Abstract
The dengue virus genome is a dynamic molecule that adopts different conformations in the infected cell. Here, using RNA folding predictions, chemical probing analysis, RNA binding assays, and functional studies, we identified new cis-acting elements present in the capsid coding sequence that facilitate cyclization of the viral RNA by hybridization with a sequence involved in a local dumbbell structure at the viral 3′ untranslated region (UTR). The identified interaction differentially enhances viral replication in mosquito and mammalian cells.
TEXT
Dengue virus (DENV) is a member of the Flaviviridae family that includes other important pathogens such as yellow fever virus (YFV), West Nile virus (WNV), Saint Louis encephalitis virus (SLEV), and Japanese encephalitis virus (JEV). The DENV genome is a plus-stranded RNA molecule that contains a single open reading frame flanked by highly structured 5′ and 3′ untranslated regions (UTRs) (1–3). RNA elements located within these regions are responsible for translation initiation and genome replication (4–7). The 5′ UTR is about 100 nucleotides (nt) long and includes three different elements: (i) stem-loop A (SLA), which is the promoter for viral polymerase binding and activation (8–10); (ii) stem-loop B (SLB), which contains a sequence known as 5′ upstream of the AUG region (5′ UAR) that is complementary to a sequence present at the 3′ UTR (3′ UAR) and mediates long-range RNA-RNA interactions between the ends of the genome (11); and (iii) a spacer sequence between SLA and SLB rich in U's, which functions as an enhancer of viral replication (10). The viral 3′ UTR is about 450 nucleotides long and comprises four defined domains: domain A1, which features a variable region (VR) (12); domains A2 and A3, which present two almost-identical dumbbell-like secondary structures (DB1 and DB2), which appear to work as enhancers for viral RNA replication (13–15); and domain A4, which contains a small hairpin (sHP) and the 3′ stem-loop (3′ SL), which are essential elements for viral replication (3, 16). In addition to RNA structures defined in the UTRs that play different roles during infection, important RNA elements have been described in the protein coding region. In this regard, essential sequences that mediate long-range RNA-RNA interactions known as 5′ cyclization sequence (5′ CS) and 5′ downstream of AUG region (5′ DAR) are located within the capsid coding sequence (11, 13, 17–21). Also, a hairpin known as cHP, located between 5′ CS and 5′ DAR, has been shown to be necessary for efficient RNA replication (22). The current model for viral RNA synthesis includes the interaction of the viral polymerase NS5 with the 5′-end SLA promoter and its transfer to the 3′-end initiation site by cyclization of the viral genome (9). Despite great advances in knowledge of cis-acting RNA elements in the flavivirus genomes, the molecular details and mechanisms by which many of them function during viral replication are still not well understood.
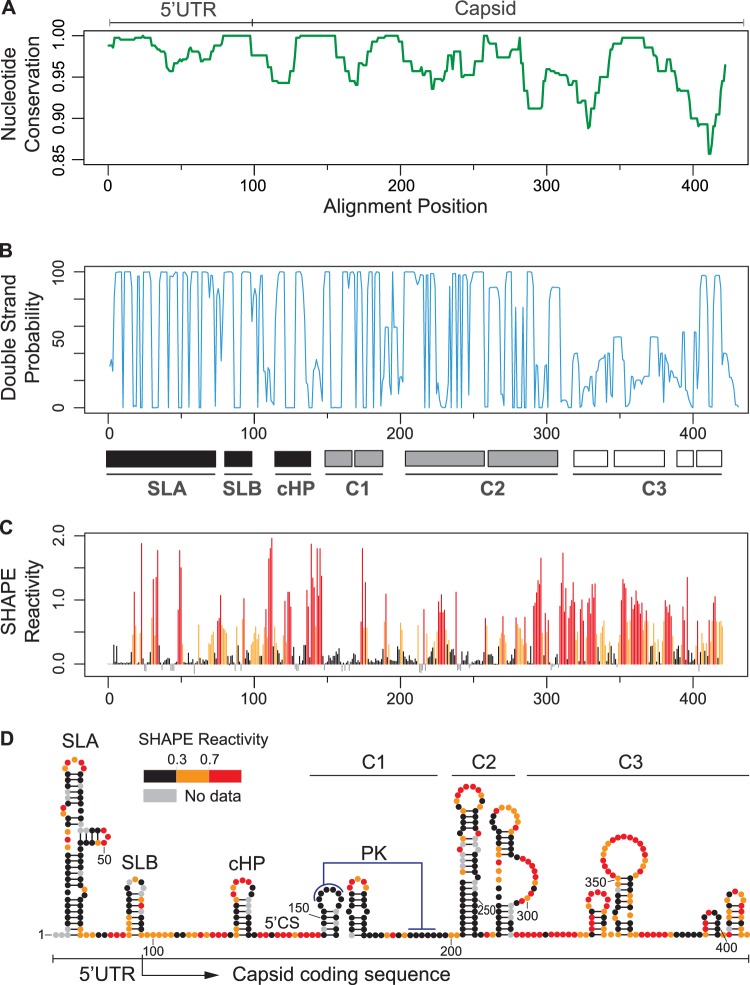
Intrigued by dual roles of RNA sequences in viral protein coding regions, we examined the complete capsid coding sequence for the presence of new cis-acting RNA elements. First, we used representative sequences of the six different genotypes of DENV type 2 (DENV2) to examine the conservation of the first 450 nucleotides of the viral genome, including the 5′ UTR and the complete capsid coding sequence. The highest nucleotide conservation was observed in the first 300 nucleotides of the viral genome (Fig. 1A). To evaluate the ability of the nucleotide sequences to form RNA secondary structures, we performed an evolutionary conservation analysis using RNAz and RNAalifold software (23–25). Interestingly, the base pairing probability plot drastically dropped around nucleotide 310 (region indicated as C3 in Fig. 1B), showing high double-strand probability for 5′ SLA, SLB, and cHP structures (black boxes) as expected but also highlighting two additional structured regions indicated as C1 and C2 (gray boxes). To confirm the RNA structure predictions, a fragment of the first 450 nt of the viral genome was probed with N-methylisotoic anhydride (NMIA) employing SHAPE (selective 2′-hydroxyl acylation analyzed by primer extension) technology (26, 27). This chemical preferentially reacts with unpaired nucleotides and poorly with those that are constrained in base pairs. The samples were resolved by capillary electrophoresis, and data were analyzed using ShapeFinder software v1.0 (28). NMIA reactivity of each nucleotide position is color coded and shown in a summary plot in Fig. 1C. The secondary structure prediction was obtained using RNAstructure software v5.4 (29–31) (Fig. 1D). A notable correlation of the pairing probability and the SHAPE reactivity was observed (compare Fig. 1B and C), showing high SHAPE reactivity downstream of nucleotide 310. Importantly, the analysis supports the presence of RNA structures within nucleotides 150 and 310 of the genome (C1 and C2 regions). For the C1 region, two small hairpins were mapped. Nucleotides located in the top of the first hairpin (GAGAA) and a downstream complementary region (UUCUC) were unreactive for NMIA, predicting the formation of a pseudoknot structure (PK) (Fig. 1D), in agreement with data recently reported using DENV4 (32). In the C2 region, two large stem-loops were predicted, while in the C3 region, low-stability structures were observed.
FIG 1.
Conserved RNA structures in the DENV capsid coding sequence. (A) Nucleotide conservation of the first 450 nucleotides of the viral genome using representative sequences of the six different DENV2 genotypes. Window size = 10. (B) Double-stranded probability of the first 450 nucleotides of the viral genome. Three regions were defined: high double-strand probability for 5′ SLA, SLB, and cHP (black boxes); C1 and C2 structures (gray boxes); and unstructured C3 region (white boxes). (C) Plot showing SHAPE reactivity at each nucleotide position of DENV2 RNA. (D) Viral RNA structures based on conservation, predictions, and SHAPE reactivity. Red notations correspond to unpaired nucleotides, and black indicates double-stranded regions, whereas residues of intermediate reactivity are noted in orange. Abbreviations: UTR, untranslated region; SLA, stem-loop A; SLB, stem-loop B; cHP, capsid hairpin; 5′ CS, 5′ cyclization sequence; PK, pseudoknot; C1, C2, and C3, RNA structures identified in the capsid coding sequence. Data shown are averages of three experiments.
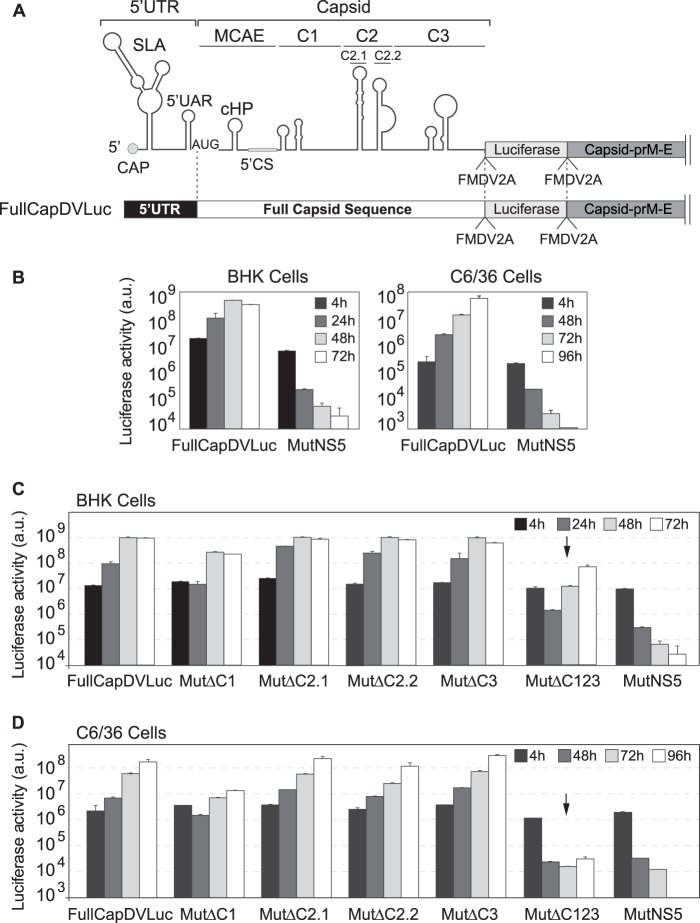
In order to study the functional significance of these RNA elements, a DENV2 system previously described (33), carrying a luciferase gene, was modified to generate a new construct that allowed manipulation of the complete coding sequence of capsid but retained the ability to produce infectious particles. This construct allowed uncoupling cis-acting signals from coding sequences of capsid. The new reporter virus (named FullCapDVLuc) contained the viral 5′ UTR, the complete capsid coding region followed by the Renilla luciferase gene, which was flanked by two foot-and-mouth disease virus 2A (FMDV2A) protease coding sequences (QLLNFDLLKLAGDVESNPGP), and the rest of the viral genome, including a second copy of the capsid protein coding sequence followed by the rest of the open reading frame and the 3′ UTR (Fig. 2A). Two FMDV2As were introduced to ensure the release of the luciferase and avoid changes in its enzymatic activity due to fusion of additional amino acids of the capsid coding sequence. In order to evaluate the ability of this virus to replicate, the in vitro-transcribed RNA was transfected into mosquito (C6/36) and mammalian (BHK) cells, and luciferase activity was measured as a function of time (Fig. 2B). A replication-impaired control (mutations in the polymerase NS5) was also constructed in the described backbone (MutNS5). The results indicate that the reporter FullCapDVLuc is fully active for translation and replication in both cell types.
FIG 2.
Functional significance of RNA structures in the capsid coding sequence. (A) Schematic representation of the DENV reporter FullCapDVLuc. FullCap and MCAE stand for full capsid coding sequence and minimal cis-acting elements, respectively. (B) Replication of new DENV reporter construct. Plots show Renilla luciferase activity as a function of time post-RNA transfection in BHK and C6/36 cells. The luciferase values are means ± standard deviations (n = 4). (C and D) Translation and replication of DENV reporter RNAs containing the indicated deletions in BHK cells (C) and C6/36 cells (D). Luciferase activity was measured as a function of time and is represented as arbitrary units (a.u.). The luciferase values are means ± standard deviations (n = 4).
To examine the requirement of the predicted RNA structures in the capsid coding sequence, we constructed a set of mutants with a deletion of each structure (MutΔC1, MutΔC2.1, MutΔC2.2, MutΔC3, and MutΔC123) in the context of the FullCapDVLuc. The mutants MutΔC2.1 and MutΔC2.2 correspond to deletions of each of the two hairpins of structure C2 (Fig. 2A). RNAs corresponding to the mutants were transfected into C6/36 and BHK cells and monitored by luciferase activity as a function of time. The mutant with simultaneous deletion of all three regions, MutΔC123, showed delayed and reduced replication in both cell lines. A reduction of about 100-fold with respect to wild-type (WT) control was observed at 48 h in BHK cells, while the reduction was more than 1,000-fold in C6/36 cells at 72 h (Fig. 2C and D, black arrows). Mutants ΔC2.1, ΔC2.2, and ΔC3 showed luciferase levels that were similar to those of the WT FullCapDVLuc in both cell lines, indicating that these structures do not play an important role in translation or RNA synthesis. On the other hand, the reporter MutΔC1 presented a delay and a 10-fold reduction in viral replication in mosquito cells, and a 6-fold reduction in mammalian cells, at 72 and 48 h, respectively (Fig. 2C and D). Although deletion of C1 showed a clear effect on viral replication, deleting each structure individually was not as drastic as the deletion of the 3 elements at the same time, suggesting an additive effect when these RNA structures are deleted collectively. For all the transfected viral RNAs, similar levels of luciferase activity were observed at 4 h, indicating that the deletions tested did not alter translation of the input RNA. The results suggest that the RNA structures present in the capsid coding sequence are important for efficient DENV amplification, with a more pronounced requirement when the virus replicates in mosquito cells than when it replicates in mammalian cells (compare Fig. 2C and D). It is important to mention that the deleted RNA structures are duplicated downstream (in the coding region of the capsid protein); thus, we cannot rule out a possible underestimation of the effect observed by RNA structure deletions due to complementation via the duplicated structures.
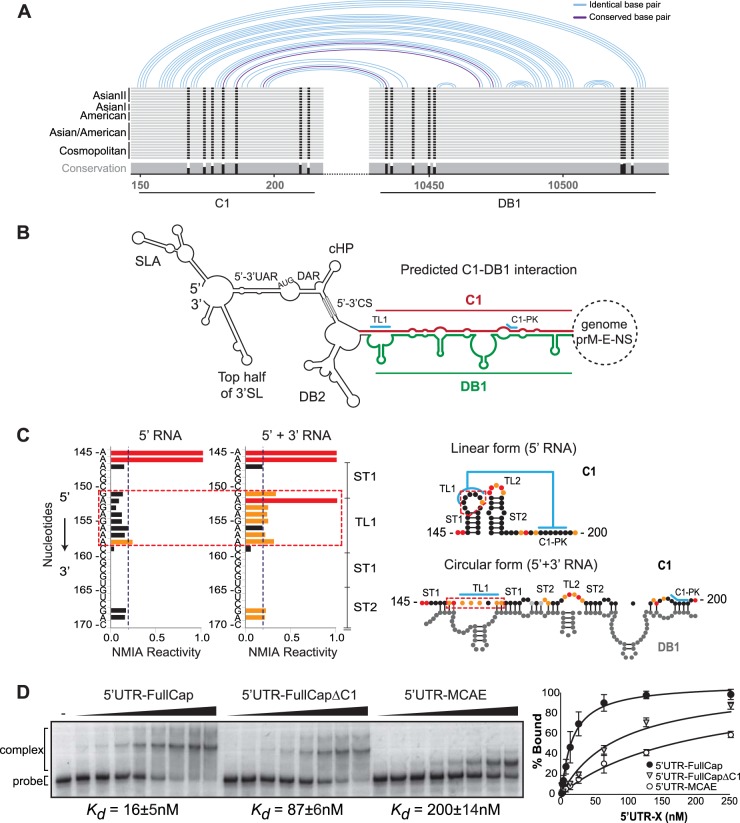
It has been suggested in a recent report that RNA elements present in the capsid coding sequence of DENV4, DCS-PK, homologous to C1, could modulate the conformation of RNA elements at the 5′ end (5′ UAR, 5′ DAR, cHP, and/or 5′ CS) during RNA cyclization (32); however, no evidence of how this RNA structure would participate during this viral process was provided. Based on the finding that the C1 region enhances viral RNA synthesis, we examined a possible direct contribution of its sequence in long-range RNA-RNA interactions. First, we evaluated conserved patterns of possible RNA-RNA interaction based on sequence alignments of the capsid coding regions and 3′ UTRs from different DENV2 genotypes. To this end, the RNAaliduplex software was used for the analysis (25), and the R-CHIE package was used to plot the data (34). The program takes two RNA sequence alignments, predicts optimal and suboptimal interactions and hybridization energies, and provides information about conserved interactions. The alignments and details of specific sequences used for this study are described in Fig. S1 in the supplemental material. Interestingly, this analysis resulted in a model that predicted a novel conserved interaction between a C1 sequence and a sequence contained in the conserved DB1 structure of the 3′ UTR (Fig. 3A and B). The predicted model is complex because it involves the interaction between sequences contained in C1 and DB1 structures, which also form local PK interactions. The PK formed between the conserved nucleotides at the loop of DB1 (GCUGU) and downstream sequence (CGACG) has been well characterized in previous studies (14, 35, 36). The proposed long-range RNA-RNA hybridization competes and disrupts not only the stem-loops in C1 and DB1 but also the two PKs. Thus, the C1-DB1 long-range hybridization and the C1-DB1 local structures represent mutually exclusive forms of the DENV genome, which may exist in equilibrium in the infected cell. We have previously described that alternative conformations of the DENV genome, linear and circular, are necessary for viral RNA replication and that a balance between these forms of the genome is crucial for DENV infectivity (37). To confirm the formation of the new predicted structures, SHAPE studies using 5′ UTR-capsid and 3′ UTR RNAs were performed. Although most of the nucleotides involved in the long-range RNA-RNA interaction are also involved in local base pairings, we detected nucleotides (151-GAGAGAAA-158) at the loop of the first hairpin of C1 (TL1) that significantly change the reactivity to NMIA in the presence or absence of the 3′ UTR (Fig. 3C, red dashed box). This observation shows that upon C1-DB1 interaction, the TL1 nucleotides (which are involved in the C1-PK) become more reactive (Fig. 3C), while the nucleotides of C1-PK do not change the reactivity, since they mostly interact with nucleotides of DB1 (Fig. 3C, circular form).
FIG 3.
Sequence in the capsid protein coding region facilitates long-range RNA-RNA interactions between the ends of the viral genome. (A) Schematic representation of the conserved patterns of long-range interactions based on alignments of genomes of different DENV2 genotypes. Details of nucleotide sequences, alignments, and predictions are described in Fig. S1 in the supplemental material. (B) Representation of the circular conformation of the DENV genome indicating the known and new complementary elements. The predicted RNA-RNA interaction between C1 (red) and DB1 (green) is indicated. (C) Analysis of 5′-end RNA secondary structure in the absence and presence of the 3′ UTR RNA (ratio of 1:5). NMIA probing profiles of an RNA corresponding to the 5′ end of the genome (5′ RNA) and the probing of the same RNA in the presence of a second RNA containing the viral 3′ UTR sequence (5′ + 3′ RNA) are shown on the left. The y axis contains the sequence and nucleotide numbers; the x axis depicts chemical reactivity. On the right, secondary structures and NMIA reactivity are indicated for the predicted 5′ end alone and the 5′-3′ hybridized form. Red dashed frames highlight a region with different probing profiles. (D) Mobility shift assays indicate that C1-DB1 hybridization stabilizes the RNA-RNA complex. The 3′ UTR RNA probe was incubated with increasing concentrations of unlabeled RNAs corresponding to molecules with viral 5′-end sequences as indicated. Mobilities of the 3′ UTR RNA probe and RNA-RNA complex as well as the apparent dissociation constants (Kd ± standard error) are indicated. On the right, the percentage of RNA probe bound was plotted as a function of unlabeled RNA concentration for the three molecules used.
To further examine whether the predicted RNA-RNA complementarity between C1 and DB1 contributes in stabilizing a complex between the ends of the viral genome, electrophoretic mobility shift assays were performed with different RNA molecules (Fig. 3D). The affinity between RNA molecules containing the sequence of the 3′ UTR and the 5′ end of the genome including the C1 sequence or not was evaluated. A uniformly 32P-labeled RNA probe corresponding to the entire DENV2 3′ UTR was in vitro transcribed and purified in a polyacrylamide gel. The radiolabeled 3′ UTR RNA was incubated with increasing concentrations of unlabeled RNAs corresponding to the 5′ UTR-FullCap (full length capsid), 5′ UTR-FullCapΔC1 (with a deletion of the C1 structure), or 5′ UTR-MCAE (minimal cis-acting elements). The complexes were analyzed in native 4% polyacrylamide gels as previously described (11). Total radioactivity for each lane was determined by quantifying the bands corresponding to the RNA-RNA complex and the free probe. For each case, the apparent dissociation constant (Kd) was estimated by nonlinear regression analysis. RNA titrations indicate a significantly higher affinity between the 5′ UTR-FullCap and the 3′ UTR (Kd = 16 ± 5 nM) than the affinities of the 5′ UTR-FullCapΔC1 (Kd = 87 ± 6 nM) and the 5′ UTR-MCAE (Kd = 200 ± 14 nM) with the 3′ UTR (Fig. 3D). These results confirm that new sequences in the capsid coding region contribute in stabilizing the RNA-RNA complex formed between the ends of the DENV genome and highlight the relevance of the C1 region.
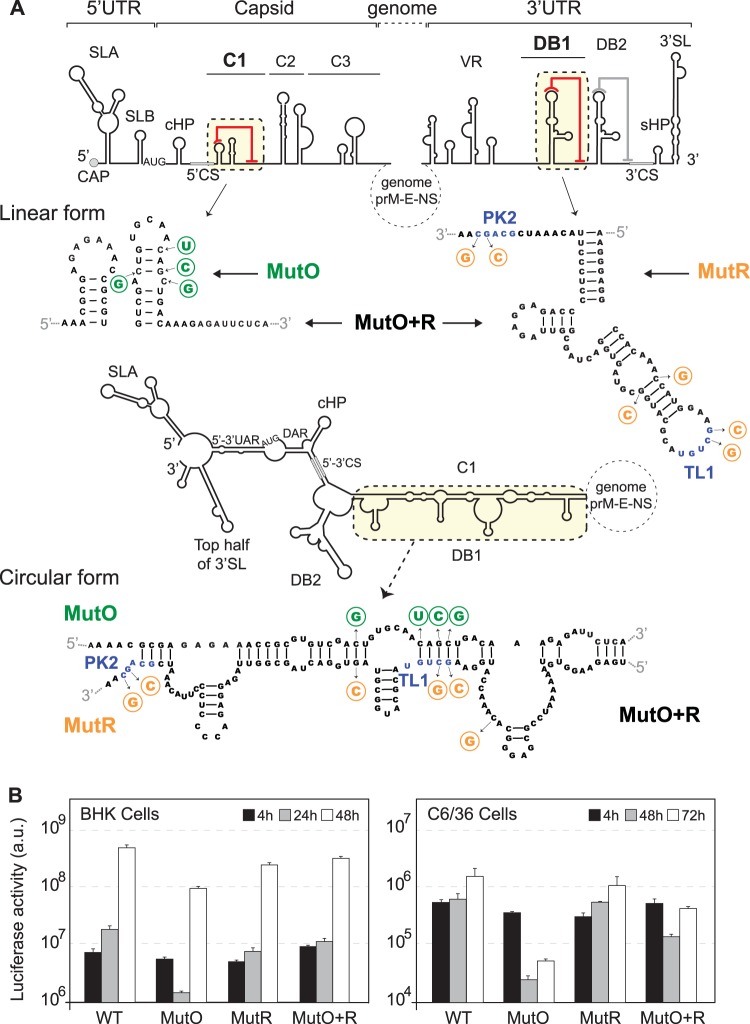
Next, we evaluated the functional significance of a possible C1-DB1 hybridization during DENV replication. To this end, we constructed recombinant viruses with nucleotide changes disrupting or restoring the predicted interactions. The design of these viruses was very complex because five different structures had to be taken into account simultaneously: C1 structure and local PK, DB1 structure and local PK, and the long-range RNA-RNA interaction (see linear and circular forms of the RNA in Fig. 4A). Mutations were incorporated along with compensations to maintain local structures. Mutant O (MutO) includes substitutions only in the 5′ end of the genome that are predicted to disrupt C1-DB1 interaction but maintain the C1 secondary structure. Mutant R (MutR) includes substitutions only in the 3′ end that are predicted to debilitate C1-DB1 interaction but maintain the local DB1 and TL1-PK2 pseudoknot structures by compensatory mutations. Finally, a reconstitution mutant (MutO+R) predicted to restore the long-range C1-DB1 interaction contained the substitutions of both MutO and MutR. For clarity, the location of the mutations is indicated in the two alternative structures (linear and circular forms, Fig. 4A). RNAs corresponding to the three mutants (MutO, MutR, and MutO+R) and the RNA of the parental virus (WT) were transfected into C6/36 and BHK cells, and replication was monitored by luciferase activity. The levels of luciferase at 4 h posttransfection were comparable for all the viruses in the two cell lines, indicating efficient translation of the input genomes. The reporter MutO showed delayed RNA replication with an 8-fold reduction of luciferase activity at 48 h in BHK cells, while in mosquito cells, replication was reduced about 50-fold (Fig. 4B). The MutR replicated slightly less than the WT virus in both cell lines. However, when the C1-DB1 interaction was reconstituted (MutO+R), the impaired replication of MutO was partially restored in both cell lines (Fig. 4B). The restitution of viral RNA replication of MutO by compensatory mutations supports the relevance of the identified long-range interaction. In addition, the data highlight a different requirement of the identified RNA elements for efficient viral replication in mosquito and mammalian cells.
FIG 4.
Relevance of C1-DB1 interaction in DENV RNA replication. (A) Design of mutation disrupting and reconstituting long-range C1-DB1 RNA-RNA interaction in the context of the viral genome. cis-acting elements in the 5′ end and the 3′ end of the genome are indicated. Nucleotide sequences of C1 and DB1 and the substitutions incorporated are shown in the two alternative conformations, linear and circular. Location, nucleotide changes, and names of mutants are indicated. MutO (green) shows substitutions in C1, and MutR (orange) shows substitutions in DB1. MutO+R contains both substitutions, restoring the interaction and maintaining C1 and DB1 structures. In addition, this mutant maintains the pseudoknot TL1/PK2 by compensatory mutations. (B) Replication of the three mutant RNAs in BHK and C6/36 cells together with the WT control. Luciferase activity was measured as a function of time and represented as arbitrary units (a.u.). The luciferase values are means ± standard deviations (n = 3).
Here, we identified sequences in the DENV genome that enhance viral RNA replication by stabilizing long-range RNA-RNA interactions, in agreement with the proposed function of genome cyclization during minus-strand RNA synthesis (9). The identified complementary sequences are located within the capsid coding region at the 5′ end of the RNA and in a well-characterized dumbbell structure at the 3′ UTR, and both form PK structures, stabilizing local RNA elements at each end of the genome. Long-distance hybridization of these RNA sequences disassembles the hairpin and dumbbell structures. This observation, together with previous studies, supports the idea that conservation of alternative (mutually exclusive) functional RNA structures is a common feature in the flavivirus genomes. For instance, in the case of the conserved 5′-3′ CS interaction, the 3′ CS lies in a region that forms a local PK with the loop sequence of the DB2 structure (located downstream of DB1) (14, 15). In the case of the 5′-3′ UAR complementary sequence, the 3′ UAR adopts local structures that are essential for viral replication, including the sHP and the base of the terminal 3′ SL (16, 21, 37–39). It is possible that conservation of competing structures in the viral genome provides a mechanism for fine tuning RNA conformations required for different viral processes. In this regard, the presence of regulatory RNA elements in protein coding sequences may be relevant for controlling functional RNA structures by translating ribosomes. In the infected cell, it is likely that RNA binding proteins modulate different viral RNA conformations. Many proteins have been previously reported as binders of the DENV RNA (40). Among them, proteins with RNA helicase and chaperone activities have been described to interact with the UTRs of flavivirus genomes (41–43). In this regard, the host helicase DDX6 was reported to bind specifically to the DENV DB1 and DB2 structures; however, the function of these interactions is still unknown (44). Another example is the host protein AUF1, which was shown to bind the WNV RNA and enhance RNA replication likely by rearranging RNA conformations (45). Moreover, the different requirements of viral RNA structures for viral RNA replication in mosquito and mammalian cells may also reflect distinct ways of controlling viral RNA conformations in different host cell environments.
A previous study has investigated the role of RNA sequences within the capsid coding region in DENV infectious particle production. These studies suggested a function of RNA elements during viral assembly in mosquito cells (46). The role of C1 in enhancing viral RNA synthesis observed in our studies limited the possibility of evaluating its function in viral assembly or encapsidation, steps that take place downstream of genome amplification. Nevertheless, we cannot rule out the possibility that C1 sequences play dual functions in viral replication and encapsidation. This possibility should be further investigated.
We propose that DENV conserves RNA sequences that participate in alternative local and long-range structures as a mechanism to modulate the viral RNA architecture for efficient replication. In addition, based on the impact of deletions and mutations of the identified viral RNA structures for replication in mosquito and mammalian cells, we propose that distinct RNA conformations may play different roles in the two host cell environments.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Richard Kinney for the dengue virus cDNA clone. We also thank the members of Andrea Gamarnik's laboratory for helpful discussions and ideas about this work.
N.G.I., C.V.F., L.G.G., and A.V.G. are members of the Argentinean Council of Investigation (CONICET). This work was supported by grants from the Agencia Argentina de Promoción Científica y Tecnológica (PICT-2010), NIH (1R01AI095175-01), and DARPA (grant HR001111C0094) to A.V.G.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.02677-14.
REFERENCES
- 1.Gamarnik AV. 2010. Role of the dengue virus 5′ and 3′ untranslated regions in viral replication, p 55–78 InHanley KA, Weaver SC (ed), Frontiers in dengue virus research. Caister Academic Press, Wymondham, Norfolk, United Kingdom. [Google Scholar]
- 2.Lindenbach BD, Thiel HJ, Rice CD. 2007. Flaviviridae: the viruses and their replication, p 1101–1152 InKnipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 5th ed, vol 1 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 3.Markoff L. 2003. 5′- and 3′-noncoding regions in flavivirus RNA. Adv Virus Res 59:177–228. doi: 10.1016/S0065-3527(03)59006-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alcaraz-Estrada SL, Yocupicio-Monroy M, del Angel RM. 2010. Insights into dengue virus genome replication. Future Virol 5:575–592. doi: 10.2217/fvl.10.49. [DOI] [Google Scholar]
- 5.Gebhard LG, Filomatori CV, Gamarnik AV. 2011. Functional RNA elements in the dengue virus genome. Viruses 3:1739–1756. doi: 10.3390/v3091739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iglesias NG, Gamarnik AV. 2011. Dynamic RNA structures in the dengue virus genome. RNA Biol 8:249–257. doi: 10.4161/rna.8.2.14992. [DOI] [PubMed] [Google Scholar]
- 7.Paranjape SM, Harris E. 2010. Control of dengue virus translation and replication. Curr Top Microbiol Immunol 338:15–34. doi: 10.1007/978-3-642-02215-9_2. [DOI] [PubMed] [Google Scholar]
- 8.Filomatori CV, Iglesias NG, Villordo SM, Alvarez DE, Gamarnik AV. 2011. RNA sequences and structures required for the recruitment and activity of the dengue virus polymerase. J Biol Chem 286:6929–6939. doi: 10.1074/jbc.M110.162289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Filomatori CV, Lodeiro MF, Alvarez DE, Samsa MM, Pietrasanta L, Gamarnik AV. 2006. A 5′ RNA element promotes dengue virus RNA synthesis on a circular genome. Genes Dev 20:2238–2249. doi: 10.1101/gad.1444206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lodeiro MF, Filomatori CV, Gamarnik AV. 2009. Structural and functional studies of the promoter element for dengue virus RNA replication. J Virol 83:993–1008. doi: 10.1128/JVI.01647-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alvarez DE, Lodeiro MF, Luduena SJ, Pietrasanta LI, Gamarnik AV. 2005. Long-range RNA-RNA interactions circularize the dengue virus genome. J Virol 79:6631–6643. doi: 10.1128/JVI.79.11.6631-6643.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shurtleff AC, Beasley DW, Chen JJ, Ni H, Suderman MT, Wang H, Xu R, Wang E, Weaver SC, Watts DM, Russell KL, Barrett AD. 2001. Genetic variation in the 3′ non-coding region of dengue viruses. Virology 281:75–87. doi: 10.1006/viro.2000.0748. [DOI] [PubMed] [Google Scholar]
- 13.Alvarez DE, De Lella Ezcurra AL, Fucito S, Gamarnik AV. 2005. Role of RNA structures present at the 3′UTR of dengue virus on translation, RNA synthesis, and viral replication. Virology 339:200–212. doi: 10.1016/j.virol.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 14.Manzano M, Reichert ED, Polo S, Falgout B, Kasprzak W, Shapiro BA, Padmanabhan R. 2011. Identification of cis-acting elements in the 3′-untranslated region of the dengue virus type 2 RNA that modulate translation and replication. J Biol Chem 286:22521–22534. doi: 10.1074/jbc.M111.234302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sztuba-Solinska J, Teramoto T, Rausch JW, Shapiro BA, Padmanabhan R, Le Grice SF. 2013. Structural complexity of dengue virus untranslated regions: cis-acting RNA motifs and pseudoknot interactions modulating functionality of the viral genome. Nucleic Acids Res 41:5075–5089. doi: 10.1093/nar/gkt203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Villordo SM, Gamarnik AV. 2013. Differential RNA sequence requirement for dengue virus replication in mosquito and mammalian cells. J Virol 87:9365–9372. doi: 10.1128/JVI.00567-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friebe P, Shi PY, Harris E. 2011. The 5′ and 3′ downstream AUG region elements are required for mosquito-borne flavivirus RNA replication. J Virol 85:1900–1905. doi: 10.1128/JVI.02037-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.You S, Falgout B, Markoff L, Padmanabhan R. 2001. In vitro RNA synthesis from exogenous dengue viral RNA templates requires long range interactions between 5′- and 3′-terminal regions that influence RNA structure. J Biol Chem 276:15581–15591. doi: 10.1074/jbc.M010923200. [DOI] [PubMed] [Google Scholar]
- 19.You S, Padmanabhan R. 1999. A novel in vitro replication system for dengue virus. Initiation of RNA synthesis at the 3′-end of exogenous viral RNA templates requires 5′- and 3′-terminal complementary sequence motifs of the viral RNA. J Biol Chem 274:33714–33722. [DOI] [PubMed] [Google Scholar]
- 20.Hahn CS, Hahn YS, Rice CM, Lee E, Dalgarno L, Strauss EG, Strauss JH. 1987. Conserved elements in the 3′ untranslated region of flavivirus RNAs and potential cyclization sequences. J Mol Biol 198:33–41. doi: 10.1016/0022-2836(87)90455-4. [DOI] [PubMed] [Google Scholar]
- 21.Khromykh AA, Meka H, Guyatt KJ, Westaway EG. 2001. Essential role of cyclization sequences in flavivirus RNA replication. J Virol 75:6719–6728. doi: 10.1128/JVI.75.14.6719-6728.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clyde K, Barrera J, Harris E. 2008. The capsid-coding region hairpin element (cHP) is a critical determinant of dengue virus and West Nile virus RNA synthesis. Virology 379:314–323. doi: 10.1016/j.virol.2008.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bernhart SH, Hofacker IL, Will S, Gruber AR, Stadler PF. 2008. RNAalifold: improved consensus structure prediction for RNA alignments. BMC Bioinformatics 9:474. doi: 10.1186/1471-2105-9-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gruber AR, Neubock R, Hofacker IL, Washietl S. 2007. The RNAz web server: prediction of thermodynamically stable and evolutionarily conserved RNA structures. Nucleic Acids Res 35:W335–W338. doi: 10.1093/nar/gkm222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hofacker IL. 2004. RNA secondary structure analysis using the Vienna RNA package. Curr Protoc Bioinformatics Chapter 12:Unit 12.2. doi: 10.1002/0471250953.bi1202s04. [DOI] [PubMed] [Google Scholar]
- 26.Weeks KM, Mauger DM. 2011. Exploring RNA structural codes with SHAPE chemistry. Acc Chem Res 44:1280–1291. doi: 10.1021/ar200051h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilkinson KA, Merino EJ, Weeks KM. 2006. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat Protoc 1:1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- 28.Vasa SM, Guex N, Wilkinson KA, Weeks KM, Giddings MC. 2008. ShapeFinder: a software system for high-throughput quantitative analysis of nucleic acid reactivity information resolved by capillary electrophoresis. RNA 14:1979–1990. doi: 10.1261/rna.1166808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bellaousov S, Reuter JS, Seetin MG, Mathews DH. 2013. RNAstructure: Web servers for RNA secondary structure prediction and analysis. Nucleic Acids Res 41:W471–W474. doi: 10.1093/nar/gkt290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deigan KE, Li TW, Mathews DH, Weeks KM. 2009. Accurate SHAPE-directed RNA structure determination. Proc Natl Acad Sci U S A 106:97–102. doi: 10.1073/pnas.0806929106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reuter JS, Mathews DH. 2010. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics 11:129. doi: 10.1186/1471-2105-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu ZY, Li XF, Jiang T, Deng YQ, Zhao H, Wang HJ, Ye Q, Zhu SY, Qiu Y, Zhou X, Qin ED, Qin CF. 2013. Novel cis-acting element within the capsid-coding region enhances flavivirus viral-RNA replication by regulating genome cyclization. J Virol 87:6804–6818. doi: 10.1128/JVI.00243-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Samsa MM, Mondotte JA, Iglesias NG, Assuncao-Miranda I, Barbosa-Lima G, Da Poian AT, Bozza PT, Gamarnik AV. 2009. Dengue virus capsid protein usurps lipid droplets for viral particle formation. PLoS Pathog 5:e1000632. doi: 10.1371/journal.ppat.1000632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lai D, Proctor JR, Zhu JY, Meyer IM. 2012. R-CHIE: a web server and R package for visualizing RNA secondary structures. Nucleic Acids Res 40:e95. doi: 10.1093/nar/gks241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Funk A, Truong K, Nagasaki T, Torres S, Floden N, Balmori Melian E, Edmonds J, Dong H, Shi PY, Khromykh AA. 2010. RNA structures required for production of subgenomic flavivirus RNA. J Virol 84:11407–11417. doi: 10.1128/JVI.01159-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olsthoorn RC, Bol JF. 2001. Sequence comparison and secondary structure analysis of the 3′ noncoding region of flavivirus genomes reveals multiple pseudoknots. RNA 7:1370–1377. doi: 10.1017/S1355838201010068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Villordo SM, Alvarez DE, Gamarnik AV. 2010. A balance between circular and linear forms of the dengue virus genome is crucial for viral replication. RNA 16:2325–2335. doi: 10.1261/rna.2120410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zeng L, Falgout B, Markoff L. 1998. Identification of specific nucleotide sequences within the conserved 3′-SL in the dengue type 2 virus genome required for replication. J Virol 72:7510–7522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Song BH, Yun SI, Choi YJ, Kim JM, Lee CH, Lee YM. 2008. A complex RNA motif defined by three discontinuous 5-nucleotide-long strands is essential for flavivirus RNA replication. RNA 14:1791–1813. doi: 10.1261/rna.993608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salazar MI, Del Angel RM, Lanz-Mendoza H, Ludert JE, Pando-Robles V. 2014. The role of cell proteins in dengue virus infection. J Proteomics 111C:6–15. doi: 10.1016/j.jprot.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 41.Li C, Ge LL, Li PP, Wang Y, Dai JJ, Sun MX, Huang L, Shen ZQ, Hu XC, Ishag H, Mao X. 2014. Cellular DDX3 regulates Japanese encephalitis virus replication by interacting with viral un-translated regions. Virology 449:70–81. doi: 10.1016/j.virol.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gomila RC, Martin GW, Gehrke L. 2011. NF90 binds the dengue virus RNA 3′ terminus and is a positive regulator of dengue virus replication. PLoS One 6:e16687. doi: 10.1371/journal.pone.0016687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ivanyi-Nagy R, Lavergne JP, Gabus C, Ficheux D, Darlix JL. 2008. RNA chaperoning and intrinsic disorder in the core proteins of Flaviviridae. Nucleic Acids Res 36:712–725. doi: 10.1093/nar/gkm1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ward AM, Bidet K, Yinglin A, Ler SG, Hogue K, Blackstock W, Gunaratne J, Garcia-Blanco MA. 2011. Quantitative mass spectrometry of DENV-2 RNA-interacting proteins reveals that the DEAD-box RNA helicase DDX6 binds the DB1 and DB2 3′ UTR structures. RNA Biol 8:1173–1186. doi: 10.4161/rna.8.6.17836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Friedrich S, Schmidt T, Geissler R, Lilie H, Chabierski S, Ulbert S, Liebert UG, Golbik RP, Behrens SE. 2014. AUF1 p45 promotes West Nile virus replication by an RNA chaperone activity that supports cyclization of the viral genome. J Virol 88:11586–11599. doi: 10.1128/JVI.01283-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Groat-Carmona AM, Orozco S, Friebe P, Payne A, Kramer L, Harris E. 2012. A novel coding-region RNA element modulates infectious dengue virus particle production in both mammalian and mosquito cells and regulates viral replication in Aedes aegypti mosquitoes. Virology 432:511–526. doi: 10.1016/j.virol.2012.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.