Abstract
Autophagy is a catabolic process conserved among eukaryotes. Under nutrient starvation, a portion of the cytoplasm is non-selectively sequestered into autophagosomes. Consequently, ribosomes are delivered to the vacuole/lysosome for destruction, but the precise mechanism of autophagic RNA degradation and its physiological implications for cellular metabolism remain unknown. We characterized autophagy-dependent RNA catabolism using a combination of metabolome and molecular biological analyses in yeast. RNA delivered to the vacuole was processed by Rny1, a T2-type ribonuclease, generating 3′-NMPs that were immediately converted to nucleosides by the vacuolar non-specific phosphatase Pho8. In the cytoplasm, these nucleosides were broken down by the nucleosidases Pnp1 and Urh1. Most of the resultant bases were not re-assimilated, but excreted from the cell. Bulk non-selective autophagy causes drastic perturbation of metabolism, which must be minimized to maintain intracellular homeostasis.
Keywords: autophagy, metabolome analysis, ribosome, RNA degradation, vacuole
See also: E Welter & Z Elazar (January 2015)
Introduction
Every cellular activity is maintained by a balance between continuous synthesis and degradation of constituents, proteins, nucleic acids, and lipids. Autophagy, a principal pathway involved in degradation of cellular components, is a membrane pathway that is highly conserved among eukaryotes (Klionsky, 2007; Ohsumi, 2014). Autophagy occurs constitutively at a low level, thereby serving as an important means for quality control by removing excessive or damaged proteins and organelles. Nutrient starvation notably stimulates autophagy, contributing to cell survival by recycling constituents and maintaining energy levels. During autophagy, a portion of the cytoplasm including organelles is sequestered into autophagosomes and delivered to the lysosome/vacuole, where the contents of autophagosomes are degraded by various hydrolytic enzymes. Bulk degradation via autophagy is principally a non-selective process; however, recently selective or preferential degradation of proteins and organelles has attracted intense attention and has been shown to play important roles on cell physiology (Reggiori et al, 2012).
To date, more than 38 autophagy-related (Atg) proteins have been identified and characterized in yeast (Ohsumi, 2014). Of these, 18 Atg proteins, Atg1–Atg10, Atg12–Atg14, Atg16–Atg18, Atg29, and Atg31, have been identified as components essential for autophagosomal formation upon starvation. Deletion of any of these proteins results in complete defect in autophagy and decreased viability under nitrogen starvation conditions. Most of these proteins are also required for selective types of autophagy, such as the cytoplasm-to-vacuole targeting (Cvt) pathway, mitophagy, and pexophagy (Suzuki, 2013). Other Atg proteins are dispensable for starvation-induced non-selective autophagy, but are specifically required for selective types of autophagy.
In rapidly growing yeast cells, the cytoplasm contains large quantities of ribosomes (2 × 105/cell) (Warner, 1999); these ribosomes contain almost 50% of all cellular proteins and 80% of total RNA. Under nutrient-rich conditions, more than 2,000 ribosomes are assembled every minute, and cellular ribosome contents correlate closely with growth rate. Upon nitrogen starvation, ribosome synthesis is immediately stopped and superfluous ribosomes are degraded. In our first paper on autophagy in yeast, we showed that most autophagosomes contain the same density of ribosomes as the cytoplasm, and occasionally contain even more ribosomes per unit volume (Takeshige et al, 1992; Fig1A). During ribosome degradation via autophagy, not only ribosomal proteins, but also a large amount of ribosomal RNAs must be degraded in the vacuole/lysosome. To date, the process of protein degradation has been extensively studied, but the molecular details of RNA degradation via autophagy remain mostly unknown. Pioneer work by Mortimore using a rat liver perfusion system showed that bulk RNA degradation is induced under amino-acid starvation (Lardeux et al, 1987; Mortimore et al, 1989). Recently, selective ribosome degradation, ribophagy, has been reported in yeast (Kraft et al, 2008). Although the mechanisms are not completely understood, it is clear that ribosomes are targets of selective autophagy as well as non-selective degradation.
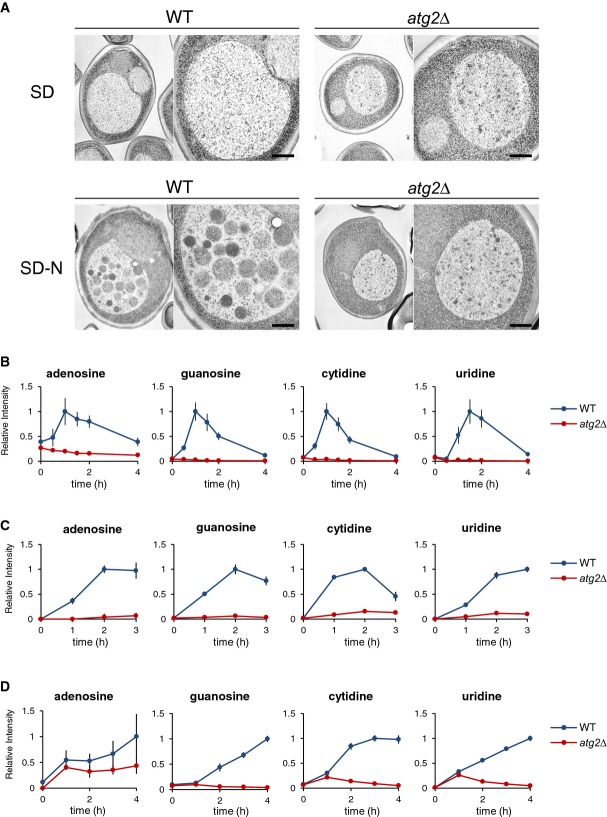
Figure 1. Dynamics of intracellular nucleosides under starvation.
- A Electron microscopic analyses to see autophagic bodies in the vacuoles under both growth and nitrogen starvation conditions. Autophagic bodies typically contain cytosolic ribosomes. pep4Δprb1Δ and pep4Δprb1Δatg2Δ cells were grown in SD to mid-log phase (OD600 = 1) and transferred to SD(-N) for 5 h. The cells were examined by transmission electron microscopy as described in Materials and Methods. Scale bar, 500 nm.
- B–D Time-dependent changes in nucleoside contents under nitrogen starvation (B), rapamycin treatment (C), and carbon starvation (D) conditions. Wild-type and atg2Δ cells were grown in SD and transferred to SD(-N), SD medium with 0.2 μM rapamycin, or SD(-C) at time 0. At the indicated time points, nucleosides were analyzed by LC/MS as described in Materials and Methods. Results are presented as normalized intensities on the basis of peak height of each metabolite in wild-type cells. All data are means of triplicate samples. The error bars represent the standard deviation.
Ribonucleases (RNases) play important roles in many aspects of RNA metabolism. Multiple types of RNase have been identified and classified according to their structural and biochemical properties. Broadly, RNases can be divided into endoribonucleases and exoribonucleases. Classical biochemical studies revealed the existence of alkaline RNases, such as the RNase T1 and RNase A families, and acid RNases, comprising the RNase T2 family (Irie, 1999). RNase T2 is an endonuclease with weak nucleobase specificity that is found in all organisms. In yeast, Rny1 is the sole T2 family RNase identified, but its biological role has not yet been determined (MacIntosh et al, 2001).
Using metabolome analysis, we investigated the dynamic change of intracellular metabolites under starvation conditions. In addition, using molecular genetic approaches tractable in yeast, we obtained a comprehensive picture of RNA degradation via autophagy. In this study, we identified the enzymes involved in the process and characterized the dynamic flow of RNA metabolism under nitrogen starvation conditions.
Results
Nitrogen starvation conditions induce a transient increase in intracellular nucleoside contents
To deepen our understanding of the physiological roles of autophagy in metabolism, we performed metabolome analysis. To minimize metabolic requirements, we derived all strains used in these experiments from the prototrophic wild-type strain X2180. Wild-type and autophagy-defective atg2Δ cells were grown in minimal synthetic defined (SD) medium to mid-log phase (optical density [OD600] about 1) and then transferred to nitrogen starvation [SD(-N)] medium to induce autophagy. Using liquid chromatography/mass spectrometry (LC/MS), we examined the time-dependent changes in the contents of intracellular metabolites. As expected, we observed that amino acids were depleted during starvation (Onodera & Ohsumi, 2005). Strikingly, contents of nucleosides (adenosine, guanosine, cytidine, and uridine) exhibited remarkable changes in wild-type cells during nitrogen starvation (Fig1B): Intracellular levels increased for up to 1 h (for adenosine, guanosine, and cytidine) or 1.5 h (for uridine), started decreasing, and subsided after 4 h of starvation. By contrast, such transient increase was not observed in atg2Δ cells at any time during nitrogen starvation.
We next examined the change of nucleosides under other autophagy-triggering conditions. Upon induction of autophagy by rapamycin, which induces the starvation response even under nutrient-rich conditions (Noda & Ohsumi, 1998), wild-type but not atg2Δ cells showed a transient increase in nucleoside levels (Fig1C). Upon induction of autophagy by carbon starvation (SD(-C)), nucleosides continued to increase in an autophagy-dependent manner (Fig1D). Therefore, all autophagy-triggering conditions tested here inevitably cause transient elevation of intracellular nucleoside levels, although the kinetics seems to be different among starvation conditions. As autophagy is intensively induced by nitrogen starvation and lasts for at least several hours, this transient generation of nucleosides strongly indicates that nucleosides are rapidly further metabolized during nitrogen starvation (see below). To follow this, we used nitrogen starvation as the representative autophagy-triggering conditions for the rest of this study.
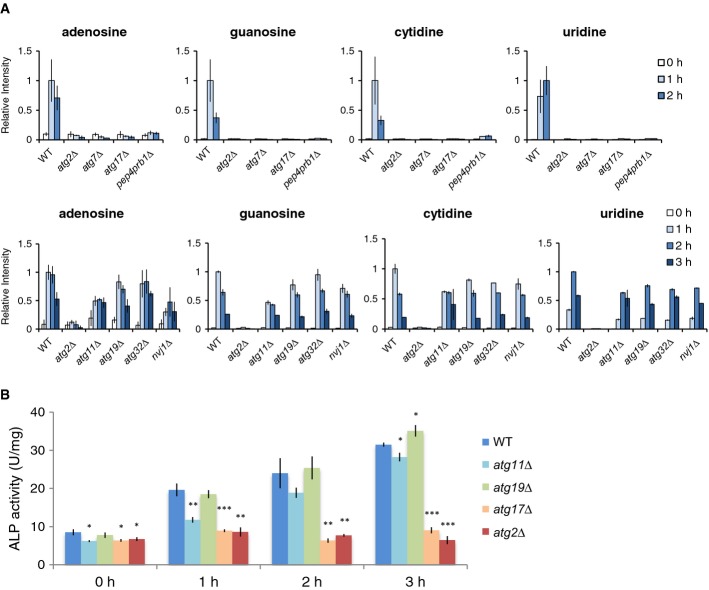
We next investigated which type of autophagy is involved in the generation of nucleosides during starvation. Mutants lacking ATG7 or ATG17, both of which are essential for starvation-induced autophagy, exhibited no accumulation of nucleosides at all, as in atg2Δ (Fig2). By contrast, deletion of ATG19, ATG32, or NVJ1, which are necessary for the Cvt pathway, the constitutive and selective transport system for vacuolar enzymes (Harding et al, 1995; Scott et al, 1997, 2001), mitophagy (Kanki et al, 2009; Okamoto et al, 2009), and the piecemeal microautophagy of the nucleus (Roberts et al, 2003), respectively, did not affect the transient increases in nucleoside levels (Fig2A). Atg11 was originally identified as an essential component for the Cvt pathway. However, recent studies suggested that although not essential, Atg11 also functions together with core autophagy components during starvation-induced autophagy (Mao et al, 2013). In agreement with this, atg11Δ cells exhibited a moderate increase in nucleoside levels as compared with wild-type cells (Fig2A). In accordance with this result, autophagic activities of atg11Δ cells were partially defective (Fig2B), which is considered to be the primary cause that led to the reduced level of nucleoside increase. Pep4 and Prb1 are the major proteases in the vacuole, and their deletions cause accumulation of autophagic bodies in the vacuole under starvation (Takeshige et al, 1992). In pep4Δprb1Δ cells, the increase in nucleoside levels was completely abolished (Fig2A), suggesting that disruption of autophagic bodies is required for generation of nucleosides. These data indicate that non-selective autophagy contributes substantially to the increase in nucleoside levels during starvation, whereas selective autophagy plays a less significant role. We discuss ribophagy below.
Figure 2. Changes in intracellular nucleoside levels under nitrogen starvation.
- Time-dependent changes in nucleoside contents under nitrogen starvation. Nucleosides in wild-type, atg2Δ, atg7Δ, atg11Δ, atg17Δ, atg19Δ, atg32Δ, nvj1Δ, and pep4prb1Δ cells under nitrogen starvation. All data are means of triplicates. The error bars represent the standard deviation.
- Alkaline phosphatase (ALP) assay (Pho8Δ60). Wild-type, atg2Δ,atg11Δ, atg17Δ, and atg19Δ cells expressing Pho8Δ60 were grown in SD to mid-log phase and transferred to SD(-N) at time 0. At the indicated time points, lysates were prepared and subjected to the ALP assay. The bars represent the standard deviation of three independent experiments. *P < 0.05; **P < 0.005; ***P < 0.0005 (paired t-test, two-tailed)
Role of Pho8, a vacuolar nucleotidase
We hypothesized that these nucleosides were derived from degradation of cytoplasmic RNAs in the vacuole and that each nucleoside was derived from the corresponding nucleotide. In our earlier electron microscopic observation of cells under nutrient starvation, we presented morphological evidence showing that cytoplasmic ribosomes are sequestered to autophagosomes and delivered to the vacuole for degradation (Takeshige et al, 1992; Fig1A). Given that ribosomes are highly abundant supramolecular structures composed of proteins and ribosomal RNAs (rRNA) at a nearly 1:1 weight ratio (Warner, 1999), it is likely that the nucleosides we detected under starvation were mostly derived from rRNAs.
We first investigated how nucleosides were generated from nucleotides. Recently, Phm8 was identified as a nucleotidase responsible for RNA degradation under starvation in yeast (Xu et al, 2013). If Phm8 functions in starvation-induced RNA degradation process by dephosphorylating nucleotides, phm8Δ cells should produce reduced levels of nucleosides and consequently accumulate higher levels of nucleotides. Contrary to our expectation, phm8Δ cells accumulated similar levels of both nucleosides and nucleotides to those observed in wild-type cells, indicating that Phm8 is not requisite for RNA degradation under nitrogen starvation (Supplementary Fig S1).
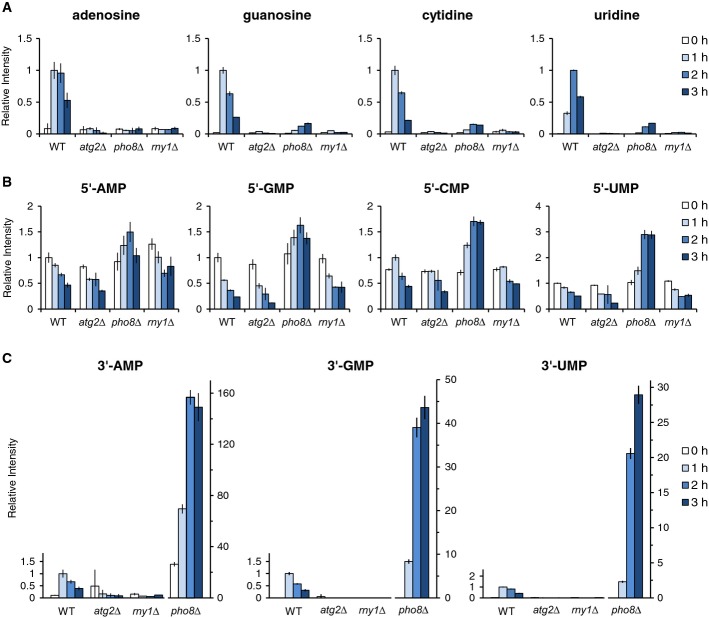
We next focused on the vacuolar non-specific phosphatase, Pho8, as a candidate phosphatase/nucleotidase (Kaneko et al, 1982; Klionsky & Emr, 1989). In pho8Δ cells, the increase of nucleosides was dramatically diminished during nitrogen starvation (Fig3A). 5′-NMP levels (5′-AMP, 5′-GMP, 5′-CMP, and 5′-UMP) were slightly higher in pho8Δ cells than those in wild-type and atg2Δ cells (Fig3B). Given that the cytosol contains considerably high levels of 5′-nucleotides and other related compounds such as NAD and UDP-glucose, a portion of them would be enwrapped into autophagosomes and delivered to the vacuoles, in which they might be retained in the absence of Pho8. Nevertheless, the accumulation of 5′-NMP in pho8Δ cells was not as remarkable as we had expected. Thus, it seems likely that the nucleosides detected here were not mainly derived from 5′-NMPs. We therefore decided to measure intracellular 3′-NMPs, which are thought to be relatively minor metabolites in normal cells (see below). Our LC/MS was capable of determining the amounts of 3′-AMP, 3′-GMP, and 3′-UMP but not 3′-CMP, which lacks a high-quality authentic standard. Just like nucleosides, 3′-NMPs exhibited transient increases in wild-type and phm8Δ cells, but not in atg2Δ cells (Fig3C and Supplementary Fig S1). As shown in Fig3C, in pho8Δ cells, 3′-NMPs accumulated at very high levels in response to nitrogen starvation. Based on these results, we concluded that the nucleosides were derived from 3′-NMPs and Pho8 is the nucleotidase responsible for autophagy-induced RNA degradation. This conclusion is consistent with previous biochemical studies showing that Pho8 is a non-specific phosphatase with broad substrate specificity and that Phm8 is a 5′-NMP-specific nucleotidase (Plankert et al, 1991; Donella-Deana et al, 1993; Xu et al, 2013).
Figure 3. Changes in intracellular nucleotide and nucleoside levels under nitrogen starvation.
- A–C Wild-type, atg2Δ, pho8Δ, and rny1Δ cells were grown in SD to mid-log phase and transferred to SD(-N) at time 0. At the indicated time points, nucleosides (A), 5′-NMPs (B), and 3′-NMPs (C) were analyzed by LC/MS as described in Materials and Methods. The results are presented as normalized intensities on the basis of the peak height of each metabolite in wild-type cells. Note that 3′-NMPs in pho8Δ are presented in a different scale. All data are means of triplicates. The error bars represent the standard deviation.
Rny1 is the nuclease responsible for RNA degradation in the vacuole
Next, we asked how 3′-NMPs are generated in the vacuole. Searching for a possible RNase, we took note of Rny1, the sole vacuolar RNase identified to date in S. cerevisiae (MacIntosh et al, 2001). Rny1 belongs to the T2 family of RNases, highly conserved endoribonucleases with weak base specificity that catalyze the cleavage of single-strand RNA through 2′,3′-cyclic phosphate intermediates, yielding mono- or oligonucleotides with a terminal 3′ phosphate group (Irie, 1999). These enzymatic characteristics suggest that Rny1 acts on RNA, but not DNA. We investigated the involvement of Rny1 in generation of 3′-NMPs during autophagy. In rny1Δ cells, the elevation of 3′-NMPs, and consequently that of nucleosides, was completely abolished (Fig3A–C). Even prolonged starvation of up to 24 h did not result in any nucleoside accumulation in rny1Δ cells (Supplementary Fig S2A). We concluded that Rny1 is the essential ribonuclease for RNA degradation into mononucleotides by autophagy.
To exclude the possibility that Rny1 and Pho8 are involved in some aspect of the autophagic process itself, such as sensing of starvation, induction of autophagy, autophagosome formation, or protein degradation, we monitored autophagy in rny1Δ and pho8Δ cells. First, we quantitatively estimated the autophagic activity of rny1Δ cells using the alkaline phosphatase (ALP) assay (Noda et al, 1995). Deletion of Rny1 did not affect autophagic activity at all (Supplementary Fig S2B). We next examined autophagy by fluorescence microscopy using GFP-Atg8 as a marker. GFP-Atg8 expressed in rny1Δ and pho8Δ cells was transported normally to the vacuoles in response to starvation (Supplementary Fig S2C). From these results, we concluded that Rny1 and Pho8 do not affect the membrane dynamics necessary for autophagy, but instead play specific roles in RNA degradation via autophagy.
RNY1 deletion causes accumulation of RNA in the vacuole
Although previous studies have suggested that Rny1 and Pho8 are vacuolar enzymes, the physiological roles of these proteins in the vacuole have not yet been well defined. Indeed, other physiological functions have been suggested for Rny1: Under standard nutrient conditions, a fraction of Rny1 is secreted from the cell through the ER and Golgi apparatus and has been proposed to play a role in scavenging nutrients from extracellular RNAs (MacIntosh et al, 2001; Shcherbik, 2013). Thompson et al suggested that Rny1 exits the vacuole to the cytosol and specifically cleaves cytosolic tRNA, as well as rRNA, in response to oxidative stress (Thompson & Parker, 2009). In the case of Pho8, although an authentic function has not been established, molecular details of its induction under phosphate starvation, delivery to the vacuole, and activation have been well studied (Kaneko et al, 1985; Klionsky & Emr, 1989). Pho8 is transported to the vacuole through the ER and the Golgi apparatus. Upon reaching the vacuole, the pro-form of Pho8 is C-terminally processed to an active form by vacuolar protease Pep4 and Prb1. We examined the expression and cellular localization of Rny1 and Pho8 under our conditions. For this purpose, we constructed cells chromosomally expressing Rny1-GFP or Pho8-GFP via their own promoters in wild-type and atg2Δ cells. Rny1 and Pho8 were localized in the vacuoles, under both growth and starvation conditions (Fig4A), and we observed no striking difference between wild-type and atg2Δ. As previously suggested, large portions of Rny1-GFP as well as Pho8-GFP were C-terminally processed and liberated from the GFP moiety in the vacuole (Fig4B) (Campomenosi et al, 2006). By estimating the total amounts of these proteins as the sum of the full-length and cleaved bands, we found that Pho8 expression was highly induced under nitrogen starvation conditions, and Rny1 expression was moderately upregulated (Fig4A and B).
Figure 4. Accumulation of RNA in the vacuole in rny1Δ cells under nitrogen starvation.
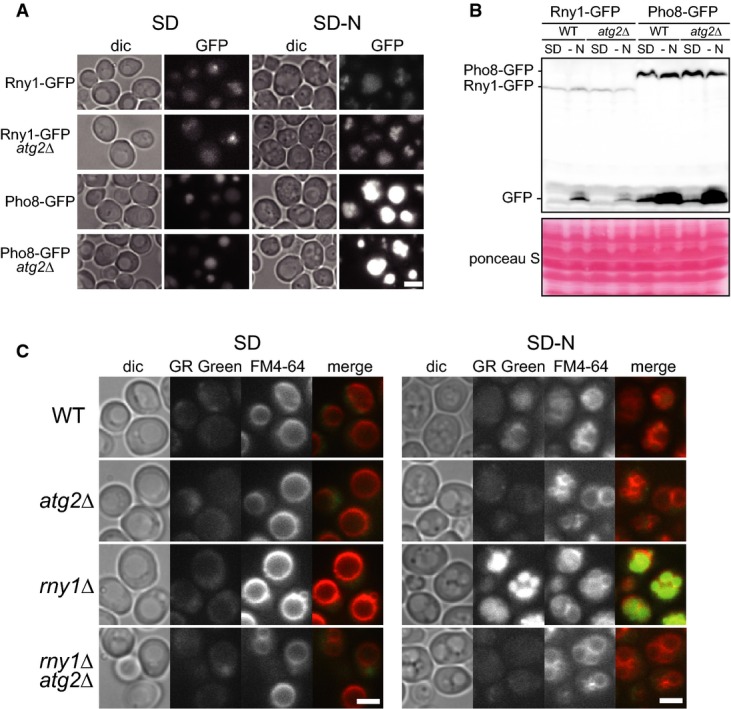
- A, B Expression and localization of Rny1 and Pho8. Wild-type cells and atg2Δ cells expressing Rny1-GFP or Pho8-GFP were grown in SD to mid-log phase and transferred to SD(-N). After 2 h of starvation, GFP-tagged proteins were observed by fluorescence microscopy (A) or analyzed by Western blot with anti-GFP antibody (B). Scale bar, 5 μm.
- C Detection of free RNA. Wild-type, atg2Δ, rny1Δ, and rny1Δatg2Δ cells grown in SD (left) or SD(-N) for 2 h (right) were stained with FM4-64 and GR Green and observed under a fluorescence microscope. Scale bar, 5 μm.
We next performed microscopic monitoring of free RNA in cells using the nucleic acid-binding fluorescent dye, GR Green. Under growing conditions, wild-type, atg2Δ, and rny1Δ cells did not show significant fluorescence signals (Fig4C). By contrast, under starvation conditions, vacuoles in wild-type cells but not atg2Δ cells were faintly but significantly stained. Strikingly, rny1Δ cells exhibited brightly stained vacuoles with the dye (Fig4C), indicating that in the absence of Rny1, free RNA was retained in the vacuole without further degradation. Deletion of ATG2 in rny1Δ cells abolished starvation-induced fluorescence signals in the vacuoles, which further supports our conclusion that starvation-induced RNA degradation in the vacuoles is completely autophagy-dependent. Based on these observations, it is clear that Rny1 functions as the vacuolar nuclease for RNA degradation during autophagy.
Ubp3- and Bre5-mediated ribophagy plays an irrelevant role in starvation-induced RNA degradation
Recently, Kraft et al (2008) reported a novel type of selective autophagy, ribophagy, which degrades ribosomes preferentially to other cytosolic proteins during nitrogen starvation. They showed that Ubp3 and Bre5, a ubiquitin deconjugation enzyme and its cofactor, mediate preferential degradation of ribosomal 60S proteins, but not 40S proteins. Therefore, we asked the question whether ribophagy plays an important role in nucleotide metabolism in our experimental conditions. We constructed ubp3Δ and bre5Δ cells and analyzed the changes in nucleotides and related metabolites. Similar levels of transient accumulation of 3′-NMPs were observed (Fig5A). However, the time course of 3′-NMP generation was slightly delayed, and the conversion of 3′-NMPs to nucleosides was also slow, resulting in reductions of nucleoside levels. It is possible that these phenotypes were caused partly by some defect in the formation of ribophagy-specific autophagosomes under our experimental conditions, or simply the slow-growth phenotype of these mutants. However, it is more likely that these proteins regulate membrane trafficking processes, as reported in a previous study (Cohen et al, 2003). In fact, we found these mutants have partial defects in delivery or processing of vacuolar enzymes, as demonstrated by the presence of pro-forms of these enzymes under both growing and starvation conditions (Supplementary Fig S3A). Furthermore, endogenous Pho8 activity was low (Supplementary Fig S3B), which may explain the lower levels of nucleoside generation in both ubp3Δ and bre5Δ cells (Fig5A). Microscopic monitoring of free RNA in rny1Δubp3Δ and rny1Δbre5Δ using GR Green also clearly showed fluorescence signals inside the vacuoles under starvation conditions, demonstrating that RNAs are transported to the vacuole in the absence of Ubp3 and Bre5 (Fig5B). At present, it is hard to estimate how many ribosomes are delivered via the ribophagy pathway, but it is clear that at least a certain number of ribosomes are delivered to the vacuole via non-selective and bulk autophagy.
Figure 5. Evaluation of ribophagy on RNA degradation under nitrogen starvation.
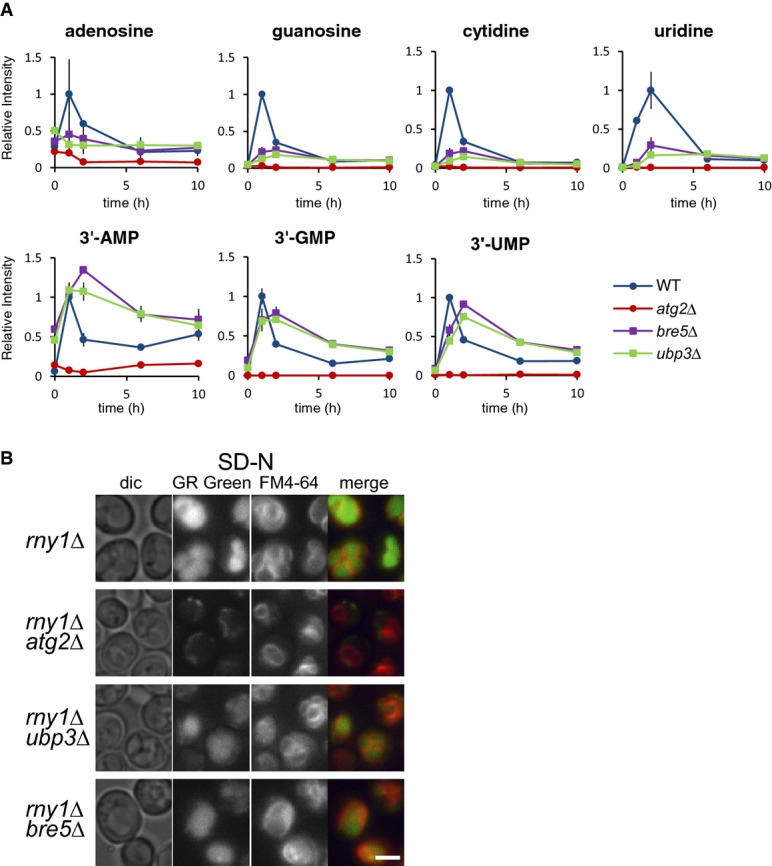
- Time-dependent change in nucleoside levels after nitrogen starvation. Wild-type, atg2Δ, ubp3Δ, and bre5Δ cells were grown in SD to mid-log phase and transferred to SD(-N) at time 0. Metabolites were analyzed by LC/MS as described in Materials and Methods. The results are presented as normalized intensities on the basis of the peak height of each metabolite in wild-type cells. All data are means of triplicates. The error bars represent the standard deviation.
- Detection of free RNA within cells. rny1Δ, rny1Δatg2Δ, rny1Δubp3Δ, and rny1Δbre5Δ cells grown in SD(-N) for 2 h were stained with FM4-64 and GR Green and observed under a fluorescence microscope. Scale bar, 5 μm.
Further breakdown of nucleosides in the cytosol
Although autophagy continues for at least several hours under nitrogen starvation conditions (Supplementary Fig S2B), intracellular 3′-NMPs and nucleosides increased only transiently, suggesting that these compounds are rapidly metabolized further and exist in a state of dynamic equilibrium. To obtain a comprehensive picture of autophagy-dependent RNA catabolism, we quantitated the intracellular concentrations of nucleotides, nucleosides, and bases under starvation with standard addition method by LC/MS (Fig6).
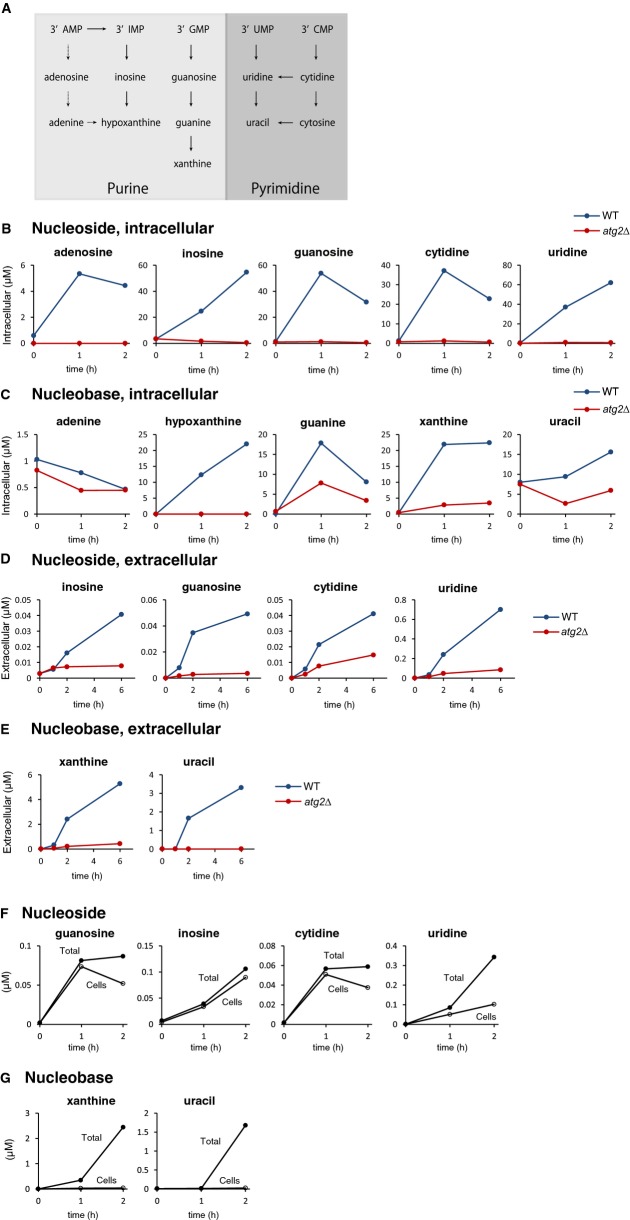
Figure 6. Quantitative analysis of nucleotides, nucleosides, and bases at their destinations.
- A Schematic representation of reactions of the nucleotide degradation pathway.
- B, C Nucleosides (B) and nucleobases (C) within the cells. Wild-type and atg2Δ were grown in SD to mid-log phase and transferred to SD(-N) at time 0. At the indicated time points, the cells were collected; their metabolites were extracted, and the nucleosides and nucleobases were analyzed by LC/MS. The level of each nucleoside and base is presented as its absolute concentration, as described in Materials and Methods and Supplementary Methods. All data are means of duplicates except the wild-type strain at 1 h.
- D, E Nucleosides (D) and nucleobases (E) in the medium. The culture medium of the above cultures was filtered, and nucleosides and nucleobases were directly analyzed by LC/MS as described in Materials and Methods and Supplementary Methods. All data are means of duplicates.
- F, G Destinations of nucleosides and bases. Based on the data from (B–E), the level of each nucleoside and base in cells (open circle) and in cells and medium (closed circle) of wild-type strain were estimated. See Supplementary Methods in detail (flow chart of the calculation procedure).
In wild-type cells at 2 h starvation, intracellular concentrations of nucleosides were calculated to be about 4 μM (adenosine), 30 μM (guanosine), 20 μM (cytidine), or 60 μM (uridine) (Fig6B). The fact that adenosine level is quite low as compared with other nucleosides might be because that most 3′-AMP was converted to 3′-IMP and then into inosine by Pho8 (Fig6A). Indeed, we found that inosine level increased to about 50 μM after 2 h of starvation in wild-type cells (Fig6B).
We also detected autophagy-dependent generation of the nucleobases guanine and uracil within the cell (Fig6C); cytosine was not detectable in our LC/MS system. The results explained why nucleosides were only transiently increased during autophagy. Significant amounts of hypoxanthine and xanthine were detected in an autophagy-dependent manner, strongly suggesting that enzymes normally working in the purine or pyrimidine salvage pathway would produce bases under starvation conditions. Since these enzymes are localized in the cytoplasm (Breker et al, 2013), nucleosides must be effectively transported from the vacuole to the cytoplasm. In the purine salvage pathway, hypoxanthine comes from either inosine or adenine, and xanthine comes from guanine (Fig6A). Pnp1 is a purine nucleoside phosphorylase that specifically acts on guanosine and inosine (Lecoq et al, 2001); the enzyme responsible for generation of adenine from adenosine has not yet been described. Urh1 is a pyrimidine nucleoside-specific hydrolase converting cytidine and uridine into cytosine and uracil, respectively (Kurtz et al, 1999), in the pyrimidine salvage pathway, during which cytidine, uridine, and cytosine are all eventually converted into uracil (Fig6A). We investigated the requirements for Pnp1 and Urh1 in nucleoside metabolism during starvation. In pnp1Δ cells, guanosine and inosine accumulated to much higher levels than in wild-type cells, whereas levels of guanine and xanthine were reduced (Supplementary Fig S4A). By contrast, in urh1Δ cells, the level of uracil was remarkably decreased (Supplementary Fig S4B). Therefore, we concluded that Pnp1 and Urh1 are the nucleosidases responsible for autophagy-induced RNA degradation, consistent with a recent study by Xu et al (2013). We further analyzed the cellular localization of Pnp1 and Urh1 by chromosomally expressing Pnp1-GFP or Urh1-GFP via their own promoters in wild-type cells. Pnp1 and Urh1 were found to be localized in the cytosol but not in the vacuole under both growth and starvation conditions (Supplementary Fig S4C), suggesting that autophagy-derived nucleosides are converted to nucleobases in the cytoplasm.
Excretion of nucleobases from cells during autophagy
We noticed that the nucleobases only transiently accumulated in the cytosol during autophagy. Further degradation of purines or pyrimidines is unlikely in S. cerevisiae, because both xanthine oxidase and the enzyme required for degradation of the pyrimidine ring are absent in this organism. Therefore, we measured both intracellular and extracellular nucleosides and bases. We found that xanthine, uracil, and four kinds of nucleosides (inosine, guanosine, cytidine, and uridine) were excreted from wild-type cells, but not atg2Δ cells, indicating that these excreted metabolites were derived from autophagic degradation (FigD and E, and Supplementary Fig S5B). The levels of these metabolites in the medium continued to rise for at least 6 h after starvation, indicating that their production by autophagy was ongoing during this period. Notably, the concentrations of xanthine and uracil secreted into the medium became very high, reaching micromolar range by 6 h.
We studied the absolute amounts of cellular and extracellular metabolites (Fig6F and G). At 2 h starvation, nucleosides were mostly retained within the cells, although a small portion of them had been excreted into the medium; by contrast, nucleobases were mostly transported to the medium. Taken together, the data indicated that cells excrete a majority of autophagy-degraded RNA as xanthine and uracil.
From the nucleobases excreted to the medium, we could estimate that a 3–4% of RNAs were catabolized per hour under nitrogen starvation conditions. About 80–85% of total cellular RNA is rRNAs and 10–15% is tRNA, whereas mRNA and other non-coding RNAs are minor. Therefore, most RNA degraded under starvation observed must be rRNA sequestered into autophagosomes, which is supported by the electron micrograph shown in Fig1A. We tried to evaluate how much and what kind of cellular RNA is targeted to the vacuole via autophagy by directly analyzing RNA (Supplementary Fig S6). The same amount of RNA from wild-type, atg2Δ, and rny1Δ cells before and after nitrogen starvation was examined by Northern blot and quantitative RT–PCR (qRT-PCR). Based on the assumption that the amount of small nuclear non-coding RNAs such as snRNA and snoRNA is not affected by autophagy, we employed them as internal references. By doing so, we observed decreases in both 25S and 18S RNAs as well as several tRNAs in wild-type cells compared to those in atg2Δ and rny1Δ cells under starvation conditions by both Northern blot and qRT-PCR analyses (Supplementary Fig S6A–C). However, we also realized that by these conventional methods, it is quite difficult to quantitate the accurate amount of RNAs and kinetics of their decrease. By contrast, as shown in this study, using metabolome analysis to quantitate the increase of degradation products such as nucleosides is far more sensitive and accurate to define bulk RNA degradation via autophagy.
Mode of RNA degradation via autophagy is conserved in fission yeast
To determine whether similar mechanisms operate in other organisms, we analyzed the intracellular metabolites in fission yeast Schizosaccharomyces pombe under both nitrogen-replete and nitrogen-starved conditions. We observed similar transient accumulations for 3′-NMPs and nucleosides, although the peaks appeared a little bit later than those of S. cerevisiae (Fig7). In Spatg1Δ cells, nucleosides levels did not change at all, indicating that autophagy-dependent RNA catabolism is well conserved between two phylogenetically distant yeast species. Thus, the principal scheme of RNA degradation may be widely conserved in higher eukaryotes.
Figure 7. RNA degradation via autophagy in S. pombe.
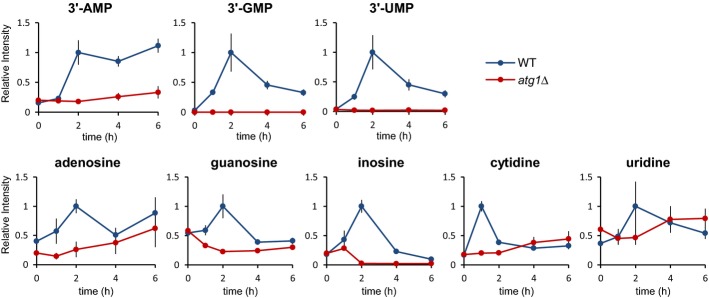
Wild-type and atg1Δ of S. pombe were grown in SD to mid-log phase and transferred to SD(-N) at time 0. At the indicated time points, the cells were collected; the metabolites in these cells were extracted and analyzed by LC/MS as described in Materials and Methods. The results are presented as normalized intensities on the basis of peak height of each metabolite in wild-type cells. All data are means of triplicates. The error bars represent the standard deviation.
Discussion
This study represents the first comprehensive characterization of autophagy-dependent bulk RNA catabolism in yeast. Under nitrogen starvation conditions, autophagy is induced, and cytoplasmic constituents including large numbers of ribosomes are delivered to the vacuole. The RNA sequestered to the vacuole via autophagosomes is first cleaved by the vacuolar ribonuclease Rny1, generating 3′ nucleotides. Next, the vacuolar non-specific phosphatase Pho8 acts as a nucleotidase by cleaving phosphate from the nucleotides, resulting in accumulation of nucleosides in the vacuole. Following transport out of the vacuole into the cytoplasm, the nucleosides are further broken down into purine and pyrimidine bases by two nucleosidases, Pnp1 and Urh1. These bases are immediately converted to the final de-aminated forms, uracil and xanthine through several enzymatic steps. These bases are not retained in the cell, but are instead excreted into the extracellular medium (Fig8). These dynamic changes in metabolites were never observed in cells defective in non-selective autophagy, but they occurred normally in cells defective in several different types of selective autophagy (Figs2A and 5A).
Figure 8. Schematic representation of the pathway for autophagy-dependent RNA degradation in yeast.
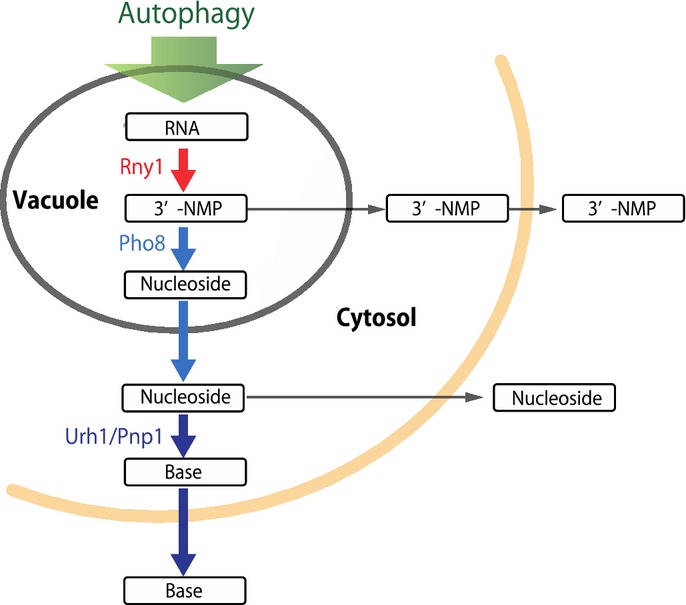
Autophagy-mediated RNA degradation pathway in S. cerevisiae. Thick arrows describe the primary process in the wild-type strain. Thin arrows indicate subordinate pathways that are not normally observed in wild-type cells. See Discussion for details.
T2-type RNase is highly conserved in almost all organisms and generally has a preference for acidic pH (Irie, 1999; Luhtala & Parker, 2010). Most of these RNases are vacuolar/lysosomal enzymes, consistent with the low pH optimum of these proteins. Therefore, it is likely that they function in the vacuole/lysosome to cleave RNA and that loss of their function may result in lysosomal dysfunction. Previous studies have suggested that homologues of Rny1 in plant and animals play critical roles in RNA turnover (Hillwig et al, 2011). Haud et al reported that loss of RNASET2 in zebrafish resulted in accumulation of undigested rRNA within lysosomes in brain cells, as revealed by microscopy (Haud et al, 2011). Henneke et al also showed that in human, mutation in the RNASET2 gene is associated with a neurological lesion, leukoencephalopathy (Henneke et al, 2009). Therefore, Rny1 homologues broadly participate in RNA catabolism in the vacuole/lysosome, and defects in these enzymes may cause serious lysosomal storage disorders. In this study, we showed that yeast T2-type ribonuclease Rny1 plays a major role in autophagy-dependent RNA degradation under starvation conditions. Consequently, 3′-NMPs are the intermediates of RNA degradation in the vacuole. At the steady state of autophagy in wild-type cells, 3′-NMP levels are quite low, suggesting that the nuclease reaction is the rate-limiting step of the whole process and that the resultant nucleotides are rapidly degraded. At present, it is not clear whether Rny1 itself produces the mononucleotides, or instead triggers RNA degradation by cleaving large RNAs into fragments. Because rRNAs possess tight secondary structure, it remains possible that some other exonuclease or RNA helicase/unwindase functions in this process. Further characterization of Rny1 and/or identification of additional protein(s) that support RNA degradation is necessary in order to understand the mechanistic basis of the first step of RNA hydrolysis in the vacuole.
3′-NMPs in the vacuole are immediately converted to nucleosides by Pho8. In pho8Δ cells, 3′-NMPs accumulated to very high levels (Fig3C and Supplementary Fig S5A). Furthermore, significant levels of nucleotides, nucleosides, and bases were detectable in the medium of pho8Δ cells after 6 h of starvation (Supplementary Fig S5B), indicating that vacuolar and plasma membranes possess transport systems for these compounds. Notably, the large quantities of nucleotides generated during the process of bulk RNA degradation are not 5′-NMPs but 3′-NMPs (Supplementary Fig S5A), possibly preventing serious perturbation of energy metabolism and signaling, as well as nucleic acid synthesis, processes in which 5′ nucleotides participate.
Pho8 was initially identified as a non-specific phosphatase, and its physiological substrates have been controversial for a long time (Kaneko et al, 1982; Donella-Deana et al, 1993). Pho8 is classified as alkaline phosphatase because it has a pH optimum above 9.0 for the artificial substrate p-nitrophenyl phosphate (Plankert et al, 1991). By contrast, Pho8 activity for fructose 2,6-bisphosphate (FBP), one of the physiological substrates of Pho8, has an acidic pH optimum (Plankert et al, 1991). A recent study showed that Pho8 plays a role in nicotinamide adenine dinucleotide metabolism, functioning as a nicotinamide mononucleotide (NMD) phosphatase in the vacuole (Lu & Lin, 2011). In this study, we proposed that Pho8 is a nucleotidase with an acidic pH optimum, similar to FBPase. In addition, we showed that Pho8 is highly induced under nitrogen starvation conditions and that it can efficiently act on 3′ nucleotides as substrates (Figs3C and 4B).
The nucleosides generated by autophagic degradation of RNA are catabolized further in the cytoplasm by two nucleosidases, Pnp1 and Urh1 [Supplementary Fig S4 and Xu et al (2013)]. Under nitrogen starvation, deamination of nucleotides, nucleosides, and bases occurs, and the resultant final products, xanthine and uracil, are excreted out of the cells. Under nitrogen starvation, the translation rate is significantly reduced due to the low abundance of amino acids, although the transcription rate is not affected much (Onodera & Ohsumi, 2005; Suzuki et al, 2011). When extracellular nitrogen sources are depleted, recycling of amino acids derived from protein degradation via autophagy is essential for survival (Kuma et al, 2004; Onodera & Ohsumi, 2005). Those amino acids are reutilized for new protein synthesis and also as an energy source. Ammonia liberated during the RNA degradation process may be reused as a nitrogen source, whereas nitrogen atoms in a purine or pyrimidine ring cannot be utilized as nitrogen sources by S. cerevisiae (Pantazopoulou & Diallinas, 2007). Therefore, superfluous bases must be excreted out of the cell, probably due to an osmotic demand as a result of elevated levels of degradation products from bulk protein and RNA. In any case, under starvation, nucleosides and nucleobases are hardly salvaged at all, but instead mostly excreted into the environment, suggesting that proper disposal of nucleotides in appropriate forms must be important for maintenance of cellular metabolic balance. Therefore, amount of the ultimate metabolites in the medium may serve as an appropriate indicator of autophagy. The fundamental mechanism of autophagy-dependent RNA degradation is likely to be conserved from yeast to human. However, the final outputs and their fates will depend on the presence and activity of metabolic enzymes, which differ among species, organs, or cell types.
For RNA degradation, it is interesting to know whether there are any specific RNA species of tRNA, mRNA, or other cytoplasmic RNAs selectively degraded by autophagy. So far, we did not find any selectivity in autophagy-mediated RNA degradation. (Supplementary Fig S6B and C). However, global analysis such as RNA-seq technique might reveal a more comprehensive assessment of changes in RNA species that are regulated by autophagy, if any.
In summary, we showed that autophagy induces drastic changes in the concentrations of cellular metabolites, not only amino acids but also nucleosides. Autophagy inevitably causes metabolic perturbations that must be overcome; failure to do so may cause various diseases including cancer. Understanding the relationship between the entire flow of cellular metabolism and autophagy will not only provide crucial insights into cell physiology, but also open new opportunities for intervention in metabolic disorders.
Materials and Methods
Yeast strains and media
Yeast strains used in this study are listed in Table1. Mutants and disruptants were generated using one-step gene replacement or gene disruption methods as described previously (Knop et al, 1999; Janke et al, 2004). All deletion and epitope-tagged constructs in this study were validated by PCR. Cells were grown in minimal synthetic defined (SD) medium (SD; 0.17% yeast nitrogen base without amino acids and ammonium sulfate (Difco), 0.5% ammonium sulfate, 2% glucose), nitrogen-depleted SD(-N) medium (SD-N; 0.17% yeast nitrogen base without amino acids and ammonium sulfate, 2% glucose), or carbon-depleted SD(-C) medium (0.17% yeast nitrogen base without amino acids and ammonium sulfate, 0.5% ammonium sulfate). All medium used was buffered with 50 mM MES/KOH.
Table 1.
Yeast strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| Saccharomyces cerevisiae | ||
| X2180-1B | MATα SUC2 mal mel gal2 CUP1 | Yeast Genetic Stock Center |
| MMY3 | X2180-1B;atg2Δ::kanMX6 | This study |
| JOY67 | X2180-1B;atg7Δ::kanMX4 | (Onodera & Ohsumi, 2005) |
| MMY7 | X2180-1B;atg11Δ::kanMX6 | This study |
| MMY9 | X2180-1B;atg17Δ::kanMX6 | This study |
| MMY10 | X2180-1B;atg19Δ::kanMX6 | This study |
| MMY198 | X2180-1B;atg32Δ::kanMX6 | This study |
| MMY279 | X2180-1B;nvj1Δ::kanMX6 | This study |
| TMK971 | X2180-1B;pep4Δ::kanMX6 prb1Δ::hphNT1 | This study |
| MMY156 | X2180-1B;pep4Δ::zeo prb1Δ::hphNT1, atg2Δ::kanMX6 | This study |
| MMY361 | X2180-1B;phm8Δ::kanMX6 | This study |
| TMK974 | X2180-1B; pho8Δ::kanMX6 | This study |
| MMY204 | X2180-1B;rny1Δ::natNT2 | This study |
| MMY375 | X2180-1B;rny1Δ::natNT2, atg2Δ::kanMX6 | This study |
| MMY337 | X2180-1B;RNY1-GFP::kanMX6 | This study |
| MMY356 | X2180-1B;RNY1-GFP::kanMX6 atg2Δ::hghNT1 | This study |
| TMK826 | X2180-1B;PHO8-GFP::kanMX6 | This study |
| MMY647 | X2180-1B;PNP1-GFP::kanMX6 | This study |
| MMY649 | X2180-1B;URH1-GFP::kanMX6 | This study |
| TMK852 | X2180-1B;PHO8-GFP::kanMX6 atg2Δ::hghNT1 | This study |
| MMY334 | X2180-1B;ubp3Δ::kanMX6 | This study |
| MMY339 | X2180-1B;bre5Δ::kanMX6 | This study |
| MMY722 | X2180-1B;ubp3Δ::kanMX6, rny1Δ::hyhNT1 | This study |
| MMY720 | X2180-1B;bre5Δ::kanMX6, rny1Δ::hyhNT1 | This study |
| MMY13 | X2180-1B;pep4Δ::kanMX6 | This study |
| MMY254 | X2180-1B;urh1Δ::kanMX6 | This study |
| MMY256 | X2180-1B;pnp1Δ::kanMX6 | This study |
| MMY20 | X2180-1B;pho8::GPDp-pho8Δ60::natNT2 | This study |
| MMY24 | X2180-1B;pho8::GPDp-pho8Δ60::natNT2 atg2Δ::kanMX6 | This study |
| MMY384 | X2180-1B;pho8::GPDp-pho8Δ60::natNT2 rny1Δ::kanMX6 | This study |
| MMY108 | X2180-1B;atg8Δ::GFP-ATG8::hphNT1 atg11Δ::kanMX6 | This study |
| MMY465 | X2180-1B;atg8Δ::GFP-ATG8::hphNT1 atg11Δ::natNT2 rny1Δ::kanMX6 | This study |
| MMY467 | X2180-1B;atg8Δ::GFP-ATG8::hphNT1 atg11Δ::natNT2 pho8Δ::kanMX6 | This study |
| MMY30 | X2180-1B;pho8::GPDp-pho8Δ60::natNT2 atg11Δ::kanMX6 | This study |
| MMY34 | X2180-1B;pho8::GPDp-pho8Δ60::natNT2 atg17Δ::kanMX6 | This study |
| MMY36 | X2180-1B;pho8::GPDp-pho8Δ60::natNT2 atg19Δ::kanMX6 | This study |
| Schizosaccharomyces pombe | ||
| JY1 | h− | (Kohda et al, 2007) |
| JV905 | h− ura4.d18 atg1::ura4+ | (Kohda et al, 2007) |
Yeast culture conditions and extraction
Cells were grown in SD medium at 30°C in a flask to an OD600 of 1. Five OD600 units of cells were collected onto a membrane filter (0.45 μm, 25 mm; Millipore, Billerica, MA, USA) and washed with 4 ml of 50 mM KCl solution precooled at 4°C. Cells were rapidly frozen in liquid nitrogen to halt metabolism and stored at −80°C until extraction. The rest of the cell culture was harvested and washed with 50 mM KCl, resuspended with an equal volume of SD(-N), SD(-C) medium, or SD medium with 0.2 μM rapamycin, and cultured at 30°C with shaking. Cells were collected at the indicated time points by filtering and stored as described above. To obtain culture fluid, 200 μl of the cell culture was filtered (0.2 μm PTFE, Millipore) and stored at −80°C until the analyses.
The cell samples were extracted according to the Bligh–Dyer method (Bligh & Dyer, 1959). Further details are described in the Supplementary Methods.
Metabolite measurement
Cell extracts were analyzed by (i) pentafluorophenylpropyl (PFPP) stationary-phase liquid chromatography coupled by electrospray ionization (ESI) (positive and negative modes) and (ii) reversed phase ion-pairing liquid chromatography with a C18 column coupled by ESI (negative mode), to a triple quadrupole mass spectrometer operated in multiple reaction monitoring (MRM) mode, with compound identities verified by precursor and product masses and retention time match to authenticated standards. Intracellular metabolite concentrations in the wild-type strain at 1 h were determined by the standard addition method, and concentrations in other samples were calculated based on fold change in ion counts relative to the wild-type strain at 1 h, multiplied by the known absolute concentration in the wild-type strain at 1 h. For medium samples, the external standard method was applied. Further details are described in the Supplementary Methods.
Western blotting
Immunoblot analyses were performed as described previously (Kushnirov, 2000). Samples corresponding to 0.5–1 OD600 units of cells were separated by SDS–PAGE followed by Western blotting. Antibodies against GFP (1:1,000, Roche), ALP (1:100, Life Technologies-Novex), Pep4, and Prb1 were used as primary antibodies. Chemiluminescence was induced by Femtoglow HRP Substrate (Michigan Diagnostics), and images were acquired by LAS-4000 imaging. The images were processed using the Multi Gauge software (Fujifilm Life Sciences).
Fluorescence microscopy
Intracellular localization of proteins was examined using an inverted fluorescence microscope as described previously (Suzuki et al, 2010). Images were acquired on an inverted fluorescence microscope (IX71; Olympus) equipped with a 150× total internal reflection fluorescence objective (UAPON 150×∼ OTIRF, NA 1.45; Olympus) and a CCD camera (ImagEM C9100-13; Hamamatsu Photonics). Images were captured using image acquisition system and analysis software (AQUACOSMOS (Hamamatsu Photonics) or MetaMorph 7.0r4 (Molecular Devices)). Images were processed in Adobe Photoshop. For RNA staining, cells grown in SD or SD(-N) medium for 2 h were incubated with GR Green (BIO-CRAFT) for 30 min at room temperature, washed three times with PBS, and visualized by fluorescence microscopy with GFP filter. Vacuoles were labeled with FM4-64 (Molecular Probes) as described previously (Vida & Emr, 1995).
Other procedures
Alkaline phosphatase assays were performed as described previously (Noda et al, 1995). Ultrastructural analysis of yeast cells was performed by Tokai-EMA Inc (Japan).
Acknowledgments
We thank Masayuki Yamamoto for providing Schizosaccharomyces pombe strains. We also thank M. Matsunami for technical assistance. This work was supported in part by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan. T.K. is a recipient of a JSPS Research Fellowship. This study represents a portion of the dissertation submitted by Hanghang Huang to Osaka University in partial fulfillment of the requirement for her PhD.
Author contributions
HH, TK, YO, and EF designed experiments and wrote the manuscript; HH, TK, TH, HT, and YN performed the experiments.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Supplementary Figures
Supplementary Information
Legends for Supplementary Figures
Review Process File
References
- Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Breker M, Gymrek M, Schuldiner M. A novel single-cell screening platform reveals proteome plasticity during yeast stress responses. J Cell Biol. 2013;200:839–850. doi: 10.1083/jcb.201301120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campomenosi P, Salis S, Lindqvist C, Mariani D, Nordstrom T, Acquati F, Taramelli R. Characterization of RNASET2, the first human member of the Rh/T2/S family of glycoproteins. Arch Biochem Biophys. 2006;449:17–26. doi: 10.1016/j.abb.2006.02.022. [DOI] [PubMed] [Google Scholar]
- Cohen M, Stutz F, Belgareh N, Haguenauer-Tsapis R, Dargemont C. Ubp3 requires a cofactor, Bre5, to specifically de-ubiquitinate the COPII protein, Sec23. Nat Cell Biol. 2003;5:661–667. doi: 10.1038/ncb1003. [DOI] [PubMed] [Google Scholar]
- Donella-Deana A, Ostojic S, Pinna LA, Barbaric S. Specific dephosphorylation of phosphopeptides by the yeast alkaline phosphatase encoded by PHO8 gene. Biochim Biophys Acta. 1993;1177:221–228. doi: 10.1016/0167-4889(93)90044-p. [DOI] [PubMed] [Google Scholar]
- Harding TM, Morano KA, Scott SV, Klionsky DJ. Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J Cell Biol. 1995;131:591–602. doi: 10.1083/jcb.131.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haud N, Kara F, Diekmann S, Henneke M, Willer JR, Hillwig MS, Gregg RG, Macintosh GC, Gartner J, Alia A, Hurlstone AF. rnaset2 mutant zebrafish model familial cystic leukoencephalopathy and reveal a role for RNase T2 in degrading ribosomal RNA. Proc Natl Acad Sci USA. 2011;108:1099–1103. doi: 10.1073/pnas.1009811107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henneke M, Diekmann S, Ohlenbusch A, Kaiser J, Engelbrecht V, Kohlschutter A, Kratzner R, Madruga-Garrido M, Mayer M, Opitz L, Rodriguez D, Ruschendorf F, Schumacher J, Thiele H, Thoms S, Steinfeld R, Nurnberg P, Gartner J. RNASET2-deficient cystic leukoencephalopathy resembles congenital cytomegalovirus brain infection. Nat Genet. 2009;41:773–775. doi: 10.1038/ng.398. [DOI] [PubMed] [Google Scholar]
- Hillwig MS, Contento AL, Meyer A, Ebany D, Bassham DC, Macintosh GC. RNS2, a conserved member of the RNase T2 family, is necessary for ribosomal RNA decay in plants. Proc Natl Acad Sci USA. 2011;108:1093–1098. doi: 10.1073/pnas.1009809108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie M. Structure-function relationships of acid ribonucleases: lysosomal, vacuolar, and periplasmic enzymes. Pharmacol Ther. 1999;81:77–89. doi: 10.1016/s0163-7258(98)00035-7. [DOI] [PubMed] [Google Scholar]
- Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno-Borchart A, Doenges G, Schwob E, Schiebel E, Knop M. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- Kaneko Y, Toh-e A, Oshima Y. Identification of the genetic locus for the structural gene and a new regulatory gene for the synthesis of repressible alkaline phosphatase in Saccharomyces cerevisiae. Mol Cell Biol. 1982;2:127–137. doi: 10.1128/mcb.2.2.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko Y, Tamai Y, Toh-e A, Oshima Y. Transcriptional and post-transcriptional control of PHO8 expression by PHO regulatory genes in Saccharomyces cerevisiae. Mol Cell Biol. 1985;5:248–252. doi: 10.1128/mcb.5.1.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell. 2009;17:98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Emr SD. Membrane protein sorting: biosynthesis, transport and processing of yeast vacuolar alkaline phosphatase. EMBO J. 1989;8:2241–2250. doi: 10.1002/j.1460-2075.1989.tb08348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol. 2007;8:931–937. doi: 10.1038/nrm2245. [DOI] [PubMed] [Google Scholar]
- Knop M, Siegers K, Pereira G, Zachariae W, Winsor B, Nasmyth K, Schiebel E. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast. 1999;15:963–972. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Kohda TA, Tanaka K, Konomi M, Sato M, Osumi M, Yamamoto M. Fission yeast autophagy induced by nitrogen starvation generates a nitrogen source that drives adaptation processes. Genes Cells. 2007;12:155–170. doi: 10.1111/j.1365-2443.2007.01041.x. [DOI] [PubMed] [Google Scholar]
- Kraft C, Deplazes A, Sohrmann M, Peter M. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol. 2008;10:602–610. doi: 10.1038/ncb1723. [DOI] [PubMed] [Google Scholar]
- Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- Kurtz JE, Exinger F, Erbs P, Jund R. New insights into the pyrimidine salvage pathway of Saccharomyces cerevisiae: requirement of six genes for cytidine metabolism. Curr Genet. 1999;36:130–136. doi: 10.1007/s002940050482. [DOI] [PubMed] [Google Scholar]
- Kushnirov VV. Rapid and reliable protein extraction from yeast. Yeast. 2000;16:857–860. doi: 10.1002/1097-0061(20000630)16:9<857::AID-YEA561>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Lardeux BR, Heydrick SJ, Mortimore GE. RNA degradation in perfused rat liver as determined from the release of [14C]cytidine. J Biol Chem. 1987;262:14507–14513. [PubMed] [Google Scholar]
- Lecoq K, Belloc I, Desgranges C, Konrad M, Daignan-Fornier B. YLR209c encodes Saccharomyces cerevisiae purine nucleoside phosphorylase. J Bacteriol. 2001;183:4910–4913. doi: 10.1128/JB.183.16.4910-4913.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu SP, Lin SJ. Phosphate-responsive signaling pathway is a novel component of NAD+ metabolism in Saccharomyces cerevisiae. J Biol Chem. 2011;286:14271–14281. doi: 10.1074/jbc.M110.217885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luhtala N, Parker R. T2 Family ribonucleases: ancient enzymes with diverse roles. Trends Biochem Sci. 2010;35:253–259. doi: 10.1016/j.tibs.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacIntosh GC, Bariola PA, Newbigin E, Green PJ. Characterization of Rny1, the Saccharomyces cerevisiae member of the T2 RNase family of RNases: unexpected functions for ancient enzymes? Proc Natl Acad Sci USA. 2001;98:1018–1023. doi: 10.1073/pnas.98.3.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao K, Chew LH, Inoue-Aono Y, Cheong H, Nair U, Popelka H, Yip CK, Klionsky DJ. Atg29 phosphorylation regulates coordination of the Atg17-Atg31-Atg29 complex with the Atg11 scaffold during autophagy initiation. Proc Natl Acad Sci USA. 2013;110:E2875–E2884. doi: 10.1073/pnas.1300064110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimore GE, Lardeux BR, Heydrick SJ. Mechanism and control of protein and RNA degradation in the rat hepatocyte: two modes of autophagic sequestration. Revis Biol Celular. 1989;20:79–96. [PubMed] [Google Scholar]
- Noda T, Matsuura A, Wada Y, Ohsumi Y. Novel system for monitoring autophagy in the yeast Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1995;210:126–132. doi: 10.1006/bbrc.1995.1636. [DOI] [PubMed] [Google Scholar]
- Noda T, Ohsumi Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J Biol Chem. 1998;273:3963–3966. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- Ohsumi Y. Historical landmarks of autophagy research. Cell Res. 2014;24:9–23. doi: 10.1038/cr.2013.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K, Kondo-Okamoto N, Ohsumi Y. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev Cell. 2009;17:87–97. doi: 10.1016/j.devcel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- Onodera J, Ohsumi Y. Autophagy is required for maintenance of amino acid levels and protein synthesis under nitrogen starvation. J Biol Chem. 2005;280:31582–31586. doi: 10.1074/jbc.M506736200. [DOI] [PubMed] [Google Scholar]
- Pantazopoulou A, Diallinas G. Fungal nucleobase transporters. FEMS Microbiol Rev. 2007;31:657–675. doi: 10.1111/j.1574-6976.2007.00083.x. [DOI] [PubMed] [Google Scholar]
- Plankert U, Purwin C, Holzer H. Yeast fructose-2,6-bisphosphate 6-phosphatase is encoded by PHO8, the gene for nonspecific repressible alkaline phosphatase. Eur J Biochem. 1991;196:191–196. doi: 10.1111/j.1432-1033.1991.tb15803.x. [DOI] [PubMed] [Google Scholar]
- Reggiori F, Komatsu M, Finley K, Simonsen A. Autophagy: more than a nonselective pathway. Int J Cell Biol. 2012;2012:219625. doi: 10.1155/2012/219625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts P, Moshitch-Moshkovitz S, Kvam E, O'Toole E, Winey M, Goldfarb DS. Piecemeal microautophagy of nucleus in Saccharomyces cerevisiae. Mol Biol Cell. 2003;14:129–141. doi: 10.1091/mbc.E02-08-0483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott SV, Baba M, Ohsumi Y, Klionsky DJ. Aminopeptidase I is targeted to the vacuole by a nonclassical vesicular mechanism. J Cell Biol. 1997;138:37–44. doi: 10.1083/jcb.138.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott SV, Guan J, Hutchins MU, Kim J, Klionsky DJ. Cvt19 is a receptor for the cytoplasm-to-vacuole targeting pathway. Mol Cell. 2001;7:1131–1141. doi: 10.1016/s1097-2765(01)00263-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbik N. Golgi-mediated glycosylation determines residency of the T2 RNase Rny1p in Saccharomyces cerevisiae. Traffic. 2013;14:1209–1227. doi: 10.1111/tra.12122. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Kondo C, Morimoto M, Ohsumi Y. Selective transport of alpha-mannosidase by autophagic pathways: identification of a novel receptor, Atg34p. J Biol Chem. 2010;285:30019–30025. doi: 10.1074/jbc.M110.143511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki SW, Onodera J, Ohsumi Y. Starvation induced cell death in autophagy-defective yeast mutants is caused by mitochondria dysfunction. PLoS ONE. 2011;6:e17412. doi: 10.1371/journal.pone.0017412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K. Selective autophagy in budding yeast. Cell Death Differ. 2013;20:43–48. doi: 10.1038/cdd.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshige K, Baba M, Tsuboi S, Noda T, Ohsumi Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J Cell Biol. 1992;119:301–311. doi: 10.1083/jcb.119.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DM, Parker R. The RNase Rny1p cleaves tRNAs and promotes cell death during oxidative stress in Saccharomyces cerevisiae. J Cell Biol. 2009;185:43–50. doi: 10.1083/jcb.200811119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vida TA, Emr SD. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol. 1995;128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- Xu YF, Letisse F, Absalan F, Lu W, Kuznetsova E, Brown G, Caudy AA, Yakunin AF, Broach JR, Rabinowitz JD. Nucleotide degradation and ribose salvage in yeast. Mol Syst Biol. 2013;9:665. doi: 10.1038/msb.2013.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures
Supplementary Information
Legends for Supplementary Figures
Review Process File