ABSTRACT
The alphaherpesvirus UL51 protein is a tegument component that interacts with the viral glycoprotein E and functions at multiple steps in virus assembly and spread in epithelial cells. We show here that pUL51 forms a complex in infected cells with another conserved tegument protein, pUL7. This complex can form in the absence of other viral proteins and is largely responsible for recruitment of pUL7 to cytoplasmic membranes and into the virion tegument. Incomplete colocalization of pUL51 and pUL7 in infected cells, however, suggests that a significant fraction of the population of each protein is not complexed with the other and that they may accomplish independent functions.
IMPORTANCE The ability of herpesviruses to spread from cell to cell in the face of an immune response is critical for disease and shedding following reactivation from latency. Cell-to-cell spread is a conserved ability of herpesviruses, and the identification of conserved viral genes that mediate this process will aid in the design of attenuated vaccines and of novel therapeutics. The conserved UL51 gene of herpes simplex virus 1 plays important roles in cell-to-cell spread and in virus assembly in the cytoplasm, both of which likely depend on specific interactions with other viral and cellular proteins. Here we identify one of those interactions with the product of another conserved herpesvirus gene, UL7, and show that formation of this complex mediates recruitment of UL7 to membranes and to the virion.
INTRODUCTION
Human herpesviruses can reactivate from latency at internal anatomical sites and be shed from epithelial surfaces long after initial infection and establishment of an adaptive immune response (1–8). This suggests that the ability to spread from cell to cell in the presence of immune effectors is a conserved property of herpesviruses. Within the alphaherpesviruses, where this phenomenon has been the most extensively studied, the evidence suggests that cell-to-cell spread (CCS) between epithelial cells in vitro has two mechanistic components. The first of these is a virion trafficking component in which virions are targeted from the site of secondary envelopment to the junctional surfaces of cells, where they have access to adjacent cells in a compartment that is sterically protected from immune effectors in the medium (9–11). The second component is a specialized entry process that may involve gE binding to specific CCS receptors and that also may involve different patterns of receptor usage by the virus entry apparatus (12–15). In alphaherpesviruses, both mechanistic components involve the complex between the envelope glycoproteins gE and gI. However, since gE and gI are encoded only by alphaherpesvirus genomes, any mechanism for CCS that is conserved among all herpesviruses is likely to involve other, more widely conserved viral factors.
Virus genome-encoded proteins that are specifically required for epithelial CCS fall into two categories: those required for virus entry into cells regardless of the mode of virus egress and those that are apparently required for trafficking of virus components or virions to junctional surfaces of cells. The four viral glycoproteins gB, gD, gH, and gL, which are required for virus entry, are in the first category (16–19), while gE and gI are in the second category. gE and gI form a heterodimeric complex that is required for efficient CCS in the nervous system in vivo (20–24). It is also required for CCS in cultured neuronal cells and in epithelial and fibroblast cells that form well-defined cell junctions (20, 24–26). Trafficking of gE to cell junctions and its function in CCS are regulated by other viral proteins that are conserved among the herpesviruses. A complex of pUL11, pUL16, and pUL21 must form on the gE cytoplasmic tail in order for it to localize to junctions and to function properly in CCS (27–30), and in some cell types, gE localization is also sensitive to mutations in the nuclear egress factor pUL34 (31). Herpes simplex virus 1 (HSV-1) pUL51 also interacts with gE, and expression of a dominant negative pUL51 fusion protein disrupts gE localization (32). However, the effect of pUL51 or pUL34 mutation on CCS in Vero cells is far more severe than that of gE deletion, suggesting that additional interactions are involved.
Alphaherpesvirus pUL34, pUL11, and pUL51 are all membrane-associated proteins, but none of them is displayed on the exterior surface of the infected cell or on the virion (33–42), suggesting that they function by interacting with viral and/or cellular factors in the cytosol or on cytoplasmic membranes and are likely to aid trafficking of virions and/or viral proteins, including gE, for cell-to-cell spread. pUL34, pUL11, and pUL51 also are important in virus assembly, and at least for pUL34 and pUL11, these functions involve specific interactions with other viral proteins and their recruitment to virus assembly sites on the nuclear or internal cytoplasmic membrane, respectively (27, 28, 34, 43–51). In addition, the human cytomegalovirus (HCMV) homolog of pUL51, pUL71, aids in the organization of specialized regions of the cytoplasm where cytoplasmic envelopment occurs, further suggesting a function in trafficking of viral or cellular proteins (52).
Alphaherpesvirus pUL51 and its cytomegalovirus homolog, pUL71, are classified as tegument proteins but are associated with membranes (40, 41, 53). At least for HSV-1, membrane association is by way of a palmitoyl group added at a highly conserved cysteine residue near the N terminus of the protein (42). The involvement of pUL51 in both virus assembly and CCS suggested that it, too, makes specific interactions with other viral proteins for assembly and perhaps with other cellular trafficking factors for CCS. In an effort to identify other interaction partners for pUL51, we affinity purified pUL51 from infected cells and discovered a novel interaction with another conserved herpesvirus gene product, pUL7.
MATERIALS AND METHODS
Cells and viruses.
HEp-2 and Vero cells and a UL51-complementing cell line (described in reference 32) were maintained as previously described (34). HaCaT cells (gift of David Johnson) were maintained in Dulbecco modified Eagle medium (DMEM) with high glucose supplemented with 10% fetal bovine serum. The properties of HSV-1(F), HSV-1 with UL51 with a FLAG tag (UL51-FLAG), and HSV-1 with UL51 with a C-terminal truncation of amino acids 73 to 244 (UL51Δ73-244) have been previously described (34, 54).
Construction of recombinant mutant viruses.
The recombinant virus with UL51 with a C-terminal truncation of amino acids 167 to 244 (UL51Δ167-244) was constructed using an HSV-1(F) bacterial artificial chromosome (BAC) and the methods of Tischer et al. (55), as previously described (31). The sequences of the primers used for virus construction are available upon request. The proper structure of the recombinant BACs was determined by sequencing of the UL51 gene region. The structures of the altered UL51 genes are indicated in Fig. 1.
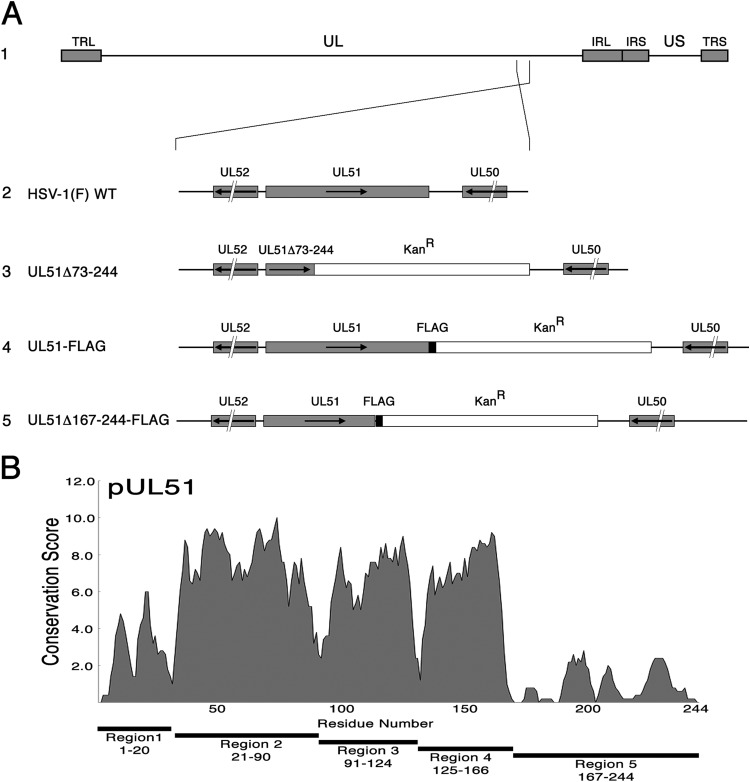
FIG 1.
Construction of recombinant viruses. (A) Schematic diagram of the HSV-1(F) genome (line 1) and of the recombinant viruses used in this study. TRL, terminal repeat long region; UL, unique long region; IRL, internal repeat long region; IRS, internal repeat short region; US, unique short region; TRS, terminal repeat short region. Line 2, structures of the wild-type sequences in the regions of UL51 and US8; line 3, the UL51Δ73-244 virus carries a stop codon and a kanamycin resistance cassette in place of the sequences coding for amino acids 73 to 244 of pUL51; line 4, the UL51-FLAG virus carries a FLAG tag at the C terminus of UL51 followed by a kanamycin resistance cassette; line 5, the UL51Δ167-244FLAG virus was constructed as described in Materials and Methods and carries a FLAG-tagged truncated pUL51 that lacks approximately the last third of the protein. (B) UL51 sequence conservation. The plot shows the conservation of the biochemical properties of amino acids using all available herpesvirus pUL51 homologous sequences aligned using the program MUSCLE (87). Each residue position receives a conservation score, and scores were averaged over a sliding 5-amino-acid-residue window.
Recombinant virus was reconstituted by transfection of BAC DNA into Vero cells. Virus containing deletions in the UL51 gene sequence were amplified on UL51-complementing cells to minimize selection for phenotypic revertants.
Plasmids and transfection.
Plasmids that express UL51-FLAG, UL7, and gE from the human cytomegalovirus major immediate early promoter were constructed by PCR amplification of the desired coding sequence and ligation into pcDNA3. The gE-coding sequence was amplified using the primers 5′-AGCTGAATTCCCATGGATCGCGGGGCGGTG-3′ and 5′-GATCCTCGAGTTACCAGAAGACGGACGAATCGGAGG-3′ and then cut with EcoRI and XhoI restriction enzymes and ligated into the EcoRI and XhoI sites of pcDNA3. The UL7-coding sequence was amplified using the primers 5′-GATCGAATTCATGGCCGCCGCGACG-3′ and 5′-CTAGTCTAGATCAACAAAACTGATAAAACAGCGACGACGTCTG-3′ and then cut with EcoRI and XbaI restriction enzymes and ligated into the EcoRI and XbaI sites of pcDNA3. UL51-FLAG was amplified from the UL51-FLAG recombinant BAC using the primers 5′-AGCTGAATTCCCATGGCTTCTCTTCTCGGGGCTATATG-3′ and 5′-GATCCTCGAGCTACTTATCGTCATCGTCTTTGTAGTCTTGACCC-3′ and then cut with EcoRI and XhoI and ligated into the EcoRI and XhoI sites of pcDNA3. Plasmids were transfected into Vero cells using the Lipofectamine reagent according to the manufacturer's protocol.
Indirect immunofluorescence.
Immunofluorescence for colocalization was performed as previously described using either 1:500 anti-UL51 rabbit antiserum (a gift of Joel Baines), 1:500 anti-UL7 rabbit antiserum (a gift of Yasushi Kawaguchi), 1:1,000 mouse monoclonal anti-gE (Virusys), or 1:1,000 mouse monoclonal anti-FLAG M2 antibody (Sigma) (45, 56). Quantitative colocalization analysis was performed using ImageJ software. Ten randomly selected cells that showed clear concentrations of gE staining were chosen from each condition. A line was drawn across each cell that included the concentration(s) of gE, and a profile of pixel intensities for both gE and pUL7 staining was obtained. Pixel intensity values from each profile were entered as data sets for determination of a Pearson correlation coefficient, as implemented at http://www.socscistatistics.com/tests/pearson.
Immunopurification.
pUL51-FLAG was purified from Vero or HEp-2 cells that had been infected with 5 PFU/cell of wild-type or recombinant tagged HSV-1 for 16 h. Infected cell monolayers from 100-mm cultures were washed with 5 ml of phosphate-buffered saline (PBS), and then the cells were scraped into 3 ml of PBS and pelleted at 110 × g for 10 min. The cell pellets were resuspended in 1.5 ml coimmunoprecipitation (co-IP) buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1× Sigma protease inhibitor cocktail), transferred to microcentrifuge tubes, and incubated on ice for 3 min. Nuclei and other cellular debris were pelleted by centrifugation at 10,000 rpm in a microcentrifuge for 10 min, and the supernatant was transferred to a fresh tube. After removal of a fraction of the sample as a lysate control, 15 μl of an anti-FLAG magnetic bead suspension (Sigma) was added to the remainder of each sample, and the tubes were placed in an end-over-end rotator at 4°C overnight. The magnetic beads were separated from the lysate using a magnetic separator, and the supernatant containing unbound proteins was discarded. Magnetic beads were washed three times each with 1.5 ml of co-IP buffer, and then bound proteins were eluted with three washes with co-IP buffer containing 100 μg/ml competitor 3× FLAG peptide (Sigma).
Immunoprecipitation.
Proteins were immunoprecipitated from HEp-2 cells that had been infected with 5 PFU/cell of wild-type HSV-1(F) for 16 h. Infected cell monolayers from 100-mm cultures were washed with 5 ml of PBS, and the cells were then scraped into 3 ml of PBS and pelleted at 110 × g for 10 min. The cell pellets were resuspended in 0.75 ml co-IP buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1× Sigma protease inhibitor cocktail), transferred to microcentrifuge tubes, and incubated on ice for 3 min. Nuclei and other cellular debris were pelleted by centrifugation at 10,000 rpm in a microcentrifuge for 10 min, and the supernatant was transferred to a fresh tube. After removal of a fraction of the sample as a lysate control, 1 μl of nonimmune rabbit serum, rabbit anti-UL51 antiserum, or anti-UL7 antiserum was added to the remainder of each sample, and the tubes were incubated at 4°C overnight. On the following day, 30 μl of a 50% suspension of protein G agarose (Thermo/Pierce) was added and samples were incubated in an end-over-end rotator for 6 h. The beads were separated from the lysate by centrifugation in a microcentrifuge at 1,000 rpm for 1 min, and the supernatant containing unbound proteins was discarded. The beads were washed five times each with 0.5 ml of co-IP buffer, and then bound proteins were eluted with two washes with 50 μl of 0.1 M glycine, pH 2.5. Eluents were neutralized with 1 M Tris base.
Peptide sequencing.
Proteins eluted following anti-FLAG purification were separated on SDS-polyacrylamide gels under reducing and denaturing conditions. The gels were stained with Coomassie stain, and a few bands of interest were excised for in-gel tryptic digestion. Following the procedure described by Shevchenko et al. (57), the quality of the digest supernatant was determined by matrix-assisted laser desorption ionization–time of flight (TOF) analysis on an Autoflex III TOF/TOF apparatus (Bruker) prior to lyophilization. Peptides from digested samples were analyzed by nano-liquid chromatography-tandem mass spectrometry (MS/MS) using a Dionex 3000 UHP nano-rapid separation liquid chromatography series high-pressure liquid chromatography system (Thermo-Electron) and a linear ion-trap mass spectrometer (Thermo LTQ/XL; Thermo Electron). MS/MS spectra were acquired in a data-dependent acquisition mode that automatically selected and fragmented the six most intense peaks from each MS spectrum.
Peptide RAW data sets were refined to a centroid list using the Distiller (version 2.4) program (MatrixScience) and matched to protein sequences in the Swiss-Prot and TrEMBl databases of 23 July 2012 with the Spectrum Mill Proteomics workbench (revision A.03.02.060; Agilent Technologies), accepting carbamidomethyl cysteine as a fixed modification and methionine oxidation as the single variable modification (58). Peptide and MS/MS mass tolerances were set at 1.8 and 0.4 Th, respectively. Taxonomy files were restricted to Homo sapiens and human herpesvirus 1, with common contaminants being included. A target/decoy search strategy was used, with mass tolerances being set to 1.8 Th for peptide precursor ions and 0.4 Th for fragment ion data (59). Search requirements were set to a maximum local peptide false discovery rate cutoff score of 0.5%, at least four y- or b-ion pairs, and at least two unique peptides. The resulting peptide alignments were manually examined and accepted only if peptides from the TREM-1sv standard protein with identical sequences revealed a consistent fragmentation pattern.
Immunoblotting.
Nitrocellulose sheets bearing proteins of interest were blocked in 5% nonfat milk plus 0.2% Tween 20 for at least 2 h. The membranes were probed with either a rabbit polyclonal anti-UL51 (1:1,000), rabbit polyclonal anti-UL7, rabbit polyclonal anti-gE (a kind gift of H. Friedman) (1:500), or mouse anti-FLAG M2 (1:1,000) (Sigma/Aldrich) monoclonal antibody, followed by reaction with alkaline phosphatase-conjugated secondary antibody.
Virion purification.
Cell-associated virions from wild-type and UL51 mutant HSV-1 were purified from infected HEp-2 cells by modification of the method of Spear and Roizman (60) as previously described (61). Briefly, cells from three confluent T150 flasks that had been infected with 5 PFU/cell of virus for 24 h were washed twice with PBS and then scraped into 20 ml PBS. Cells were then pelleted at 800 × g for 10 min in a clinical centrifuge, and the supernatant was discarded. The cell pellet was resuspended in 3.2 ml of 1 mM Na2HPO4-NaH2PO4, pH 7.0, buffer and allowed to swell on ice for 15 min. Cells were lysed with five strokes in a Dounce homogenizer with a tight pestle and then immediately adjusted to 50 mM sucrose. Large cell fragments, nuclei, and mitochondria were pelleted by centrifugation at 15,000 × g for 10 min in a Sorvall SS34 rotor. The supernatant, containing smaller membrane structures, virions, and soluble proteins, was then centrifuged at 30,000 rpm in a Beckman SW60Ti rotor for 1 h to pellet virions and other membrane structures. The supernatant was discarded, and the pellet was resuspended in 0.5 ml of 1 mM Na2HPO4-NaH2PO4, pH 7.0, buffer, sonicated for 5 s at power level 1 with a Fisher sonic Dismembrator 50, and then layered onto a 10-ml linear 15 to 30% dextran T10 gradient and centrifuged at 20,000 rpm for 1 h in a Beckman Sw40Ti rotor to separate virions and less dense cellular membrane structures. The gradient was fractionated by dripping from the bottom of the tube. Equal aliquots of each fraction were separated by SDS-PAGE, and proteins were detected by immunoblotting.
Culture supernatant (released) virions from wild-type and UL51 mutant HSV-1 were purified from infected HaCaT cell cultures by modification of the method of Lippé (62). Eight 150-mm cultures of confluent HaCaT cells were infected with 5 PFU/cell of each virus for 24 h in medium containing 1% heat-inactivated calf serum. Extracellular medium was pooled and centrifuged at 300 × g for 10 min to pellet the large debris, and the supernatant was filtered through a 0.45-μm-pore-size filter. Virus was pelleted by centrifugation at 20,000 × g for 30 min. The pellet was resuspended in 1.0 ml of DMEM, treated with 250 U DNase I for 30 min at 4°C, sonicated for 10 s at power level 1, and then layered over a 5 to 15% continuous Ficoll 400 gradient prepared in DMEM and centrifuged at 26,000 × g for 2 h in a Beckman Sw40Ti rotor as described by Szilagyi and Cunningham (63). The gradient was fractionated by dripping from the bottom of the tube. Equal aliquots of each fraction were titrated by plaque assay. Aliquots of each fraction were separated by SDS-PAGE, and proteins were detected by immunoblotting.
RESULTS
pUL7 copurifies with pUL51.
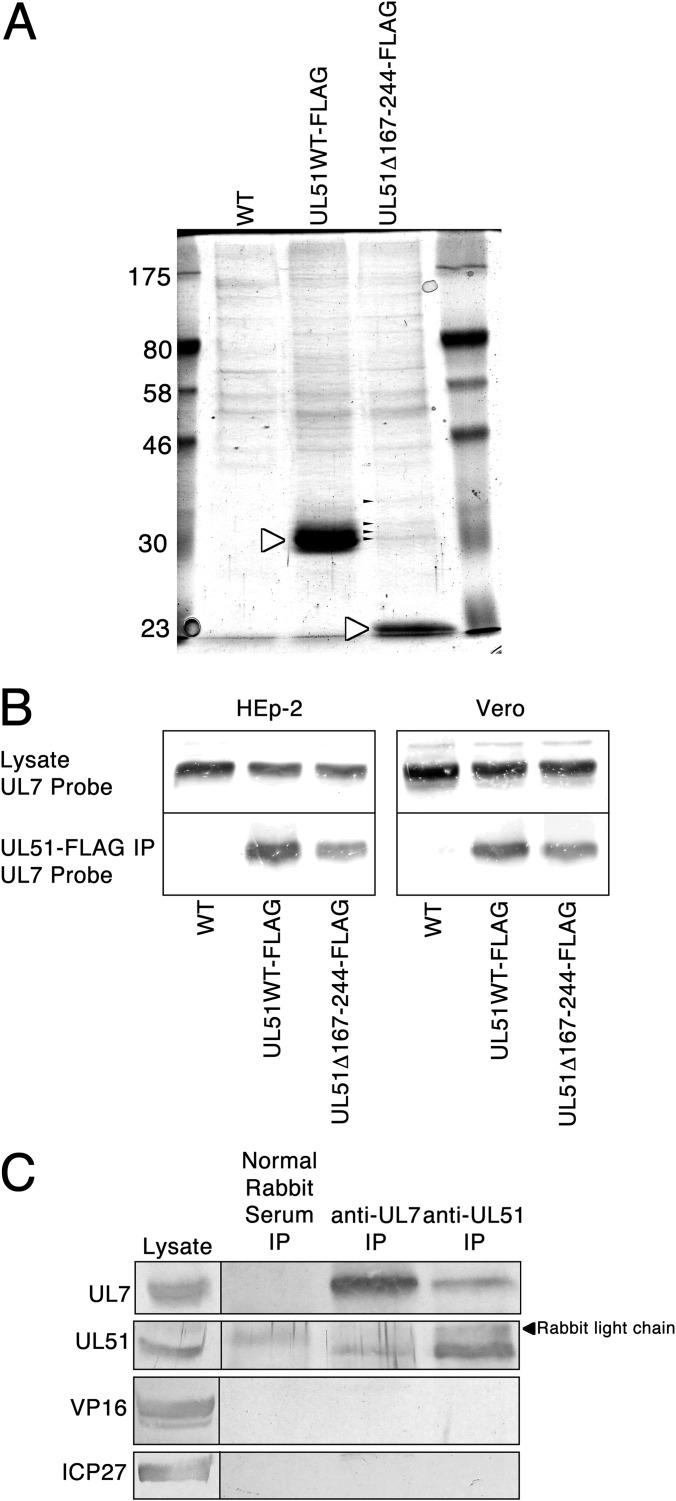
HSV-1 pUL51 has documented functions in nuclear egress, cytoplasmic envelopment, virus release, and epithelial CCS (32, 51). The protein has no known or predicted enzymatic activity and so likely accomplishes its functions by interaction with other viral or cellular factors. In order to identify some of those factors, recombinant viruses in which full-length or C-terminally truncated pUL51 was tagged at the C terminus with a FLAG epitope were generated. The C-terminal truncation (amino acids 167 to 244) was designed to correspond to a region of very low sequence similarity between pUL51 homologs from all herpesviruses (Fig. 1B), suggesting that the truncated protein might retain most or all conserved interactions of pUL51. FLAG-tagged UL51 protein from HEp-2 cells infected with wild-type (WT) virus (which carries no FLAG tag and so serves as a background control) (UL51WT-FLAG) or FLAG-tagged UL51 with a C-terminal truncation of amino acids 167 to 244 (UL51Δ167-244-FLAG) was purified on anti-FLAG magnetic beads, separated by SDS-PAGE, and stained with Coomassie brilliant blue (Fig. 2A). Both full-length pUL51 and truncated pUL51 were efficiently purified (white arrowheads). The most obvious accompanying proteins were seen in purification of the truncated pUL51 (small black arrowheads), but they were not obvious in purification of full-length pUL51 because they partially comigrated with it. Excision of these bands and sequencing by tandem mass spectrometry revealed a high representation of peptides from the HSV-1 pUL7 protein.
FIG 2.
Copurification and coimmunoprecipitation of pUL7 with pUL51. (A) Coomassie brilliant blue-stained SDS-polyacrylamide gel containing proteins purified from infected HEp-2 cells using anti-FLAG magnetic beads. White arrowheads, FLAG-tagged pUL51 proteins; small black arrowheads, copurifying proteins that partially comigrated with full-length pUL51. Numbers on the left are molecular sizes (in kilodaltons). (B) Immunoblot of proteins from Vero or HEp-2 cells infected with the viruses indicated below each lane. (Top) pUL7 present in the lysate from infected cells; (bottom) pUL7 present in eluents from anti-FLAG purification. (C) Immunoblot of proteins from HEp-2 cells infected with wild-type virus. The leftmost lane contains proteins from cell lysates. The immunoprecipitating (IP) antibody is indicated above each lane, and the antibody used for probing of the blot is indicated to the left of each panel.
In order to confirm copurification of pUL7 with pUL51, FLAG affinity-purified proteins from infected Vero and HEp-2 cells were separated by SDS-PAGE, blotted, and probed with rabbit antiserum directed against pUL7 (Fig. 2B). pUL7 is present in equivalent amounts in lysates from cells infected with each of the three viruses but is purified only from cells infected with recombinant viruses carrying FLAG-tagged pUL51. Copurification was observed from both Vero and HEp-2 cells, indicating that the pUL51/pUL7 interaction is most likely not cell specific.
In order to ensure that interaction with pUL7 is not an artifactual result of epitope tagging, wild-type, untagged protein was immunoprecipitated from HEp-2 cells using antibodies to either pUL51 or pUL7 (Fig. 2C). pUL51 was coimmunoprecipitated with anti-pUL7 and vice versa. This interaction was specific, since neither protein was precipitated by nonimmune rabbit serum and since neither antibody coimmunoprecipitated VP16 (a highly abundant viral protein that, like pUL51 and pUL7, is a component of the virion tegument) or ICP27 (an abundant protein found in both the cytoplasm and nucleus).
pUL51 is required for recruitment of pUL7 to cytoplasmic membranes in both infected and transfected cells.
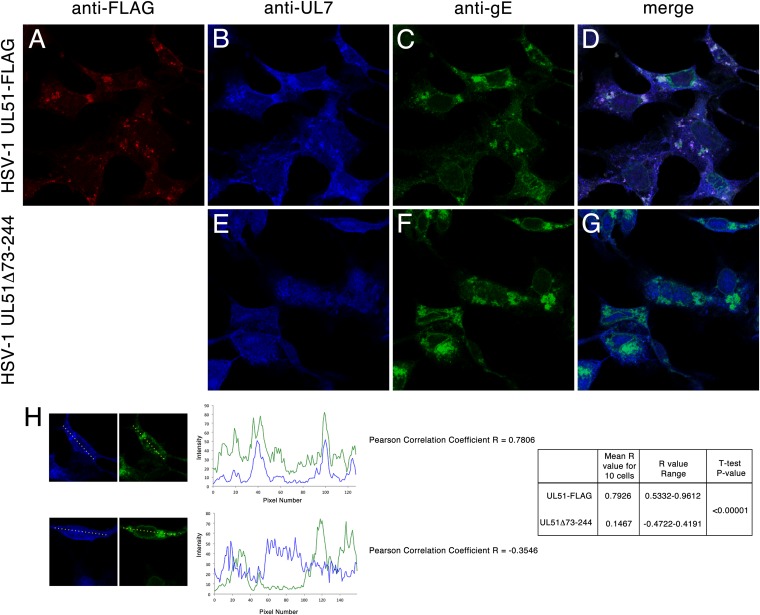
The observation that pUL51 and pUL7 interact during copurification and coimmunoprecipitation suggested that they form a complex in the infected cell. This further suggested that the proteins should colocalize in infected cells. Immunofluorescence assay of Vero cells infected with UL51WT-FLAG virus using antibodies directed against FLAG, pUL7, and gE showed that all three proteins partially colocalized (Fig. 3). As shown previously, pUL51 was largely found on cytoplasmic membranes that also contained gE (Fig. 3A, C, and D) (32). gE, however, was found on cytoplasmic membranes that did not contain pUL51 and was also present on the nuclear membrane, where pUL51 was absent. Some pUL7 was found diffusely localized in both the nucleoplasm and cytoplasm, but some of it was also concentrated on cytoplasmic membranes, where it colocalized with pUL51 and gE (Fig. 3B and D). The concentration of pUL7 on cytoplasmic membranes and colocalization with gE depended on expression of pUL51, since neither occurred in cells infected with HSV-1 UL51Δ73-244, which is missing most of the UL51 protein-coding sequence (Fig. 3E to G). Quantitative analysis of colocalization of gE and pUL7 in 10 randomly selected cells that showed concentrations of gE in the cytoplasm showed that gE and pUL7 localization was positively correlated in all infected cells that also expressed pUL7 (mean R value, 0.7926). None of the cells analyzed gave R values of less than 0.5. In contrast, pUL7 localization was not correlated with gE localization in cells infected with UL51Δ73-244 virus (mean R value, 0.1467). None of the cells analyzed gave an R value of greater than 0.5.
FIG 3.
Localization of pUL51, pUL7, and gE in infected cells. Vero cells infected for 14 h with HSV-1 UL51WT-FLAG (A to D) or HSV-1 UL51Δ73-244 (E to G) were fixed and probed with anti-FLAG antibody to detect pUL51 (A and D), anti-UL7 antiserum (B, D, E, and G), and mouse monoclonal antibody directed against gE (C, D, F, and G). Confocal images of single z-sections taken near the center of the nucleus (i.e., where the nuclear cross-section is the largest) with a 60× oil objective are shown. Negative controls (not shown) were normal rabbit serum for anti-UL7 rabbit antiserum and uninfected cells for anti-gE and anti-FLAG and showed no fluorescence at the laser and detector settings used to obtain these images. (H) Quantitation of gE and pUL7 colocalization. gE and pUL7 fluorescent intensities were measured across the linear profiles of 10 randomly selected cells, as described in Materials and Methods. Representative profiles from one cell infected with UL51-FLAG (top) and UL51Δ73-244 (bottom) are shown along with the profile plots and derived Pearson correlation coefficients. Aggregate results from 10 profiles are shown at the right.
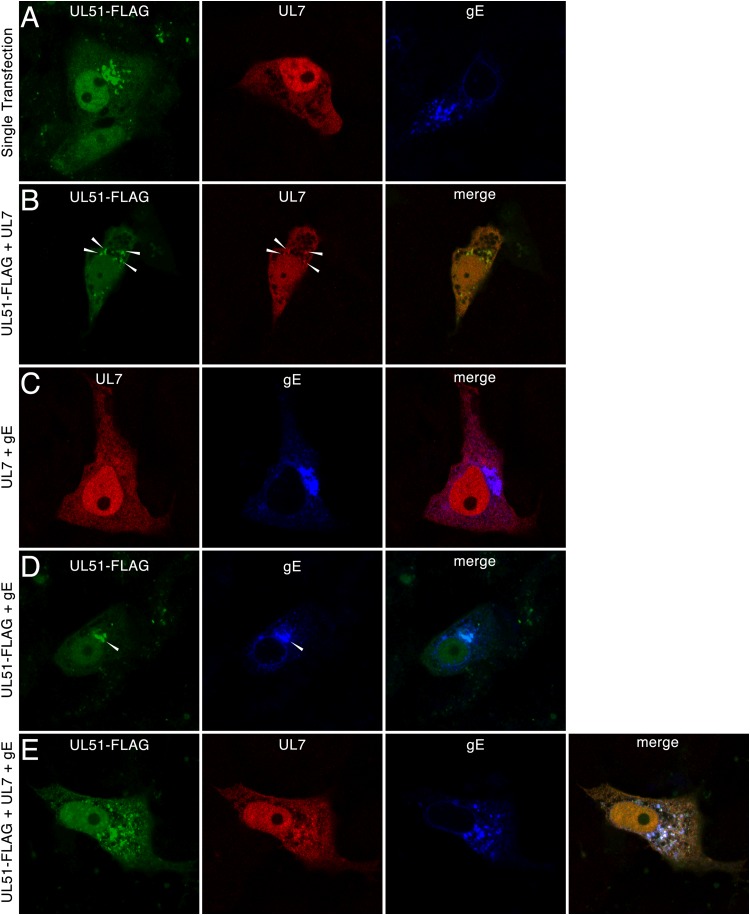
To determine whether pUL51, pUL7, and gE can interact in the absence of other viral proteins, Vero cells were transfected with plasmids carrying pUL51-FLAG, pUL7, and gE, and protein localizations were determined by immunofluorescence (Fig. 4). pUL51 expressed in the absence of any other viral gene was concentrated on cytoplasmic membranes previously show to be Golgi apparatus membranes (42), although some protein was also found diffusely distributed in both the cytoplasm and the nucleoplasm (Fig. 4A). pUL7 expressed alone was found diffusely distributed in the cytoplasm and nucleoplasm and showed no tendency to concentrate on membranes (Fig. 4A). Coexpression of pUL51 and pUL7, however, resulted in recruitment of some pUL7 to sites of pUL51 concentration (Fig. 4B, arrowheads). pUL7 showed no tendency to colocalize with gE when these two proteins were expressed alone (Fig. 4C), suggesting that interaction with pUL51 is specifically responsible for membrane recruitment of pUL7. pUL51 and gE also colocalized when these two proteins were expressed in the absence of pUL7 (Fig. 4D, arrowheads). Since both pUL51 and gE can independently localize to the Golgi apparatus, however, this does not necessarily imply interaction during coexpression. Expression of all three proteins resulted in a localization pattern very similar to that seen in infected cells, with partial colocalization of all three proteins on cytoplasmic membranes. This colocalization was observed in cells expressing low, moderate, and large amounts of all three proteins, suggesting that colocalization was not an artifact of overexpression, and the images shown were chosen, in part, because the fluorescent staining intensities were similar to those seen in infected cells.
FIG 4.
Localization of pUL51, pUL7, and gE in transfected cells. Vero cells were transfected with plasmids for 24 h and then fixed and probed by indirect immunofluorescence. Images of single z-sections taken near the center of the nucleus with a 60× oil objective are shown. Primary antibodies are indicated at the tops of the panels, and the plasmids used for transfection are indicated to the left of each row. White arrowheads, areas of colocalization between pUL51 and pUL7.
Incorporation of pUL7 into virions depends upon pUL51.
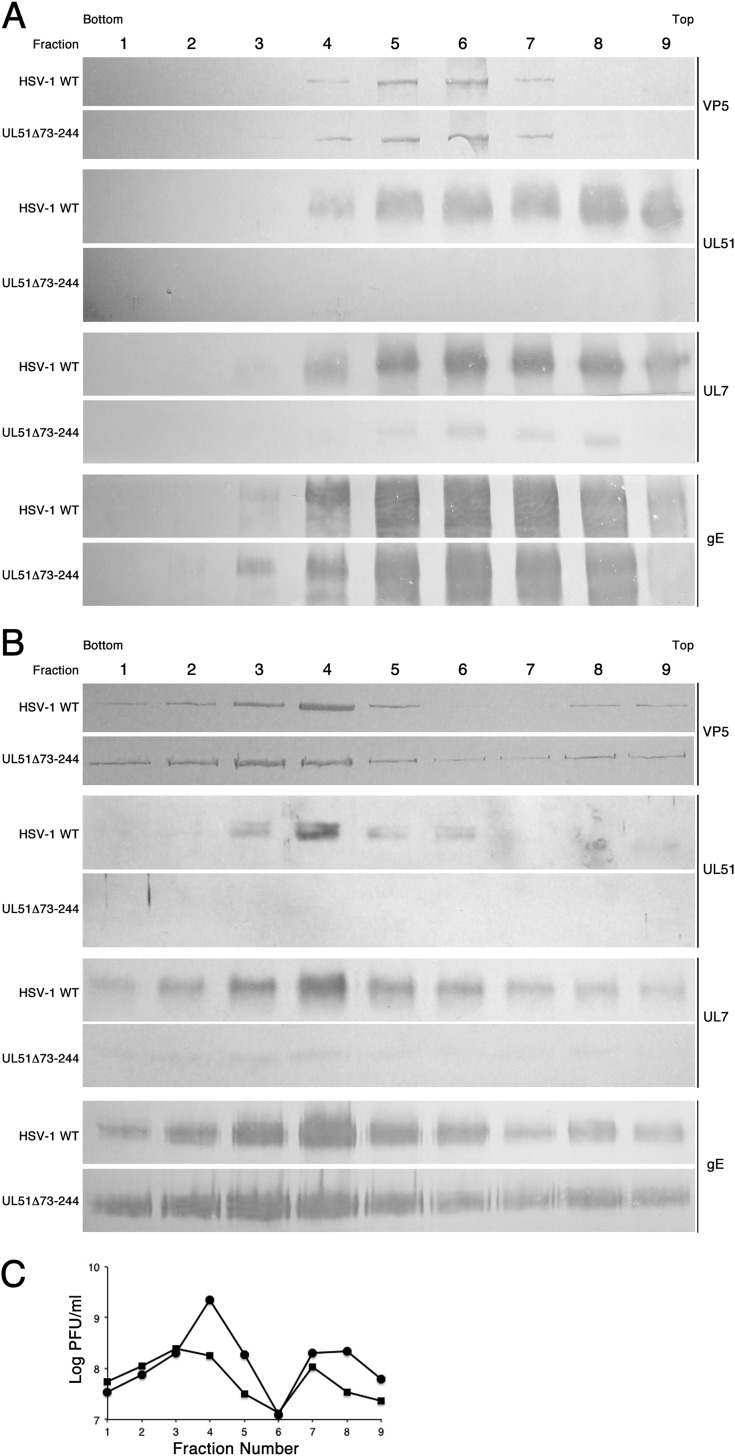
pUL7 and pUL51 are both components of the virus tegument (40, 41, 53, 64–67). Their interaction suggested that one might be responsible for recruitment of the other to sites of virion assembly on Golgi apparatus or endosomal membranes. The observation that pUL7 recruitment to cytoplasmic membranes depended upon expression of pUL51 (Fig. 3) suggested that pUL51 might also be required for recruitment of pUL7 to the virion tegument. Virions were purified from whole-cell membrane fractions and from culture supernatants, as described in Materials and Methods (Fig. 5). Cytoplasmic membranes from HEp-2 cells infected with either HSV-1(F) or HSV-1 UL51Δ73-244 were fractionated on dextran gradients, and fractions were assayed by immunoblotting for VP5 (to identify virions) and for pUL51, pUL7, and gE (Fig. 5A). Virions were concentrated in fractions 5 and 6 of the gradient. About half of the pUL51 in the gradient peaked in the same fractions as VP5, but a significant amount was also found in the lighter fractions, consistent with pUL51 association with cytoplasmic membranes other than the virion envelope. As expected, pUL51 was not detectable in fractions from cells infected with UL51 deletion virus. In fractions from wild-type virus-infected cells, pUL7 cosedimented with pUL51 and was abundant in the virion fractions. In contrast, the amount of pUL7 in the entire gradient and, specifically, in the virion fractions was greatly reduced in the UL51 deletion virus-infected cells. The near absence of pUL7 in the gradient overall is consistent with the hypothesis that pUL51 is required for the recruitment of pUL7 to cytoplasmic membranes. The greatly diminished amount of pUL7 specifically in virion fractions from UL51 deletion virus-infected cells indicated that pUL51 expression was necessary for the efficient incorporation of pUL7 into the tegument. However, pUL7 was not entirely absent from the virion fractions in the UL51 deletion virus gradient, perhaps indicating that it can be recruited at a very low efficiency by interaction with other viral factors. pUL51 also interacts with gE, but in contrast to pUL7, incorporation of gE into the virion envelope appeared to be unaffected by deletion of UL51. In order to confirm these observations in mature virions that had been released from infected cells, virions were purified from the culture supernatants of infected HaCaT cells and probed for VP5, pUL51, pUL7, and gE (Fig. 5B) or titers were determined by plaque assay to determine infectivity (Fig. 5C). As expected, VP5, pUL51, and gE all peaked in the same fractions as those with virion infectivity in both wild-type and UL51 deletion virus infections. pUL7 was greatly diminished (but, again, not completely absent) in the virion fractions from cells infected with pUL51 deletion virus.
FIG 5.
Recruitment of pUL7 to virions by pUL51. (A) Immunoblots of virion gradient fractions from fractionation of cytoplasmic membranes of cells infected with wild-type or UL51Δ73-244 virus are shown. (B) Immunoblots of virion gradient fractions from fractionation cell culture supernatants of cells infected with wild-type or UL51Δ73-244 virus are shown. For panels A and B, the infecting virus is indicated to the left of each panel. The antibody used for immunoblot probing is indicated to the right. (C) Plot of infectivity in the gradients shown in panel B.
DISCUSSION
The UL51 and UL7 genes are conserved in all herpesviruses, where they accomplish at least roughly similar functions. Homologs of both have been reported to participate in cytoplasmic envelopment in the alpha- and betaherpesviruses (50–52, 64, 67–70). pUL51 representatives from all three herpesvirus subfamilies (pUL51 in HSV and pseudorabies virus [PrV], pUL71 in human cytomegalovirus [HCMV], and BSRF1 in Epstein-Barr virus [EBV]) have been reported to be virion tegument components (40, 41, 64–67). pUL7 has also been identified to be a tegument component in HSV, PrV, and HCMV (where the homolog is UL103), but not in EBV (53, 65, 66).
Here, we show that pUL51 and pUL7 of HSV-1 form a complex in infected cells that can be purified from infected cell lysates and from which either protein can be coprecipitated with antibody directed against the other. We also show that pUL51 and pUL7 are partially colocalized in infected cells and that pUL51 is required for recruitment of a portion of the infected cell pUL7 to cytoplasmic membranes. Furthermore, recruitment of pUL7 to membranes by pUL51 occurs in cells that do not express other viral proteins. Finally, pUL51 is largely responsible for recruitment of pUL7 to the virion tegument.
We have previously shown that, in addition to its previously described function in cytoplasmic envelopment, pUL51 has cell type-specific functions in virus growth and CCS in epithelial cells (32). Its importance in CCS may be due to a function in trafficking virion components or perhaps virions themselves to the junctional surfaces of cells (32). The growth phenotypes of HSV-1 isolates with UL51 and UL7 deletions are notably similar. In both cases, single-step growth is inhibited 10- to 100-fold and the deletion viruses form minute plaques (32, 71), suggesting functions in both assembly and CCS. Furthermore, like pUL51, pUL7 from PrV and UL103 from HCMV are required for efficient cytoplasmic envelopment (64, 68). This suggests the possibility that the two proteins perform their principal growth and spread functions as a complex. pUL51 and pUL7 do not completely colocalize in infected cells, however, showing that some fraction of the population of each protein is not complexed with the other. This suggests the possibility that the uncomplexed fraction of each population may pursue independent functions.
HSV-1 pUL7 has previously been shown to interact with the mitochondrial adenine nucleotide transporter 2 (ANT2) in pulldown and coimmunoprecipitation assays (71). We did not observe colocalization between either pUL51 or pUL7 and mitochondrial markers in infected cells (not shown) at 16 h after infection. Nonetheless, interactions between pUL7 and ANT2 at earlier or later times might provide an avenue for pUL7 function independent of pUL51.
Viral proteins are thought to be recruited to the tegument by either direct or indirect interactions with the capsid or by direct or indirect interactions with the cytoplasmic extensions of proteins embedded in the virion envelope (reviewed in reference 72). In fact, interaction chains that connect the exterior face of the capsid to the exterior face of the membranes for secondary envelopment may be important for mediating both secondary envelopment and incorporation of tegument proteins into the virion. For example, the large tegument protein VP1/2 (pUL36) binds to pUL25 and pUL17 at the capsid penton vertices and recruits pUL37 (73–77). Interaction between pUL37 and the virion envelope proteins gK and pUL20 assists with secondary envelopment (78). pUL16 also associates both with capsids and with pUL11, which is embedded in membranes by way of its myristoyl group, and this association may also assist with secondary envelopment (27, 48, 49, 77, 79–83). Tegument proteins that are not necessary for envelopment may be incorporated by direct or indirect interaction with proteins that assist in assembly. pUL16, for example, is required for recruitment of VP22, which is, in turn, required for recruitment of ICP0 and ICP4 to the virion tegument (84, 85). Our results suggest that the same paradigm governs incorporation of pUL7 into the HSV virion.
HSV-1 pUL7 is a 296-amino-acid protein that lacks a putative transmembrane sequence or motifs for fatty acylation. Its association with membranes, therefore, is likely mediated by direct or indirect interaction with a membrane-associated protein. Our results suggest that this protein is pUL51. Furthermore, HSV-1 pUL7 is salt extractable from detergent-treated virions, suggesting that it is a peripheral tegument protein (66). This is consistent with its recruitment to the virion by interaction with pUL51 protein that is associated with the interior of the viral envelope by way of its palmitoyl group.
We also observed low-level incorporation into the virions during infection with HSV-1 UL51Δ73-244, suggesting that pUL7 has an alternative route for recruitment to the virion. Nozawa et al. found that pUL7 was weakly associated with capsids purified from wild-type virus-infected cells (86). Association with capsids may, therefore, provide a redundant, albeit inefficient mechanism for pUL7 virion incorporation and may account for the inefficient incorporation of pUL7 into virions that was observed when pUL51 was not expressed (Fig. 5). Bridging of the capsid and membrane by way of a pUL7/pUL51 interaction might also provide a means for this complex to assist with cytoplasmic capsid envelopment.
ACKNOWLEDGMENTS
We are grateful to Harvey Friedman for the gift of anti-gE antiserum, to Yasushi Kawaguchi for UL7 antiserum, and to Keith Jarosinski and members of the R. J. Roller laboratory for helpful discussions and critical readings of the manuscript.
Mass spectrometry analysis was performed in the Roy J. Carver Charitable Trust-supported CCOM Proteomics Facility at the University of Iowa. This work was supported by NIH grant AI097212 (to R.J.R.).
REFERENCES
- 1.Arbuckle JH, Medveczky PG. 2011. The molecular biology of human herpesvirus-6 latency and telomere integration. Microbes Infect 13:731–741. doi: 10.1016/j.micinf.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beaulieu BL, Sullivan JL. 2002. Epstein-Barr virus, p 479–494 InRichman DD, RJ Whitley, Hayden FG (ed), Clinical virology, 2nd ed ASM Press, Washington, DC. [Google Scholar]
- 3.Di Luca D, Mirandola P, Ravaioli T, Dolcetti R, Frigatti A, Bovenzi P, Sighinolfi L, Monini P, Cassai E. 1995. Human herpesviruses 6 and 7 in salivary glands and shedding in saliva of healthy and human immunodeficiency virus positive individuals. J Med Virol 45:462–468. doi: 10.1002/jmv.1890450418. [DOI] [PubMed] [Google Scholar]
- 4.Gershon AA, Gershon MD. 2013. Pathogenesis and current approaches to control of varicella-zoster virus infections. Clin Microbiol Rev 26:728–743. doi: 10.1128/CMR.00052-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giffin L, Damania B. 2014. KSHV: pathways to tumorigenesis and persistent infection. Adv Virus Res 88:111–159. doi: 10.1016/B978-0-12-800098-4.00002-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Griffiths PD, Emery VC. 2002. Cytomegalovirus, p 433–461 InRichman DD, RJ Whitley, Hayden FG (ed), Clinical virology, 2nd ed ASM Press, Washington, DC. [Google Scholar]
- 7.Sacks SL, Griffiths PD, Corey L, Cohen C, Cunningham A, Dusheiko GM, Self S, Spruance S, Stanberry LR, Wald A, Whitley RJ. 2004. HSV shedding. Antiviral Res 63(Suppl 1):S19–S26. doi: 10.1016/j.antiviral.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 8.Widmer IC, Erb P, Grob H, Itin P, Baumann M, Stalder A, Weber R, Cathomas G. 2006. Human herpesvirus 8 oral shedding in HIV-infected men with and without Kaposi sarcoma. J Acquired Immune Defic Syndr 42:420–425. doi: 10.1097/01.qai.0000226790.31463.e6. [DOI] [PubMed] [Google Scholar]
- 9.Mingo RM, Han J, Newcomb WW, Brown JC. 2012. Replication of herpes simplex virus: egress of progeny virus at specialized cell membrane sites. J Virol 86:7084–7097. doi: 10.1128/JVI.00463-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson DC, Webb M, Wisner TW, Brunetti C. 2001. Herpes simplex virus gE/gI sorts nascent virions to epithelial cell junctions, promoting virus spread. J Virol 75:821–833. doi: 10.1128/JVI.75.2.821-833.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farnsworth A, Johnson DC. 2006. Herpes simplex virus gE/gI must accumulate in the trans-Golgi network at early times and then redistribute to cell junctions to promote cell-cell spread. J Virol 80:3167–3179. doi: 10.1128/JVI.80.7.3167-3179.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wisner T, Brunetti C, Dingwell K, Johnson DC. 2000. The extracellular domain of herpes simplex virus gE is sufficient for accumulation at cell junctions but not for cell-to-cell spread. J Virol 74:2278–2287. doi: 10.1128/JVI.74.5.2278-2287.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Polcicova K, Goldsmith K, Rainish BL, Wisner TW, Johnson DC. 2005. The extracellular domain of herpes simplex virus gE is indispensable for efficient cell-to-cell spread: evidence for gE/gI receptors. J Virol 79:11990–12001. doi: 10.1128/JVI.79.18.11990-12001.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collins WJ, Johnson DC. 2003. Herpes simplex virus gE/gI expressed in epithelial cells interferes with cell-to-cell spread. J Virol 77:2686–2695. doi: 10.1128/JVI.77.4.2686-2695.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rauch DA, Rodriguez N, Roller RJ. 2000. Mutations in herpes simplex virus glycoprotein D distinguish entry of free virus from cell-cell spread. J Virol 74:11437–11446. doi: 10.1128/JVI.74.24.11437-11446.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cai JS, Jang HK, Izumiya Y, Tsushima Y, Kato K, Damiani AM, Miyazawa T, Kai C, Takahashi E, Mikami T. 1999. Identification and structure of the Marek's disease virus serotype 2 glycoprotein M gene: comparison with glycoprotein M genes of Herpesviridae family. J Vet Med Sci 61:503–511. doi: 10.1292/jvms.61.503. [DOI] [PubMed] [Google Scholar]
- 17.Forrester A, Farrell H, Wilkinson G, Kaye J, Davis-Poynter N, Minson T. 1992. Construction and properties of a mutant herpes simplex virus type 1 with glycoprotein H coding sequences deleted. J Virol 66:341–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ligas MW, Johnson DC. 1988. A herpes simplex virus mutant in which glycoprotein D sequences are replaced by β-galactosidase sequences binds to but is unable to penetrate into cells. J Virol 62:1486–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roop C, Hutchinson L, Johnson DC. 1993. A mutant herpes simplex virus type 1 unable to express glycoprotein L cannot enter cells and its particles lack glycoprotein H. J Virol 67:2285–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dingwell KS, Brunetti CR, Hendricks RL, Tang Q, Tang M, Rainbow AJ, Johnson DC. 1994. Herpes simplex virus glycoproteins E and I facilitate cell-to-cell spread in vivo and across junctions of cultured cells. J Virol 68:834–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dingwell KS, Doering LC, Johnson DC. 1995. Glycoproteins E and I facilitate neuron-to-neuron spread of herpes simplex virus. J Virol 69:7087–7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson DC, Feenstra V. 1987. Identification of a novel herpes simplex virus type 1-induced glycoprotein which complexes with gE and binds immunoglobulin. J Virol 61:2208–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson DC, Frame MC, Ligas MW, Cross AM, Stow ND. 1988. Herpes simplex virus immunoglobulin G Fc receptor activity depends on a complex of two viral glycoproteins, gE and gI. J Virol 62:1347–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McGraw HM, Awasthi S, Wojcechowskyj JA, Friedman HM. 2009. Anterograde spread of herpes simplex virus type 1 requires glycoprotein E and glycoprotein I but not US9. J Virol 83:8315–8326. doi: 10.1128/JVI.00633-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Balan P, Davis-Poynter N, Bell S, Atkinson H, Browne H, Minson T. 1994. An analysis of the in vitro and in vivo phenotypes of mutants of herpes simplex virus type 1 lacking glycoproteins gG, gE, gI or the putative gJ. J Gen Virol 75:1245–1258. doi: 10.1099/0022-1317-75-6-1245. [DOI] [PubMed] [Google Scholar]
- 26.Dingwell KS, Johnson DC. 1998. The herpes simplex virus gE-gI complex facilitates cell-to-cell spread and binds to components of cell junctions. J Virol 72:8933–8942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chadha P, Han J, Starkey JL, Wills JW. 2012. Regulated interaction of tegument proteins UL16 and UL11 from herpes simplex virus. J Virol 86:11886–11898. doi: 10.1128/JVI.01879-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han J, Chadha P, Meckes DG Jr, Baird NL, Wills JW. 2011. Interaction and interdependent packaging of tegument protein UL11 and glycoprotein E of herpes simplex virus. J Virol 85:9437–9446. doi: 10.1128/JVI.05207-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han J, Chadha P, Starkey JL, Wills JW. 2012. Function of glycoprotein E of herpes simplex virus requires coordinated assembly of three tegument proteins on its cytoplasmic tail. Proc Natl Acad Sci U S A 109:19798–19803. doi: 10.1073/pnas.1212900109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yeh PC, Han J, Chadha P, Meckes DG Jr, Ward MD, Semmes OJ, Wills JW. 2011. Direct and specific binding of the UL16 tegument protein of herpes simplex virus to the cytoplasmic tail of glycoprotein E. J Virol 85:9425–9436. doi: 10.1128/JVI.05178-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haugo AC, Szpara ML, Parsons L, Enquist LW, Roller RJ. 2011. Herpes simplex virus type 1 pUL34 plays a critical role in cell-to-cell spread of virus in addition to its role in virus replication. J Virol 85:7203–7215. doi: 10.1128/JVI.00262-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roller RJ, Haugo AC, Yang K, Baines JD. 2014. The herpes simplex virus 1 UL51 gene product has cell type-specific functions in cell-to-cell spread. J Virol 88:4058–4068. doi: 10.1128/JVI.03707-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klupp BG, Granzow H, Mettenleiter TC. 2001. Effect of the pseudorabies virus US3 protein on nuclear membrane localization of the UL34 protein and virus egress from the nucleus. J Gen Virol 82:2363–2371. [DOI] [PubMed] [Google Scholar]
- 34.Roller RJ, Zhou Y, Schnetzer R, Ferguson J, DeSalvo D. 2000. Herpes simplex virus type 1 UL34 gene product is required for viral envelopment. J Virol 74:117–129. doi: 10.1128/JVI.74.1.117-129.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shiba C, Daikoku T, Goshima F, Takakuwa H, Yamauchi Y, Koiwai O, Nishiyama Y. 2000. The UL34 gene product of herpes simplex virus type 2 is a tail-anchored type II membrane protein that is significant for virus envelopment. J Gen Virol 81:2397–2405. [DOI] [PubMed] [Google Scholar]
- 36.Baines JD, Jacob RJ, Simmerman L, Roizman B. 1995. The herpes simplex virus 1 UL11 proteins are associated with cytoplasmic and nuclear membranes and with nuclear bodies of infected cells. J Virol 69:825–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baird NL, Yeh PC, Courtney RJ, Wills JW. 2008. Sequences in the UL11 tegument protein of herpes simplex virus that control association with detergent-resistant membranes. Virology 374:315–321. doi: 10.1016/j.virol.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loomis JS, Bowzard JB, Courtney RJ, Wills JW. 2001. Intracellular trafficking of the UL11 tegument protein of herpes simplex virus type 1. J Virol 75:12209–12219. doi: 10.1128/JVI.75.24.12209-12219.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.MacLean CA, Clark B, McGeoch DJ. 1989. Gene UL11 of herpes simplex virus type 1 encodes a virion protein which is myristylated. J Gen Virol 70:3147–3157. doi: 10.1099/0022-1317-70-12-3147. [DOI] [PubMed] [Google Scholar]
- 40.Daikoku T, Ikenoya K, Yamada H, Goshima F, Nishiyama Y. 1998. Identification and characterization of the herpes simplex virus type 1 UL51 gene product. J Gen Virol 79:3027–3031. [DOI] [PubMed] [Google Scholar]
- 41.Lenk M, Visser N, Mettenleiter TC. 1997. The pseudorabies virus UL51 gene product is a 30-kilodalton virion component. J Virol 71:5635–5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nozawa N, Daikoku T, Koshizuka T, Yamauchi Y, Yoshikawa T, Nishiyama Y. 2003. Subcellular localization of herpes simplex virus type 1 UL51 protein and role of palmitoylation in Golgi apparatus targeting. J Virol 77:3204–3216. doi: 10.1128/JVI.77.5.3204-3216.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fuchs W, Klupp BG, Granzow H, Osterrieder N, Mettenleiter TC. 2002. The interacting UL31 and UL34 gene products of pseudorabies virus are involved in egress from the host-cell nucleus and represent components of primary enveloped but not mature virions. J Virol 76:364–378. doi: 10.1128/JVI.76.1.364-378.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klupp BG, Granzow H, Mettenleiter TC. 2000. Primary envelopment of pseudorabies virus at the nuclear membrane requires the UL34 gene product. J Virol 74:10063–10073. doi: 10.1128/JVI.74.21.10063-10073.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reynolds AE, Ryckman BJ, Baines JD, Zhou Y, Liang L, Roller RJ. 2001. UL31 and UL34 proteins of herpes simplex virus type 1 form a complex that accumulates at the nuclear rim and is required for envelopment of nucleocapsids. J Virol 75:8803–8817. doi: 10.1128/JVI.75.18.8803-8817.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reynolds AE, Wills EG, Roller RJ, Ryckman BJ, Baines JD. 2002. Ultrastructural localization of the herpes simplex virus type 1 UL31, UL34, and US3 proteins suggests specific roles in primary envelopment and egress of nucleocapsids. J Virol 76:8939–8952. doi: 10.1128/JVI.76.17.8939-8952.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baines JD, Roizman B. 1992. The UL11 gene of herpes simplex virus 1 encodes a function that facilitates nucleocapsid envelopment and egress from cells. J Virol 66:5168–5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kopp M, Granzow H, Fuchs W, Klupp BG, Mundt E, Karger A, Mettenleiter TC. 2003. The pseudorabies virus UL11 protein is a virion component involved in secondary envelopment in the cytoplasm. J Virol 77:5339–5351. doi: 10.1128/JVI.77.9.5339-5351.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Loomis JS, Courtney RJ, Wills JW. 2003. Binding partners for the UL11 tegument protein of herpes simplex virus type 1. J Virol 77:11417–11424. doi: 10.1128/JVI.77.21.11417-11424.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Klupp BG, Granzow H, Klopfleisch R, Fuchs W, Kopp M, Lenk M, Mettenleiter TC. 2005. Functional analysis of the pseudorabies virus UL51 protein. J Virol 79:3831–3840. doi: 10.1128/JVI.79.6.3831-3840.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nozawa N, Kawaguchi Y, Tanaka M, Kato A, Kato A, Kimura H, Nishiyama Y. 2005. Herpes simplex virus type 1 UL51 protein is involved in maturation and egress of virus particles. J Virol 79:6947–6956. doi: 10.1128/JVI.79.11.6947-6956.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schauflinger M, Fischer D, Schreiber A, Chevillotte M, Walther P, Mertens T, von Einem J. 2011. The tegument protein UL71 of human cytomegalovirus is involved in late envelopment and affects multivesicular bodies. J Virol 85:3821–3832. doi: 10.1128/JVI.01540-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Varnum SM, Streblow DN, Monroe ME, Smith P, Auberry KJ, Paša-Tolić L, Wang D, Camp DG, Rodland K, Wiley S, Britt W, Shenk T, Smith RD, Nelson JA. 2004. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J Virol 78:10960–10966. doi: 10.1128/JVI.78.20.10960-10966.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ejercito PM, Kieff ED, Roizman B. 1968. Characteristics of herpes simplex virus strains differing in their effect on social behavior of infected cells. J Gen Virol 2:357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- 55.Tischer BK, von Einem J, Kaufer B, Osterrieder N. 2006. Two-step Red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques 40:191–197. doi: 10.2144/000112096. [DOI] [PubMed] [Google Scholar]
- 56.Bjerke SL, Cowan JM, Kerr JK, Reynolds AE, Baines JD, Roller RJ. 2003. Effects of charged cluster mutations on the function of herpes simplex virus type 1 UL34 protein. J Virol 77:7601–7610. doi: 10.1128/JVI.77.13.7601-7610.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. 2006. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protocols 1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 58.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20:3551–3567. doi:. [DOI] [PubMed] [Google Scholar]
- 59.Elias JE, Haas W, Faherty BK, Gygi SP. 2005. Comparative evaluation of mass spectrometry platforms used in large-scale proteomics investigations. Nat Methods 2:667–675. doi: 10.1038/nmeth785. [DOI] [PubMed] [Google Scholar]
- 60.Spear PG, Roizman B. 1972. Proteins specified by herpes simplex virus. V. Purification and structural proteins of the herpesvirion. J Virol 9:143–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roller RJ, Roizman B. 1992. The herpes simplex virus 1 RNA binding protein US11 is a virion component and associates with ribosomal 60S subunits. J Virol 66:3624–3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lippé R. 2014. Characterization of extracellular HSV-1 virions by proteomics, p 181–190 InDiefenbach RJ, Fraefel C (ed), Herpes simplex virus, vol 1144 Springer, New York, NY. [DOI] [PubMed] [Google Scholar]
- 63.Szilagyi JF, Cunningham C. 1991. Identification and characterization of a novel non-infectious herpes simplex virus-related particle. J Gen Virol 72:661–668. doi: 10.1099/0022-1317-72-3-661. [DOI] [PubMed] [Google Scholar]
- 64.Fuchs W, Granzow H, Klopfleisch R, Klupp BG, Rosenkranz D, Mettenleiter TC. 2005. The UL7 gene of pseudorabies virus encodes a nonessential structural protein which is involved in virion formation and egress. J Virol 79:11291–11299. doi: 10.1128/JVI.79.17.11291-11299.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Johannsen E, Luftig M, Chase MR, Weicksel S, Cahir-McFarland E, Illanes D, Sarracino D, Kieff E. 2004. Proteins of purified Epstein-Barr virus. Proc Natl Acad Sci U S A 101:16286–16291. doi: 10.1073/pnas.0407320101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Loret S, Lippe R. 2012. Biochemical analysis of infected cell polypeptide (ICP)0, ICP4, UL7 and UL23 incorporated into extracellular herpes simplex virus type 1 virions. J Gen Virol 93:624–634. doi: 10.1099/vir.0.039776-0. [DOI] [PubMed] [Google Scholar]
- 67.Womack A, Shenk T. 2010. Human cytomegalovirus tegument protein pUL71 is required for efficient virion egress. mBio 1(5):e00282–10. doi: 10.1128/mBio.00282-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ahlqvist J, Mocarski E. 2011. Cytomegalovirus UL103 controls virion and dense body egress. J Virol 85:5125–5135. doi: 10.1128/JVI.01682-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Meissner CS, Suffner S, Schauflinger M, von Einem J, Bogner E. 2012. A leucine zipper motif of a tegument protein triggers final envelopment of human cytomegalovirus. J Virol 86:3370–3382. doi: 10.1128/JVI.06556-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Messerle M, Rapp M, Lucin P, Koszinowski UH. 1995. Characterization of a conserved gene block in the murine cytomegalovirus genome. Virus Genes 10:73–80. doi: 10.1007/BF01724298. [DOI] [PubMed] [Google Scholar]
- 71.Tanaka M, Sata T, Kawaguchi Y. 2008. The product of the herpes simplex virus 1 UL7 gene interacts with a mitochondrial protein, adenine nucleotide translocator 2. Virol J 5:125. doi: 10.1186/1743-422X-5-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mettenleiter TC, Klupp BG, Granzow H. 2009. Herpesvirus assembly: an update. Virus Res 143:222–234. doi: 10.1016/j.virusres.2009.03.018. [DOI] [PubMed] [Google Scholar]
- 73.Cardone G, Newcomb WW, Cheng N, Wingfield PT, Trus BL, Brown JC, Steven AC. 2012. The UL36 tegument protein of herpes simplex virus 1 has a composite binding site at the capsid vertices. J Virol 86:4058–4064. doi: 10.1128/JVI.00012-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Coller KE, Lee JI, Ueda A, Smith GA. 2007. The capsid and tegument of the alphaherpesviruses are linked by an interaction between the UL25 and VP1/2 proteins. J Virol 81:11790–11797. doi: 10.1128/JVI.01113-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Desai P, Sexton GL, Huang E, Person S. 2008. Localization of herpes simplex virus type 1 UL37 in the Golgi complex requires UL36 but not capsid structures. J Virol 82:11354–11361. doi: 10.1128/JVI.00956-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Klupp BG, Fuchs W, Granzow H, Nixdorf R, Mettenleiter TC. 2002. Pseudorabies virus UL36 tegument protein physically interacts with the UL37 protein. J Virol 76:3065–3071. doi: 10.1128/JVI.76.6.3065-3071.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vittone V, Diefenbach E, Triffett D, Douglas MW, Cunningham AL, Diefenbach RJ. 2005. Determination of interactions between tegument proteins of herpes simplex virus type 1. J Virol 79:9566–9571. doi: 10.1128/JVI.79.15.9566-9571.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jambunathan N, Chouljenko D, Desai P, Charles A-S, Subramanian R, Chouljenko VN, Kousoulas KG. 2014. Herpes simplex virus 1 protein UL37 interacts with viral glycoprotein gK and membrane protein UL20 and functions in cytoplasmic virion envelopment. J Virol 88:5927–5935. doi: 10.1128/JVI.00278-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kopp M, Granzow H, Fuchs W, Klupp B, Mettenleiter TC. 2004. Simultaneous deletion of pseudorabies virus tegument protein UL11 and glycoprotein M severely impairs secondary envelopment. J Virol 78:3024–3034. doi: 10.1128/JVI.78.6.3024-3034.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Leege T, Fuchs W, Granzow H, Kopp M, Klupp BG, Mettenleiter TC. 2009. Effects of simultaneous deletion of pUL11 and glycoprotein M on virion maturation of herpes simplex virus type 1. J Virol 83:896–907. doi: 10.1128/JVI.01842-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Meckes DG, Wills JW. 2007. Dynamic interactions of the UL16 tegument protein with the capsid of herpes simplex virus. J Virol 81:13028–13036. doi: 10.1128/JVI.01306-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Oshima S, Daikoku T, Shibata S, Yamada H, Goshima F, Nishiyama Y. 1998. Characterization of the UL16 gene product of herpes simplex virus type 2. Arch Virol 143:863–880. doi: 10.1007/s007050050338. [DOI] [PubMed] [Google Scholar]
- 83.Yeh PC, Meckes DG Jr, Wills JW. 2008. Analysis of the interaction between the UL11 and UL16 tegument proteins of herpes simplex virus. J Virol 82:10693–10700. doi: 10.1128/JVI.01230-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Elliott G, Hafezi W, Whiteley A, Bernard E. 2005. Deletion of the herpes simplex virus VP22-encoding gene (UL49) alters the expression, localization, and virion incorporation of ICP0. J Virol 79:9735–9745. doi: 10.1128/JVI.79.15.9735-9745.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Starkey JL, Han J, Chadha P, Marsh JA, Wills JW. 2014. Elucidation of the block to herpes simplex virus egress in the absence of tegument protein UL16 reveals a novel interaction with VP22. J Virol 88:110–119. doi: 10.1128/JVI.02555-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nozawa N, Daikoku T, Yamauchi Y, Takakuwa H, Goshima F, Yoshikawa T, Nishiyama Y. 2002. Identification and characterization of the UL7 gene product of herpes simplex virus type 2. Virus Genes 24:257–266. doi: 10.1023/A:1015332716927. [DOI] [PubMed] [Google Scholar]
- 87.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]