeable under certain conditions, further broadening the scope of such c-type cytochromes (Fu et al. 2014). Thus, it is possible that multiple small c-type cytochromes may mediate electron transfer from the bc1 complex to the cbb3-HCO, a scenario reported in Rhodobacter sphaeroides before (Daldal et al. 2001).
Based on our data, we propose two mechanisms that underlie enhanced nitrite resistance of the cbb3-HCO in the presence of overabundant ScyA. One involves with electron transfer. As ScyA is a minor player in transferring electrons from the bc1 complex/CymA to the cbb3-HCO, its loss does not significantly compromise the activity of the cbb3-HCO. This is evident with the phenotypes of the scyA mutant, with respect to aerobic growth, nitrite susceptibility, and the activity revealed by the Nadi assay. When in excess, ScyA may be able to increase the electron turnover rate of the cbb3-HCO, resulting in the augment of the nitrite resistance. This notion gains support clearly from in vivo data. Perhaps more importantly, it is also in line with in vitro results, at least to some extent. The in vitro experiment was conducted with NADH, a condition under which ScyA likely exists in its reduced form. Hence, electron transfer may occur from ScyA to the cbb3-HCO. The other involves direct interaction with the cbb3-HCO. The E. coli cytochrome bd oxidase, which confers NO resistance to E. coli, has a NO dissociation rate faster than both the cytochrome bo oxidase and aa3-HCO(Mason et al. 2009). Given that the in vitro data presented in Figure7 resemble those reported in the E. coli study, it is therefore possible that ScyA in excess helps alter the biochemical properties of the cbb3-HCO, leading to its increased resistance to nitrite. How these intricate processes are spatially and temporally coordinated are currently under our investigation.
Our results presented here and before demonstrated that the cytochrome bc1 complex is essential to the cbb3-HCO for reducing oxygen (Zhou et al. 2013; Fu et al. 2014). So, why does the strain devoid of the complex exhibit an enhanced activity of the cbb3-HCO? We suggest a mechanism illustrated in Figure8. In S. oneidensis, there are two ways to supply electrons to the cbb3-HCO for oxygen respiration: the major one is through the bc1 complex, which may couple with a yet unidentified cytochrome c, and the minor one is mediated by ScyA. Both CymA and the bc1 complex may serve as the electron donors for these c-type cytochromes as ScyA has been shown to be able to interact with CymA (Coursolle and Gralnick 2010; Schütz et al. 2011). However, contribution from CymA as the electron source is negligible. Neither of these two electron mediators is required for the Nadi assay: α-naphthol + N′,N′-dimethyl-p-phenylenediamine (DMPD) + O2 → Indophenol Blue + H2O because DMPD serves as the electron donor (Marrs and Gest 1973). However, oxygen is the substrate for both the Nadi reaction and oxygen respiration, which requires electrons from the bc1 complex and/or cytochrome c electron mediators. As a consequence, the two processes compete with each other for oxygen. When electrons are no longer delivered to the cbb3-HCO, the enzyme exclusively reacts with the Nadi reagents, exhibiting the highest activities. It is worth mentioning that it is still uncertain of whether or not there exists a yet unidentified cytochrome c mediating electron transfer from the cytochrome bc1 complex to the cbb3-HCO. Efforts to test this are underway.
Figure 8.
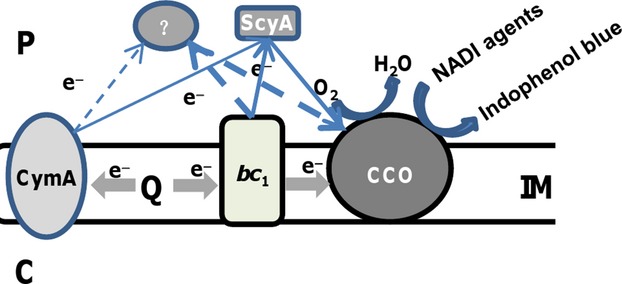
Model: electron transfer to the cbb3-HCO in Shewanella oneidensis. The cytochrome bc1 complex may transfer electrons to the cbb3-HCO directly or more likely via an undetermined cytochrome c as the major mediator and ScyA as the minor, allowing oxygen reduction. Contribution from CymA as the electron source is extremely limited. According to the Nadi reaction (α-naphthol + DMPD + O2 → Indophenol Blue + H2O), oxygen is required for both oxygen respiration and the Nadi reaction. Solid and dash lines represent established and suggested electron pathways, respectively. C, IM, and P represent cytoplasm, inner-membrane, and periplasm, respectively.
Acknowledgments
This research was supported by Major State Basic Research Development Program (973 Program: 2010CB833803), National Natural Science Foundation of China (31270097), and Doctoral Fund of Ministry of Education of China (20130101110142) to H. G.
Conflict of Interest
None declared.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Figure S1. Characteristics of recombinant ScyA. (A) SDS-PAGE analysis of purified recombinant ScyA by heme-staining. ScyA-His6 was extracted from the S. oneidensis wild-type carrying pHGE-ScyA-His6. M represents protein marker. (B) Functional analysis of recombinant ScyA and mutant proteins.
References
- Atkinson SJ, Mowat CG, Reid GA. Chapman SK. An octaheme c-type cytochrome from Shewanella oneidensis can reduce nitrite and hydroxylamine. FEBS Lett. 2007;581:3805–3808. doi: 10.1016/j.febslet.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Borisov VB, Gennis RB, Hemp J. Verkhovsky MI. The cytochrome bd respiratory oxygen reductases. Biochim. Biophys. Acta. 2011;1807:1398–1413. doi: 10.1016/j.bbabio.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretschger O, Obraztsova A, Sturm CA, Chang IS, Gorby YA, Reed SB, et al. Current production and metal oxide reduction by Shewanella oneidensis MR-1 wild type and mutants. Appl. Environ. Microbiol. 2007;73:7003–7012. doi: 10.1128/AEM.01087-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschmann S, Warkentin E, Xie H, Langer JD, Ermler U. Michel H. The structure of cbb3 cytochrome oxidase provides insights into proton pumping. Science. 2010;329:327–330. doi: 10.1126/science.1187303. [DOI] [PubMed] [Google Scholar]
- Chen H, Luo Q, Yin J, Gao T. Gao H. Evidence for the requirement of CydX in function but not assembly of the cytochrome bd oxidase in Shewanella oneidensis. Biochim. Biophys. Acta. 2014 doi: 10.1016/j.bbagen.2014.10.005. doi: 10.1016/j.bbagen.2014.10.005. [DOI] [PubMed] [Google Scholar]
- Cordova CD, Schicklberger MFR, Yu Y. Spormann AM. Partial functional replacement of CymA by SirCD in Shewanella oneidensis MR-1. J. Bacteriol. 2011;193:2312–2321. doi: 10.1128/JB.01355-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coursolle D. Gralnick JA. Modularity of the Mtr respiratory pathway of Shewanella oneidensis strain MR-1. Mol. Microbiol. 2010;77:995–1008. doi: 10.1111/j.1365-2958.2010.07266.x. [DOI] [PubMed] [Google Scholar]
- Coursolle D. Gralnick JA. Reconstruction of extracellular respiratory pathways for iron(III) reduction in Shewanella oneidensis strain MR-1. Front. Microbiol. 2012;3:56. doi: 10.3389/fmicb.2012.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daldal F, Mandaci S, Winterstein C, Myllykallio H, Duyck K. Zannoni D. Mobile cytochrome c2 and membrane-anchored cytochrome cy are both efficient electron donors to the cbb3- and aa3-type cytochrome c oxidases during respiratory growth of Rhodobacter sphaeroides. J. Bacteriol. 2001;183:2013–2024. doi: 10.1128/JB.183.6.2013-2024.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Noe JC, Paik S. Kitten T. An improved arbitrary primed PCR method for rapid characterization of transposon insertion sites. J. Microbiol. Methods. 2005;63:89–94. doi: 10.1016/j.mimet.2005.02.011. [DOI] [PubMed] [Google Scholar]
- Dibrova DV, Cherepanov DA, Galperin MY, Skulachev VP. Mulkidjanian AY. Evolution of cytochrome bc complexes: from membrane-anchored dehydrogenases of ancient bacteria to triggers of apoptosis in vertebrates. Biochim. Biophys. Acta. 2013;1827:1407–1427. doi: 10.1016/j.bbabio.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Wang J, Fu H, Zhou G, Shi M. Gao H. A Crp-dependent two-component system regulates nitrate and nitrite respiration in Shewanella oneidensis. PLoS ONE. 2012;7:e51643. doi: 10.1371/journal.pone.0051643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn AK, Karr EA, Wang Y, Batton AR, Ruby EG. Stabb EV. The alternative oxidase (AOX) gene in Vibrio fischeri is controlled by NsrR and upregulated in response to nitric oxide. Mol. Microbiol. 2010;77:44–55. doi: 10.1111/j.1365-2958.2010.07194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekici S, Pawlik G, Lohmeyer E, Koch H-G. Daldal F. Biogenesis of cbb3-type cytochrome c oxidase in Rhodobacter capsulatus. Biochim. Biophys. Acta. 2012;1817:898–910. doi: 10.1016/j.bbabio.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firer-Sherwood M, Pulcu G. Elliott S. Electrochemical interrogations of the Mtr cytochromes from Shewanella: opening a potential window. J. Biol. Inorg. Chem. 2008;13:849–854. doi: 10.1007/s00775-008-0398-z. [DOI] [PubMed] [Google Scholar]
- Fonseca BM, Paquete CM, Neto SE, Pacheco I, Soares CM. Louro RO. Mind the gap: cytochrome interactions reveal electron pathways across the periplasm of Shewanella oneidensis MR-1. Biochem. J. 2013;449:101–108. doi: 10.1042/BJ20121467. [DOI] [PubMed] [Google Scholar]
- Fredrickson JK, Romine MF, Beliaev AS, Auchtung JM, Driscoll ME, Gardner TS, et al. Towards environmental systems biology of Shewanella. Nat. Rev. Microbiol. 2008;6:592–603. doi: 10.1038/nrmicro1947. [DOI] [PubMed] [Google Scholar]
- Fu H, Chen H, Wang J, Zhou G, Zhang H, Zhang L, et al. Crp-dependent cytochrome bd oxidase confers nitrite resistance to Shewanella oneidensis. Environ. Microbiol. 2013;15:2198–2212. doi: 10.1111/1462-2920.12091. [DOI] [PubMed] [Google Scholar]
- Fu H, Jin M, Ju L, Mao Y. Gao H. Evidence for function overlapping of CymA and the cytochrome bc1 complex in the Shewanella oneidensis nitrate and nitrite respiration. Environ. Microbiol. 2014 doi: 10.1111/1462-2920.12457. doi: 10.1111/1462-2920.12457. [DOI] [PubMed] [Google Scholar]
- Gao H, Wang Y, Liu X, Yan T, Wu L, Alm E, et al. Global transcriptome analysis of the heat shock response of Shewanella oneidensis. J. Bacteriol. 2004;186:7796–7803. doi: 10.1128/JB.186.22.7796-7803.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao HC, Yang ZMK, Wu LY, Thompson DK. Zhou J Z. Global transcriptome analysis of the cold shock response of Shewanella oneidensis MR-1 and mutational analysis of its classical cold shock proteins. J. Bacteriol. 2006;188:4560–4569. doi: 10.1128/JB.01908-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Wang X, Yang Z, Palzkill T. Zhou J. Probing regulon of ArcA in Shewanella oneidensis MR-1 by integrated genomic analyses. BMC Genom. 2008;9:42. doi: 10.1186/1471-2164-9-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Yang ZK, Barua S, Reed SB, Romine MF, Nealson KH, et al. Reduction of nitrate in Shewanella oneidensis depends on atypical NAP and NRF systems with NapB as a preferred electron transport protein from CymA to NapA. ISME J. 2009;3:966–976. doi: 10.1038/ismej.2009.40. [DOI] [PubMed] [Google Scholar]
- Gao H, Barua S, Liang Y, Wu L, Dong Y, Reed S, et al. Impacts of Shewanella oneidensis c-ype cytochromes on aerobic and anaerobic respiration. Microb. Biotechnol. 2010a;3:455–466. doi: 10.1111/j.1751-7915.2010.00181.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Wang X, Yang Z, Chen J, Liang Y, Chen H, et al. Physiological roles of ArcA, Crp, and EtrA and their interactive control on aerobic and anaerobic respiration in Shewanella oneidensis. PLoS ONE. 2010b;5:e15295. doi: 10.1371/journal.pone.0015295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon EH, Pike AD, Hill AE, Cuthbertson PM, Chapman SK. Reid GA. Identification and characterization of a novel cytochrome c(3) from Shewanella frigidimarina that is involved in Fe(III) respiration. Biochem. J. 2000;349:153–158. doi: 10.1042/0264-6021:3490153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralnick JA, Vali H, Lies DP. Newman DK. Extracellular respiration of dimethyl sulfoxide by Shewanella oneidensis strain MR-1. Proc. Natl. Acad. Sci. USA. 2006;103:4669–4674. doi: 10.1073/pnas.0505959103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidelberg JF, Paulsen IT, Nelson KE, Gaidos EJ, Nelson W C. Read TD. Genome sequence of the dissimilatory metal ion-reducing bacterium Shewanella oneidensis. Nat. Biotechnol. 2002;20:1118–1123. doi: 10.1038/nbt749. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Dong Y, Luo Q, Li N, Wu G. Gao H. Protection from oxidative stress relies mainly on derepression of OxyR-dependent KatB and Dps in Shewanella oneidensis. J. Bacteriol. 2014;196:445–458. doi: 10.1128/JB.01077-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin M, Jiang Y, Sun L, Yin J, Fu H, Wu G, et al. Unique organizational and functional features of the cytochrome c maturation system in Shewanella oneidensis. PLoS ONE. 2013;8:e75610–e75610. doi: 10.1371/journal.pone.0075610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranz RG, Richard-Fogal C, Taylor J-S. Frawley ER. Cytochrome c biogenesis: mechanisms for covalent modifications and trafficking of heme and for heme-iron redox control. Microbiol. Mol. Biol. Rev. 2009;73:510–528. doi: 10.1128/MMBR.00001-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Laz S, Kpebe A, Bauzan M, Lignon S, Rousset M. Brugna M. A biochemical approach to study the role of the terminal oxidases in aerobic respiration in Shewanella oneidensis MR-1. PLoS ONE. 2014;9:e86343. doi: 10.1371/journal.pone.0086343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leys D, Tsapin AS, Nealson KH, Meyer TE, Cusanovich MA. Beeumen JJV. Structure and mechanism of the flavocytochrome c fumarate reductase of Shewanella putrefaciens MR-1. Nat. Struct. Mol. Biol. 1999;6:1113–1117. doi: 10.1038/70051. [DOI] [PubMed] [Google Scholar]
- Leys D, Meyer TE, Tsapin AS, Nealson KH, Cusanovich MA. Van Beeumen JJ. Crystal structures at atomic resolution reveal the novel concept of “electron-harvesting” as a role for the small tetraheme cytochrome c. J. Biol. Chem. 2002;277:35703–35711. doi: 10.1074/jbc.M203866200. [DOI] [PubMed] [Google Scholar]
- Lukat P, Rudolf M, Stach P, Messerschmidt A, Kroneck PMH, Simon J, et al. Binding and reduction of sulfite by cytochrome c nitrite reductase. Biochemistry. 2008;47:2080–2086. doi: 10.1021/bi7021415. [DOI] [PubMed] [Google Scholar]
- Luo Q, Dong Y, Chen H. Gao H. Mislocalization of Rieske protein PetA predominantly accounts for the aerobic growth defect of tat mutants in Shewanella oneidensis. PLoS ONE. 2013;8:e62064. doi: 10.1371/journal.pone.0062064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier T. Myers C. The outer membrane protein Omp35 affects the reduction of Fe(III), nitrate, and fumarate by Shewanella oneidensis MR-1. BMC Microbiol. 2004;4:23. doi: 10.1186/1471-2180-4-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marritt SJ, Lowe TG, Bye J, McMillan DGG, Shi L, Fredrickson J, et al. A functional description of CymA, an electron-transfer hub supporting anaerobic respiratory flexibility in Shewanella. Biochem. J. 2012;444:465–474. doi: 10.1042/BJ20120197. [DOI] [PubMed] [Google Scholar]
- Marrs B. Gest H. Genetic mutations affecting the respiratory electron-transport system of the photosynthetic bacterium Rhodopseudomonas capsulata. J. Bacteriol. 1973;114:1045–1051. doi: 10.1128/jb.114.3.1045-1051.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason MG, Shepherd M, Nicholls P, Dobbin PS, Dodsworth KS, Poole RK, et al. Cytochrome bd confers nitric oxide resistance to Escherichia coli. Nat. Chem. Biol. 2009;5:94–96. doi: 10.1038/nchembio.135. [DOI] [PubMed] [Google Scholar]
- McMillan DGG, Marritt SJ, Firer-Sherwood MA, Shi L, Richardson DJ, Evans SD, et al. Protein–protein interaction regulates the direction of catalysis and electron transfer in a redox enzyme complex. J. Am. Chem. Soc. 2013;135:10550–10556. doi: 10.1021/ja405072z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer TE, Tsapin AI, Vandenberghe I, De Smet L, Frishman D, Nealson KH, et al. Identification of 42 possible cytochrome c genes in the Shewanella oneidensis genome and characterization of six soluble cytochromes. OMICS. 2004;8:57–77. doi: 10.1089/153623104773547499. [DOI] [PubMed] [Google Scholar]
- Nissen S, Liu X, Chourey K, Hettich RL, Wagner DD, Pfiffner SM, et al. Comparative c-type cytochrome expression analysis in Shewanella oneidensis strain MR-1 and Anaeromyxobacter dehalogenans strain 2CP-C grown with soluble and insoluble oxidized metal electron acceptors. Biochem. Soc. Trans. 2012;40:1204–1210. doi: 10.1042/BST20120182. [DOI] [PubMed] [Google Scholar]
- Reese MG. Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput. Chem. 2001;26:51–56. doi: 10.1016/s0097-8485(01)00099-7. [DOI] [PubMed] [Google Scholar]
- Richardson DJ, Butt JN, Fredrickson JK, Zachara JM, Shi L, Edwards MJ, et al. The ‘porin–cytochrome’ model for microbe-to-mineral electron transfer. Mol. Microbiol. 2012;85:201–212. doi: 10.1111/j.1365-2958.2012.08088.x. [DOI] [PubMed] [Google Scholar]
- Richter K, Schicklberger M. Gescher J. Dissimilatory reduction of extracellular electron acceptors in anaerobic respiration. Appl. Environ. Microbiol. 2012;78:913–921. doi: 10.1128/AEM.06803-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romine MF, Carlson TS, Norbeck AD, McCue LA. Lipton MS. Identification of mobile elements and pseudogenes in the Shewanella oneidensis MR-1 genome. Appl. Environ. Microbiol. 2008;74:3257–3265. doi: 10.1128/AEM.02720-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum MA, Bar HY, Beg QK, Segrè D, Booth J, Cotta MA, et al. Transcriptional analysis of Shewanella oneidensis MR-1 with an electrode compared to Fe(III)citrate or oxygen as terminal electron acceptor. PLoS ONE. 2012;7:e30827. doi: 10.1371/journal.pone.0030827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuetz B, Schicklberger M, Kuermann J, Spormann AM. Gescher J. Periplasmic electron transfer via the c-type cytochromes MtrA and FccA of Shewanella oneidensis MR-1. Appl. Environ. Microbiol. 2009;75:7789–7796. doi: 10.1128/AEM.01834-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schütz B, Seidel J, Sturm G, Einsle O. Gescher J. Investigation of the electron transport chain to and the catalytic activity of the diheme cytochrome c peroxidase CcpA of Shewanella oneidensis. Appl. Environ. Microbiol. 2011;77:6172–6180. doi: 10.1128/AEM.00606-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slauch JM. Silhavy TJ. Genetic analysis of the switch that controls porin gene expression in Escherichia coli K-12. J. Mol. Biol. 1989;210:281–292. doi: 10.1016/0022-2836(89)90330-6. [DOI] [PubMed] [Google Scholar]
- Song H. Niederweis M. Uptake of sulfate but not phosphate by Mycobacterium tuberculosis is slower than that for Mycobacterium smegmatis. J. Bacteriol. 2012;194:956–964. doi: 10.1128/JB.06132-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Jin M, Ding W, Yuan J, Kelly J. Gao H. Posttranslational modification of flagellin FlaB in Shewanella oneidensis. J. Bacteriol. 2013;195:2550–2561. doi: 10.1128/JB.00015-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Dong Y, Shi M, Jin M, Zhou Q, Luo ZQ, et al. Two residues predominantly dictate functional difference in motility between Shewanella oneidensis flagellins FlaA and FlaB. J. Biol. Chem. 2014;289:14547–14559. doi: 10.1074/jbc.M114.552000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor P, Pealing SL, Reid GA, Chapman SK. Walkinshaw MD. Structural and mechanistic mapping of a unique fumarate reductase. Nat. Struct. Mol. Biol. 1999;6:1108–1112. doi: 10.1038/70045. [DOI] [PubMed] [Google Scholar]
- Thomas PE, Ryan D. Levin W. An improved staining procedure for the detection of the peroxidase activity of cytochrome P-450 on sodium dodecyl sulfate polyacrylamide gels. Anal. Biochem. 1976;75:168–176. doi: 10.1016/0003-2697(76)90067-1. [DOI] [PubMed] [Google Scholar]
- Tsapin AI, Vandenberghe I, Nealson KH, Scott JH, Meyer TE, Cusanovich MA, et al. Identification of a small tetraheme cytochrome c and a flavocytochrome c as two of the principal soluble cytochromes c in Shewanella oneidensis strain MR1. Appl. Environ. Microbiol. 2001;67:3236–3244. doi: 10.1128/AEM.67.7.3236-3244.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong SM. Mekalanos JJ. Genetic footprinting with mariner-based transposition in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA. 2000;97:10191–10196. doi: 10.1073/pnas.97.18.10191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Wang J, Tang P, Chen H. Gao H. Genetic and molecular characterization of flagellar assembly in Shewanella oneidensis. PLoS ONE. 2011;6:e21479. doi: 10.1371/journal.pone.0021479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H, Buschmann S, Langer JD, Ludwig B. Michel H. Biochemical and biophysical characterization of the two isoforms of cbb3-type cytochrome c oxidase from Pseudomonas stutzeri. J. Bacteriol. 2014;196:472–482. doi: 10.1128/JB.01072-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Wei B, Shi M. Gao H. Functional assessment of EnvZ/OmpR two-component system in Shewanella oneidensis. PLoS ONE. 2011;6:e23701. doi: 10.1371/journal.pone.0023701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Fu H, Wang J, Sun L, Jiang Y, Zhang L, et al. Impacts of nitrate and nitrite on physiology of Shewanella oneidensis. PLoS One. 2013;8:e62629. doi: 10.1371/journal.pone.0062629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G, Yin J, Chen H, Hua Y, Sun L. Gao H. Combined effect of loss of the caa3 oxidase and Crp regulation drives Shewanella to thrive in redox-stratified environments. ISME J. 2013;7:1752–1763. doi: 10.1038/ismej.2013.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Characteristics of recombinant ScyA. (A) SDS-PAGE analysis of purified recombinant ScyA by heme-staining. ScyA-His6 was extracted from the S. oneidensis wild-type carrying pHGE-ScyA-His6. M represents protein marker. (B) Functional analysis of recombinant ScyA and mutant proteins.


