Abstract
We analyzed the diversity of bacterial epibionts and trophic ecology of a new species of Kiwa yeti crab discovered at two hydrothermal vent fields (E2 and E9) on the East Scotia Ridge (ESR) in the Southern Ocean using a combination of 454 pyrosequencing, Sanger sequencing, and stable isotope analysis. The Kiwa epibiont communities were dominated by Epsilon- and Gammaproteobacteria. About 454 sequencing of the epibionts on 15 individual Kiwa specimen revealed large regional differences between the two hydrothermal vent fields: at E2, the bacterial community on the Kiwa ventral setae was dominated (up to 75%) by Gammaproteobacteria, whereas at E9 Epsilonproteobacteria dominated (up to 98%). Carbon stable isotope analysis of both Kiwa and the bacterial epibionts also showed distinct differences between E2 and E9 in mean and variability. Both stable isotope and sequence data suggest a dominance of different carbon fixation pathways of the epibiont communities at the two vent fields. At E2, epibionts were putatively fixing carbon via the Calvin-Benson-Bassham and reverse tricarboxylic acid cycle, while at E9 the reverse tricarboxylic acid cycle dominated. Co-varying epibiont diversity and isotope values at E2 and E9 also present further support for the hypothesis that epibionts serve as a food source for Kiwa.
Keywords: East Scotia Ridge, epibionts, hydrothermal vent, Kiwa sp., microbial diversity, stable isotopes
Introduction
Hydrothermal vents are highly productive ecosystems, where primary production occurs through chemoautotrophic microbial production. They support dense communities of metazoans, some of which are in endo- or episymbiotic relationships with microbes. The spatial distribution of organisms within these habitats is governed by temperature and composition of chemically reduced fluids emitted from the seafloor as well as topology and substrate type (Cuvelier et al. 2009; Takai et al. 2009; Podowski et al. 2010; Flores et al. 2011; Petersen et al. 2011; Beinart et al. 2012). Metazoan communities at hydrothermal vents vary globally and a series of biogeographical provinces has been proposed (Moalic et al. 2012; Rogers et al. 2012). In areas closest to high temperature venting, alvinellid polychaetes (North Pacific), alvinochoncid gastropods (western Pacific), rimicarid shrimp (Atlantic Ocean), and the decapod Shinkaia crosnieri (western Pacific) are the dominant metazoan fauna in their respective biogeographical provinces (Tunnicliffe et al. 2003; Moalic et al. 2012). All these species are known to be in symbiotic relationships with either endo- or epibiotic bacteria.
In 2010, the first hydrothermal vent fields south of the polar front were discovered on the East Scotia Ridge (ESR) (Rogers et al. 2012). The vent fields are situated ca. 440 km apart on segments E2 and E9 of the ESR at a similar depth, E2 at ca. 2600 m and E9 at ca. 2400 m. Both have a number of actively venting black smoker chimneys, where temperatures of up to 353°C (E2) and 383°C (E9) have been measured (Rogers et al. 2012). The dominant fauna at both sites is a new species of anomuran crab (Fig.1A and B). Morphological and molecular analyses identified the anomuran crab as a new species of yeti crab within the genus Kiwa (Roterman et al. 2013), hereafter referred to as Kiwa sp. nov. ESR, which has ventral setae covered in filamentous epibiotic bacteria (Rogers et al. 2012). Kiwa sp. nov. ESR is found in densities of up to 4000 individuals m−2 (Marsh et al. 2012), much higher than the other two known Kiwaids, Kiwa hirsuta (Macpherson et al. 2005), found at Pacific-Antarctic Ridge hydrothermal vents, and Kiwa puravida (Thurber et al. 2011), found at cold methane seeps off the Coast of Costa Rica. Given the high abundance of species like Kiwa sp. nov. ESR (Marsh et al. 2012), Rimicaris exoculata (>1120 ind. m−2, Copley et al. 2007) and S. crosnieri (>560 ind. m−2, Tokeshi 2011) the contribution of epibiont communities to the overall biological production of hydrothermal vents food webs is likely to be high.
Figure 1.
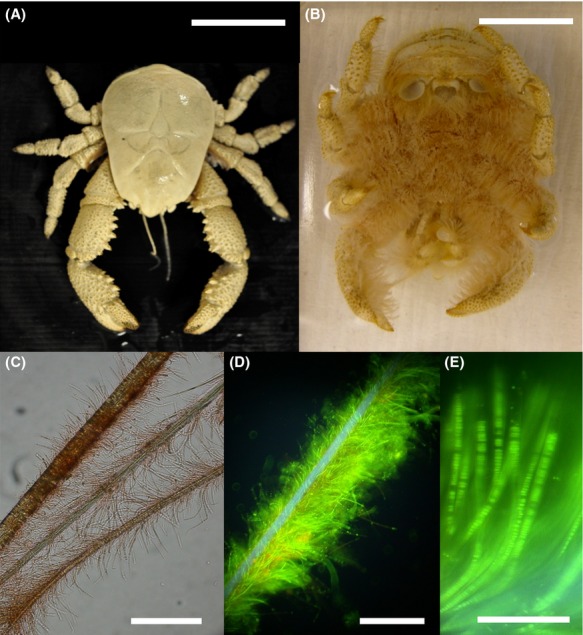
(A) Kiwa sp. nov. East Scotia Ridge (ESR), (B) ventral view, showing the setae covering the ventral side. (C) individual setae covered with epibiotic bacteria, (D and E) SYBR green staining of epibiotic bacteria. Scale bars: (A, B) – 5 cm, (C, D) – 100 μm, (E) – 50 μm.
Chemolithoautotrophic bacteria can fix carbon via different pathways, which are largely conserved within a phylogenetic group (reviewed by Hügler and Sievert 2011). Within the Epsilonproteobacteria, all autotrophic members are believed to use the reductive tricarboxylic acid (rTCA) cycle (Campbell and Cary 2004; Hügler et al. 2005; Takai et al. 2005), whereas the Calvin-Benson-Bassham (CBB) cycle is typical for autotrophic Gammaproteobacteria (Hügler and Sievert 2011 and references therein). Differences in the carbon fixation pathways are reflected in the stable carbon isotope (δ13C) signatures of the microbial primary producers resulting in δ13C values less than -20‰ and greater than −16‰ for organic carbon fixed via CBB and rTCA cycle, respectively (Hügler and Sievert 2011). As a result of minimal trophic discrimination in δ13C between food source and consumer, a metazoan consumer's δ13C values reflect those of their carbon source (Hügler and Sievert 2011). Spatial variability in δ13C can therefore reveal the relative importance of different food sources within and among locations when individuals consume food sources which have widely different δ13C values (Bearhop et al. 2004). The phylogenetic composition of the epibiont community, that is, for example, dominated by either Gammaproteobacteria or Epsilonproteobacteria, in combination with the δ13C values can be an indication of the carbon fixation pathway utilized by epibionts, and thus the relative contribution of either the CBB or rTCA cycles to sustaining the metazoan consumer.
In order to investigate epibiont-Kiwa interactions, vent fluids, epibionts, and Kiwa sp. nov. ESR were collected concurrently at various sites within the E2 and E9 vent fields. Preliminary chemical and physical analysis has revealed that the vent fields have contrasting vent fluid end-member chemistries and there are also detectable differences within E9 between the northern (E9N) and southern (E9S) parts of the vent field (Rogers et al. 2012). The major aims of this study were to: (1) examine epibiont diversity of the newly discovered Kiwa sp. nov. ESR at E2 and E9; (2) compare the Kiwa sp. nov. ESR epibiont community to that harbored by other hydrothermal decapods, which occupy a similar ecological niche; (3) examine the stable isotope signature of Kiwa sp. nov. ESR to assess whether the degree of isotopic variability was consistent with epibiont diversity.
Experimental Procedures
Sampling and sample preparation
Samples were collected during cruise JC42 on board the RRS James Cook in January–February 2010 using the remotely operated vehicle (ROV) Isis. Specimen were collected at various sites (as listed in Table2 and Table S2) at the E2 and E9 vent fields in the ESR. For DNA extraction and sequencing, the Kiwa sp. nov. ESR specimen were either fixed in 100% ethanol and then stored at −80°C or frozen directly at −80°C until used. Carapace length of specimens (CL) was measured from the base of the rostrum to the posterior-lateral margin of the carapace.
Table 2.
Details of the Kiwa sp. nov. ESR specimen, which were used for 454 analysis of the epibionts
| Sample | OTUs | Sequences | Coverage | Gender | Carapace length (cm) | Sample site | Shannon |
|---|---|---|---|---|---|---|---|
| 121_E2 | 283 | 2513 | 0.938 | Male | 9 | E2 – Anemone field | 3.196 |
| 122_E2 | 226 | 2522 | 0.956 | Male | 7.5 | E2 – Anemone field | 2.891 |
| 123_E2 | 329 | 2020 | 0.907 | Female | 5.5 | E2 – Anemone field | 4.287 |
| 124_E2 | 372 | 2317 | 0.916 | Female | 5.5 | E2 – Anemone field | 4.392 |
| 125_E2 | 285 | 2735 | 0.944 | Female | 6.5 | E2 – Anemone field | 2.932 |
| 2091_E2 | 168 | 2412 | 0.960 | Female | 5.5 | E2 – crab city | 2.387 |
| 2092_E2 | 178 | 1987 | 0.951 | Female | 4.5 | E2 – crab city | 2.715 |
| SLE2 | 281 | 4019 | 0.970 | Female | 7 | E2 – crab city | 2.825 |
| 2391_E9 | 210 | 2894 | 0.963 | Juvenile | 3 | E9 South – Marshland | 2.958 |
| 2392_E9 | 134 | 1800 | 0.961 | Juvenile | 3 | E9 South – Marshland | 2.619 |
| 2393_E9 | 52 | 549 | 0.949 | Juvenile | 3 | E9 South – Marshland | 2.331 |
| SL1_E9 | 94 | 1264 | 0.960 | Male | 5.5 | E9 North – B&W | 1.717 |
| SL2_E9 | 51 | 769 | 0.964 | Male | 6 | E9 North – B&W | 1.269 |
| SL5_E9 | 66 | 1161 | 0.974 | Juvenile | 3 | E9 North – B&W | 1.360 |
| SLE9 | 187 | 5221 | 0.987 | Juvenile | 3 | E9 North – B&W | 2.177 |
| Total | 34,183 |
ESR, East Scotia Ridge; OTUs, operational taxonomic units.
Microscope analysis
For SYBR green staining (Fig.1D and E), epibiont-covered setae of an ethanol-fixed Kiwa sp. nov. ESR were cut off with sterilized scissors and stained with a 200× solution of SYBR Green I (Sigma, St. Louis, MO) for 1 h at room temperature in the dark. The setae were washed with ddH2O and then analyzed with an epifluorescence microscope.
DNA extraction
A patch of setae covered with epibiotic bacteria was cut off from the ventral side of the Kiwa sp. nov. ESR with sterilized scissors and DNA was extracted using a phenol/chloroform protocol. See supplementary data S1.
PCR, cloning and Sanger sequencing
The 16S rRNA gene was amplified with the universal bacterial primers 1492R (Stackebrand and Liesack 1993) and 27F (Lane 1991). The PCR conditions were 94°C, 3 min, followed by 30 cycles of 94°C, 45 sec, 50°C, 30 sec, 72°C, 90 sec, and a final extension of 5 min at 72°C, using MyTaq polymerase (Bioline, Cambridge, UK). PCR products were cloned using TOPO TA cloning (Invitrogen/Life Technologies, Carlsbad, CA, USA) and sequenced bi-directionally (LGC Genomics, Berlin, Germany). Sequences have been submitted to Genbank (accession numbers KF438845–KF439049). Clone libraries for Sanger sequencing were generated from the epibionts of one E2 Kiwa sp. nov. ESR collected at Crab City and one E9 Kiwa sp. nov. ESR collected at Black and White.
454 sequencing
Primers 786Fm (Roesch et al. 2007) and 1194R (Maeda et al. 2003) were used to amplify part of the 16S rRNA gene. PCR conditions were 94°C, 3 min, followed by 30 cycles of 94°C, 45 sec, 50°C, 30 sec, 72°C, 60 sec, and a final extension of 5 min at 72°C using MyTaq polymerase (Bioline, UK Cambridge). Primer 786Fm was coupled with the fusion primer A plus a barcode to allow multiplexing, while primer 1194R was coupled with fusion primer B of the LibL chemistry. Sequencing was done from the 786R end, that is, sequences start just before V5 region. Sequencing was done by LGC Genomics (Berlin, Germany) on a Roche454 FLX Titanium sequencer 454 Life Sciences, Branford, CT. Sequence data have been submitted to Genbank, Biosample accession number SAMN02189919.
Sequence analysis
Sanger sequences were quality checked, assembled and trimmed with Geneious (www.geneious.com) and further analyzed with arb (Ludwig et al. 2004). Principal coordinate analysis was done with Fast unifrac (Hamady et al. 2009), based on a maximum likelihood tree generated in arb. Only sequences >1300 bp were used.
454 sequence data were analyzed with mothur v18.0 (Schloss et al. 2009) and arb. For further details see supplementary data S1.
The P (phylogenetic) test (Martin 2002) was used to test for significant differences between the epibiont communities from different sampling sites. The P-test uses a phylogenetic tree of the sample sequences and a parsimony approach to estimate the number of sequence changes necessary to explain the different distribution of sequences between samples. This tool is incorporated in the Fast unifrac website.
Stable isotope analysis
Muscle was removed from the chelipeds of Kiwa sp. nov. ESR, frozen at −80°C before being freeze dried and ground to a homogeneous powder for carbon and nitrogen SIA. Approximately 0.7 mg of powder was weighed into tin capsules and isotopic ratios were then measured by continuous-flow isotope ratio mass spectrometry using a Costech Elemental Analyzer interfaced with Thermo Finnigan Delta Plus XP (Natural Environment Research Council, Life Sciences Mass Spectrometry Facility, SUERC, East Kilbride, UK). Two laboratory standards were analyzed every ten samples in each analytical sequence. These alternated between paired alanine standards, of differing δ13C and δ15N, and an internal laboratory gelatin standard. Stable isotope ratios were expressed in delta (δ) notation as parts per mil (‰). All internal standards are traceable to the following international standards v-PDB (Pee Dee Belemnite) and AIR (atmospheric nitrogen). An external reference material of white fish muscle was also analyzed (δ13C, n = 24, −18.94‰ ± SD 0.09; δ15N, n = 24, 13.11‰ ± SD 0.38).
Isotope data analysis
Kiwa sp. nov. ESR isotopic niches were examined using the dispersion of δ13C and δ15N values in xy-space by calculating sample size corrected standard ellipse areas (SEAc) (Jackson et al. 2011) using the SIAR package (Parnell et al. 2010) implemented in the R statistical package version 3.0.1. See Jackson et al. (2011) and Data S1 for more details.
The relationships between δ13C and CL were analyzed by linear regression. Regression diagnostics were examined to assess normality (qq-plots) and homogeneity of variance (standardized residuals vs. fitted values).
Chemistry
Samples were taken and processed as described in Rogers et al. 2012 and James et al. 2014. Briefly, samples were taken by ROV using titanium major samplers linked to an inductively coupled temperature probe, and chloride and time sensitive parameters such as H2S and gases were fixed and analyzed on board. Major cations were analyzed by inductively coupled plasma – optical emission spectroscopy (ICP-OES) at the National Oceanography Centre Southampton after acidification, storage at 4°C and dilution. Results for each analyte from each vent chimney were corrected for seawater mixing by extrapolating to zero magnesium as is the convention, except for pH where the lowest measured result is reported.
Results
Physical and chemical properties of the vent fluids
The maximum temperatures of the vent fluids measured were 353°C at E2 vent and 383°C at E9 (Table1). Both vent fields had a similar pH of ca. 3, but differed in their chemistry. Recent volcanic activity possibly led to very low chloride and high H2S concentrations at E9 and particularly the southern (E9S) part of the E9 vent field (James et al. 2014). Vent fluids in the northern part of the E9 vent field (E9N) were hotter and slightly lower in H2S than E9S (Table1). However, these differences were very small in comparison with the differences to the E2 vent field, which had roughly seawater chloride concentrations, lower H2S, and a lower Fe:Mn ratio, all characteristic of sub-surface cooling. Despite the differences in the end-member vent fluids, within the diffuse flow areas around the chimneys, where most Kiwa were situated, concentrations of various chemicals as well as physical properties were very similar between E2 and E9 (Table1).
Table 1.
Location and properties of the sample sites (average of 2 measured values is shown, unless the values differed by more than 20%, in which case both values are shown)
| E2-Anemone field | E2-Crab city | E9 South-Marshland | E9 North-Black&White | Seawater1 | |
|---|---|---|---|---|---|
| Latitude | 56°5.31 S | 56°4.80 S | 60°2.79 S | 60°02.57 S | |
| Longitude | 30°19.07 W | 30°18.60 W | 29°58.71 W | 29°58.92 W | |
| Depth (m) | 2597 | 2641 | 2402 | 2400 | |
| Measurements in the diffuse flow near Kiwa | |||||
| pH | 7.59 | 6.39 | 7.37/5.91 | 5.96 | 7.5–8.4 |
| Temperature (°C) | 3.5 | 20 | 5/20 | 11 | ∼0 |
| H2S (mmol/L) | 0 | 0.12 | 0/0.1 | 0.1 | <0.000002 |
| SO4 (mmol/L) | 27.2 | 26.5 | 26.7 | 27.6 | 28 |
| Cl (mmol/L) | 531 | 541 | 525 | 539 | 546 |
| Mg (mmol/kg) | 52.7 | 49.9 | 51.5 | 52.0 | 52.7 |
| Mn (μmol/kg) | 9.76 | 92.9 | 0.82/18.9 | 4.18 | 0.00036 |
| Fe (μmol/kg) | 2.17 | 4.13 | 1.2/5.4 | 5.57 | 0.0005 |
| E2–Dog'sHead | E2-Sepia | E9 South – Ivory Tower | E9 North- Black&White | |
|---|---|---|---|---|
| Measurements in the end-member fluid of the nearest vent chimney | ||||
| pH | 3.02 | 3.05 | 3.08 | 3.42 |
| Temperature (°C) | 351 | 353 | 348 | 383 |
| H2S (mmol/L) | 6.7 | 7.1 | 11 | 9.5 |
| Cl (mmol/L) | 536 | 532 | 220 | 98.2 |
| Mn (μmol/kg) | 2050 | 2060 | 508 | 199 |
| Fe (μmol/kg) | 1280 | 1010 | 2520 | 800 |
Average values in seawater. Source: http://www.mbari.org/chemsensor/pteo.htm).
Microscopic analysis of epibionts
The setae of the Kiwa sp. nov. ESR were densely covered in bacteria, with a mostly filamentous cell morphology (Fig.1C–E). There were no obvious differences in cell morphology or cell density of the epibionts of E2 and E9 Kiwa sp. nov. ESR specimen. However, it should be noted that at E2 several crabs were observed further away (within 100 m) from actively venting chimneys. These individuals had notably fewer epibionts. They were mostly females and the majority of them were carrying eggs (Marsh 2014). All sequencing and isotope analysis were carried out on specimen collected in close proximity to the vents.
Identification of the epibionts by Sanger sequencing
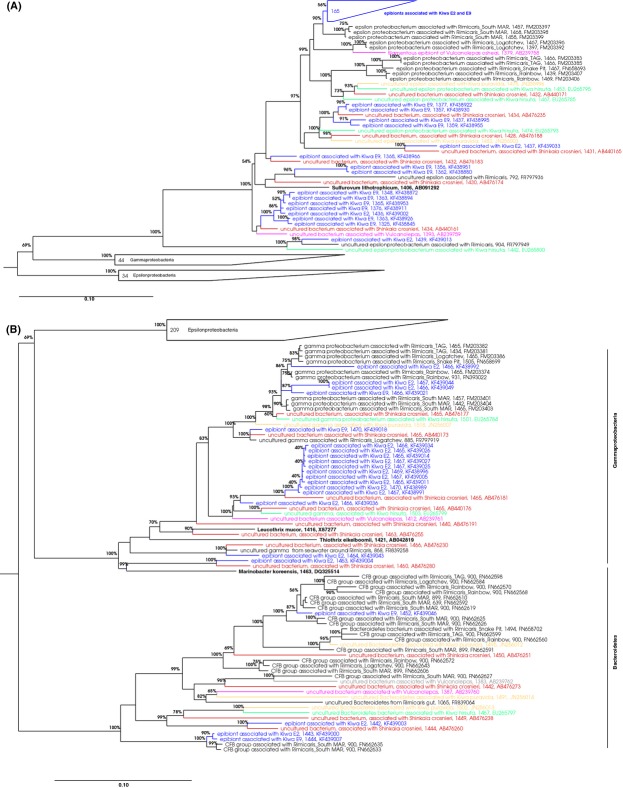
Sanger sequencing was used for a detailed phylogenetic classification of the dominant epibionts based on full length 16S rRNA sequences, while the 454 sequencing approach (see below) was used to compare the relative abundances of dominant phylogenetic groups and potential presence of rare taxa among epibionts from different Kiwa sp. nov. ESR specimen, sites, and vent fields. DNA was extracted from the epibionts of one E2 and one E9 Kiwa and clone libraries of the 16S rRNA gene were generated and sequenced. A total of 45 sequences from E2 and 165 sequences from E9 were analyzed (Table S1 and Fig.2). At E2, there was a roughly even distribution between Gamma- and Epsilonproteobacteria, as well as a few representatives from the Bacteroidetes. In contrast, at E9 the vast majority of sequences (97%) was assigned to the Epsilonproteobacteria. Most of these sequences clustered into three distinct phylotypes (i.e., with sequences within a phylotype showing a sequence identity of ≥97%). Two of the phylotypes were found at both E2 and E9, the third one was only found at E2 (Fig.2A).
Figure 2.
Neighbor joining tree of 16S rRNA sequences of Kiwa sp. nov. ESR (blue), Kiwa hirsuta (green), Kiwa puravida (yellow), Shinkaia crosnieri (red), Rimicaris exoculata (black), and Vulcanolepas sp. (pink). (A) Epsilonproteobacteria, (B) Gammaproteobacteria and Bacteroidetes. The tree was calculated within arb using neighbor joining with the Felsenstein correction using only sequences >1300 nt. Shorter sequences were then added by parsimony within arb. The length of each sequence is shown in the tree, followed by the Genbank accession number. Bootstrap values were calculated with the arb parsimony interactive tool. Only bootstrap values >50% are shown.
Within the Epsilonproteobacteria, all sequences from E2 and E9 fell into the Campylobacterales-Sulfurovum cluster, with Sulfurovum lithotrophicum as the closest cultured relative. The majority of E2 and E9 sequences within the Epsilonproteobacteria (87%) clustered into one distinct phylotype, which has a moderate sequence identity with S. lithotrophicum of 93.3%. More closely related to this dominant phylotype, with a sequence identity of 98.6%, are several sequences from uncultured epibionts associated with R. exoculata derived from the South vent field in the Mid Atlantic Ridge (South MAR) (Petersen et al. 2010). The dominant Kiwa sp. nov. ESR epibiont and R. exoculata South MAR epibiont form a separate cluster from other epibionts of various species of vent fauna, such as those associated with the other two currently described yeti crabs K. hirsuta and K. puravida, the more distantly related crab S. crosnieri, the stalked barnacle Vulcanolepas osheai, as well as R. exoculata epibionts from other MAR vent fields (Fig.2A).
Within the Gammaproteobacteria, sequences were assigned to the Thiotrichales-Thiothrix-Leucothrix cluster, with 81.8–89.9% sequence identity with Leucothrix mucor as the closest cultured relative. The dominant phylotype of E2 epibiont sequences has epibionts of S. crosnieri (94.6% sequence identity) and K. hirsuta (94.9%) as the closest uncultured relatives. Most of the other E2 and E9 epibiont sequences are more closely related to R. exoculata epibionts, including ones from the South MAR location, than to epibionts of other vent species (Fig.2B).
Within the Bacteroidetes, three of the four retrieved Kiwa sp. nov. ESR epibiont sequences have R. exoculata epibionts as the closest relative, with two of the sequences showing high similarity (98.3%) to South MAR epibionts (Fig.2B).
Comparison of E2 and E9 Kiwa sp. nov. ESR epibionts based on 454 pyrosequencing data
16S rDNA amplicon sequencing was carried out with epibionts from 15 individuals (eight from E2 and seven from E9). This generated a data set of 34,183 sequences after trimming and quality control. Kiwa sp. nov. ESR were collected at two different sites at E2 (Anemone field and Crab city) and two sites at E9 (E9S-Marshland and E9N-Black&White) (Table2 and Fig.3).
Figure 3.
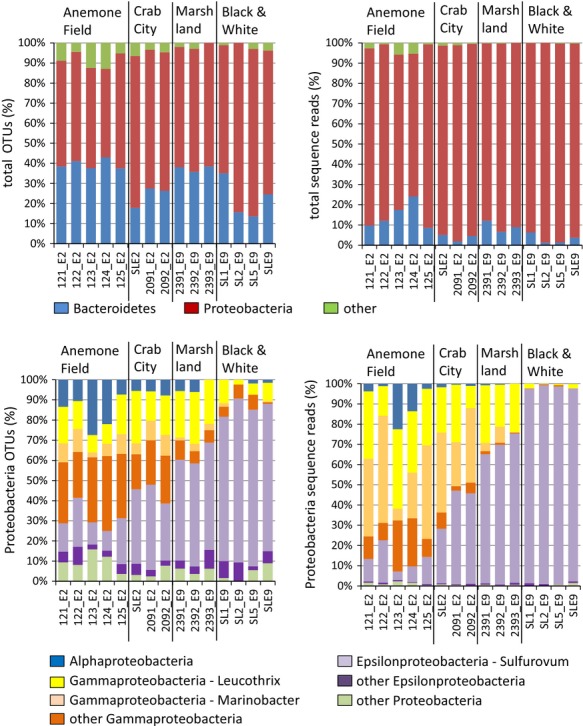
Phylogenetic analysis of 454 sequencing data of epibionts from 15 Kiwa sp. nov. ESR individuals. OTUs are defined at a cutoff of 0.03. ESR, East Scotia Ridge; OTUs, operational taxonomic units.
Overall Shannon diversity was higher in the E2 samples than the E9 samples. As expected based on the Sanger sequencing results, Proteobacteria and Bacteroidetes were the dominant phyla at all sites, with Proteobacteria accounting for 70.6–98.7% of all sequence reads and 44.1–84.3% of all operational taxonomic units (OTUs) (at 0.03 cutoff) and Bacteroidetes accounting for 1.3–24.1% of sequence reads and 13.6–43.0% of OTUs (Fig.3 and Table S2). The striking contrast between the Epsilonproteobacteria-dominated E9 epibionts and the Epsilon-/Gammaproteobacteria mix of E2 epibionts found with Sanger sequencing was supported with the increased replication that was undertaken with the 454 data for a larger number of samples derived from four different sampling sites. There was a high consistency of the phylogenetic composition of samples from the same site. Across the samples from the E9N Black & White site 94.4% (standard deviation 2.8) of all sequence reads were identified as Epsilonproteobacteria. At the E9S sampling site Marshland the proportion of Epsilonproteobacteria was lower (63.1%, SD 4.8) and Gammaproteobacteria were more abundant (26.7%, SD 3.2). At E2 the ratio was shifted more toward Gammaproteobacteria, with 55.7% (SD 7.0) and 64.5% (SD 9.0) at the E2 sites Crab City and Anemone Field compared to just 38.1% (SD 8.7) and 10.3% (SD 5.4) of Epsilonproteobacteria at these sites.
Notable is the appearance of up to 17.3% Alphaproteobacteria in the samples from E2 – Anemone Field. Within the Epsilonproteobacteria, sequences were almost exclusively assigned to the Campylobacterales-Sulfurovum cluster, while there was a split between Thiotrichales-Thiothrix-Leucothrix and Alteromonadales-Marinobacter within the Gammaproteobacteria. No Marinobacter sequences were found with the Sanger sequencing approach, although this may be due to the limited number of clones analyzed. In contrast to the Epsilon- and Gammaproteobacteria, where the majority of epibionts clustered into one group (represented by several phylotypes), the Bacteriodetes epibionts were widespread across several orders and multiple families with only distant similarities (<85%) to cultured species, but high similarities (>95%) to epibionts of other vent fauna, particularly R. exoculata. The most common Bacteroidetes families found among the Kiwa ESR epibionts were Saprospiraceae, Chitinophagaceae, and Flavobacteriaceae.
Principal coordinates analysis (PCoA) (Fig.4) confirmed the split between E2 and E9 epibiont communities and also showed a strong clustering of epibionts from Kiwa sp. nov. ESR collected at the same site. The epibiont community of samples from each site was significantly different (P < 0.001) from the epibiont communities of samples from the other sites (P-test, Martin 2002).
Figure 4.
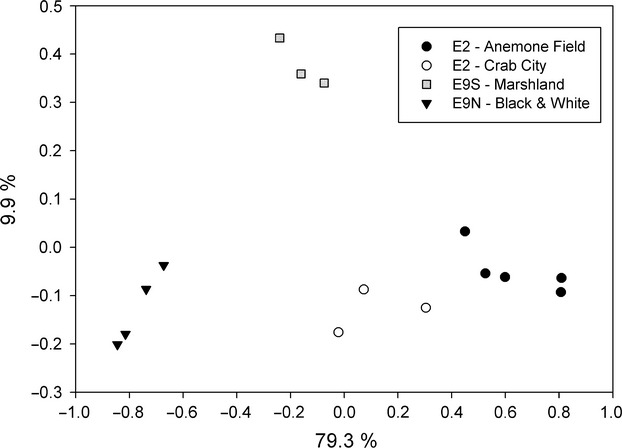
PCoA of Kiwa sp. nov. East Scotia Ridge (ESR) epibionts of 15 specimen at four E2 and E9 sampling sites based on 454 data.
Isotopic niches of Kiwa sp. nov. ESR at E2 and E9
The isotopic niche area (‰2), as defined by a sample size SEAc, overlapped by only 18% between female and male Kiwa sp. nov. ESR at E2 (Fig.5). Bayesian inference showed an 80% probability that males had a greater isotopic niche area than females at E2. Neither E2 male nor female isotopic niche area overlapped with the niche area of males collected from E9 (Fig.5). Bayesian inference indicated that there was a > 98% probability that E2 males and female isotopic niche area were greater than E9 male Kiwa sp. nov. ESR.
Figure 5.
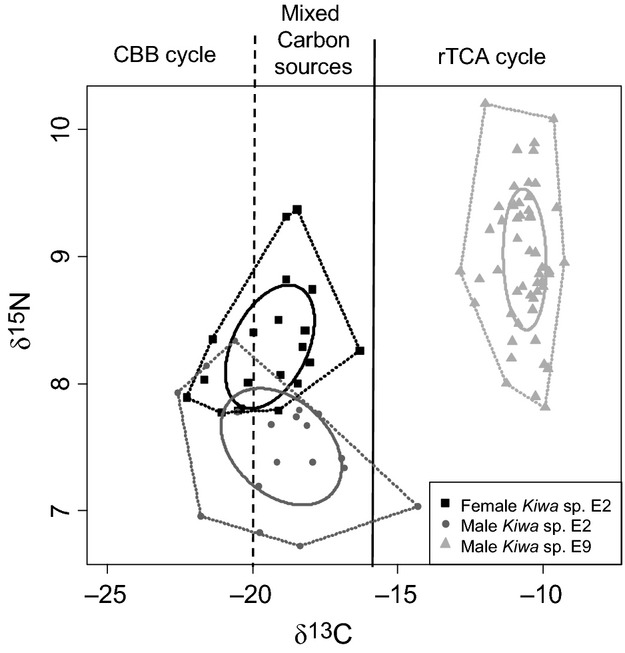
Isotopic niche of Kiwa sp. nov. East Scotia Ridge (ESR) δ13C and δ15N values for female (squares) and male (circles) Kiwa sp. nov. ESR from E2 and male (triangle) Kiwa sp. nov. ESR from E9 with their respective standard ellipse area (solid line) with a boundary plotted (dotted line) around the extreme points to aid visualization. The dashed and solid vertical lines represent the range of δ13C values for different carbon fixation pathways: <−20‰ for the Calvin-Benson-Bassham (CBB) cycle, >−16‰ for the reductive tricarboxylic acid (rTCA) cycle and −20‰ to −16‰ for mixing of the two carbon sources.
Trends related to the size of the host
Within the E2 vent field, the sampled female Kiwa sp. nov. ESR were significantly smaller (CL) than the males (t-test, n = 37, P < 0.01). The population at E9 was dominated by males and juveniles of undetermined sex and we were not able to collect females for stable isotope or epibiont sequence analysis. A comparison of male Kiwa sp. nov. ESR at E2 and E9 showed that the E2 individuals were significantly larger than those at E9 (t-test, n = 69, P < 0.01). At the same time, the Shannon diversity index (Table2) of the epibionts was higher at E2 than at E9 (t-test, n = 15 [including male, female and juvenile hosts], P < 0.01).
δ13C values increased with CL in male E9 Kiwa and male E2 Kiwa, but not in female E2 Kiwa (Fig.6).
Figure 6.
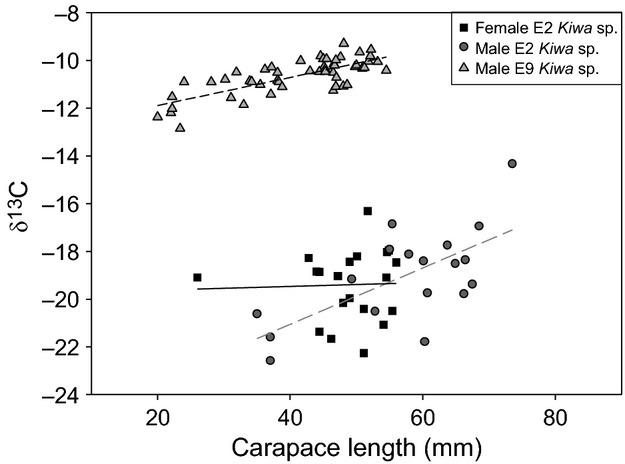
Plots of δ13C against carapace length (mm) for Kiwa sp. nov. East Scotia Ridge (ESR) for samples collected from E2 female (squares and solid line), E2 male (circles and long dash line), and E9 male (triangles and short dash line) Kiwa. In male Kiwa sp. nov. ESR there was an increase in δ13C with carapace length (mm) (CL) in E2 (δ13C = −25.78 + 0.118 CL; F1,16 = 12.93, r2adj = 0.41, P < 0.01) and E9 (δ13C = −13.06 + 0.059 CL; F1,50 = 67.81, r2adj = 0.58, P < 0.001). However, female Kiwa sp. nov. ESR did not increase in δ13C with CL (δ13C = −21.52 + 0.118 CL; F1,17 = 0.2428, r2adj = −0.04, P = 0.69).
Discussion
Phylogenetic diversity of the epibiont communities
The vast majority of the Epsilonproteobacteria among the Kiwa sp. nov. ESR epibionts fell into the Sulfurovum cluster. Members of the genus Sulfurovum are considered to be important primary producers in chemosynthetic systems (Campbell et al. 2006) and have been found living on various species of vent fauna close to active venting (Goffredi et al. 2008; Goffredi 2010; Petersen et al. 2010; Tsuchida et al. 2011). A recent metatranscriptomic analysis of a Sulfurovum dominated biofilm at Loki's Castle vent field on the MAR (Dahle et al. 2013) showed expression of genes involved in respiration with sulfur species, hydrogen, formate, nitrate, and oxygen. Particularly common were transcripts for cbb3-type cytochrome c oxidase, which supports microaerobic respiration, as well as transcripts of the sox gene cluster, responsible for oxidation of sulfur compounds. The presence of transcripts of the ATP citrate lyase and other key enzymes suggested C-fixation via the reverse tricarboxylic acid (rTCA) cycle.
Within the Gammaproteobacteria, the genera Leucothrix and Marinobacter were identified as the closest relatives (albeit with only moderate sequence identity) for a large proportion of the epibiont sequences at E2. Marinobacter is abundant in hydrothermal vent environments, particularly within metal sulfide deposits (Rogers et al. 2003; Kaye et al. 2011) and in water samples among mussels in the Logatchev vent field at MAR (Perner et al. 2013). To our knowledge, Marinobacter has not previously been found as a common component of epibiont communities. Physiological studies on Marinobacter isolates from the Juan de Fuca Ridge identified them as iron oxidizing, microaerophilic and obligate chemolithoautotrophs and suggested a role in the weathering of metal sulfide deposits (Edwards et al. 2003).
Leucothrix is also commonly found at hydrothermal vents, including the epibiont communities of K. hirsuta, S. crosnieri, and R. exoculata. The closest cultured relative, L. mucor, has been shown to be a chemolithoheterotroph, oxidizing thiosulfate (Grabovich et al. 1999). However, a study on R. exoculata showed evidence for the cbbM gene in the Leucothrix-containing epibiont community in the mouthparts of the shrimp (Hügler et al. 2011). The cbbM gene codes for RubisCO form II and is thus indicative for the capability of autotrophic growth via the CBB cycle. This was confirmed in a recent study of the metagenome of R. exoculata epibionts, which showed the presence of a complete set of genes for the CBB cycle in the Leucothrix-like epibionts (Jan et al. 2014). Therefore, a mixotrophic life-style of Leucothrix-like epibionts in vent systems has been suggested (Hügler et al. 2011).
Sequence identity of the Kiwa sp. nov. ESR epibionts with both Sulfurovum and Leucothrix is only moderate and deductions on the metabolic capabilities of the epibionts therefore uncertain. The closest relatives among uncultured bacteria for the dominant Epsilonproteobacteria phylotype as well as Gammaproteobacteria and Bacteroidetes sequences of the Kiwa sp. nov. ESR epibionts are not epibionts of other Kiwa species, but rather R. exoculata epibionts from the South MAR vent field. Petersen et al. (2010) analyzed R. exoculata epibionts from several vent fields along MAR and found a correlation between genetic distance of the epibionts and geographic distance. Interestingly, in that study the South MAR R. exoculata epibionts were shown to form a separate group from R. exoculata epibionts from other MAR vent fields. The geographic distance between the South MAR vent field and E9-ESR is very similar to the distance between South MAR and Rainbow (the northernmost MAR vent field analyzed by Petersen et al.), ca. 8500 km along the ridges. This points to a genetic link between the epibionts at ESR and South MAR with potentially several currently unknown vent sites along the ridges as stepping stones. The geographic distance along the edges of tectonic plates with known or unknown hydrothermal activity is greater between Kiwa sp. nov. ESR and either K. hirsuta, K. puravida or S. crosnieri than between Kiwa sp. nov. ESR and R. exoculata South MAR. This could explain why the Kiwa sp. nov. ESR epibionts were shown to be genetically more similar to R. exoculata South MAR epibionts than epibionts of any of the other vent species analyzed here.
Epibionts as food source for Kiwa sp. nov. ESR
The role of epibionts as a food source has been discussed for several vent and seep species, such as S. crosnieri (Watsuji et al. 2010; Tsuchida et al. 2011), K. puravida (Thurber et al. 2011), K. hirsuta (Goffredi et al. 2008) and R. exoculata (Polz and Cavanaugh 1995; Petersen et al. 2010; Ponsard et al. 2013). Evidence for this is based on isotopic signatures of epibionts and hosts (Van Dover 2002; Suzuki et al. 2005), uptake and transfer of isotope labeled inorganic carbon (Ponsard et al. 2013), fatty acid analysis (Suzuki et al. 2005; Thurber et al. 2011) as well as behavioral observations like the “dancing for food” (i.e., waving the epibiont-covered arms in nutrient rich seep fluid) described by Thurber et al. (2011) and combing epibionts toward the mouth (Thurber et al. 2011; Tsuchida et al. 2011).
Assuming that epibionts serve as a food source, differences in the epibiont community composition have implications for the δ13C values of associated bulk organic carbon of both epibionts and host. Gamma- and Epsilonproteobacteria fix carbon via different metabolic pathways, the CBB and rTCA cycle, respectively, resulting in isotopically distinct δ13C values (Hügler et al. 2011). Organic carbon produced via the rTCA cycle at hydrothermal vents tends to have values >−16‰, while values between −20‰ and −30‰ are believed indicative of the CBB cycle (Hügler et al. 2011).
We found clear differences in isotopic niches of Kiwa between E2 and E9 (Fig.5), which indicated that there were differences in the food source that was assimilated. At the same time, phylogenetic sequence analysis of the epibionts also showed striking differences between E2 and E9 (Fig.3). At E9, the δ13C values of the Epsilonproteobacteria, which dominated the epibiont community, (−9.9‰ SD 0.3, n = 5) indicate carbon fixation via the rTCA cycle (Reid et al. 2013). Furthermore, the δ13C values of E9 Kiwa (−10.6‰ SD 0.7, n = 52) also lie within the range reported for rTCA cycle (Reid et al. 2013) and therefore suggest a direct trophic link between the epibionts and Kiwa sp. nov. ESR.
At E2, the mean δ13C values for epibionts (−18.9‰ SD 5.3, n = 5) and host (−19.2‰ SD 1.7, n = 38) suggest a direct trophic link between epibionts and host, but they are slightly higher than the reported values for carbon fixation exclusively via CBB. It is likely that the δ13C values are the result of assimilation of a mix of CBB and rTCA fixed carbon. Following the hypothesis that the epibionts serve as a food source, both the lower mean and the broader range of δ13C values in E2 Kiwa compared to E9 Kiwa (Figs.5, 6) can be seen as an indication of a more diverse food source that reflects the spatial differences in the epibiont community associated with the ventral setae. Epibiont sequence data, showing a mix of Gamma- and Epsilonproteobaceria at E2 Kiwa, confirmed this. Taken together, the co-varying isotope values and epibiont diversity provide strong support for the food source hypothesis.
There was no evidence for Kiwa sp. nov. ESR preying on other vent animals. Grazing on bacterial mats around the vents could be another potential food source for the crabs. Unfortunately we do not have any sequence data for bacterial mats at ESR. Isotope analysis of rock scrapings at E9 and particulate suspended material at E2 (Reid et al. 2013) does not suggest a trophic link between bacteria from these substrates and Kiwa sp. nov. ESR, although it should be noted that the number of samples analyzed was too small to allow conclusive statistical analysis.
Differences in epibiont community composition within and between the ESR vent fields
Regional variations in the community composition of the epibionts are likely the result of a combination of factors. We propose that the main driving forces, in decreasing order of importance, are:
1. Geology and geochemistry of the vents. Geologically, E2 is characterized by more mature chimney structures and there are indications of sub-surface cooling, which can influence the chemical composition of the vent fluids (Seyfried and Ding 1993). E9, on the other hand, appears to be hydrothermally more active, possibly following a recent volcanic eruption, and is characterized by nascent chimneys and extremely low Cl- concentrations.
Differences in the chemical environmental conditions at E2 and E9 could account for some of the phylogenetic differences in the epibiont communities. It should be noted that the exact environmental parameters within the diffuse flow at the point where the Kiwa sp. nov. ESR were sampled were difficult to determine, as the location of the temperature probe, the Ocean Test Equipment (OTE) bottles for taking water samples and the robotic arms for collecting Kiwa sp. nov. ESR are located on different positions on the ROV and variation can occur at spatial scales of <1 m. The values given in Table1 for the diffuse flow therefore serve as a guideline. More conclusive are the measurements of the end-member fluids, which show distinct differences in the overall chemistry between E2 and E9. These fluids are diluted with seawater in sub-surface mixing before emission into the deep ocean in the diffuse flow areas.
The E9 vent field, and particularly E9N, is characterized by higher H2S and lower Cl- concentrations compared to E2 (Table1 and Rogers et al. 2012). This may benefit the sulfur oxidizer Sulfurovum. E2, on the other hand, has overall lower H2S concentrations, higher concentrations of Fe and especially high concentrations of Mn (Table1 and James et al. 2014). Members of the genus Marinobacter have been shown to be Fe oxidizers (Edwards et al. 2003) and at least one species, Marinobacter manganoxydans, is known to oxidize Mn and is able to tolerate extreme Mn concentrations (Wang et al. 2012).
2. Size (age) and moulting stage of the host. Kiwa at E2 were on average larger and had a more diverse epibiont community than Kiwa at E9, where a large proportion of the population consisted of juveniles. Using size as a proxy for age, this could be an indication that the epibiont community composition changes with age. This has been described in R. exoculata (Guri et al. 2012) and more recently in K. puravida (Goffredi et al. 2014). In both cases, a shift from a Gammaproteobacteria dominated to an Epsilonproteobacteria-dominated epibiont community was observed from egg to adult. We did not analyze any eggs or larvae of Kiwa sp. nov. ESR, so may only see later stages of a possible shift. However, the clear increase of δ13C values with size, within both E2 and E9 (Fig.6), indicates a change in the food source of Kiwa sp. nov. ESR and therefore putatively a change of their epibionts. As δ13C increases with size, this would suggest that more carbon is being assimilated that was fixed via the rTCA cycle. Length-based trends in δ13C are also found in other caridean vent shrimps (Polz et al. 1998; Van Dover 2002). For example, R. exoculata increases in δ13C by 6–7‰ with increasing length as it changes from a photosynthetic diet during larval and juvenile stages to a chemosynthetic diet as an adult (Polz et al. 1998; Vereshchaka et al. 2000; Van Dover 2002). The δ13C-length trends in R. exoculata are in contrast to the gradual increase in δ13C with size observed in Kiwa sp. nov. ESR sp. Even though Kiwa sp. nov. ESR δ13C-length relationships varied with site, the trends still suggest they were not driven by a dilution of photosynthetic primary production over the size range sampled but are reflective of a change in the utilization of carbon within the hydrothermal vent. Despite the 454 sequencing sample size not being large enough to draw reliable conclusions regarding a correlation between microbial diversity and size, the much greater sample size for stable isotope analysis (n = 90) would suggest that changes in epibiont communities with size or gender is a feasible hypothesis.
The Kiwa moulting cycle may also have an effect on the epibiont community composition. After each moulting, epibiotic bacteria have to recolonize their host. In Rimicaris, this recolonization involves a succession of different bacteria (Corbari et al. 2007; Guri et al. 2012). At E2, Kiwa generally had a visibly “dirtier” carapace than Kiwa at E9 and showed some necrosis of the setae, which is indicative of a later stage of the moulting cycle (Sven Thatje, pers. comm. 2013). This could account for the stark contrast between the Epsilonproteobacteria-dominated E9 epibionts and the Gamma-/Epsilonproteobacteria mix at E2.
3. Differential behavior of male and female Kiwa sp. nov. ESR. At E2, the δ13C values of Kiwa sp. nov. ESR showed a broader range than at E9 as well as a distinct split between males and females (Figs.5, 6).
This may be due to changes in behavior and location throughout the life cycle of the crabs (Marsh 2014). While juveniles (particularly at E9) were generally found sitting in diffuse flow regions at the bottom of actively venting chimneys, larger, and especially large male Kiwa sp. nov. ESR were often observed climbing toward the top of a chimney (Marsh et al. 2012), where they are likely to be exposed to different chemical and physical environmental conditions. At E2, females were frequently observed further away from active venting and outside areas of diffuse flow, resulting again in different environmental conditions. A large number of these females were carrying eggs (Sven Thatje, pers. comm 2010). At the same time, these individuals had noticeably fewer epibionts (K. Zwirglmaier, pers. obs. 2010), underlining the dependence of the epibionts on the environmental conditions in close proximity to the vents. No sequence data are available for these off-site individuals and at this point it is unknown, whether adult Kiwa sp. nov. ESR regularly move from these extreme locations (off-site or on top of chimney) to the more temperate conditions within the diffuse flow.
Summary and Conclusion
This study presents the first description of the epibiont community of a new species of Kiwa from the first hydrothermal vent system discovered in the Antarctic. High throughput sequencing analysis of the epibionts of 15 individuals revealed strong regional differences between the two studied vent fields, which were also reflected in δ13C values. The combination of the sequencing results with δ13C values of both the hosts and the epibionts provided insights into the putative carbon fixation pathways of the epibionts and the symbiotic relationship with their hosts and supports the hypothesis that the epibionts serve as a food source. Near full-length 16S rRNA Sanger sequencing helped to clarify the phylogenetic affiliation of the major groups identified with the shorter 454 sequences and showed that most phylotypes of the Kiwa sp. nov. ESR epibionts are closely related to R. exoculata epibionts from the South MAR vent field. The major Epsilonproteobacteria epibionts of Kiwa sp. nov. ESR form a subgroup together with the R. exoculata South MAR epibionts that is separate from epibiont communities of other vent fauna.
Future work on the metagenome and/or metatranscriptome in combination with further (geo)chemical analysis of the vent fields is required to further describe the epibiotic relationship of the microorganisms and the host and define their role in the nutrient cycles of this vent system.
Acknowledgments
The authors thank the ChEsSO consortium PI Prof. Paul Tyler, the PSO Alex Rogers, the Master, and crew of the RRS James Cook during cruise JC42, as well as the team of technicians of the ROV Isis. We would also like to thank Rona McGill of SUERC for undertaking the isotope ratio mass spectrometry sample analysis and Sven Thatje, University of Southampton, for valuable discussions on Kiwa sp. nov. ESR anatomy and behavior. NERC funded the study through the ChEsSO – Chemosynthetically driven ecosystems south of the Polar Front: biogeography and ecology consortium grant (grant number NE/D01249x/1, consortium PI: Prof. Paul Tyler), sample analysis through Life Sciences Mass Spectrometry Facilities grant LSMSFBRIS043_04/10_R_09/10 and studentships NE/F010664/1 (W. D. K. R.) and NE/H524922 (J. A. H.). The authors declare that there are no conflicts of interest.
Conflict of Interest
None declared.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Data S1. Supplementary information.
Table S1. Comparison of the phylogenetic association of near full-length (>1300 nt) 16S rRNA sequences of epibionts of Kiwa sp. nov ESR at E2 and E9.
Table S2. 454 sequence data. (A) % sequence reads, (B) % OUT at 0.03 cutoff.
Table S3. Details of Kiwa sp. nov ESR specimen collected for stable isotope analysis.
References
- Bearhop S, Adams CE, Waldron S, Fuller RA. Macleod H. Determining trophic niche width: a novel approach using stable isotope analysis. Anim. Ecol. 2004;73:1007–1012. [Google Scholar]
- Beinart RA, Sanders JG, Faure B, Sylva SP, Lee RW, Becker EL, et al. Evidence for the role of endosymbionts in regional-scale habitat partitioning by hydrothermal vent symbioses. Proc. Natl Acad. Sci. USA. 2012;109:E3241–E3250. doi: 10.1073/pnas.1202690109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell BJ. Cary SC. Abundance of reverse tricarboxylic acid cycle genes in free-living microorganisms at deep-sea hydrothermal vents. Appl. Environ. Microbiol. 2004;70:6282–6289. doi: 10.1128/AEM.70.10.6282-6289.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell BJ, Engel AS, Porter ML. Takai K. The versatile epsilon-proteobacteria: key players in sulphidic habitats. Nat. Rev. Microbiol. 2006;4:458–468. doi: 10.1038/nrmicro1414. [DOI] [PubMed] [Google Scholar]
- Copley JTP, Jorgensen PBK. Sohnt RA. Assessment of decadal-scale ecological change at a deep Mid-Atlantic hydrothermal vent and reproductive time-series in the shrimp Rimicaris exoculata. J. Mar. Biol. Assoc. U.K. 2007;87:859–867. [Google Scholar]
- Corbari L, Zbinden M, Cambon-Bonavita MA, Gaill F. Compère P. Bacterial symbionts and mineral deposits in the branchial chamber of the hydrothermal vent shrimp Rimicaris exoculata: relationship to moult cycle. Aquat. Biol. 2007;1:225–238. [Google Scholar]
- Cuvelier D, Sarrazin J, Colaco A, Copley J, Desbruyeres D, Glover AG, et al. Distribution and spatial variation of hydrothermal faunal assemblages at Lucky Strike (Mid-Atlantic Ridge) revealed by high-resolution video image analysis. Deep Sea Res. Part I Oceanogr. Res. Pap. 2009;56:2026–2040. [Google Scholar]
- Dahle H, Roalkvam I, Thorseth IH, Pedersen RB. Steen IH. The versatile in situ gene expression of an epsilonproteobaceria dominated biofilm from a hydrothermal chimney. Environ. Microbiol. Rep. 2013;5:282–290. doi: 10.1111/1758-2229.12016. [DOI] [PubMed] [Google Scholar]
- Edwards KJ, Rogers DR, Wirsen CO. McCollom TM. Isolation and characterization of novel psychrophilic, neutrophilic, Fe-oxidizing, chemolithoautotrophic α- and γ-proteobacteria from the deep sea. Appl. Environ. Microbiol. 2003;69:2906–2913. doi: 10.1128/AEM.69.5.2906-2913.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores GE, Campbell JH, Kirshtein JD, Meneghin J, Podar M, Steinberg JI, et al. Microbial community structure of hydrothermal deposits from geochemically different vent fields along the Mid-Atlantic Ridge. Environ. Microbiol. 2011;13:2158–2171. doi: 10.1111/j.1462-2920.2011.02463.x. [DOI] [PubMed] [Google Scholar]
- Goffredi SK. Indigenous ectosymbiotic bacteria associated with diverse hydrothermal vent invertebrates. Environ. Microbiol. Rep. 2010;2:479–488. doi: 10.1111/j.1758-2229.2010.00136.x. [DOI] [PubMed] [Google Scholar]
- Goffredi SK, Jones WJ, Erhlich H, Springer A. Vrijenhoek RC. Epibiotic bacteria associated with the recently discovered yeti crab, Kiwa hirsuta. Environ. Microbiol. 2008;10:2623–2634. doi: 10.1111/j.1462-2920.2008.01684.x. [DOI] [PubMed] [Google Scholar]
- Goffredi SK, Gregory A, Jones WJ, Morella NM. Sakamoto RI. Ontogenetic variation in epibiont community structure in the deep-sea yeti crab, Kiwa puravida: convergence among crustaceans. Mol. Ecol. 2014;23(6):1457–1472. doi: 10.1111/mec.12439. , and 2014 Mar; . doi: 10.1111/mec.12439 . Epub 2013 Aug 16. [DOI] [PubMed] [Google Scholar]
- Grabovich MY, Muntyan MS, Lebedeva VY, Ustiyan VS. Dubinina GA. Lithoheterotrophic growth and electron transfer chain components of the filamentous gliding bacterium Leucothrix mucor DSM 2157 during oxidation of sulfur compounds. FEMS Microbiol. Lett. 1999;178:155–161. [Google Scholar]
- Guri M, Durand L, Cueff-Gauchard V, Zbinden M, Crassous P, Shillito B, et al. Acquisition of epibiotic bacteria along the life cycle of the hydrothermal shrimp Rimicaris exoculata. ISME J. 2012;6:597–609. doi: 10.1038/ismej.2011.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamady M, Lozupone C. Knight R. Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J. 2009;4:17–27. doi: 10.1038/ismej.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hügler M. Sievert SM. Beyond the Calvin cycle: autotrophic carbon fixation in the ocean. Ann. Rev. Mar. Sci. 2011;3:261–289. doi: 10.1146/annurev-marine-120709-142712. [DOI] [PubMed] [Google Scholar]
- Hügler M, Wirsen CO, Fuchs G, Taylor CD. Sievert SM. Evidence for autotrophic CO2 fixation via the reductive tricarboxylic acid cycle by members of the ε subdivision of proteobacteria. J. Bacteriol. 2005;187:3020–3027. doi: 10.1128/JB.187.9.3020-3027.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hügler M, Petersen JM, Dubilier N, Imhoff JF. Sievert SM. Pathways of carbon and energy metabolism of the epibiotic community associated with the deep-sea hydrothermal vent shrimp Rimicaris exoculata. PLoS One. 2011;6:e16018. doi: 10.1371/journal.pone.0016018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AL, Inger R, Parnell AC. Bearhop S. Comparing isotopic niche widths among and within communities: SIBER – Stable Isotope Bayesian Ellipses in R. J. Anim. Ecol. 2011;80:595–602. doi: 10.1111/j.1365-2656.2011.01806.x. [DOI] [PubMed] [Google Scholar]
- James RH, Green DRH, Stock MJ, Alker BJ, Banerjee NR, Cole C, et al. Composition of hydrothermal fluids and mineralogy of associated chimney material on the East Scotia Ridge back-arc spreading centre. Geochim. Cosmochim. Acta. 2014;139:47–71. , 15 August 2014, [Google Scholar]
- Jan C, Petersen JM, Werner J, Teeling H, Huang S, Glöckner FO, et al. The gill chamber epibiosis of deep-sea shrimp Rimicaris exoculata: an in-depth metagenomic investigation and discovery of zetaproteobacteria. Environ. Microbiol. 2014;16(9):2723–2738. doi: 10.1111/1462-2920.12406. 2014 Sep; . doi: 10.1111/1462-2920.12406 Epub 2014 Mar 4. [DOI] [PubMed] [Google Scholar]
- Kaye JZ, Sylvan JB, Edwards KJ. Baross JA. Halomonas and Marinobacter ecotypes from hydrothermal vent, subseafloor and deep-sea environments. FEMS Microbiol. Ecol. 2011;75:123–133. doi: 10.1111/j.1574-6941.2010.00984.x. [DOI] [PubMed] [Google Scholar]
- Lane DJ. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York, NY: John Wiley & Sons; 1991. pp. 115–147. , eds. . [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar A, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macpherson E, Jones W. Segonzac M. A new squat lobster family of Galatheoidea (Crustacea, Decapoda, Anomura) from the hydrothermal vents of the Pacific-Antarctic Ridge. Zoosystema. 2005;27:709–723. [Google Scholar]
- Maeda H, Fujimoto C, Haruki Y, Maeda T, Kokeguchi S, Petelin M, et al. Quantitative real-time PCR using TaqMan and SYBR Green for Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis, Prevotella intermedia, tetQ gene and total bacteria. FEMS Immunol. Med. Microbiol. 2003;39:81–86. doi: 10.1016/S0928-8244(03)00224-4. [DOI] [PubMed] [Google Scholar]
- Marsh L. 2014. Controls on faunal microdistribution and reproductive development in deep-sea chemosynthetic environments in the Antarctic . University of Southampton, Ocean and Earth Science, Doctoral thesis. Doctoral thesis University of Southampton, Ocean and Earth Science.
- Marsh L, Copley JT, Huvenne VAI, Linse K, Reid WDK, Rogers AD, et al. Microdistribution of faunal assemblages at deep-sea hydrothermal vents in the Southern Ocean. PLoS One. 2012;7:e48348. doi: 10.1371/journal.pone.0048348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AP. Phylogenetic approaches for describing and comparing the diversity of microbial communities. Appl. Environ. Microbiol. 2002;68:3673–3682. doi: 10.1128/AEM.68.8.3673-3682.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moalic Y, Desbruyeres D, Duarte CM, Rozenfeld AF, Bachraty C. Arnaud-Haond S. Biogeography revisited with network theory: retracing the history of hydrothermal vent communities. Syst. Biol. 2012;61:127–137. doi: 10.1093/sysbio/syr088. [DOI] [PubMed] [Google Scholar]
- Parnell AC, Inger R, Bearhop S. Jackson AL. Source partitioning using stable isotopes: coping with too much variation. PLoS One. 2010;5:e9672. doi: 10.1371/journal.pone.0009672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perner M, Gonnella G, Hourdez S, Böhnke S, Kurtz S. Girguis P. In situ chemistry and microbial community compositions in five deep-sea hydrothermal fluid samples from Irina II in the Logatchev field. Environ. Microbiol. 2013;15:1551–1560. doi: 10.1111/1462-2920.12038. [DOI] [PubMed] [Google Scholar]
- Petersen JM, Ramette A, Lott C, Cambon-Bonavita M-A, Zbinden M. Dubilier N. Dual symbiosis of the vent shrimp Rimicaris exoculata with filamentous gamma- and epsilonproteobacteria at four Mid-Atlantic Ridge hydrothermal vent fields. Environ. Microbiol. 2010;12:2204–2218. doi: 10.1111/j.1462-2920.2009.02129.x. [DOI] [PubMed] [Google Scholar]
- Petersen JM, Zielinski FU, Pape T, Seifert R, Moraru C, Amann R, et al. Hydrogen is an energy source for hydrothermal vent symbioses. Nature. 2011;476:176–180. doi: 10.1038/nature10325. [DOI] [PubMed] [Google Scholar]
- Podowski EL, Ma SF, Luther GW, Wardrop D. Fisher CR. Biotic and abiotic factors affecting distributions of megafauna in diffuse flow on andesite and basalt along the Eastern Lau Spreading Center, Tonga. Mar. Ecol. Prog. Ser. 2010;418:25–45. [Google Scholar]
- Polz MF. Cavanaugh CM. Dominance of one bacterial phylotype at a Mid-Atlantic Ridge hydrothermal vent site. Proc. Natl Acad. Sci. USA. 1995;92:7232–7236. doi: 10.1073/pnas.92.16.7232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polz M, Robinson J, Cavanaugh C. Va Dover C. Trophic ecology of massive shrimp aggregations at a Mid-Atlantic Ridge hydrothermal vent site. Limnol. Oceanogr. 1998;43:1631–1638. [Google Scholar]
- Ponsard J, Cambon-Bonavita M-A, Zbinden M, Lepoint G, Joassin A, Corbari L, et al. Inorganic carbon fixation by chemosynthetic ectosymbionts and nutritional transfers to the hydrothermal vent host-shrimp Rimicaris exoculata. ISME J. 2013;7:96–109. doi: 10.1038/ismej.2012.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid WDK, Sweeting CJ, Wigham BD, Zwirglmaier K, Hawkes JA, McGill RAR, et al. Spatial differences in East Scotia Ridge hydrothermal vent food webs: influences of chemistry, microbiology and predation on trophodynamics. PLoS One. 2013;8:e65553. doi: 10.1371/journal.pone.0065553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesch LFW, Fulthorpe RR, Riva A, Casella G, Hadwin AKM, Kent AD, et al. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 2007;1:283–290. doi: 10.1038/ismej.2007.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers DR, Santelli CM. Edwards KJ. Geomicrobiology of deep-sea deposits: estimating community diversity from low-temperature seafloor rocks and minerals. Geobiology. 2003;1:109–117. [Google Scholar]
- Rogers AD, Tyler PA, Connelly DP, Copley JT, James R, Larter RD, et al. The discovery of new deep-sea hydrothermal vent communities in the Southern Ocean and implications for biogeography. PLoS Biol. 2012;10:e1001234. doi: 10.1371/journal.pbio.1001234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roterman CN, Copley JT, Linse K, Tyler PA. Rogers AD. The biogeography of the yeti crabs (Kiwaidae) with notes on the phylogeny of the Chirostyloidea (Decapoda: Anomura) Proc. R. Soc. B Biol. Sci. 2013;280:20130718. doi: 10.1098/rspb.2013.0718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyfried WE., Jr Ding K. The effect of redox on the relative solubilities of copper and iron in Cl-bearing aqueous fluids at elevated temperatures and pressures: an experimental study with application to subseafloor hydrothermal systems. Geochim. Cosmochim. Acta. 1993;57:1905–1917. [Google Scholar]
- Stackebrand E. Liesack W. Nucleic acids and classification. In: Goodfellow M, O'Donnell AG, editors; Handbook of new bacterial systematics. London: Academic Press; 1993. pp. 152–189. , eds., and . [Google Scholar]
- Suzuki Y, Sasaki T, Suzuki M, Nogi Y, Miwa T, Takai K, et al. Novel chemoautotrophic endosymbiosis between a member of the epsilonproteobacteria and the hydrothermal vent gastropod Alviniconcha aff. hessleri (Gastropoda: Provannidae) from the Indian Ocean. Appl. Environ. Microbiol. 2005;71:5440–5450. doi: 10.1128/AEM.71.9.5440-5450.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai K, Campbell BJ, Cary SC, Suzuki M, Oida H, Nunoura T, et al. Enzymatic and genetic characterization of carbon and energy metabolisms by deep-sea hydrothermal chemolithoautotrophic isolates of epsilonproteobacteria. Appl. Environ. Microbiol. 2005;71:7310–7320. doi: 10.1128/AEM.71.11.7310-7320.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai K, Nunoura T, Horikoshi K, Shibuya T, Nakamura K, Suzuki Y, et al. Variability in microbial communities in black smoker chimneys at the NW caldera vent field, Brothers volcano, Kermadec arc. Geomicrobiol J. 2009;26:552–569. [Google Scholar]
- Thurber AR, Jones WJ. Schnabel K. Dancing for food in the deep sea: bacterial farming by a new species of Yeti crab. PLoS One. 2011;6:e26243. doi: 10.1371/journal.pone.0026243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokeshi M. Spatial structures of hydrothermal vents and vent-associated megafauna in the back-arc basin system of the Okinawa Trough, western Pacific. J. Oceanogr. 2011;67:651–665. [Google Scholar]
- Tsuchida S, Suzuki Y, Fujiwara Y, Kawato M, Uematsu K, Yamanaka T, et al. Epibiotic association between filamentous bacteria and the vent-associated galatheid crab, Shinkaia crosnieri (Decapoda: Anomura) J. Mar. Biol. Assoc. U.K. 2011;91:23–32. [Google Scholar]
- Tunnicliffe V, Juniper SK. Sibuet M. Reducing environments of the deep-sea floor. In: Tyler PA, editor; Ecosystems of the deep ocean. Amsterdam: Elsevier Science; 2003. pp. 81–110. , ed., and . [Google Scholar]
- Van Dover C. Trophic relationships among invertebrates at the Kairei hydrothermal vent field (Central Indian Ridge) Mar. Biol. 2002;141:761–772. [Google Scholar]
- Vereshchaka A, Vinogradov G, Yu Lein A, Dalton S. Dehairs F. Carbon and nitrogen isotopic composition of the fauna from the Broken Spur hydrothermal vent field. Mar. Biol. 2000;136:11–17. [Google Scholar]
- Wang H, Li H, Shao Z, Liao S, Johnstone L, Rensing C, et al. Genome sequence of deep-sea manganese-oxidizing bacterium Marinobacter manganoxydans MnI7-9. J. Bacteriol. 2012;194:899–900. doi: 10.1128/JB.06551-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watsuji T-O, Nakagawa S, Tsuchida S, Toki T, Hirota A, Tsunogai U, et al. Diversity and function of epibiotic microbial communities on the galatheid crab, Shinkaia crosnieri. Microbes Environ. 2010;25:288–294. doi: 10.1264/jsme2.me10135. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Supplementary information.
Table S1. Comparison of the phylogenetic association of near full-length (>1300 nt) 16S rRNA sequences of epibionts of Kiwa sp. nov ESR at E2 and E9.
Table S2. 454 sequence data. (A) % sequence reads, (B) % OUT at 0.03 cutoff.
Table S3. Details of Kiwa sp. nov ESR specimen collected for stable isotope analysis.