ABSTRACT
Sarcocystis neurona is a member of the coccidia, a clade of single-celled parasites of medical and veterinary importance including Eimeria, Sarcocystis, Neospora, and Toxoplasma. Unlike Eimeria, a single-host enteric pathogen, Sarcocystis, Neospora, and Toxoplasma are two-host parasites that infect and produce infectious tissue cysts in a wide range of intermediate hosts. As a genus, Sarcocystis is one of the most successful protozoan parasites; all vertebrates, including birds, reptiles, fish, and mammals are hosts to at least one Sarcocystis species. Here we sequenced Sarcocystis neurona, the causal agent of fatal equine protozoal myeloencephalitis. The S. neurona genome is 127 Mbp, more than twice the size of other sequenced coccidian genomes. Comparative analyses identified conservation of the invasion machinery among the coccidia. However, many dense-granule and rhoptry kinase genes, responsible for altering host effector pathways in Toxoplasma and Neospora, are absent from S. neurona. Further, S. neurona has a divergent repertoire of SRS proteins, previously implicated in tissue cyst formation in Toxoplasma. Systems-based analyses identified a series of metabolic innovations, including the ability to exploit alternative sources of energy. Finally, we present an S. neurona model detailing conserved molecular innovations that promote the transition from a purely enteric lifestyle (Eimeria) to a heteroxenous parasite capable of infecting a wide range of intermediate hosts.
IMPORTANCE
Sarcocystis neurona is a member of the coccidia, a clade of single-celled apicomplexan parasites responsible for major economic and health care burdens worldwide. A cousin of Plasmodium, Cryptosporidium, Theileria, and Eimeria, Sarcocystis is one of the most successful parasite genera; it is capable of infecting all vertebrates (fish, reptiles, birds, and mammals—including humans). The past decade has witnessed an increasing number of human outbreaks of clinical significance associated with acute sarcocystosis. Among Sarcocystis species, S. neurona has a wide host range and causes fatal encephalitis in horses, marine mammals, and several other mammals. To provide insights into the transition from a purely enteric parasite (e.g., Eimeria) to one that forms tissue cysts (Toxoplasma), we present the first genome sequence of S. neurona. Comparisons with other coccidian genomes highlight the molecular innovations that drive its distinct life cycle strategies.
INTRODUCTION
The coccidia are a large clade of protozoan parasites within the phylum Apicomplexa. In addition to a single definitive host species in which the parasite undergoes its sexual cycle, a subgroup of coccidia, the members of the family Sarcocystidae (Sarcocystis, Toxoplasma, and Neospora) have evolved the ability to infect a broad range of intermediate hosts (1, 2). To drive this transition, the members of the family Sarcocystidae produce infectious tissue cysts surrounded by glycosylated cyst walls. Different species and even strains exhibit distinct patterns of organ tropism, with Toxoplasma forming cysts in any organ, whereas Sarcocystis cysts are largely restricted to muscle. Ingestion of tissue cysts through predation or scavenging by the definitive host propagates the life cycle (e.g., felids for Toxoplasma, canids for Neospora, and humans for two Sarcocystis species) (3).
To survive and persist in their respective hosts, apicomplexan parasites have evolved a variety of molecular strategies. These include a group of specialized proteins that facilitate parasite entry, egress, and colonization, as well as molecular decoys that modulate host immune signaling (4, 5). The majority of these proteins localize to exocytic organelles (micronemes, rhoptries, and dense granules) that discharge in a highly coordinated program of invasion (5). For Toxoplasma gondii, initial host recognition and attachment are performed by members of the SAG1-related sequence (SRS) family. This is followed by secretion of the microneme (MIC) proteins that strengthen host cell attachment and result in the formation of a “moving junction” that provides the motive force required to penetrate the host cell. The moving junction is further controlled by a set of proteins known as rhoptry neck (RON) proteins that facilitate invasion (for a review, see reference 6). Subsequently, rhoptry (ROP) proteins and dense-granule (GRA) proteins are secreted into the host cytosol to interact with cellular targets to protect the (now) intracellular parasite from clearance. The parasite is further protected through encasement within a parasitophorous vacuole (PV) that is, interestingly, absent from the schizont form of Sarcocystis.
Recent genomic comparisons of Toxoplasma, Neospora, and Hammondia (the closest extant relative of T. gondii) have identified a series of ROP proteins whose expression targets host-specific immune signaling pathways. ROP5, ROP16, and ROP18 have all been shown to affect parasite virulence and contribute to host specialization in the mouse model (4, 5). Recently, an expanded repertoire of SRS proteins, previously implicated in host range expansion of T. gondii, were also identified in Neospora (7). Modeling of T. gondii metabolism also identified strain-specific differences in growth potential, establishing metabolism as an evolutionary factor capable of influencing host adaptation (8). To complement the recently generated Eimeria genome sequences and understand the transition from a purely enteric, monoxenous life cycle (e.g., Eimeria) to a heteroxenous one that includes the formation of tissue cysts, we sequenced the genome of Sarcocystis neurona.
Sarcocystosis, caused by parasites within the genus Sarcocystis, is typically asymptomatic but can be associated with myositis, diarrhea, or infection of the central nervous system (CNS). The genus is ancient (relative age, 246 to 500 million years based on small-subunit RNA sequences), diverse (more than 150 catalogued species), highly successful (all vertebrates are susceptible hosts, including fish, birds, reptiles, and mammals), and prevalent (cattle exhibit a 90% infection rate worldwide) (9). Interestingly, Sarcocystis species are not structurally similar; for example, S. neurona sporozoites, like T. gondii, lack the crystalloid body present in other coccidia, including S. cruzi of cattle (10). Sarcocystis species typically have a two-host predator-prey life cycle, with one host supporting asexual multiplication while the other acts as the definitive host, supporting a sexual cycle that results in sporocyst shedding in feces. Humans are definitive hosts of S. suihominis and S. hominis and can be infected by S. nesbitti, with associated sequelae, including muscular sarcocystosis. Opossums are the definitive hosts of S. neurona (11), a species that has a broad intermediate-host range, including raccoons, cats, skunks, and more recently a variety of mustelids, pinnipeds, and cetaceans (12–15). S. neurona produces tissue cysts, typically in muscle and occasionally in the CNS (16, 17). Horses are considered aberrant hosts, in which the parasite typically multiplies as schizonts in the CNS but fails to encyst. Unabated destruction of neural tissue can be fatal to horses and many other hosts, and the disease was called equine protozoal myeloencephalitis before the etiologic protozoan S. neurona was identified and named in 1991 (2). With migration of opossums to the west coast of North America during the last century (14) the S. neurona host range expanded to cause epizootics in sea otters, harbor seals, and harbor porpoises (18). S. neurona is now being monitored for its potential as an emerging disease threat. Here, we sequenced and performed a systems-based analysis of the genome of type II S. neurona strain SO SN1, isolated from a southern sea otter that died of protozoal encephalitis (19), which represents the most common genotype infecting animals throughout the United States.
RESULTS
The S. neurona SO SN1 genome is more than twice the size of the T. gondii genome.
Combining 7,020,033 reads from the 454 Life Sciences sequencing platform with 529,830,690 reads from the Illumina Hi-Seq sequencing platform, we generated 47,722 Mbp of sequence data from S. neurona SO SN1 DNA. By integrating a variety of assembly algorithms (see Materials and Methods), these data were assembled into 116 genomic scaffolds with a combined size of 127 Mbp, over twice the size of the Neospora caninum and T. gondii genomes (Table 1 is a summary of the genome statistics obtained). An additional 3.1 Mbp of sequence is encoded in 2,950 unscaffolded contigs (each greater than 500 bp in length). The assembly N50 value was 3,117,290 bp, with a maximum scaffold length of 9,217,112 bp. To help annotation efforts, we generated an additional 59,622,019 reads from S. neurona RNA. From these data, we predict a complement of 7,093 genes, 5,853 of which are supported by RNA-Seq evidence (see Table S1 in the supplemental material). The number of genes predicted is comparable to that of the related coccidia T. gondii and Eimeria tenella. Comparisons of gene orderings reveal blocks of syntenic relationships between homologous genes in S. neurona and T. gondii, with the longest such block aligning 43 genes on scaffold 1 of S. neurona and chromosome IX of T. gondii (Fig. 1A). Chromosome-wide synteny was not observed, suggesting a significant level of genome rearrangement between S. neurona and T. gondii. A more detailed view of the largest syntenic region reveals the extent of gene order preservation but also reveals differences in the structures of individual genes (Fig. 1B).
TABLE 1 .
Genome statistics
| Parameter | Statistic |
|---|---|
| Genome size (bp) | 130,222,184 |
| Genome GC% | 51.5 |
| No. of scaffolds | 116 |
| Total scaffolded length (bp) | 127,077,592 |
| No. of contigs in scaffolds | 11,452 |
| Scaffold N50 (bp) | 3,117,920 |
| No. of unscaffolded contigs | 2,950 |
| Contig N50 (bp) | 20,915 |
| Total no. of bp in gaps | 12,350,913 |
| No. of genes | 7,093 |
| Mean gene length (bp) | 9,121 |
| Mean no. of exons | 5.5 |
| Mean coding size (no. of amino acid residues) | 856 |
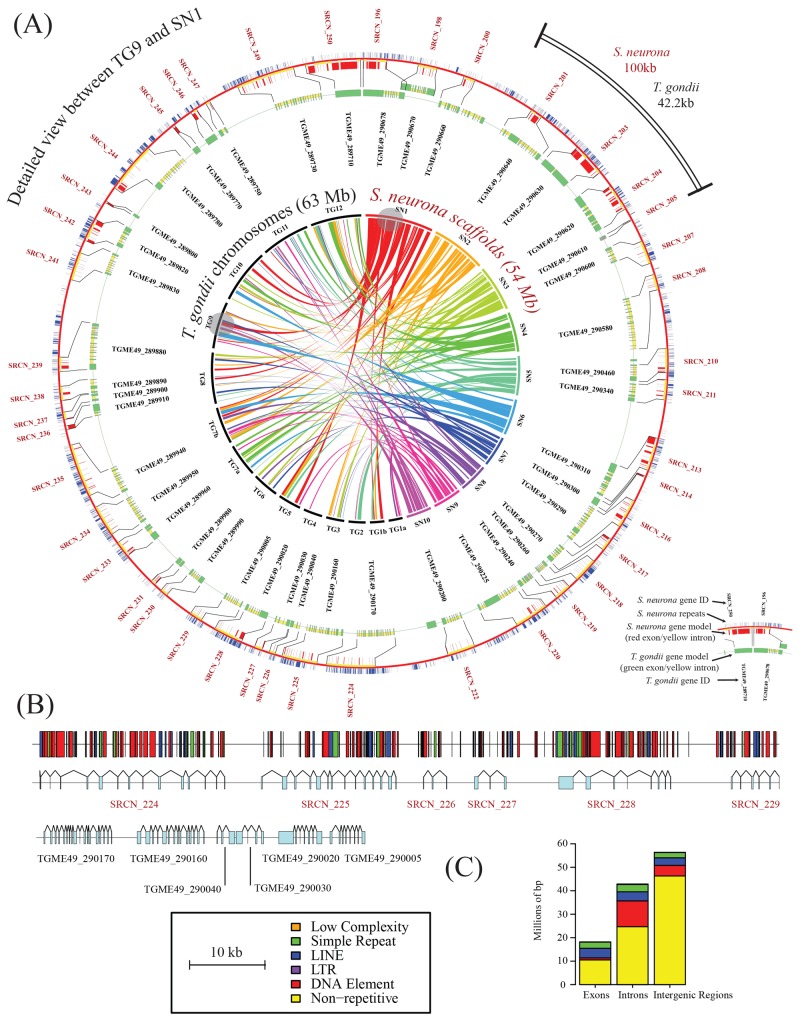
FIG 1 .
Architecture and syntenic relationships of the S. neurona genome. (A) Circos representations (50) of syntenic relationships between the genomes of S. neurona and T. gondii ME49. The inner circle shows syntenic relationships among the 10 largest S. neurona genomic scaffolds (maximum size, 9.2 Mb; minimum size, 3.5 Mb) and the 14 chromosomes of T. gondii. Bandwidths indicate alignment length, and colors represent the S. neurona scaffold of origin for the gene clusters. Grey circles indicate the largest regions of genomic synteny between S. neurona scaffold SO SN1 and T. gondii chromosome 9. The outer circles show a detailed view of synteny indicated by the grey circles. Red and green bars indicate exons of S. neurona and T. gondii, respectively, and yellow and blue bars indicate intronic and repeat regions, respectively. (B) Detailed view of the synteny map shown in panel A revealing larger introns in S. neurona relative to those in T. gondii and the relative positioning of repetitive elements. (C) Incidence of repeats in different genomic regions as defined by RepeatModeler (20).
Given similarities in gene numbers and exon lengths, we next determined the source of the additional sequence associated with the S. neurona genome. Comparisons of intron numbers and lengths show that S. neurona possesses a similar number of exons per gene (5.50 in T. gondii, 5.93 in N. caninum, and 5.44 in S. neurona) but that the average length of the introns in S. neurona (1,437.5 bp) is roughly triple that of T. gondii and N. caninum (497.5 and 465.2 bp, respectively). Further, comparisons of intergenic regions reveal these to be larger in S. neurona (8,495 ± 222 bp) than in T. gondii and E. tenella (2,381 ± 72 and 2,934 ± 268 bp, respectively). To identify factors responsible for the increased intra- and intergenic region sizes, we performed a systematic analysis of repetitive regions across representative apicomplexans with the software tool RepeatModeler (20). This analysis revealed that the S. neurona genome is rich in repeats largely associated with long interspersed nucleotide element (LINE) and DNA element sequences (class I and II transposons, respectively). Mapping of the repeats to scaffolds revealed that many of the repeats are associated with genes (Fig. 1B). Further comparisons of introns, exons, and intergenic regions showed clear differences in the repeat type based on the genomic context (Fig. 1C). DNA element-type repeats were enriched in intronic regions and virtually absent from exons, suggesting evolutionary pressure against the integration of DNA elements within coding regions. Conversely, LINE-like repeats were equally distributed across exons, introns, and intergenic regions. In total, 31 Mbp of the S. neurona genome had repetitive sequences, compared to 17.9 Mbp of the E. tenella genome and 2.5 Mbp of the T. gondii genome (Fig. 2A).
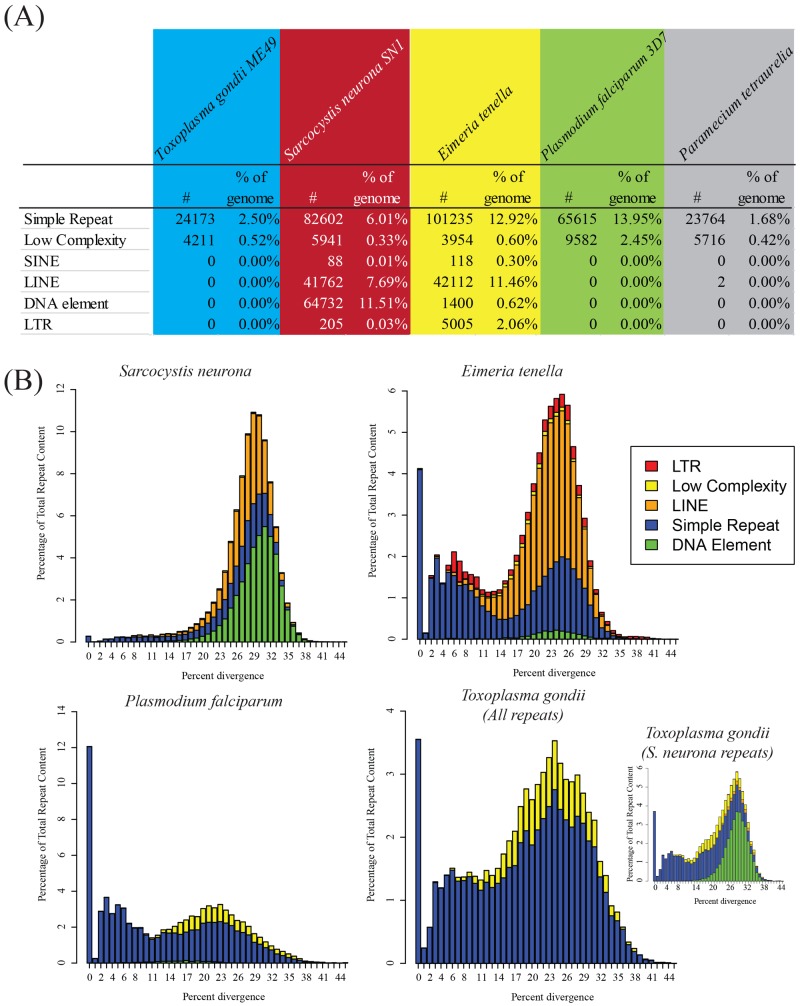
FIG 2 .
Repeat incidence and diversity in selected alveolate genomes. (A) Incidence of repeats, as defined by RepeatModeler (20), in a selected group of alveolate genomes. While E. tenella is rich in LTR elements, S. neurona is rich in DNA elements. Neither type of element is abundant in other alveolates. (B) Diversity of different repeat families within coccidian genomes. Bar graphs indicate the relative abundance of each repeat class as a function of Kimura divergence from the consensus repeat sequence. Note that DNA elements were not initially detected in the T. gondii genome; however, subsequent searches revealed the presence of DNA elements predicted from the S. neurona genome (indicated in the inset bar chart of S. neurona repeats).
S. neurona displays a diverse set of repetitive elements.
The repetitive sequences present in S. neurona are extraordinarily diverse, with 203 families of repeats discovered with RepeatModeler, compared to 101 families in E. tenella and 5 in Plasmodium falciparum. The majority of simple repeats within the S. neurona genome belong to more diverse families, unlike other apicomplexan parasites, where simple repeats are largely composed of short repeats (e.g., CAGn in E. tenella [21]). For example, 33 simple repeat families in S. neurona were composed of consensus sequences with an average length of 287 bp. The average length of the simple repeats was 105 bp in S. neurona, compared to 48 and 68 bp in P. falciparum and E. tenella, respectively.
Type II transposons, or DNA elements with 64,732 members, represent the largest family of repeats present in S. neurona, totaling 14.6 Mbp (11.5%) of the genome, considerably more than in E. tenella (Fig. 2A). All of the DNA elements identified belong to the “cut-and-paste” families of transposons, which propagate through genomes through excision and insertion of DNA intermediates. The most abundant family of DNA elements belonged to the CACTA-Mirage-Chapaev family of transposons, although a minority of Mutator-like elements was also identified. Active DNA transposons contain transposase genes; however, we were unable to detect any such gene within the S. neurona genome, suggesting that these DNA elements are ancient and degraded. Supporting this view, we found that the ratio of transversions to transitions in alignments of repetitive sequences to DNA repeat families was almost exactly 2:1, the statistically expected rate of mutation in the absence of evolutionary pressure.
Given the relative lack of repeats in the T. gondii genome, we explored whether the repeats identified in S. neurona are less active than those identified in E. tenella. When repeats are active, it is possible to identify clades of repeats with significant sequence similarity. Pairwise sequence alignments of members of five families of repeats (Fig. 2B) were highly divergent, indicating that the LINEs and DNA elements are no longer active in S. neurona. Further, the LINEs in S. neurona are more diverse and therefore likely to be more ancient than those in E. tenella. Interestingly, E. tenella, T. gondii, and P. falciparum all feature a bimodal distribution of simple repeats that is lacking in S. neurona. Finally, all three of the coccidian genomes analyzed here displayed similar distributions of sequence divergence of DNA elements, albeit with slightly different means (22.5, 28.1, and 27.8% for E. tenella, S. neurona, and S. neurona-like DNA repeats in T. gondii, respectively). This suggests that while DNA elements are no longer active in these genomes, they did remain active for slightly longer within E. tenella. From these analyses, we conclude that the maintenance of large numbers of LINEs and DNA elements in S. neurona (and E. tenella), even though they are inactive, likely plays a functional role, since T. gondii has removed most of these elements from its genome.
The S. neurona apicoplast genome is well conserved with other Apicomplexa.
In addition to its nuclear genome, the apicoplast genome of S. neurona SO SN1 was studied by reference mapping to the assembled S. neurona SN3 apicoplast sequence (see Fig. S1 in the supplemental material). Both organellar genome architectures are highly similar to those of Toxoplasma (GenBank accession no. U87145.2) and P. falciparum (22). There are, however, a few key differences. As in Toxoplasma, both Sarcocystis apicoplast sequences are missing open reading frame A (ORFA). However, unlike Toxoplasma, both S. neurona sequences show a loss of rpl36 and a loss of one copy of tRNA-Met (from the tRNA cluster between rps4 and rpl4). S. neurona also has a feature first observed in the Piroplasmida that is not seen in Toxoplasma, namely, a division of the RNA polymerase C2 gene into two distinct genes (23). Both S. neurona apicoplast genome sequences uniquely have the insertion of a fragment of rps4 between ORFG and one copy of the large-subunit rRNA in common (see Fig. S1 in the supplemental material). The insert was verified in S. neurona SN3 via PCR and sequencing across this region (see Fig. S2, S3, and S4 in the supplemental material). The rps4 fragment insertion appears to be very recent because the S. neurona SN3 fragment insert is identical in sequence to the corresponding region in the full-length rps4 gene. Comparison of the S. neurona SO SN1 and SN3 nucleotide sequences to each other reveals a few indels but no single-nucleotide polymorphisms (see Text S1 in the supplemental material). Indels, when present, occur in up to one-third of the reads for the locus. The dominant sequence is identical to that determined for SN3. Each S. neurona apicoplast genome was sequenced to greater than 200× coverage.
The S. neurona genome encodes many novel genes and identifies many coccidian-specific innovations.
InParanoid predictions suggest that S. neurona has more orthologs in common with T. gondii than with E. tenella, supporting a closer evolutionary relationship (3,169 versus 1,759 groups of orthologs, respectively) (Fig. 3A). Consistent with previous gene studies, we identified 1,285 (18%) S. neurona genes with no detectable homology (BLAST score, <50) to any known gene, suggesting either a high degree of gene innovation or significant sequence divergence from remote homologs. Among the conserved genes, 715 (10%) were conserved (possessing orthologs) in both Cryptosporidium parvum and either P. falciparum or Theileria annulata, identifying a large collection of proteins that could be amenable to broad-spectrum drug development. These include members of a variety of ATPase genes, heat shock proteins, DEAD/DEAH helicases, proteins with EF-hand domains, and protein kinases. In addition, we identified 1,285 (18%) genes with homology only within the family Sarcocystidae, representing potential drug targets against tissue cyst-forming coccidia. A majority (55%) of these are annotated as “hypothetical proteins” in the ToxoDB resource (24). Of the proteins that are annotated, AP2 domain transcription factors, rhoptry kinase (ROPK) and neck proteins, and zinc fingers, as well as proteins with RNA recognition motifs, are prevalent.
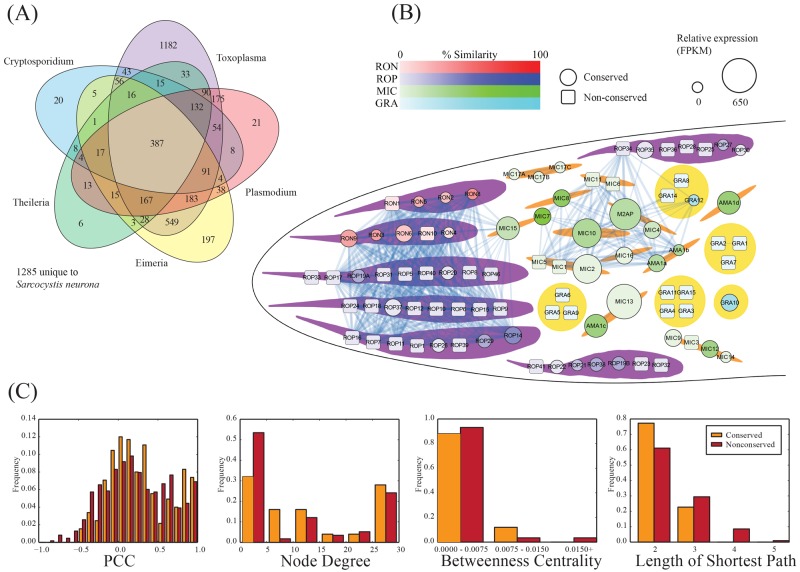
FIG 3 .
Coexpression network for T. gondii invasion-associated genes. (A) Ortholog distribution of S. neurona genes. Orthologs were predicted by using the InParanoid pipeline (51). (B) Network of T. gondii proteins involved in the invasion process. Nodes indicate genes, colored by family or location, with size indicating the relative expression of the S. neurona ortholog as determined through RNA-Seq expression. Square nodes indicate the absence of an ortholog in S. neurona. Links between nodes indicate significant coexpression (Pearson correlation coefficient [PCC], >0.8). Two main clusters of proteins are observed, one involving batteries of rhoptry proteins (ROPs and RONs) and one involving microneme proteins (MICs). (C) Network statistics associated with the invasion network.
The S. neurona attachment and invasion machinery is broadly conserved with T. gondii.
The process of host cell invasion by apicomplexan parasites is a rapid and complex process that relies on a coordinated cascade of interactions between the invading parasite and the host cell. To orchestrate these processes, apicomplexans have evolved families of invasion proteins that are broadly conserved but nevertheless exhibit unique lineage-specific innovations (25). To identify S. neurona gene models involved in invasion relative to T. gondii, we constructed an invasion protein coexpression network (Fig. 3B) in which pairs of T. gondii proteins are linked if they exhibit significant coexpression with S. neurona (Pearson correlation coefficient, >0.8), as has been done for other organisms (26–28). This network provides a scaffold onto which conservation and expression data from S. neurona are mapped to yield insights into evolutionary and functional relationships. Consistent with previous studies, we found that conserved proteins (those that have an ortholog in common with S. neurona) tend to have more correlated expression (high Pearson correlation coefficients) and more connections (high node degree) and are better connected within the network (shorter average path lengths and higher betweenness) than their nonconserved counterparts (Fig. 3C). These findings highlight the potential importance of conserved proteins to the function of the invasion machinery. Within our T. gondii invasion network, we identified two main clusters of highly correlated genes associated with key invasion events. The first involves proteins associated with the micronemes (MIC proteins) which strengthen host cell attachment and play a major role in the formation of the moving junction that forms a specific interface, facilitating invasion. The second involves proteins associated with the rhoptries (RON and ROP proteins), an organelle that is absent from the merozoite stage of all Sarcocystis species, including S. neurona (29).
The genome analyses identified nine previously reported S. neurona orthologs of T. gondii MICs (MIC7, MIC8, MIC10, MIC12, MIC13, MIC14, MIC15, MIC16, and M2AP) (30). We also identified potential homologs of MIC2, MIC4, and MIC9 that had not been annotated through the gene model prediction pipeline. MIC4 has previously been shown to form part of a heterocomplex with MIC1 and MIC6 (31). The absence of the latter two homologs from S. neurona suggests that MIC4 likely mediates the more important functional role. MIC7, MIC8, MIC9, and MIC12 are relatively unique in T. gondii with the possession of epidermal growth factor-like domains, suggesting a potential role in ligand binding. MIC10, together with MIC11 (absent from S. neurona) is thought to be involved in the organization of organellar contents. Also secreted by the microneme is apical membrane antigen 1 (AMA1), which functions to link the inner membrane complex (IMC) to the host cell via interactions with RON proteins that together make up the moving junction (6, 32). Our searches revealed two loci, separated by approximately 80 kb on the S. neurona assembly’s largest scaffold, homologous to T. gondii AMA1 protein TGME49_315730. Interestingly, the reading frames of the two S. neurona paralogs (SnAMA1a and SnAMA1b) occur in opposite directions, suggesting an inverted duplication. Supporting this, two inverted repeats >100 bp in length and with >70% identity were identified ~20,000 bp apart, separating the two paralogs. While the region upstream of the paralogs appears repeat rich, containing simple repeats, as well as LINEs and DNA elements, the region downstream of the second paralog is uncharacteristically repeat poor, with no repetitive sequences in ~14,000 bp of sequence. T. gondii possesses additional paralogs of the AMA1 protein, including TGME49_300130. Again, S. neurona appears to possess these two additional AMA1 paralogs (SnAMA1c and SnAMA1d), but in this case, they are present on two different scaffolds. AMA1 has been shown to interact remarkably strongly with RON2, RON4, and RON5 (32).
In general, RONs were well conserved in S. neurona and T. gondii, with RON2, RON3, RON5, and RON8 orthologs displaying significant sequence similarity across their entire length. Three paralogs of T. gondii RON3 (TgRON3) were identified on a single scaffold, suggesting a tandem duplication, two of which appear to be expressed as predicted by the RNA-Seq data. Putative S. neurona orthologs of RON4 and RON6 were identified through manual inspection of sequence alignments. A pattern of conservation and divergence was observed for a putative ortholog of TgRON9. In T. gondii, RON9 and RON10 form a stable complex distinct from the AMA1-RON2/4/5/8 complex, with disruption of either gene leading to the retention of its partner in the endoplasmic reticulum, followed by degradation. This complex does not play a role in T. gondii invasion and virulence but, because of its conservation with C. parvum, has been linked to interactions involving epithelial cells (33). While an S. neurona protein could be aligned over ~25% of the TgRON9 sequence, it was found to lack a single copy of the 22 copies of the PAEENAEEPKQAEEQANASQSSET motif associated with the T. gondii protein. No homologs to RON1 or RON10 could be identified.
Another critical organelle required for host invasion is the IMC, which additionally confers stability and shape on the cell and is thought to mediate critical roles in cytokinesis and host cell egress (34). We identified 20 putative S. neurona IMC orthologs, with additional evidence of a further six (see Table S2A in the supplemental material). Only TgGAP70, TgAlv6, and TgAlv7 appear to lack homologs.
Molecular modeling reveals that SnAMA1a is capable of intimately coordinating SnRON2.
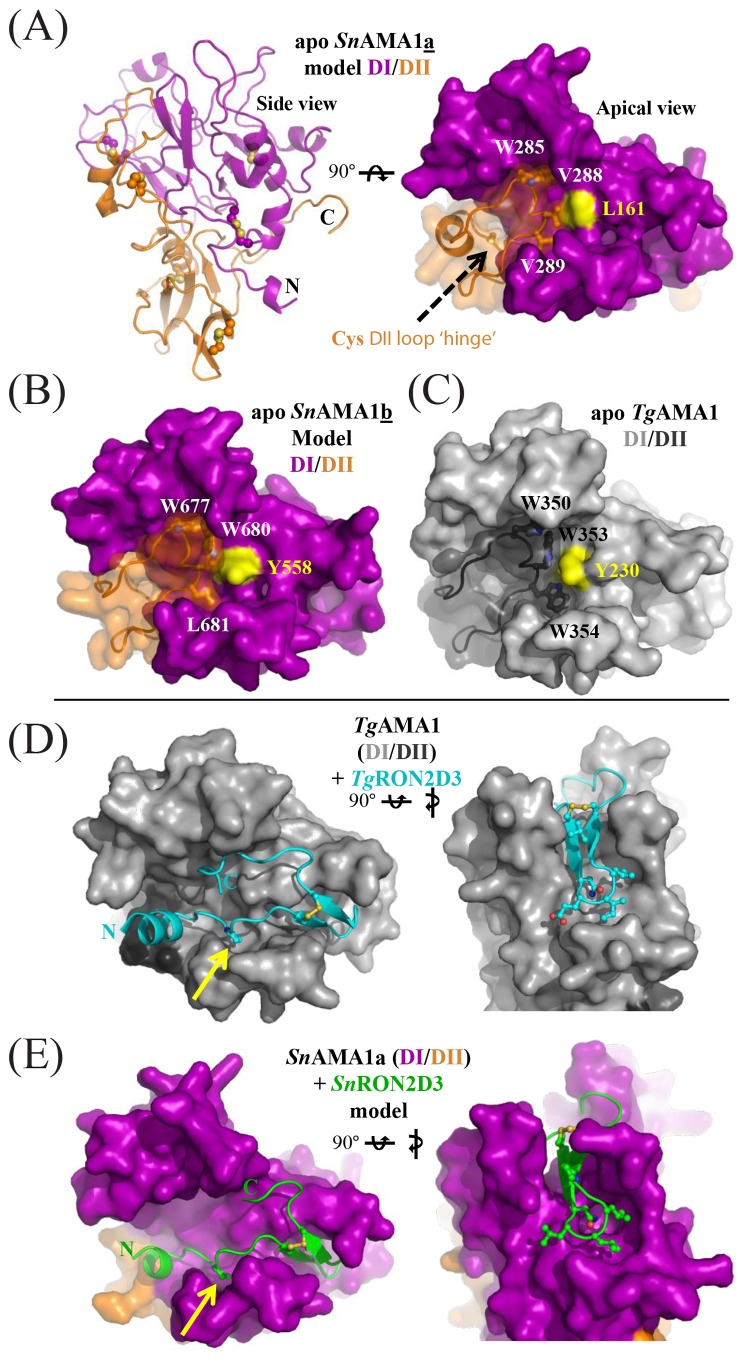
To examine if S. neurona AMA1 homologs can bind S. neurona RON2 homologs, we generated structural models of SnAMA1a and SnAMA1b, which show the highest sequence identity (49 and 44%, respectively) with TgAMA1. Both models possess a PAN-like domain architecture for DI and DII (SnAMA1a, Fig. 4A) consistent with homologs from other apicomplexans (35, 36). A key feature of DII is an extended loop that packs into the groove of DI and regulates RON2 binding in related AMA1 proteins (37). In the SnAMA1a model, a cysteine pair localized within the DII loop is a novel feature of AMA1s and may serve as a hinge to regulate loop displacement and, consequently, RON2 binding (Fig. 4A). Furthermore, the SnAMA1a DII loop appears to be loosely anchored within the DI groove via a Val-Val pair surrounding a central Leu (Fig. 4A). This is in contrast to TgAMA1 (Fig. 4C), where a Trp-Trp pair surround a central Tyr, and the SnAMA1b model (Fig. 4B), where a Trp-Leu pair surround a central Tyr (Fig. 4B). These models suggest that AMA1 paralogs in S. neurona employ divergent strategies that control DII loop dynamics and govern access to the ligand-binding groove. Focusing on the interaction with RON2, removal of the apical segment of the DII loop from the model of SnAMA1a (mimicking the mature binding surface) led to a pronounced groove similar to the RON2 binding surface observed in TgAMA1 (Fig. 4D). Indeed, an energy-minimized docked model revealed that SnRON2 domain 3 was accommodated in a U-shaped conformation (SnRON2D3; Fig. 4E) with an overall topology conserved with respect to the TgAMA1 costructure with a synthetic TgRON2D3 peptide (Fig. 4D) (37). Key features of the TgAMA1-TgRON2sp interface appear to be conserved at the SnAMA1-SnRON2D3 interface, including a RON2 proline residue that occupies an AMA1 pocket exposed by displacement of the DII loop (Fig. 4D and E, yellow arrow) and a reliance on hydrophobic interactions to engage AMA1.
FIG 4 .
SnAMA1a and SnAMA1b accessorize the canonical AMA1 DI and DII domains with unique features but maintain an apical surface capable of coordinating SnRON2D3. (A) Secondary-structure (left) and surface representations of SnAMA1a DI (purple) and DII (orange); five conserved disulfides and two extra cysteines in the DII loop are highlighted as ball-and-stick structures. Two cysteines predicted to form a disulfide at the DII loop hinge are shown as a ball-and-stick structure (black arrow). Residues anchoring the DII loop are labeled and surround a central Leu residue colored yellow. (B) Surface representations of SnAMA1b colored and labeled as for SnAMA1a. (C) Surface representation of TgAMA1 (Protein Data Bank [PDB] accession no. 2x2z); DI, light grey; DII, dark grey. (D) Complementary views of the TgAMA1-TgRON2sp costructure (PDB accession no. 2y8t) with TgAMA1 colored light grey (DI) or dark grey (DII) and TgRON2sp colored cyan. (E) Complementary views of the SnAMA1a-SnRON2D3 costructure model, with SnAMA1 colored as in panel A and SnRON2D3 in green. Residues making up the RON2 cystine loop tip are shown as ball-and-stick structures to highlight shape complementarity.
Overall, modeling of apo SnAMA1a and SnAMA1b, in combination with the complex of SnAMA1a with SnRON2D3, supports the hypothesis that these two proteins can form an intimate binary complex, as observed in related apicomplexan homologs (37, 38). Of note, both SnAMA1a and SnAMA1b exhibited relatively low levels of expression in the merozoite stage sampled (10.4 and 6.7 fragments per kilobase of exon model per million mapped reads [FPKM], respectively) compared to SnAMA1c and SnAMA1d (63.1 and 83.9 FPKM, respectively), perhaps reflecting a stage-specific role for each AMA1-RON2 pairing.
Proteins involved in host regulation in T. gondii are not well conserved in S. neurona.
In addition to RONs, rhoptries also secrete a battery of ROP proteins, the products of a group of genes displaying high levels of correlated expression (Fig. 3B). ROP proteins are secreted into the host cytosol to interact with host cell targets, manipulating pathways that protect the intracellular parasite against clearance. To identify putative S. neurona ROP homologs, we used previously published hidden Markov models (HMMs) (39). In addition to the eight SnROPKs reported previously (39), our phylogenetic analysis identified seven new ROPK orthologs, including: ROP20, ROP26, ROP33, ROP34, and ROP45, as well as two SnROPKs that appear unique to S. neurona (Fig. 5A). RNA-Seq data support the expression of six of these (ROP14, ROP21, ROP27, ROP30, ROP35, and ROP37) during the merozoite stage, despite this stage’s lack of rhoptries and the ability of schizonts to develop in host cell cytoplasm in the absence of a PV (3).
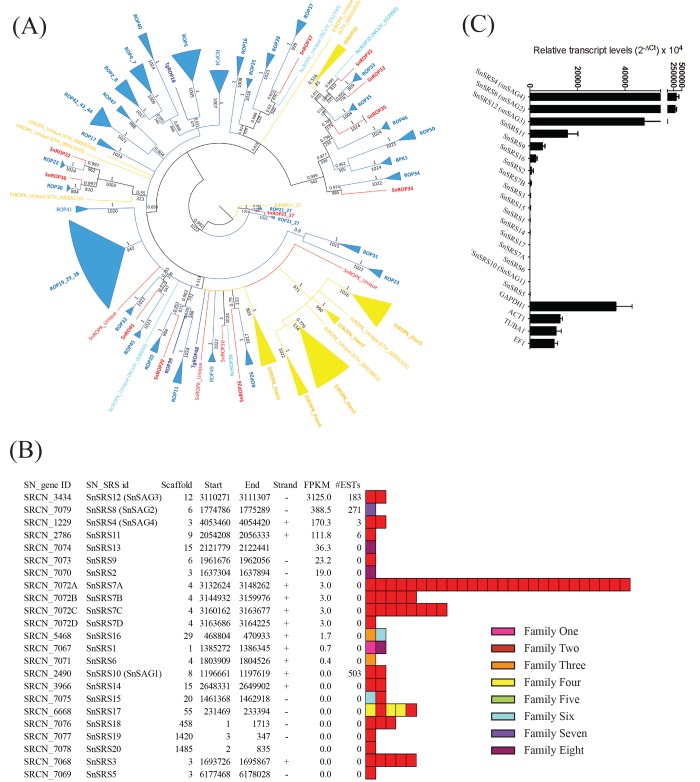
FIG 5 .
Coccidian-specific protein families implicated in virulence and host range determination. (A) Maximum-likelihood-based phylogenetic tree of the ROPK family. Values indicate the bootstrap support (of 1,000 replicates). S. neurona members are red. T. gondii members are dark blue. N. caninum members are cyan. T. gondii and N. caninum clades are blue. E. tenella members and clades are yellow. (B) Summary of the 23 SRS family members identified in the S. neurona genome. Relative expression in the S. neurona merozoite stage are provided as FPKM values, and domain architectures are indicated. #ESTs, number of expressed sequence tags. (C) The expression of the SnSRS-encoding genes was assessed by TaqMan qPCR. Genes were sorted in descending order by their expression levels.
Overall, S. neurona contains a smaller complement of ROPKs (n = 15) than E. tenella (n = 27) and a considerably smaller set than T. gondii (n = 55) and N. caninum (n = 44), both of which feature distinct lineage-specific expansions. However, despite its lower number of ROPKs, S. neurona was found to have more ROPKs in common with T. gondii and N. caninum than with E. tenella; only two of the ROPKs in the three tissue cyst-forming coccidia are conserved with E. tenella (ROP21/27 and ROP35), Importantly, we did not find S. neurona homologs corresponding to T. gondii ROPK proteins implicated in murine virulence (ROP5 and ROP18), modulation of STAT3 and STAT6 signaling (ROP16), or mitogen-activated protein (MAP) kinase signaling (ROP38), which suggests that S. neurona’s success and pathogenesis are not dependent on the inactivation of these host-specific pathways and may explain, in part, why this parasite is not infectious in rodents. No information is available regarding the functional role of S. neurona ROPKs. However, six are likely to be active kinases since they retain key “catalytic triad” residues critical for protein kinase function (SnROP21/27, SnROP30, SnROP33, SnROP34, SnROP35). Further, five are likely to be pseudokinases (SnROP20, SnROP22, SnROP26, SnROP36, SnROP37) that have been shown to act as cofactors of the active kinases (e.g., SnROP5 to SnROP18).
Finally, only two dense-granule (GRA) protein homologs of T. gondii, GRA10 and GRA12, were identified. The discovery of the latter is surprising, given that it has not been annotated in the N. caninum genome. In addition, like the ROPKs, the majority of the GRA proteins encoded by T. gondii that specifically target host immune signaling pathways to alter parasite pathogenesis are not encoded by S. neurona. These include GRA6, which regulates the activation of the host transcription factor nuclear factor of activated T cells (NFAT4); GRA15, which regulates NF-κB activation (40); GRA24, which promotes nuclear translocation of host cell p38a MAP kinase (41); and the phosphoprotein GRA25, which alters CXCL1 and CCL2 levels to regulate immune responses and control parasite replication (42). These data indicate either that a different suite of GRA proteins facilitate Sarcocystis host and niche specialization or that Sarcocystis does not require an expanded repertoire of GRA proteins during merozoite replication since it replicates in the host cytosol and is not contained within a PV, like T. gondii or N. caninum.
S. neurona encodes a distinct set of SRS proteins.
The SRS proteins exist as a developmentally regulated superfamily of parasite surface adhesins within the tissue cyst-forming coccidia that promote host cell attachment and modulate host immunity to regulate parasite growth and virulence. In previous work, we identified 109 and 246 SRS proteins in the T. gondii and N. caninum genomes, respectively (4, 7). Applying our previously generated HMMs, we identified a more restricted set of only 23 SRS-encoding genes in the S. neurona genome. Twenty of the 23 SRS-encoding genes were distributed across 11 of the major scaffolds, but unlike the SRS-encoding genes in T. gondii and N. caninum, only one genomic locus (SnSRS7 on scaffold 4) existed as a tandem array of duplicated paralogs. The 23 SRS-encoding genes were made up of 75 putative SRS domains (Fig. 5B). Of note, 63 (84%) of these 75 domains were associated with family 2 (fam2) domains, including SRS7A, which contained 26 fam2 domains. In general, each SRS protein possessed either one or two SRS domains, with individual domains classifiable into one of the eight previously defined families, although no fam5 domains were identified. The 26-fam2-domain SRS7A protein genomic locus also contained several gaps bordered by nucleic acids with which a high number of reads could be aligned. This might indicate repetitive elements that could promote domain expansion within this locus through ectopic recombination. Interestingly, the SRS7A protein fam2 domains possessed the highest sequence similarity to the 13 fam2 domains encoded by TgSRS44, a protein previously implicated as an integral structural constituent of the T. gondii cyst wall (43). TgSRS44 also possesses a mucin domain, which has been shown to be highly glycosylated and is thought to protect the cyst from immune recognition and/or dehydration. However, we did not identify any mucin domains in our S. neurona homolog. Only four SRS proteins possessed either fam7 or fam8 domains (one and three copies, respectively), in contrast to T. gondii and N. caninum, where the majority of SRS proteins possess one or the other of these two fam domains. Previous work suggested that the relative expansion of fam7 and fam8 domains in T. gondii and N. caninum is linked to their role in host specificity (7). Other noteworthy features include unique combinations of a fam1 domain with a fam8 domain (SnSRS1), and a fam3 domain with a fam6 domain (SnSRS16), which likely promote specific cell recognition events for S. neurona.
RNA-Seq data identified at least seven SRS proteins expressed in merozoites, which was confirmed by TaqMan reverse transcription-PCR (Fig. 5C). The three most abundantly expressed SnSRS proteins were SnSRS12 (SnSAG3), SnSRS8 (SnSAG2), and SnSRS4 (SnSAG4), as has been observed previously (44). Importantly, the SO SN1 strain did not express SnSRS10 (SnSAG1). SnSRS10 is a highly immunogenic protein and is the major surface antigen expressed on the SN3 strain (45), which explains the high number of SN3-derived expressed sequence tags that mapped to SnSRS10, which was transcriptionally silent in this study (Fig. 5).
Reconstruction and analysis of S. neurona metabolism reveal the potential to exploit alternative sources of energy.
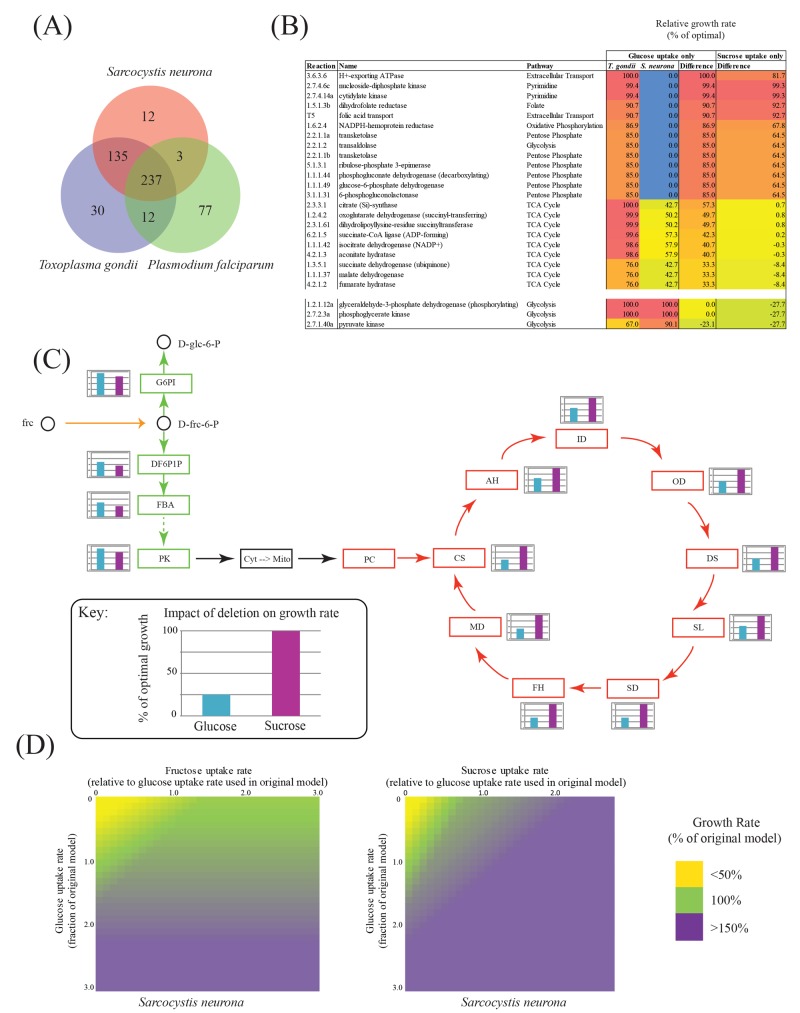
S. neurona has 372 metabolic enzymes (unique enzyme classification [EC] numbers, excluding those involved in nonmetabolic reactions) in common with T. gondii but is missing 42 enzymes and has an additional 13 enzymes that are expressed by RNA-Seq in the merozoite stage (Fig. 6A; see Text S1 in the supplemental material). Our analyses predict putative T. gondii orthologs for 12 of these enzymes, including the fatty acid elongation genes very-long-chain 3-oxoacyl coenzyme A synthase (EC 2.3.1.199; TGME49_205350) and very-long-chain (3R)-3-hydroxyacyl acyl carrier protein dehydratase (EC 4.2.1.134; TGME49_311290). Only threonine ammonia-lyase (EC 4.3.1.19) is unique and adds functionality to S. neurona.
FIG 6 .
Metabolic reconstruction and analysis of S. neurona based on iCS382. (A) Overlap in enzyme predictions for genes from S. neurona, T. gondii, and P. falciparum. (B) Species-specific differences in growth rates of single-reaction knockouts. Only reactions that show a growth rate difference of 20% between T. gondii and S. neurona are shown. (C) Impact of deletion of reactions involved in glycolysis and the TCA cycle on S. neurona growth under conditions of exclusive glucose or sucrose uptake. d-glc-6-P, d-glucose-6-phosphate; G6PI, glucose-6-phosphate isomerase; d-frc-6-P, d-fructose-6-phosphate; DF6P1P, diphosphate-fructose-6-phosphate 1-phosphotransferase; FBA, fructose-bisphosphate aldolase; PK, pyruvate kinase; Cyt, cytosol; Mito, mitochondrion; PC, pyruvate carboxylase; CS, citrate synthase; AH, aconitate hydratase; ID, isocitrate dehydrogenase (NADP+); OD, oxoglutarate dehydrogenase (succinyl-transferring); DS, dihydrolipoyllysine-residue succinyltransferase; SL, succinate-CoA ligase (ADP-forming); SD, succinate dehydrogenase (ubiquinone); FH, fumarate hydratase; MD, malate dehydrogenase; frc, fructose. (D) Relationship between fructose, glucose, and sucrose import and growth.
We incorporated these differences into our previously published metabolic reconstruction of T. gondii named iCS382 (8) and performed flux balance analyses of both iCS382 and the modified S. neurona reconstruction. Scaling the iCS382 model to produce a doubling time of 11.8 h with glucose as the sole energy source (see Text S1 in the supplemental material), we show that S. neurona has a slightly longer doubling time of 13.8 h. Single-reaction knockouts identified 22 reactions whose deletion resulted in a significantly greater impact on S. neurona than on T. gondii (>20% maximal growth rate difference) (Fig. 6B; see Table S2 in the supplemental material). Critical reactions include members of the pentose phosphate and glycolysis pathways, the tricarboxylic acid (TCA) cycle, and two members of the pyrimidine biosynthetic pathway, nucleoside-diphosphate kinase (EC 2.7.4.6) and cytidylate kinase (EC 2.7.4.14). Conversely, we identified only a single reaction, catalyzed by pyruvate dehydrogenase, whose deletion had a significantly greater impact on T. gondii than on S. neurona (>20% maximal growth rate difference).
The S. neurona annotation effort predicted a gene for alpha-glucosidase (EC 3.2.1.20) (see Text S1 and Fig. S4 in the supplemental material). Since conversion of sucrose to fructose and glucose by alpha-glucosidase would add functionality to the metabolic reconstruction, we tested in silico for its potential impact on growth. S. neurona was predicted to grow faster in the presence of sucrose and the absence of glucose than in the presence of glucose and the absence of sucrose (doubling time of 11.4 h versus 13.8 h). This is due, in part, to an increase in the concentration of fructose-6-phosphate caused by the action of hexokinase (EC 2.7.1.1, Fig. 6C). Consequently, under conditions of sucrose uptake, knockout of enzymes involved in glycolysis has a greater impact on the growth rate than conditions of glucose uptake (see Table S2 in the supplemental material). When we examined the impact of combining access to different carbohydrates, our simulations suggested that S. neurona has the capacity to significantly enhance its growth by utilizing fructose, with an even greater effect when sucrose is used as an additional energy source (Fig. 6D). For example, while fructose supplementation alters parasite growth to 120%, supplementation with sucrose extends parasite growth to 180% of its original rate. Interestingly, glucose-6-phosphate isomerase, the enzyme responsible for the conversion of glucose-6-phosphate to fructose-6-phosphate, operates in the reverse direction under glucose or sucrose uptake conditions. When only sucrose is available, more glucose-6-phosphate is produced from the conversion of fructose-6-phosphate, which is predicted to feed into other pathways (e.g., the pentose phosphate pathway), resulting in the elevated production of NADPH and an increased growth rate. Importantly, glycolysis is utilized more when sucrose is available, so there is less reliance on the TCA cycle. Furthermore, the breakdown of sucrose makes fructose available for the synthesis of other key metabolites (e.g., branched-chain amino acids), decreasing the parasite’s dependency on the TCA cycle for their production. Hence, the deletion of individual TCA cycle reactions has a greater impact on the growth rate in the presence of glucose than in the presence of sucrose (Fig. 6C).
DISCUSSION
Coccidian parasites represent a major clade within the phylum Apicomplexa, and the genomes of three species, E. tenella, T. gondii, and N. caninum, have already been sequenced (7, 21). S. neurona is the first genome in the genus Sarcocystis to be sequenced. The 127-Mbp genome is more than twice the size of other sequenced coccidian genomes, largely because of a high proportion of repetitive LINEs and DNA elements. The organization of the S. neurona genome into 116 genomic scaffolds produces the first molecular karyotype, or physical linkage map, which should greatly facilitate future genetic and comparative genomic studies of this important genus. Sarcocystis chromosomes do not condense, nor have they been resolved by pulse-field gel electrophoresis. Our comparative genomic, transcriptomic, and metabolic flux data analyses show that the invasion machinery is largely conserved among the coccidia but that the tissue cyst-forming coccidia have evolved families of dense-granule (GRA), ROPK, and surface-associated SRS adhesins that promote their ability to persist chronically in cyst-like structures or disrupt the induction of sterilizing immunity, representing novel molecular strategies that facilitate their transition from largely enteric pathogens within a single host (Eimeria) to heteroxenous pathogens that cycle between a definitive host and an intermediate host(s) (Sarcocystis).
Genome comparisons reveal that S. neurona has more orthologs in common with T. gondii than with E. tenella (3,169 versus 1,759 orthologs, respectively), supporting the notion that the Eimeria lineage is more divergent. However, S. neurona is also quite distinct from T. gondii; it possesses only limited genomic synteny, restricted to only dozens of genes, and additionally encodes 1,285 (18%) genes with no detectable homology to any other species. As in E. tenella, LINEs and DNA elements are present in S. neurona, but the DNA elements are significantly expanded in S. neurona, partially accounting for its increased genome size. The presence of the LINEs and DNA elements, however, is not associated with gene model misannotation, since LINEs are as frequently associated with T. gondii orthologs (13.3%) as they are with unique genes (11.6%), indicating that they may drive genome innovations within S. neurona (46). We did not find any examples of the coronavirus-like long terminal repeat (LTR) element previously associated with the E. tenella genome (21), strengthening the suggestion that this element was acquired by horizontal gene transfer within that lineage.
The Sarcocystis invasion machinery was largely conserved within the coccidia, and the construction of the S. neurona interaction network based on gene expression data identified two main clusters of conservation, one composed largely of MIC, AMA1, and RON proteins required for the mechanics of cell attachment and invasion and another composed of a limited set of ROP and GRA proteins thought to alter host immune effector function. However, the complement of the latter ROP and GRA proteins is greatly reduced compared to that of other tissue cyst-forming coccidia such as Toxoplasma or Neospora. While mouse models have shown ROP5 and ROP18, which are absent from S. neurona, to impact virulence in Toxoplasma, the lack of a suitable such model, i.e., immunocompetent mice, for S. neurona means that little is known about its strain virulence determinants. Additionally, all strains induce fatal encephalitis in immunodeficient mice, irrespective of the dose. Only two ROPKs were conserved with E. tenella, implying specialization in the ROPK machinery required for the different life cycles. Hence, the reduced complement of ROPKs within the S. neurona genome likely underscores the important role the expanded repertoire of ROPKs plays in promoting Toxoplasma and Neospora host and niche adaptation among the susceptible hosts in which these parasites establish transmissible infections. Likewise, the distinctive set of ROPKs previously reported for E. tenella and thought to map to the sporozoite rhoptry (47) might suggest a specialized role for these proteins during the initial establishment of infection.
Consistent with a transition from a strictly enteric coccidian pathogen to a tissue-invasive one capable of establishing long-term, chronic infection by encystment within host cells, S. neurona expresses a distinct surface antigen coat of SRS proteins that promote parasite recognition, attachment, and long-term encystment within host cells to promote transmissibility of infection. In comparison to T. gondii and N. caninum, however, the S. neurona SRS protein repertoire is surprisingly small and less divergent (25); there is a dramatic reduction in the number of SRS proteins composed of fam7 and fam8 domains, with the vast majority of the 23 SnSRS-encoding genes composed of fam2 domains. Previous studies of T. gondii suggest that proteins composed of the former domains modulate host immune responses and mediate critical roles in parasite virulence. Our data suggest that with only a single copy of a fam7- and fam8-containing SRS protein, S. neurona has evolved other mechanisms for control of immune activation and/or that such control is not required for the successful transmission of this highly prevalent protozoan pathogen. The latter point is consistent with observed differences between the S. neurona and Toxoplasma/Neospora life cycles. Sarcocystis spp., once encysted, undergo a terminal commitment to their gamont stage, requiring access to their definitive host to complete their life cycle. In contrast, both Toxoplasma and Neospora are capable of recrudescing their infection after encystation, and expansion of fam7 and fam8 domain SRS proteins capable of altering host protective immunity may function to increase the cyst burden or alter intermediate-host behavior, promoting transmission of the parasite to the definitive host to complete its life cycle. Alternatively, Sarcocystis spp. are exclusively restricted to sexual development within the intestine of the definitive host, whereas Toxoplasma infection of its felid definitive host results in both sexual development and asexual expansion of infection, so the expanded repertoire of fam7 and fam8 domain SRS proteins may promote dissemination of infection to a wide range of tissue and cell types and vaccinate the definitive host against reinfection.
The sheer dominance of the fam2 domain proteins among the limited repertoire of SnSRS proteins suggests that they play a critical functional role in the life cycle. A recent study (43) found that TgSRS44 (CST1), a T. gondii SRS protein with 13 tandemly repeated fam2 domains, is an important structural constituent of the cyst wall, suggesting that the emergence of fam2 SnSRS domain-containing proteins in the common ancestor of S. neurona and T. gondii likely provided the parasite with the ability to form cysts, thereby extending its host range and promoting the transition to a heteroxenous (two-host) life cycle. Strains of S. neurona are known to exhibit important differences in the immunodominant SnSRS-encoding genes that they possess. SnSRS10 (SnSAG1) is encoded by an immunodominant gene present and expressed abundantly in some S. neurona isolates but absent from others (45). While the type II SO SN1 strain sequenced encodes SnSRS10, it does not express it during merozoite growth (Fig. 5), whereas SN3, another type II isolate, highly expresses this protein. While the mechanism of gene regulation within the SnSRS family has yet to be elucidated, it may influence the host range, the capacity to promote coinfection, and/or pathogenicity among the broad intermediate-host range of S. neurona, much the same way differential expression of TgSRS2 alters the parasite load and the pathology of T. gondii infection in mice (4). Importantly, a high prevalence of coinfection with different genetic types of S. neurona within intermediate hosts would promote outcrossing during sexual reproduction. Outcrossing in Toxoplasma has previously been shown to produce progeny possessing altered biological potentials, including virulence and a capacity to cause outbreaks (48), which has recently also been established for S. neurona (15).
Finally, regulation of energy production has likewise evolved as a strategy for parasites to extend their host range, by tuning growth in relation to the host burden or carrying capacity (8). We found only a limited number of differences between the enzyme complements of T. gondii and S. neurona. Notably, S. neurona possesses 13 enzymes not present in T. gondii and a homolog of an alpha-glucosidase (EC 3.2.1.20) that preferentially gives S. neurona the potential to use alternative carbon sources to help drive growth. Hence, our flux balance analysis showed that S. neurona is less reliant on the TCA cycle when it is grown in the presence of sucrose and that sucrose supplementation can increase parasite growth to 180% of its original rate, a capability that may be important for allowing the parasite to exploit new host niches. These findings serve to highlight subtle differences in pathway utilization that the two parasites may have adopted to optimize their distinct life cycle strategies.
Together, our data support a model in which, following the split with the Eimeria lineage, the ancestor of Sarcocystis and Toxoplasma gained the ability to invade intermediate hosts and form tissue cysts. This transition required the evolution of SRS family proteins as structural constituents of the cyst wall, as well as immune evasion molecules protecting the parasite from sterilizing immunity. Subsequently, while the Sarcocystis lineage abandoned the use of the PV during its schizont stage in the intermediate host, committing the parasite to its sexual cycle after encystation, the Toxoplasma lineage maintained the use of a PV during intermediate-host infection. The use of the PV could conceivably shield the parasite from the host developing an effector memory CD8 T cell response that is naturally induced by the presence of parasite antigens in the cytosol of infected host cells. This, in turn, allows Toxoplasma to recrudesce postencystation and, aided by an expanded repertoire of ROPK, GRA, and SRS proteins, provides further opportunities to increase the cyst burden and extend its host range. In addition to addressing questions of host range and specificity, we expect that the availability of this resource will help drive the development of novel therapeutics that are urgently required for these devastating pathogens. Further, reference genome mapping will facilitate genus-wide and population studies that focus on questions of host specialization and virulence mechanisms. The latter, for example, might be expected to inform on the spate of fatal infections in marine mammals to resolve at the genome level the genetic basis of the emergence of these disease-producing strains. Key to these studies will be the generation of robust expression data sets that allow the identification of critical proteins associated with distinct stages of the parasite’s life cycle.
MATERIALS AND METHODS
Culturing of parasites, extraction of DNA/RNA, and sequencing.
S. neurona strain SO SN1 was isolated from a southern sea otter (19) and obtained from Patricia Conrad, University of California, Davis, CA. S. neurona parasites were maintained in MA-104 cells as described previously (49). Genomic DNA was extracted from frozen pellets of S. neurona SO SN1 by proteinase K digestion and subsequence phenol-chloroform extraction. Five libraries were prepared: two Roche 454 Shotgun GS-Titanium libraries prepared in accordance with the Rapid Library Preparation Method Manual (Roche), a Roche 454 8-kb paired-end GS-Titanium Library prepared in accordance with the Paired-End Library Preparation Method Manual with the modification of setting up four circularization reactions to increase the final library yield, an Illumina 2- to 3-kb mate pair library synthesized with the TruSeq DNA sample prep kit (Illumina) and run on an Illumina GA IIx, and a Nextera 8- to 15-kb mate pair library prepared in accordance with the manufacturer’s recommendations and run on an Illumina HiSeq 2000. These sequencing efforts generated 7,020,033 shotgun reads, 5,919,255 shotgun reads, 1,100,788 paired-end reads, 128,614,194 mate pair reads, and 136,301,151 mate pair reads, respectively. S. neurona SO SN1 RNA was isolated from merozoites with the RNeasy minikit (Qiagen), snap-frozen, and stored at −80°C. A single TruSeq v2 RNA library (mRNA enriched) was prepared for Illumina sequencing by the standard Illumina protocol and used to generate 59,622,019 reads. For further details of genome assembly and annotation, as well as bioinformatics and experimental analyses, see Text S1 in the supplemental material.
Nucleotide sequence accession number.
Further information on the S. neurona genome project, including sequence files, is available through the bioproject repository at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/bioproject/252030) using the accession number SRP052925.
SUPPLEMENTAL MATERIAL
Materials and methods showing the details of S. neurona metabolic reconstruction and apicoplast single-nucleotide polymorphism differences. Download
Organization of the apicoplast genome sequence and comparison to T. gondii. Gene names are as indicated. Red and blue colors indicate the coding strand. Differences with T. gondii are indicated on the outside of the circle. White circles within genes denote in-frame UGA codons. The S. neurona apicoplast genome, like that of Toxoplasma, uses an alternate genetic code. Download
PCR amplification of rps4 fragment. (A) Primer combinations used for PCR amplification of the rps4 fragment insert. (B) PCR was performed with Verbatim high-fidelity DNA polymerase (Thermo Fisher Scientific Inc., Pittsburgh, PA). Three 25-μl PCRs (S. neurona SN3 genomic DNA, S. neurona apicoplast DNA, or no DNA) were set up for each of the primer pairs. The cycling conditions included initial denaturation at 95°C for 3 min; 35 cycles of denaturation at 95°C for 30 s, annealing as shown in the table, and extension at 68°C for 1 min; and a final extension at 68°C for 2 min. The amplified PCR products were analyzed on 1.5% agarose gels. The 635-bp product amplified with primer pair F1-R3 was purified with a PCR purification kit (Qiagen, Valencia, CA) and sequenced in both directions at the Advanced Genetic Technologies Center, University of Kentucky. (C) Electrophoretic analysis of rps4 fragment insert PCRs. The PCR product sequence confirms the presence of the rps4 fragment insert. In a multiple-sequence alignment of the PCR products, the fragment insert and the original gene show no mutations of the rps4 gene fragment insert from the original gene, suggesting that it is a relatively recent event. Download
Alignment of the rps4 gene and the rps4 fragment insert. The multiple-sequence alignment is composed of four sequences, the rps4 gene, the rps4 fragment insert, and the sequence from each strand of the 635-bp PCR product. Highlighted in yellow is the alignment of the rps4 insert. The single nucleotide highlighted in blue is an ambiguity in the length of the homopolymer run in the rps4 insert. Download
Conservation of rpoC2. (A) Diagram of the rpoC2 gene across four apicomplexans. (B) Three-frame translation of the rpoC2a and rpoC2b gap. Highlighted in yellow is the stop codon of rpoC2a. Highlighted in green is the hypothetical start codon of rpoC2b. Download
Resolving the annotation of EC 3.2.1.20 and EC 3.2.1.84 in apicomplexan homologs. Enzymes with EC 3.2.1.20 and EC 3.2.1.84 form part of the glycosyl-hydrolase 31 family. The enzyme corresponding to EC 3.2.1.20 is glucosidase I, a single chain composed of the catalytic subunit. The enzyme corresponding to EC 3.2.1.84 is glucosidase II, a heterodimer composed of the catalytic alpha subunit and the regulatory beta subunit. A multiple-sequence alignment of all of the SwissProt annotated EC 3.2.1.20 and EC 3.2.1.84 sequences (nonredundant at 90% identity) along with apicomplexan homologs (generated with ProbCons) is shown (right panel) with active-site residues indicated by red blocks. The maximum-likelihood tree for this multiple-sequence alignment generated with PhyML with 1,024 bootstrap replicates is also shown (left panel). The evidence used for annotation of the sequences in SwissProt is indicated by colored asterisks. At the bottom, a multiple-sequence alignment of only the apicomplexan homologs is shown. Black bars correspond to regions with 100% identity, and regions of decreasing greyness correspond to decreasing identity levels. The region similar to EC 3.2.1.20 and the alpha subunit of EC 3.2.1.84 is enclosed in a red box. No homolog of the beta subunit of EC 3.2.1.84 was found in any of the apicomplexan genomes. Download
ORFs, annotations, and expression.
(A) List of Sarcocystis neurona IMC proteins. (B) Changes in enzyme:gene assignments for the application of new constraints in iCS382. (C) Predicted impact of single reaction knockouts on parasite growth.
PCR primers used to confirm SRS expression.
Metabolic reconstruction of S. neurona
ACKNOWLEDGMENTS
This study was financially supported by the Canadian Institute for Health Research (CIHR-MOP 84556 to J.P. and M.E.G.) and the Intramural Research Program of the NIH and NIAID (M.E.G.). M.J.B. was supported by a discovery grant from the Natural Sciences and Engineering Research Council of Canada (NSERC). M.E.G. is a scholar of the Canadian Institute for Advanced Research (CIFAR) Integrated Microbial Biodiversity Program. D.K.H. and J.C.K. were supported by a grant from the USDA NIFA (2009-65109-05918). D.K.H. was additionally supported by the Amerman Family Equine Research Fund.
We gratefully acknowledge the assistance of J. Wendte, M. Quinones, A. Kennard, A. Khan, D. Bruno, S. Anzick, C. Martens, and E. Dahlstrom in DNA sequencing and assembly. We also thank Patricia Conrad for the SO SN1 strain and thank the University of Toronto SciNet facility for the use of its computing resources.
Footnotes
Citation Blazejewski T, Nursimulu N, Pszenny V, Dangoudoubiyam S, Namasivayam S, Chiasson MA, Chessman, Tonkin M, Swapna LS, Hung SS, Bridgers J, Ricklefs SM, Boulanger MJ, Dubey JP, Porcella SF, Kissinger JC, Howe DK, Grigg ME, Parkinson J. 2015. Systems-based analysis of the Sarcocystis neurona genome identifies pathways that contribute to a heteroxenous life cycle. mBio 6(1):e02445-14. doi:10.1128/mBio.02445-14.
REFERENCES
- 1.Cowper B, Matthews S, Tomley F. 2012. The molecular basis for the distinct host and tissue tropisms of coccidian parasites. Mol Biochem Parasitol 186:1–10. doi: 10.1016/j.molbiopara.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 2.Dubey JP, Lindsay DS, Saville WJ, Reed SM, Granstrom DE, Speer CA. 2001. A review of Sarcocystis neurona and equine protozoal myeloencephalitis (EPM). Vet Parasitol 95:89–131. doi: 10.1016/S0304-4017(00)00384-8. [DOI] [PubMed] [Google Scholar]
- 3.Dubey JP, Speer CA, Fayer R. 1988. Sarcocystosis of animals and man. CRC Press, Boca Raton, FL. [Google Scholar]
- 4.Wasmuth JD, Pszenny V, Haile S, Jansen EM, Gast AT, Sher A, Boyle JP, Boulanger MJ, Parkinson J, Grigg ME. 2012. Integrated bioinformatic and targeted deletion analyses of the SRS gene superfamily identify SRS29C as a negative regulator of Toxoplasma virulence. mBio 3(6):e00321-12. doi: 10.1128/mBio.00321-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sibley LD. 2011. Invasion and intracellular survival by protozoan parasites. Immunol Rev 240:72–91. doi: 10.1111/j.1600-065X.2010.00990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Besteiro S, Dubremetz JF, Lebrun M. 2011. The moving junction of apicomplexan parasites: a key structure for invasion. Cell Microbiol 13:797–805. doi: 10.1111/j.1462-5822.2011.01597.x. [DOI] [PubMed] [Google Scholar]
- 7.Reid AJ, Vermont SJ, Cotton JA, Harris D, Hill-Cawthorne GA, Könen-Waisman S, Latham SM, Mourier T, Norton R, Quail MA, Sanders M, Shanmugam D, Sohal A, Wasmuth JD, Brunk B, Grigg ME, Howard JC, Parkinson J, Roos DS, Trees AJ, Berriman M, Pain A, Wastling JM. 2012. Comparative genomics of the apicomplexan parasites Toxoplasma gondii and Neospora caninum: coccidia differing in host range and transmission strategy. PLoS Pathog. 8:e1002567. doi: 10.1371/journal.ppat.1002567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song C, Chiasson MA, Nursimulu N, Hung SS, Wasmuth J, Grigg ME, Parkinson J. 2013. Metabolic reconstruction identifies strain-specific regulation of virulence in Toxoplasma gondii. Mol Syst Biol 9:708. doi: 10.1038/msb.2013.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levine ND. 1986. The taxonomy of Sarcocystis (Protozoa, Apicomplexa) species. J Parasitol 72:372–382. doi: 10.2307/3281676. [DOI] [PubMed] [Google Scholar]
- 10.Lindsay DS, Mitchell SM, Vianna MC, Dubey JP. 2004. Sarcocystis neurona (Protozoa: Apicomplexa): description of oocysts, sporocysts, sporozoites, excystation, and early development. J Parasitol 90:461–465. doi: 10.1645/GE-230R. [DOI] [PubMed] [Google Scholar]
- 11.Fenger CK, Granstrom DE, Langemeier JL, Stamper S, Donahue JM, Patterson JS, Gajadhar AA, Marteniuk JV, Xiaomin Z, Dubey JP. 1995. Identification of opossums (Didelphis virginiana) as the putative definitive host of Sarcocystis neurona. J Parasitol 81:916–919. doi: 10.2307/3284040. [DOI] [PubMed] [Google Scholar]
- 12.Dubey JP, Chapman JL, Rosenthal BM, Mense M, Schueler RL. 2006. Clinical Sarcocystis neurona, Sarcocystis canis, Toxoplasma gondii, and Neospora caninum infections in dogs. Vet Parasitol 137:36–49. doi: 10.1016/j.vetpar.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 13.Dubey JP, Saville WJ, Stanek JF, Lindsay DS, Rosenthal BM, Oglesbee MJ, Rosypal AC, Njoku CJ, Stich RW, Kwok OC, Shen SK, Hamir AN, Reed SM. 2001. Sarcocystis neurona infections in raccoons (Procyon lotor): evidence for natural infection with sarcocysts, transmission of infection to opossums (Didelphis virginiana), and experimental induction of neurologic disease in raccoons. Vet Parasitol 100:117–129. doi: 10.1016/S0304-4017(01)00500-3. [DOI] [PubMed] [Google Scholar]
- 14.Miller MA, Conrad PA, Harris M, Hatfield B, Langlois G, Jessup DA, Magargal SL, Packham AE, Toy-Choutka S, Melli AC, Murray MA, Gulland FM, Grigg ME. 2010. A protozoal-associated epizootic impacting marine wildlife: mass-mortality of southern sea otters (Enhydra lutris nereis) due to Sarcocystis neurona infection. Vet Parasitol 172:183–194. doi: 10.1016/j.vetpar.2010.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wendte JM, Miller MA, Lambourn DM, Magargal SL, Jessup DA, Grigg ME. 2010. Self-mating in the definitive host potentiates clonal outbreaks of the apicomplexan parasites Sarcocystis neurona and Toxoplasma gondii. PLoS Genet 6:e1001261. doi: 10.1371/journal.pgen.1001261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dubey JP, Saville WJ, Lindsay DS, Stich RW, Stanek JF, Speert CA, Rosenthal BM, Njoku CJ, Kwok OC, Shen SK, Reed SM. 2000. Completion of the life cycle of Sarcocystis neurona. J Parasitol 86:1276–1280. doi: 10.1645/0022-3395(2000)086[1276:COTLCO]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 17.Miller MA, Barr BC, Nordhausen R, James ER, Magargal SL, Murray M, Conrad PA, Toy-Choutka S, Jessup DA, Grigg ME. 2009. Ultrastructural and molecular confirmation of the development of Sarcocystis neurona tissue cysts in the central nervous system of southern sea otters (Enhydra lutris nereis). Int J Parasitol 39:1363–1372. doi: 10.1016/j.ijpara.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gibson AK, Raverty S, Lambourn DM, Huggins J, Magargal SL, Grigg ME. 2011. Polyparasitism is associated with increased disease severity in Toxoplasma gondii-infected marine sentinel species. PLoS Negl Trop Dis 5:e1142. doi: 10.1371/journal.pntd.0001142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller MA, Crosbie PR, Sverlow K, Hanni K, Barr BC, Kock N, Murray MJ, Lowenstine LJ, Conrad PA. 2001. Isolation and characterization of Sarcocystis from brain tissue of a free-living southern sea otter (Enhydra lutris nereis) with fatal meningoencephalitis. Parasitol Res 87:252–257. doi: 10.1007/s004360000340. [DOI] [PubMed] [Google Scholar]
- 20.Smit AFA, Hubley R. 2008–2010, posting date RepeatModeler Open-1.0. Institute for Systems Biology, Seattle, WA. http://www.repeatmasker.org. [Google Scholar]
- 21.Reid AJ, Blake DP, Ansari HR, Billington K, Browne HP, Dunn M, Hung SS, Kawahara F, DM-S, Malas TB, Mourier TB, Nagra H, Nair M, Otto TD, Rawlings ND, Rivailler P, Sanchez-Flores A, Sanders M, Subramaniam C, Tay Y-LL, Wu X, Dear PH, Doerig C, Gruber A, Ivens AC, Parkinson J, Shirley MW, Wan K-L, Berriman M, Tomley FM, Pain A. 2014. Genomic analysis of the causative agents of coccidiosis in domestic chickens. Genome Res 24:1676–1685. doi: 10.1101/gr.168955.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson RJ, Denny PW, Preiser PR, Rangachari K, Roberts K, Roy A, Whyte A, Strath M, Moore DJ, Moore PW, Williamson DH. 1996. Complete gene map of the plastid-like DNA of the malaria parasite Plasmodium falciparum. J Mol Biol 261:155–172. doi: 10.1006/jmbi.1996.0449. [DOI] [PubMed] [Google Scholar]
- 23.Brayton KA, Lau AO, Herndon DR, Hannick L, Kappmeyer LS, Berens SJ, Bidwell SL, Brown WC, Crabtree J, Fadrosh D, Feldblum T, Forberger HA, Haas BJ, Howell JM, Khouri H, Koo H, Mann DJ, Brayton KA, Lau AO, Herndon DR, Hannick L, Kappmeyer LS, Berens SJ, Bidwell SL, Brown WC, Crabtree J, Fadrosh D, Feldblum T, Forberger HA, Haas BJ, Howell JM, Khouri H, Koo H, Mann DJ, Norimine J, Paulsen IT, Radune D, Ren Q, Smith RK Jr, Suarez CE, White O, Wortman JR, Knowles DP Jr, McElwain TF, Nene VM. 2007. Genome sequence of Babesia bovis and comparative analysis of apicomplexan hemoprotozoa. PLoS Pathog 3:1401-1413. doi: 10.1371/journal.ppat.0030148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aurrecoechea C, Barreto A, Brestelli J, Brunk BP, Cade S, Doherty R, Fischer S, Gajria B, Gao X, Gingle A, Grant G, Harb OS, Heiges M, Hu S, Iodice J, Kissinger JC, Kraemer ET, Li W, Pinney DF, Pitts B, Roos DS, Srinivasamoorthy G, Stoeckert CJ Jr., Wang H, Warrenfeltz S. 2013. EuPathDB: the eukaryotic pathogen database. Nucleic Acids Res 41:D684–D691. doi: 10.1093/nar/gks1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wasmuth J, Daub J, Peregrín-Alvarez JM, Finney CA, Parkinson J. 2009. The origins of apicomplexan sequence innovation. Genome Res 19:1202–1213. doi: 10.1101/gr.083386.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jordan IK, Katz LS, Denver DR, Streelman JT. 2008. Natural selection governs local, but not global, evolutionary gene coexpression networks in Caenorhabditis elegans. BMC Syst Biol 2:96. doi: 10.1186/1752-0509-2-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peregrín-Alvarez JM, Xiong X, Su C, Parkinson J. 2009. The modular organization of protein interactions in Escherichia coli. PLOS Comput Biol 5:e1000523. doi: 10.1371/journal.pcbi.1000523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu G, Cabrera A, Kono M, Mok S, Chaal BK, Haase S, Engelberg K, Cheemadan S, Spielmann T, Preiser PR, Gilberger TW, Bozdech Z. 2010. Transcriptional profiling of growth perturbations of the human malaria parasite Plasmodium falciparum. Nat Biotechnol 28:91–98. doi: 10.1038/nbt.1597. [DOI] [PubMed] [Google Scholar]
- 29.Speer CA, Dubey JP. 2001. Ultrastructure of schizonts and merozoites of Sarcocystis neurona. Vet Parasitol 95:263–271. doi: 10.1016/S0304-4017(00)00392-7. [DOI] [PubMed] [Google Scholar]
- 30.Hoane JS, Carruthers VB, Striepen B, Morrison DP, Entzeroth R, Howe DK. 2003. Analysis of the Sarcocystis neurona microneme protein SnMIC10: protein characteristics and expression during intracellular development. Int J Parasitol 33:671–679. doi: 10.1016/S0020-7519(03)00031-6. [DOI] [PubMed] [Google Scholar]
- 31.Reiss M, Viebig N, Brecht S, Fourmaux M-N, Soete M, Di Cristina M, Dubremetz JF, Soldati D. 2001. Identification and characterization of an Escorter for two secretory adhesins in Toxoplasma gondii. J Cell Biol 152:563–578. doi: 10.1083/jcb.152.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boothroyd JC, Dubremetz JF. 2008. Kiss and spit: the dual roles of Toxoplasma rhoptries. Nat Rev Microbiol 6:79–88. doi: 10.1038/nrmicro1800. [DOI] [PubMed] [Google Scholar]
- 33.Lamarque MH, Papoin J, Finizio AL, Lentini G, Pfaff AW, Candolfi E, Dubremetz JF, Lebrun M. 2012. Identification of a new rhoptry neck complex RON9/RON10 in the Apicomplexa parasite Toxoplasma gondii. PLoS One 7:e32457. doi: 10.1371/journal.pone.0032457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kono M, Herrmann S, Loughran NB, Cabrera A, Engelberg K, Lehmann C, Sinha D, Prinz B, Ruch U, Heussler V, Spielmann T, Parkinson J, Gilberger TW. 2012. Evolution and architecture of the inner membrane complex in asexual and sexual stages of the malaria parasite. Mol Biol Evol 29:2113–2132. doi: 10.1093/molbev/mss081. [DOI] [PubMed] [Google Scholar]
- 35.Crawford J, Tonkin ML, Grujic O, Boulanger MJ. 2010. Structural characterization of apical membrane antigen 1 (AMA1) from Toxoplasma gondii. J Biol Chem 285:15644–15652. doi: 10.1074/jbc.M109.092619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tonkin ML, Crawford J, Lebrun ML, Boulanger MJ. 2013. Babesia divergens and Neospora caninum apical membrane antigen 1 structures reveal selectivity and plasticity in apicomplexan parasite host cell invasion. Protein Sci 22:114–127. doi: 10.1002/pro.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tonkin ML, Roques M, Lamarque MH, Pugnière M, Douguet D, Crawford J, Lebrun M, Boulanger MJ. 2011. Host cell invasion by apicomplexan parasites: insights from the co-structure of AMA1 with a RON2 peptide. Science 333:463–467. doi: 10.1126/science.1204988. [DOI] [PubMed] [Google Scholar]
- 38.Vulliez-Le Normand B, Tonkin ML, Lamarque MH, Langer S, Hoos S, Roques M, Saul FA, Faber BW, Bentley GA, Boulanger MJ, Lebrun M. 2012. Structural and functional insights into the malaria parasite moving junction complex. PLoS Pathog. 8:e1002755. doi: 10.1371/journal.ppat.1002755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Talevich E, Kannan N. 2013. Structural and evolutionary adaptation of rhoptry kinases and pseudokinases, a family of coccidian virulence factors. BMC Evol Biol 13:117. doi: 10.1186/1471-2148-13-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosowski EE, Lu D, Julien L, Rodda L, Gaiser RA, Jensen KD, Saeij JP. 2011. Strain-specific activation of the NF-kappaB pathway by GRA15, a novel Toxoplasma gondii dense granule protein. J Exp Med 208:195–212. doi: 10.1084/jem.20100717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Braun L, Brenier-Pinchart MP, Yogavel M, Curt-Varesano A, Curt-Bertini RL, Hussain T, Kieffer-Jaquinod S, Coute Y, Pelloux H, Tardieux I, Sharma A, Belrhali H, Bougdour A, Hakimi MA. 2013. A Toxoplasma dense granule protein, GRA24, modulates the early immune response to infection by promoting a direct and sustained host p38 MAPK activation. J Exp Med 210:2071–2086. doi: 10.1084/jem.20130103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shastri AJ, Marino ND, Franco M, Lodoen MB, Boothroyd JC. 2014. Gra25 is a novel virulence factor of Toxoplasma gondii and influences the host immune response. Infect Immun 82:2595–2605. doi: 10.1128/IAI.01339-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tomita T, Bzik DJ, Ma YF, Fox BA, Markillie LM, Taylor RC, Kim K, Weiss LM. 2013. The Toxoplasma gondii cyst wall protein CST1 is critical for cyst wall integrity and promotes bradyzoite persistence. PLoS Pathog. 9:e1003823. doi: 10.1371/journal.ppat.1003823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Howe DK, Gaji RY, Mroz-Barrett M, Gubbels MJ, Striepen B, Stamper S. 2005. Sarcocystis neurona merozoites express a family of immunogenic surface antigens that are orthologues of the Toxoplasma gondii surface antigens (SAGs) and SAG-related sequences. Infect Immun 73:1023–1033. doi: 10.1128/IAI.73.2.1023-1033.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Howe DK, Gaji RY, Marsh AE, Patil BA, Saville WJ, Lindsay DS, Dubey JP, Granstrom DE. 2008. Strains of Sarcocystis neurona exhibit differences in their surface antigens, including the absence of the major surface antigen SnSAG1. Int J Parasitol 38:623–631. doi: 10.1016/j.ijpara.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 46.Toll-Riera M, Radó-Trilla N, Martys F, Albà MM. 2012. Role of low-complexity sequences in the formation of novel protein coding sequences. Mol Biol Evol 29:883–886. doi: 10.1093/molbev/msr263. [DOI] [PubMed] [Google Scholar]
- 47.Oakes RD, Kurian D, Bromley E, Ward C, Lal K, Blake DP, Reid AJ, Pain A, Sinden RE, Wastling JM, Tomley FM. 2013. The rhoptry proteome of Eimeria tenella sporozoites. Int J Parasitol 43:181–188. doi: 10.1016/j.ijpara.2012.10.024. [DOI] [PubMed] [Google Scholar]
- 48.Grigg ME, Bonnefoy S, Hehl AB, Suzuki Y, Boothroyd JC. 2001. Success and virulence in Toxoplasma as the result of sexual recombination between two distinct ancestries. Science 294:161–165. doi: 10.1126/science.1061888. [DOI] [PubMed] [Google Scholar]
- 49.Miller MA, Sverlow K, Crosbie PR, Barr BC, Lowenstine LJ, Gulland FM, Packham A, Conrad PA. 2001. Isolation and characterization of two parasitic protozoa from a Pacific harbor seal (Phoca vitulina richardsi) with meningoencephalomyelitis. J Parasitol 87:816–822. doi: 10.1645/0022-3395(2001)087[0816:IACOTP]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 50.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. 2009. Circos: an information aesthetic for comparative genomics. Genome Res 19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Berglund AC, Sjölund E, Ostlund G, Sonnhammer EL. 2008. InParanoid 6: eukaryotic ortholog clusters with inparalogs. Nucleic Acids Res 36:D263–D266. doi: 10.1093/nar/gkm1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Materials and methods showing the details of S. neurona metabolic reconstruction and apicoplast single-nucleotide polymorphism differences. Download
Organization of the apicoplast genome sequence and comparison to T. gondii. Gene names are as indicated. Red and blue colors indicate the coding strand. Differences with T. gondii are indicated on the outside of the circle. White circles within genes denote in-frame UGA codons. The S. neurona apicoplast genome, like that of Toxoplasma, uses an alternate genetic code. Download
PCR amplification of rps4 fragment. (A) Primer combinations used for PCR amplification of the rps4 fragment insert. (B) PCR was performed with Verbatim high-fidelity DNA polymerase (Thermo Fisher Scientific Inc., Pittsburgh, PA). Three 25-μl PCRs (S. neurona SN3 genomic DNA, S. neurona apicoplast DNA, or no DNA) were set up for each of the primer pairs. The cycling conditions included initial denaturation at 95°C for 3 min; 35 cycles of denaturation at 95°C for 30 s, annealing as shown in the table, and extension at 68°C for 1 min; and a final extension at 68°C for 2 min. The amplified PCR products were analyzed on 1.5% agarose gels. The 635-bp product amplified with primer pair F1-R3 was purified with a PCR purification kit (Qiagen, Valencia, CA) and sequenced in both directions at the Advanced Genetic Technologies Center, University of Kentucky. (C) Electrophoretic analysis of rps4 fragment insert PCRs. The PCR product sequence confirms the presence of the rps4 fragment insert. In a multiple-sequence alignment of the PCR products, the fragment insert and the original gene show no mutations of the rps4 gene fragment insert from the original gene, suggesting that it is a relatively recent event. Download
Alignment of the rps4 gene and the rps4 fragment insert. The multiple-sequence alignment is composed of four sequences, the rps4 gene, the rps4 fragment insert, and the sequence from each strand of the 635-bp PCR product. Highlighted in yellow is the alignment of the rps4 insert. The single nucleotide highlighted in blue is an ambiguity in the length of the homopolymer run in the rps4 insert. Download
Conservation of rpoC2. (A) Diagram of the rpoC2 gene across four apicomplexans. (B) Three-frame translation of the rpoC2a and rpoC2b gap. Highlighted in yellow is the stop codon of rpoC2a. Highlighted in green is the hypothetical start codon of rpoC2b. Download
Resolving the annotation of EC 3.2.1.20 and EC 3.2.1.84 in apicomplexan homologs. Enzymes with EC 3.2.1.20 and EC 3.2.1.84 form part of the glycosyl-hydrolase 31 family. The enzyme corresponding to EC 3.2.1.20 is glucosidase I, a single chain composed of the catalytic subunit. The enzyme corresponding to EC 3.2.1.84 is glucosidase II, a heterodimer composed of the catalytic alpha subunit and the regulatory beta subunit. A multiple-sequence alignment of all of the SwissProt annotated EC 3.2.1.20 and EC 3.2.1.84 sequences (nonredundant at 90% identity) along with apicomplexan homologs (generated with ProbCons) is shown (right panel) with active-site residues indicated by red blocks. The maximum-likelihood tree for this multiple-sequence alignment generated with PhyML with 1,024 bootstrap replicates is also shown (left panel). The evidence used for annotation of the sequences in SwissProt is indicated by colored asterisks. At the bottom, a multiple-sequence alignment of only the apicomplexan homologs is shown. Black bars correspond to regions with 100% identity, and regions of decreasing greyness correspond to decreasing identity levels. The region similar to EC 3.2.1.20 and the alpha subunit of EC 3.2.1.84 is enclosed in a red box. No homolog of the beta subunit of EC 3.2.1.84 was found in any of the apicomplexan genomes. Download
ORFs, annotations, and expression.
(A) List of Sarcocystis neurona IMC proteins. (B) Changes in enzyme:gene assignments for the application of new constraints in iCS382. (C) Predicted impact of single reaction knockouts on parasite growth.
PCR primers used to confirm SRS expression.
Metabolic reconstruction of S. neurona