Abstract
Abnormal choline metabolism is emerging as a metabolic hallmark that is associated with oncogenesis and tumour progression. Following transformation, the modulation of enzymes that control anabolic and catabolic pathways causes increased levels of choline-containing precursors and breakdown products of membrane phospholipids. These increased levels are associated with proliferation, and recent studies emphasize the complex reciprocal interactions between oncogenic signalling and choline metabolism. Because choline-containing compounds are detected by non-invasive magnetic resonance spectroscopy (MRS), increased levels of these compounds provide a non-invasive biomarker of transformation, staging and response to therapy. Furthermore, enzymes of choline metabolism, such as choline kinase, present novel targets for image-guided cancer therapy.
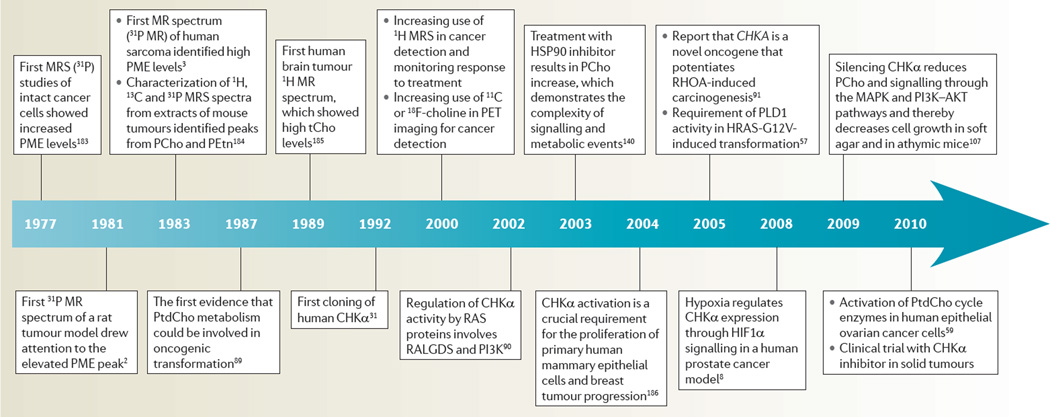
The complexity and adaptability of cancer cells and their ability to recruit and modulate host stromal cells to create a tumour microenvironment that enables survival has led to the disease continuing to confound conventional treatments and molecular targeting. Because of their genomic and phenotypic plasticity, cancer cells have a multitude of redundant pathways and networks, as well as a genomic and proteomic diversity that highlights the need for personalized medicine in this era of ‘omics’ and disease fingerprinting. A handful of common hallmarks of cancer1 have emerged that cut through this variation. Activated choline metabolism, which is characterized by increased phosphocholine (PCho) and total choline-containing compounds (tCho) and which we refer to as the cholinic phenotype, is a fairly new metabolic hallmark that was discovered mostly owing to the introduction of magnetic resonance spectroscopy (MRS) studies of tumours in the 1980s2,3 (TIMELINE). Initially, the increased PCho levels in cancer cells were interpreted as a requirement for the high rate of cell proliferation4. In subsequent studies, non-malignant breast or prostate epithelial cells that were induced to proliferate as rapidly as cancer cells using growth hormones still exhibited significantly lower levels of PCho and tCho levels, identifying malignant transformation rather than just cell proliferation as the cause of abnormal choline metabolism in cancers5. The tumour microenvironment also influences choline metabolism. Tumours have abnormal physiological environments such as hypoxia and acidic extracellular pH6. In studies with perfused mammalian cells, an acidic extracellular pH was found to significantly increase glycerophosphocholine (GPC) levels and to decrease PCho levels7. Hypoxia was also found to increase tCho and PCho levels in a human prostate cancer xenograft model that was genetically engineered to express green fluorescent protein under the control of a hypoxia response element8. A close correlation was observed between regions of high tCho levels and hypoxia in vivo8, which was subsequently identified to be due to the regulation of choline kinase-α (CHKα) expression by the transcription factor hypoxia-inducible factor 1 (HIF1)8. The conditioned growth medium from cultured cancer cells was also found to increase PCho levels when used to culture human vascular endothelial cells9, suggesting that cancer cells can influence stromal cells in the tumour microenvironment.
Timeline. Deregulated choline metabolism in cancer.
CHKα, choline kinase-α; HSP90, heat shock protein 90; MRS, magnetic resonance spectroscopy; PCho, phosphocholine; PET, positron emission tomography; PEtn, phosphoethanolamine; PLD1, phospholipase D1; PME, phosphomonoester; PtdCho, phosphatidylcholine; RALGDS, RAL GTPase guanine nucleotide dissociation stimulator; tCho, total choline-containing compounds.
Because of the ability of non-invasive clinically available imaging techniques such as MRS and positron emission tomography (PET) (BOX 1; FIG. 1) to detect these changes in choline metabolism, the cholinic phenotype is being exploited for radiological diagnosis, prognosis, monitoring response and the development of novel treatments. In fact, early 31P MRS studies showed a close association between phosphomonoesters (PMEs) and phosphodiesters (PDEs) with therapeutic response2,3, and paved the way for later studies with 1H MRS to study choline metabolism. The search for biomarkers to detect cancer and to non-invasively monitor the response to treatment has led to several clinical studies evaluating the non-invasive detection of tCho for these objectives. Similar to studies in cancer cells10, human tumour xenograft models10 and excised tumour tissue11,12, clinical studies have confirmed the activation of choline metabolism in human tumours in vivo10,13.
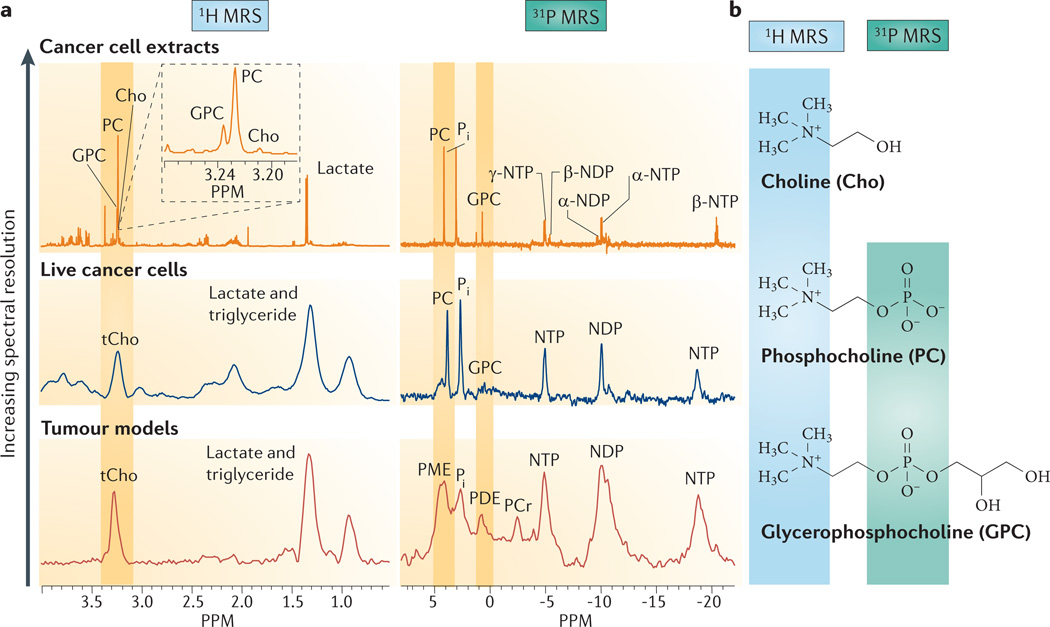
Box 1. Detecting deregulated cancer choline metabolism with MRS.
Choline metabolism in cells and tumour xenografts has been extensively investigated with 1H, 13C and 31P magnetic resonance spectroscopy (MRS). In 1H MR spectra obtained in vivo, the total choline-containing compounds (tCho) signal in tumours is detected as a single peak observed between 3.2 ppm and 3.3 ppm (see part a of the figure). This peak consists of choline (Cho), phosphocholine (PCho) and glycerophosphocholine (GPC), which can be resolved in high-resolution 1H MR spectra of tissue and cell extracts, or in high-resolution magic angle spinning (MAS) 1H MR spectra of tissue biopsy samples (see part a of the figure). These peaks arise from the nine chemically equivalent protons in the highlighted choline-N(CH3)3 groups of Cho, PCho and GPC (see part b of the figure).
In vivo31P MR spectra contain signals from phosphomonoesters(PMEs)that consist of the overlapping peaks from membrane precursors PCho and phosphoethanolamine (PEtn), and from phosphodiesters (PDEs) that consist of the overlapping peaks from membrane breakdown products GPC and glycerophosphoethanolamine (GPE) (see part a of the figure). PCho and PEtn can also be formed during the breakdown of phosphatidylcholine (PtdCho) and phosphatidylethanol-amine (PtdEtn). Individual PCho, PEtn, GPC and GPE peaks are detected in high-resolution 31P MR spectra (see part a of the figure). These PCho and GPC peaks arise from the 31P nuclei in the highlighted phosphate groups (see part b of the figure). Increased levels of PMEs were evident in the very first MR spectrum of a human sarcoma, which was acquired in 1983 (REF. 3). Because of its low sensitivity, 31P MRS has limited application in human studies.
13C MRS studies of choline metabolism are carried out to track the metabolism of exogenously supplied 13C-labelled choline in cells143 or in vivo174. The incorporation of 13C-labelled choline into different metabolites in the choline phospholipid metabolic pathways can be used to calculate flux rates from the rate of enrichment of the metabolite pool. Future studies might demonstrate the value of hyperpolarized Cho for monitoring Cho uptake and phosphorylation with greater sensitivity.
Figure is modified, with permission, from REF. 175 © 2011 Elsevier.
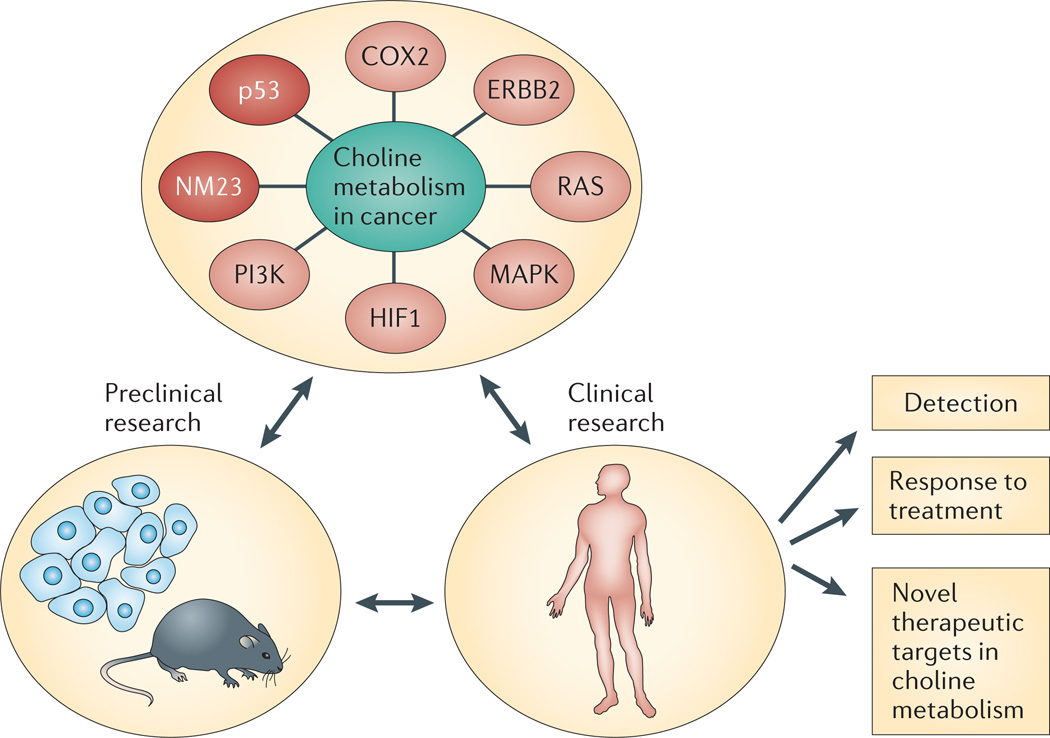
Figure 1. Overview of deregulated choline metabolism in cancer.
Magnetic resonance spectroscopy (MRS) studies with cells, animal models and human studies have revealed deregulated choline metabolism in cancer. Oncogenic pathways (shown in light red), tumour suppressor pathways (dark red) and molecules that are associated with choline metabolism (green) in cancer are shown in the upper circle. These studies have led to the use of MRS for cancer detection and for following the treatment-induced changes in total choline-containing compounds (tCho) to determine response to therapy. The association of choline metabolism with cancer has also led to the development of inhibitors that target enzymes in the pathway, and a Phase I clinical trial with a choline kinase-α (CHKα) inhibitor has been initiated. COX2, cyclooxygenase 2; HIF1, hypoxia-inducible factor 1.
Phosphocholine is both a precursor and a breakdown product of phosphatidylcholine (PtdCho), which, together with other phospholipids such as phosphatidylethanolamine (PtdEtn) and neutral lipids, forms the characteristic bilayer structure of cellular membranes and regulates membrane integrity14. The synthesis of PtdCho and PtdEtn, which are the most abundant phospholipids in the cell membrane, was first described by Kennedy and Weiss in 1956 (REF. 15) and this de novo biosynthesis of PtdCho and PtdEtn is termed the Kennedy pathway. The high-energy intermediates cytidine diphosphate (CDP)-choline and CDP-ethanolamine are required to synthesize PtdCho and PtdEtn. These two mirroring pathways, one that uses choline and the other that uses ethanolamine, are called the CDP-choline and CDP-ethanolamine pathways, respectively. The biosynthesis and hydrolysis of PtdCho mediates mitogenic signal transduction events in cells, and products of choline phospholipid metabolism, such as PCho16, diacylglycerol (DAG)17,18 and arachidonic acid metabolites17, may function as second messengers that are essential for this mitogenic activity. Growth factor stimulation18, cytokines19, oncogenes5 or requirements for eicosanoid production17 also regulate choline phospholipid metabolism (FIGS 1,2). Underlying these phenotypic alterations in choline metabolism, is a network of transporter systems and enzymes involved in choline phospholipid metabolism that are deregulated in cancer cells.
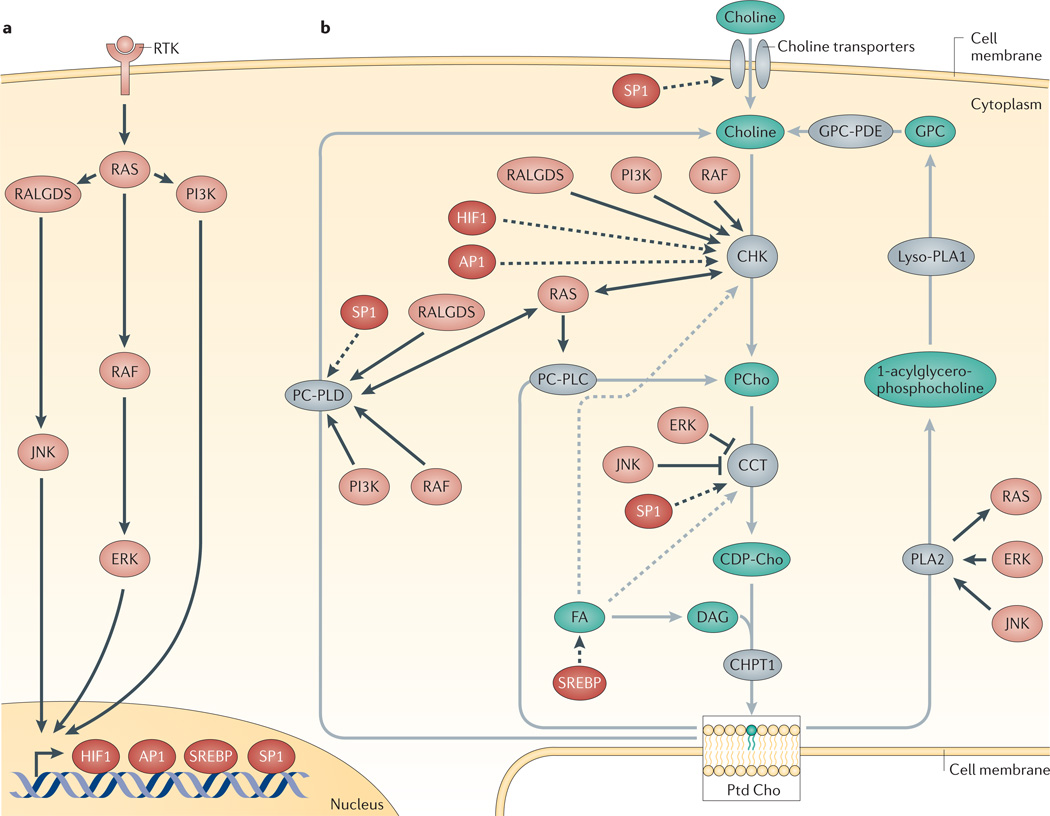
Figure 2. Control of choline metabolism by oncogenic signalling pathways.
Oncogenic signalling pathways (part a) that interact with the choline metabolic pathway (part b) are shown. Grey arrows represent the choline metabolism pathway, proteins in grey catalyse the reaction that is depicted by the corresponding grey arrow. Dashed grey arrows indicate the regulation of enzyme activity in the choline metabolism pathway. Black arrows indicate connections to the oncogenic signalling pathways shown in part a. Solid black arrows indicate increased or decreased enzyme activity, dashed black arrows indicate increased gene transcription. AP1, activator protein 1; CHK, choline kinase; CHPT1, diacylglycerol cholinephosphotransferase 1; CCT, CTP:phosphocholine cytidylyltransferase; DAG, diacylglycerol; FA, fatty acid; GPC, glycerophosphocholine; HIF1, hypoxia-inducible factor 1; JNK, JUN N-terminal kinase; PC-PLC, phosphatidylcholine-specific phospholipase C; PC-PLD, phosphatidylcholine-specific phospholipase D; PLA2, phosphatidylcholine-specific phospholipase A2; PCho, phosphocholine; PtdCho, phosphatidylcholine; RALGDS, RAL GTPase guanine nucleotide dissociation stimulator; RTK, receptor tyrosine kinase; SREBP, sterol regulatory element binding protein.
This Review highlights the causes and consequences of the cholinic phenotype within the context of radiological applications, the molecular mechanisms and the interconnected networks underlying this phenotype, and the possibility of targeting this metabolic hallmark of cancer.
Key enzymes in choline metabolism in cancer
The increased PCho and tCho levels that have been detected in human cancers are caused by interplay between multiple enzymes, which are at the core of choline metabolism (FIG. 3) and which constitute the biosynthetic and catabolic pathways of PtdCho. Here, we review the different enzymes that are involved in choline metabolism. Reported enzyme expression levels and activities in human cancers are compared with those in normal tissue of the same origin and are summarized in TABLE 1.
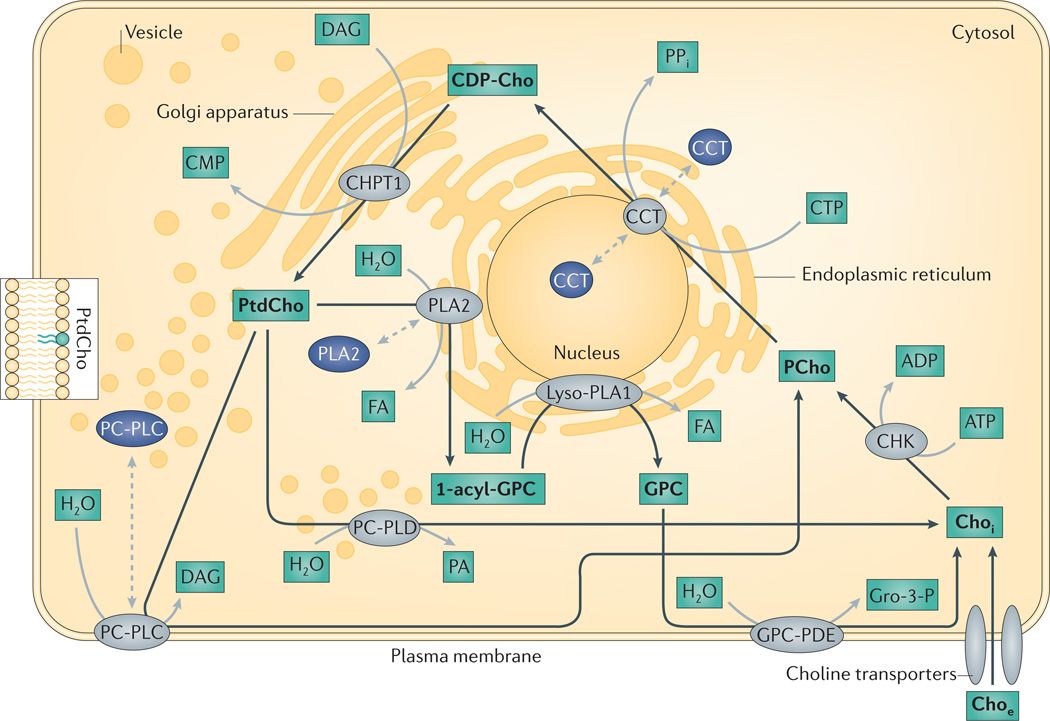
Figure 3. The major enzymes involved in choline phospholipid metabolism in the cell.
Enzymes shown in grey indicate active choline cycle enzymes, which are shown in the organelle in which they are active. Enzymes shown in dark blue indicate the location of choline cycle enzymes that are deactivated by translocation to a different organelle. Black arrows represent the choline metabolism pathway, proteins in grey catalyse the reaction that is depicted by the corresponding black arrow and choline cycle metabolites are shown in bold. Dashed grey arrows show translocation to different subcellular locations, which can deactivate (dark blue) or activate (grey) the enzyme. CCT, CTP: phospho-choline cytidylyltransferase; CDP-Cho, cytidine diphosphate-choline; CHKα, choline kinase-α; Choe, extracellular free choline; Choi, intracellular free choline; CHPT1, diacylglycerol cholinephosphotransferase 1; CMP, cytidine monophosphate; CTP, cytidine triphosphate; FA, fatty acid; GPC, glycerophosphocholine; GPC-PDE, glycerophospho-choline phosphodiesterase; Gro-3-P, glycerol-3-phosphate; Lyso-PLA1, lyso-phospholipase A1; PCho, phosphocholine; PC-PLC, phosphatidylcholine-specific phospholipase C; PC-PLD, phosphatidylcholine-specific phospholipase D; PLA2, cytoplasmic phosphatidylcholine-specific phospholipase A2; PPi, diphosphate.
Table 1.
Choline metabolite concentrations and choline enzyme expression and activity levels in human cancers
| Cancer site |
PCho |
GPC |
tCho |
Enzyme expression |
Enzyme activity |
|||
|---|---|---|---|---|---|---|---|---|
| Normal | Cancer | Normal | Cancer | Normal | Cancer | |||
| Bladder | ND | ND | ND | ND | ND | ND | CHKα ↑176 | CHKα ↑176 |
| Brain | 0.48 ±0.08 (REF. 177) |
0.91 ±0.20 (REF. 177) |
0.86 ±0.11 (REF. 177) |
0.68 ±0.17 (REF. 177) |
0.64* ±0.10 (REF. 178) and 1.79‡ ±0.24 (REF. 177) |
1.19§ ±0.30 (REF. 178) and 1.88║ ±0.33 (REF. 177) |
ND | ND |
| Breast | 0.03 ±0.03 (REF. 42) |
0.79 ±0.55 (REF. 42) |
0.04 ±0.04 (REF. 42) |
0.28 ±0.20 (REF. 42) |
0.07 ±0.07 (REF. 42) |
1.07 ±0.75 (REF. 42) |
CHKα ↑38, CHT1 ↑28 OCT2 ↑28 and PLD1 ↑61 |
CT ↑28 CHKα ↑41, CCT inconclusive28 and PLD ↑60,61 |
| Colon | ND | ND | ND | ND | ND | ND | CTL1 ↑24,27 CHKα ↑38,40, CCT ↑50 and PLD2 ↑64,65 |
CT ↑27, CHKα ↑40, CCT ↑50, PLD2 ↑64 and PC-PLC↓,50 |
| Liver | 0.17 ±0.11 (REF 49) |
1.36 ±0.50 (REF 49) |
2.46 ±0.37 (REF 49) |
0.59 ±0.15 (REF 49) |
ND | ND | CCT ↑48 | CT ↑179, CCT ↑48 and PC-PLC ↑78 |
| Lung | Increased in tumour tissue versus normal tissue180,181 |
Increased in tumour tissue versus normal tissue181 |
Increased in tumour tissue versus normal tissue180,181 |
CHKα ↑38,39 CTL1 ↑26 and OCT3 ↑26 |
CHKα ↑39 | |||
| Ovary | Increased in tumour tissue versus normal tissue59 |
Increased in tumour tissue versus normal tissue59 |
Increased in tumour tissue versus normal tissue59 |
CHKα ↑59 OCT3 ↓9, CTL3 ↑59 CCT ↓59 and PC-PLC↑59 |
CHKα ↑59 PC-PLD ↑59 and PC-PLC ↑59 |
|||
| Prostate | 0.02 ±0.07 (REF. 182) |
0.39 ±0.40 (REF. 182) |
0.29 ±0.26 (REF. 182) |
0.57 ±0.87 (REF. 182) |
0.31 ±0.33 (REF. 182) |
0.96 ±1.27 (REF. 182) |
CHKα ↑38 | CT ↑29 |
CCT, CTP:phosphocholine cytidylyltransferase; CHKα, choline kinase-α; CHT1, choline transporter 1; CT, choline transport; CTL, choline transporter-like; GPC, glycerophosphocholine; ND, not determined; OCT, organic cation transporter; PCho, phosphocholine; PC-PLC, phosphatidylcholine-specific phospholipase C; PLD, phospholipase D1; tCho, total-choline-containing compounds.
Cortex.
White matter.
Glioblastoma.
Astrocytoma grade IV.
Choline transporters
Choline is an essential nutrient that is derived from the diet, with plasma concentrations of about 10 µM. The enhanced transport of free extracellular choline into cancer cells (FIG. 3) has been identified as a dominant cause for the cholinic phenotype, as it can be a rate-limiting step in PCho formation under some circumstances20. Four different types of choline-transporting transmembrane systems have been implicated in cancer. These are high-affinity choline transporters (CHTs), choline transporter-like proteins (CTLs), organic cation transporters (OCTs) and organic cation/carnitine transporters (OCTNs). Subtypes of each transporter have increased expression levels in cancer cells (TABLE 1).
High-affinity choline transport has been attributed to CHT1 (also known as SLC5A7)21, which has a Km (Michaelis constant) for choline of less than 10 µM. CTLs are responsible for intermediate-affinity, sodium-independent choline transport22. The CTL family consists of at least six genes, and all gene products undergo complex alternative splicing23. SLC44A1 (which encodes CTL1)23 exists as two splice variants. It is ubiquitously expressed in human tissues24 and supplies choline for phospholipid biosynthesis22,24. Both OCTs and OCTNs are part of the SLC22 family and carry out low-affinity choline transport in addition to the transportation of other organic cations25. Human OCTs exist as three subtypes and translocate organic cations in an electrogenic, sodium-independent and reversible manner25. There are two human OCTN isoforms25. OCTN1 (also known as SLC22A4) functions in a sodium-independent manner25; whereas OCTN2 (also known as SLC22A5) functions as a sodium-dependent co-transporter for certain zwitterions, and as a sodium-independent transporter of organic cations25.
CTL1 was shown by microarray analysis to be expressed in cancers of the central nervous system (CNS), ovary, breast and prostate, as well as in leukaemia, and to be highly expressed in melanoma, and renal and colon cancer24. Human pulmonary adenocarcinoma tissues highly overexpress CTL1 and somewhat overexpress OCT3 (also known as SLC22A3) compared with matched normal tissues, as shown by immunohistochemistry26. In lung adenocarcinoma cell lines, enhanced choline uptake mostly depends on CTL1, and only partially on OCT3, OCTN1 and OCTN2 (REF. 26). However, in human HT-29 colon carcinoma cells, enhanced choline transport is predominantly mediated by CTL1, and to a lesser extent by CTL2 (also known as SLC44A2) and CTL4 (also known as SLC44A4)27. Increased choline transport20 and elevated mRNA expression of SLC5A7 (which encodes CHT1) and SLC22A2 (which encodes OCT2)28 was observed in breast cancer cells compared with mammary epithelial cells. Choline transport in PC-3 prostate cancer cells had a facilitative component that was characterized by sodium dependence and intermediate affinity, as well as non-facilitative components, although the transporters involved were not identified29.
More molecular characterization is required to decipher the exact roles of the different transporters and how they lead to the enhanced choline transport that is observed in cancer cells. As a common theme so far, CTL1 is overexpressed in multiple types of cancer. However, it is also possible that different types of cancer use different choline transport mechanisms depending on their tissue of origin.
Choline kinase
CHK was first described in 1953 by Wittenberg and Kornberg30, and first cloned in 1992 by Hosaka et al.31. It catalyses the phosphorylation of free choline using ATP as a phosphate donor, thereby producing PCho, and CHK can take on a rate-limiting, regulatory role in PtdCho biosynthesis under some circumstances32. CHK is primarily located in the cytoplasm of cells from various tissues, and its enzymatic properties have been extensively studied (reviewed in REF. 32). At least three isoforms of CHK exist in mammalian cells, and these are encoded by two genes: choline kinase-α (CHKA) and choline kinase-β (CHKB)32. The two functional isoforms, CHKα1 and CHKα2, are derived from CHKA by alternative splicing32. Homodimeric or heterodimeric forms of CHK are enzymatically active32. Both CHKα1 and CHKα2 homodimers display a dual choline and ethanolamine kinase activity, with a lower Km for choline, whereas the CHKβ homodimer predominantly has ethanolamine kinase activity, and the CHKα–CHKβ heterodimer has an intermediate substrate specificity33,34. The proportions of different homodimer and heterodimer populations are tissue-specific33, and studies with knockout mice demonstrate that Chka loss, but not Chkb loss, is embryonically lethal35,36. Taken together, knockout studies suggest that CHKβ cannot compensate for the loss of CHKα, which is necessary to sustain PtdCho biosynthesis35,36. Although the activity of yeast choline kinase (CKI) can be upregulated by protein kinase A (PKA)-dependent phosphorylation at both Ser30 and Ser85, there has been only one report indicating that PKA-mediated phosphorylation partially regulates human CHK37. Thus, the upregulation of CHK activity in cancer probably results from an increase in CHKα expression, which would lead to a higher proportion of CHKα–CHKα dimers in cancer cells and in turn a higher CHK activity level than CHKα–CHKβ heterodimers or CHKβ–CHKβ homodimers.
Overexpression of CHKα, but not of CHKβ, has been reported in several human tumour-derived cell lines of multiple origins, as well as in biopsy samples of lung, colon and prostate carcinomas, among others, which were compared with matched normal tissue from the same patient34,38,39 (TABLE 1). These findings, combined with the studies in knockout mice, indicate that CHKα, but not CHKβ, is essential for PtdCho biosynthesis, which is required for the uncontrolled growth of cancer cells. In addition, the activity of CHKα was shown to increase in human colon cancers40 and in human breast carcinomas compared with normal breast tissue41. Increased enzymatic activity and overexpression of CHKα correlated with advanced histological tumour grade and negative oestrogen receptor status in breast carcinomas41, which is consistent with the increased levels of PCho and tCho that are observed in breast cancers42. By contrast, no significant correlation was found with age, tumour size, progesterone receptor status, vascular invasion and histological tumour type, or with expression of p53, ERBB2 or Ki-67, in these breast tumours41. These clinical findings, combined with a large body of work in cancer cells, suggest that CHKα expression and activity is directly associated with increased cancer cell proliferation and malignancy, making it a potential prognostic marker of some cancers, such as non-small-cell lung cancer39. CHKα expression and activity levels have not yet been investigated in metastases.
CTP:phosphocholine cytidylyltransferase
CTP:phosphocholine cytidylyltransferase (CCT) catalyses the synthesis of CDP-choline and PPi from PCho and cytidine triphosphate (CTP) (FIG. 3). CDP-choline is the most highly activated Cho intermediate of the Kennedy pathway, and it is directly used by diacylglycerol cholinephosphotransferase 1 (CHPT1) to form the membrane lipid PtdCho. First described in 1957 by Kennedy et al.43, CCT catalyses the ratelimiting step in PtdCho synthesis in normal cells, and it catalyses the main rate-limiting and regulatory step in PtdCho biosynthesis in cancer cells44, but CHK32, and choline transporters20, can take on an important regulatory role in some cases. CCT exists as an inactive soluble form and as an active lipid-bound form in the nuclear membrane45. Four highly homologous isoforms of CCT exist in mammalian cells: CCTα (also known as PCYT1A), which is ubiquitously expressed and active as a homodimer45; and CCTβ1, CCTβ2 and CCTβ3, which are splice variants of PCYT1B that exhibit tissue-specific expression46. Much of the work on CCT was carried out before the different isoforms were identified; nonetheless, it is clear that the control of CCT activity is complex and that it involves multiple factors that modulate membrane localization, phosphorylation and transcription of CCTs47. All of these factors have been linked to oncogenic signalling.
Hepatocarcinogenesis was demonstrated to be associated with increased CCT activity and mRNA expression, whereas phosphatidylethanolamine N-methyltransferase (PEMT) activity and mRNA expression were decreased, suggesting a reciprocal relationship between these two enzymes48. PEMT converts PtdEtn to PtdCho and can therefore replenish cellular PtdCho levels at the expense of PtdEtn48. An increase in CCT activity would therefore result in the depletion of cellular PCho levels. However, 31P MRS has frequently demonstrated that liver tumours contain significantly increased cellular PCho levels compared with normal liver49. Because several enzymes, other than CCT, such as CHK and PtdCho-specific phospholipase C (PC-PLC), can increase intracellular PCho levels, these enzymes may counteract a possible depletion of cellular PCho levels by increased CCT activity in liver tumorigenesis (TABLE 1). CCT expression and activity were also increased in colon cancer, which resulted in elevated PtdCho levels in colon cancer50. Taken together, CTL1, CHKα and CCT expression and activity are increased in colon cancer, which results in elevated PCho and PtdCho levels, thereby facilitating enhanced cell growth and proliferation (TABLE 1).
PtdCho-specific phospholipase D
PtdCho-specific phospholipase D (PC-PLD) was discovered in plants in 1947 (REF. 51), but it only attracted widespread attention when experiments in mammalian cell cultures revealed that PLD1 and PLD2 were rapidly activated in response to extracellular stimuli (reviewed in REF. 52). PLD1 and PLD2 hydrolyse PtdCho to phosphatidic acid and Cho, which is one of the breakdown pathways in PtdCho metabolism that directly produces intracellular free Cho (FIG. 3). PLD1 and PLD2 occur as three splice variants and share 50% sequence homology53,54. PLD1 is predominantly localized to Golgi membranes, whereas PLD2 is associated with both Golgi membranes and the plasma membrane55. Both PLD enzymes were also found in the cytoplasm and a variety of other organelles, and their location may depend on the cell type and physiological state of the cell56. PLD1 and PLD2 are crucial in cell proliferation, survival signalling, cell transformation and tumour progression57.
Elevated activity of PC-PLD has been observed in gastric58 and ovarian cancers59, as well as in epithelial ovarian cancer cell lines59. PLD1 activity and expression were increased in breast cancers60,61, as well as several human breast cancer62 and melanoma63 cell lines. PLD2 activity and expression were elevated in colorectal64,65 and renal66 cancers. A C1814T polymorphism occurs in PLD2, resulting in T577I substitution, which is associated with colorectal cancer but which does not affect PLD2 activity67. PLD1 or PLD2 expression were able to transform cells that overexpressed a tyrosine kinase such as epidermal growth factor receptor (EGFR) or SRC, suggesting a contribution of PLD1 or PLD2 to malignant progression in cells with elevated tyrosine kinase activity68 (TABLE 1).
The overexpression of PLD1 and PLD2 in mouse fibroblasts caused neoplastic transformation, resulting in upregulated matrix metalloproteinase 9 (MMP9) activity, anchorage-independent growth in soft agar and the formation of undifferentiated sarcoma in nude mice69. Multidrug-resistant colon and breast cancer cells exhibited a higher level of PLD2 activity and expression than the parental drug-sensitive cells70. A correlation between increased PC-PLD activity and loss of oestrogen receptor expression in breast cancer cells has been observed, suggesting that PC-PLD could provide a survival signal that is normally provided by oestrogen in a developing tumour71. PC-PLD activity has also been implicated in tumour invasion and may play an important part in the metastasis of cancer cells72,73. High levels of PC-PLD activity correlate with high invasive potential in human breast cancer cells71, and elevated PC-PLD activity was shown to correlate with increased protease secretion, which is commonly observed in invasive cancer cells72–75.
PtdCho-specific phospholipase C
PC-PLC catalyses the hydrolysis of PtdCho, thereby producing PCho and DAG (FIG. 3). DAG is a second messenger that induces protein kinase C (PKC) activation, which phosphorylates multiple target proteins and which is involved in several signal transduction cascades. PtdCho hydrolysis produces a longer and more sustained action of DAG than phosphatidylinositol cleavage by PC-PLC76. To date, no mammalian PC-PLC isoforms have been sequenced or cloned, and studies investigating the putative mammalian PC-PLC enzyme have relied on detecting PC-PLC activity and/or protein expression with rabbit polyclonal antibodies against bacterial PC-PLC from Bacillus cereus. PC-PLC translocates from a perinuclear cytosolic area to the plasma membrane following activation by growth factors, insulin or phorbol esters77 (FIG. 3).
A function for PC-PLC has been demonstrated in hepatocarcinogenesis78, in which the activity of calcium-dependent PC-PLC, but not of calcium-independent PC-PLC, increased significantly in a rat model of N-nitrosodiethylamine-induced hepatocarcinoma78. Stable transfection of NIH3T3 fibroblasts with bacterial PC-PLC led to chronically elevated levels of DAG and PCho, and a transformed phenotype in these cells that was mediated by RAF1 and PKCζ79. In ovarian cancer cells, the elevated PCho levels, which are linked to ovarian carcinogenesis, are partially caused by PC-PLC activation80. In breast cancer cells, PC-PLC accumulates on the plasma membrane of HER2 (also known as ERBB2)-overexpressing cells and associates with HER2 in lipid raft domains81.
Other choline-related enzymes
Additional enzymes, such as PtdCho-specific phospholipase A2 (PC-PLA2), lysophospholipase, GPC phosphodiesterase (GPC-PDE; also known as GPCPD1) and CHPT1, may also be important in establishing the cholinic phenotype by altering the metabolite levels of PCho, GPC and Cho (FIG. 3). PC-PLA2 exists as 19 isoforms, complicating its investigation. Ovarian cancer lines contained unaltered or decreased expression of PC-PLA2 isoforms, which was accompanied by decreased overall PC-PLA2 activity59. The expression of cytosolic phospholipase A2 (PLA2; also known as PLA2G4A) was decreased in breast cancer cells82, and may modulate the cellular MRS-detected GPC levels83. Sphingomyelinases, which catalyse the cleavage of the phosphodiester bond in sphingomyelin to form ceramide and PCho, are involved in cancer pathogenesis84, and may potentially contribute to elevated PCho levels that constitute the cholinic phenotype. Inhibition of CHKα has been reported to increase ceramide levels, and therefore sphingomyelinases may have a role in the action of CHKα inhibitors as antitumour drugs85.
Oncogenic regulation of choline metabolism
The first indication that choline metabolism could be regulated by oncogenic signalling came from early work showing that PCho levels increased following growth factor stimulation of NIH3T3 fibroblasts86. Subsequent studies (discussed below) have demonstrated the complex and often reciprocal interactions (FIG. 2) between oncogenic signalling pathways and the enzymes that are involved in choline metabolism.
The RAS pathway
Each of the enzymes involved in choline metabolism is affected by the RAS pathway (FIG. 2). Activation of CHK and elevated PCho levels were initially reported in serum-stimulated, KRAS- or HRAS-transformed NIH3T3 fibroblasts87–89. Detailed investigations indicate that CHK activation downstream of oncogenic HRAS occurs via combined RAL GTPase guanine nucleotide dissociation stimulator (RALGDS) and PI3K signalling90. Activation of CHK could also be mediated by the small GTPase RHOA and its effector RHO-associated coiled-coil-containing protein kinases (ROCKs), which are associated with transformation and metastasis, and which are overexpressed in various cancers91.
Oncogenic HRAS controls the transcription of CCT through the activation of the MAPK pathways92. MAPK signalling also modulates the expression of sterol regulatory element binding proteins (SREBPs). SREBPs, in turn, can induce CCT expression, but are more likely to lead to increased CCT activity by enhancing fatty acid synthesis, which promotes membrane translocation and thus the activation of CCT93. By contrast, other studies indicate that RAS signalling can result in the inhibition of CCT. Phosphorylation, and thus inhibition, of CCT is mediated by ERK94 and JUN N-terminal kinase (JNK)95. Accordingly, oncogenic signalling can reduce CCT activity and can increase PCho levels92,94,96. Elevated CCT expression could lead to enhanced PtdCho synthesis, enabling rapid cell proliferation and tumour growth. However, the importance of reduced CCT activity in cell transformation remains unclear.
The activation of PC-PLD was first reported in serum-stimulated HRAS- or KRAS-transformed NIH3T3 cells97. Recent work has demonstrated the complex oncogenic regulation of PC-PLD with two positive regulatory pathways that are mediated by RALGDS and PI3K, and two negative feedback mechanisms that are mediated by RAF and RALGDS71,98. In turn, PC-PLD was identified as a regulator of cell transformation and tumour progression71. This effect could be mediated by phosphatidic acid, which couples EGFR stimulation to RAS activation through SOS99, or it could be mediated through a positive feedback loop via WNT57,100.
The activation of PC-PLC in cells that express oncogenic HRAS was hypothesized in early studies as an explanation for the presence of elevated PCho levels in these18. Subsequently, increased PC-PLC activity was observed downstream of serum stimulation, or platelet-derived growth factor (PDGF) stimulation via RAF1, and was associated with cell transformation79,80,101. However, an alternative mechanism for the elevation of PCho levels downstream of PDGF has been proposed and involves increased PC-PLD expression combined with increased CHK activity102. Unfortunately, the lack of molecular biological information regarding PC-PLC has hampered more detailed studies into its role in cancer.
The expression of PLA2 is mediated through JNK and ERK signalling 103, and its activity is further induced by ERK through phosphorylation on Ser505 (REF. 104). In turn, PLA2 has been reported to mediate cell transformation downstream of HRAS105.
The PI3K–AKT pathway
Inhibiting the PI3K–AKT signalling pathway blocked choline uptake in lung adenocarcinoma cell lines, suggesting that this pathway might participate in the regulation of choline transport26. As mentioned above, PI3K–AKT signalling also affects CHK activation, which occurs via combined RALGDS and PI3K signalling90 (FIG. 2). Recent studies point to a reciprocal interaction between the enzyme and oncogenic pathways. CHKα overexpression resulted in the differential expression of multiple genes that are associated with cell cycle progression, whereas partial inhibition of CHKα resulted in cell cycle arrest or apoptosis106. Knockdown of CHKα using small interfering RNA (siRNA) led to the simultaneous attenuation of MAPK and PI3K–AKT signalling and was associated with the inhibition of cell proliferation107. A separate study indicated that knockdown of CHKα or CHKβ inhibited AKT-S473 phosphorylation independently of PI3K, and this was associated with the inhibition of cell proliferation108. Consistent with this observation, treatment with the CHKα inhibitor Mn58b inhibited tumour growth in vivo 108. Most recently, a role for CHKα in mediating the synergistic actions of EGFR and SRC in breast cancer development has also been shown109. These observations reinforce the proposed role of CHKA as an oncogene, with its overexpression contributing, through a positive feedback loop, to increased MAPK and PI3K signalling.
Transcription factors
Transcriptional control of the regulators of choline metabolism by factors associated with cancer has been most clearly demonstrated for CHK. In the liver, the binding of JUN to a distal activator protein 1 (AP1) element in the promoter region of CHKA mediates its expression, indicating a possible link between the role of AP1 in cell proliferation and transformation and CHK110 (FIG. 2). Two additional transcription factors, which are important in oncogenesis and which are likely to be involved in the control of CHK expression, are MYC and HIF1. A possible role for the proto-oncogene MYC was indicated in studies that showed elevated levels of CHK and PCho in Myc+/+ compared with Myc−/− rat fibroblasts111. In the case of HIF1, a recent study clearly identified putative hypoxia response elements within the promoter of CHKA and showed that hypoxia and HIF1 stabilization increase PCho levels8 (FIG. 2). How this effect could enhance cancer cell survival under hypoxic conditions is unclear, but this observation indicates that cells that survive in a hypoxic environment are likely to demonstrate elevated PCho levels.
With regard to other transcription factors that are associated with choline metabolism, SP1, which is involved in cell growth and which is upregulated in a variety of cancers, controls the expression of CTL1 (REF. 24), CCTα112 and PLD1 (REF. 113). Collectively, these enzymes could therefore enhance PtdCho synthesis, facilitating cell growth and proliferation.
Clinical implications
Several clinical implications arise from the deregulated choline metabolism of cancers. The most evident of these is non-invasively detecting cancer with MRS, and indeed many centres throughout the world are evaluating the use of spectroscopic imaging to assist in the diagnosis and the staging of cancer. Two additional, equally valuable, areas of application are in detecting therapeutic response and exploiting choline phospholipid metabolism to identify new targets for cancer treatment (FIG. 3).
Diagnosis and staging of cancer
Several ongoing multi-centre trials evaluating 1H MRS imaging (MRSI) of breast114, brain115 and prostate116,117 cancer, have reported an elevation of tCho levels in tumours. Increased tCho levels have been associated with aggressiveness in breast cancer118. The elevation of tCho expression is a specific biomarker of prostate cancer and correlates with Gleason score and aggressiveness117. In breast and ovarian cancers, the low PCho and high GPC levels that are observed in non-malignant cells change to high PCho and low GPC levels with malignant transformation5,59. PCho, GPC and tCho concentrations in human cancers versus normal tissue of the same origin are summarized in TABLE 1. The elevated tCho level in brain tumours has been used for detection119 and grading120, radiation therapy treatment planning121, and determining recurrence from radiation-induced necrosis and reactive astrocytosis122.
Although the spatial resolution of MRS is orders of magnitude lower than histological evaluation and staging from biopsies or excised tumours, MRS has the advantage of non-invasively detecting tumour metabolism, and can be combined with clinically approved magnetic resonance imaging (MRI) methods in the same setting. Unlike the histological evaluation of biopsies, MRS also provides information about the entire tumour.
MRS applications in the clinic are technically challenging and, unfortunately, are currently not reimbursed by healthcare providers in the United States123. This prevents patients from receiving the potential benefits of MRS, and slows down future progress in clinical MRS techniques, as, without reimbursement, there is no incentive for manufacturers to support MRS development or for radiologists to use MRS123. Nonetheless, several large multi-centre clinical trials are currently evaluating the use of MRS for the detection of tCho and other metabolites in diagnosis, staging and therapeutic response monitoring. This reflects the fairly early stages of MRS technology in terms of clinical translation111,112,114,115. However, the value of MRS in identifying tumours and predicting survival has been demonstrated111,112,114,115. Furthermore, the introduction of higher field clinical MR scanners, such as 7 Tesla and higher, although to date technically more demanding, should allow for better spatial and spectral resolution of non-invasive MRS in clinical applications, probably achieving resolution of the tCho signal into GPC, PC and Cho and further enhancing the information obtained by 1H MRS.
Recent advances in the process of dynamic nuclear polarization (DNP) and the development of commercial polarizers provide a novel method to dramatically increase the MRS signal that arises from exogenous hyperpolarized compounds and their metabolites. The use of hyperpolarized 13C MRS is now in clinical trials at the University of California in San Francisco (UCSF), USA, with very promising results. Whereas monitoring of hyperpolarized PCho synthesis from hyperpolarized choline has not yet been demonstrated in cells or tumours, some studies indicate that this approach might be feasible in the future. Synthesis of PCho from hyperpolarized 15N-labelled choline was observed in vitro using purified CHK124, and the presence of 15N choline, al though not its metabolism, was reported in vivo in rat brain125. Metabolism of hyperpolarized 13C-labelled choline to acetylcholine was also observed in vitro 126. In addition, 11C- and 18F-labelled choline and choline derivatives are being evaluated for non-invasive PET radiological diagnosis of metastatic or recurrent cancers127.
Detecting therapeutic response
Treatment with most conventional chemotherapeutic agents results in a decrease of tCho levels in responding tumours in preclinical models128 and in human tumours129,130. Studies investigating the molecular basis underlying these changes are urgently needed. These studies will provide much needed insight into how choline metabolism in cancer cells is affected by chemotherapy, and whether treatment outcome would be improved by combining conventional chemotherapy with the downregulation of enzymes in this pathway.
Because the enzymes that control choline metabolism are modulated by oncogenic signalling pathways (FIG. 2), the inhibition of this signalling results in altered choline-containing metabolite levels of Cho, PCho and GPC. Accordingly, these metabolites could provide downstream metabolic readouts of effective inhibition of these pathways. Several studies have used MRS and have demonstrated the value of this approach for the non-invasive detection of drug molecular action. The inhibition of oncogenic signalling in mutant HRAS-transfected NIH3T3 cells resulted in reduced levels of PCho, whereas PCho levels remained unchanged in the parent line131. Inhibition of PI3K or MAPK signalling also resulted in reduced PCho and tCho levels, with recent studies indicating that this effect is primarily mediated by CHKα inhibition as a result of its reduced expression downstream of HIF1 activation8,132–135. Consistent with these findings, and with the fact that p53 can regulate HRAS136, loss of p53 function also led to an increase in PCho levels137. In addition, the inhibition of HIF1α138 resulted in reduced PCho levels, and tumours deficient in HIF1β (also known as ARNT), which HIFα heterodimerizes with to form the HIF transcription factor, showed lower levels of PCho than controls139.
Other metabolic imaging studies highlight the need for further investigation. The inhibition of heat shock protein 90 (HSP90), which results in reduced signalling through both the MAPK and the PI3K pathways, leads in most cases to an unexpected increase in PCho and GPC levels140,141 and to increased uptake of the PET tracer [11C]-choline142. These effects could be mediated by the increased expression of the choline transporter CTL1 (REF. 143), but this varies across cancer types, which possibly reflects differences in genetic background144. Drug-induced inhibition of fatty acid synthesis resulted in reduced PCho levels and lower CHK activity, potentially identifying fatty acids as post-translational modulators of CHK activity, which is similar to their role in the control of CCT activity, but such an effect remains to be confirmed145. The inhibition of cyclooxygenase resulted in reduced levels of PCho and an increase in GPC levels with no concomitant changes in the expression of the enzymes that are associated with choline metabolism, but this study identified several candidate genes that could be involved in altering choline metabolism through secondary, as yet undetermined, effects146. Finally, phenylbutyrate induced increases in GPC levels, which was consistent with an as yet undetermined mechanism whereby peroxisome proliferator-activated receptor-γ (PPARγ) is responsible for increased PLA2 activity147.
Therapeutic opportunities
The increased expression and activity of CHKα make it an attractive molecular target for anticancer therapy148–151. Lacal and colleagues first suggested CHKα inhibition as a potential antitumour therapy152, and developed novel chemical compounds for this purpose152. The most promising of these compounds exhibited significant antiproliferative activity, induced the reduction of tumour growth and was specific for CHKα inhibition152,153. Overall, these novel pharmacological inhibitors targeting CHKα151 resulted in tumour growth arrest and apoptosis through the induction of a cytotoxic increase in ceramide levels85,149. A CHKα inhibitor (TCD-717) is currently undergoing Phase I clinical trials for toxicity testing in cancer patients.
Targeting CHKα results in decreased PCho and tCho levels, which can be detected non-invasively with 31P or 1H MRS149,150. Silencing of CHKα in human breast cancer cells by RNA interference (RNAi) has been demonstrated to decrease cellular 1H MRS-detectable PCho levels, while leaving human mammary epithelial cells unaffected148,150,154. A significant decrease of cell proliferation and induction of differentiation in highly invasive and metastatic human breast cancer cells and tumour xenografts was observed with CHKα downregulation148. Non-invasive 31P MRS detected a reduction in PCho levels following gene therapy with an intravenously injected lentiviral vector that delivered short hairpin RNA targeting CHKα in a breast cancer model150. Combining CHKα silencing with chemotherapeutic 5-fluorouracil treatment resulted in a more pronounced anti-proliferative and growth inhibitory effect in breast cancer cells, but not in non-malignant breast epithelial cells154. Radiolabelled choline or choline analogues can also be used as pharmacodynamic markers to monitor CHKα-targeted therapies using PET155.
Hemicholinium-3 (HC-3), which inhibits low-affinity sodium-independent choline transport156, has been proposed as a potential therapeutic anticancer agent152. However, HC-3 is not specific to choline transport and also inhibits CHK152. Radiolabelled HC-3 was evaluated as a molecular imaging agent in a preclinical study, to potentially detect prostate cancer by PET29.
Hexadecylphosphocholine (HePC; also known as miltefosine) is a synthetic alkylphosphocholine that is currently being investigated for antineoplastic therapy and it has been approved for the topical treatment of cutaneous metastases of mammary carcinomas157. Its mechanism of action involves the inhibition of CCT activity by preventing CCT translocation to the membrane158,159. This leads to elevated cellular PCho levels, and decreasing cellular PtdCho levels, and identifies a potential approach for monitoring the effects of treatment158,159.
Because PC-PLD is involved in several aspects of cell proliferation and oncogenic signalling, it could prove to be valuable as a target for therapeutic intervention in cancer. HePC was also shown to modulate PC-PLD activity160. Chronic exposure of different non-tumorigenic mammalian cell lines to HePC strongly inhibits PC-PLD activity, which may be related to its antitumour activity160. The selective oestrogen receptor modulators tamoxifen and raloxifene, which are commonly used for the chemotherapy of oestrogen receptor-positive breast cancer, were reported to differentially affect PC-PLD activity161. Multidrug resistance in human cancer cells was accompanied by increased PLD2 activity in detergent-insoluble glycolipidrich and caveolar membranes70. High levels of PC-PLD activity also conferred resistance to rapamycin, an inhibitor of mTOR, in human breast cancer cells162.
The anti-oestrogen drug tamoxifen, which is widely used for the treatment of breast cancer163, as well as some other cancers164,165, activates cellular PC-PLD and PC-PLC, thereby causing increased levels of DAG, PCho and phosphoethanolamine (PEtn), and sustained translocation and activation of PKC, which may ultimately lead to changes in cell growth166. PC-PLC inhibition reduced HER2 expression on the plasma membrane, thereby inducing antiproliferative effects, which suggests that it may provide a valuable strategy to counteract the tumorigenic effects of HER2 and thus complement HER2-targeting therapies81. The antiviral, potentially antitumoural xanthate D609 was shown to be a potent inhibitor of PC-PLC167, and significantly reduced PCho levels along with cell proliferation in ovarian cancer cells59. D609 can also inhibit sphingomyelin synthase168. Although it has shown promising results in cancer cell lines and transformed cells in vitro169, D609 exhibited little antitumour activity in in vivo animal studies170. The dual-specificity phosphatase inhibitor SC-ααδ93,4-(benzyl-(2-[(2,5-diphenyloxazole-4-carbonyl)amino] ethyl)carba-moyl)-2-decanoylaminobutyric acid, which exhibits antitumour activity in vivo, was also shown to be a potent inhibitor of PC-PLC171.
Conclusions
As a disease, cancer continues to confound the technological advances that are available in the twenty-first century. Common pathways are a rarity in this disease, but these provide the vision and hope for finding effective treatments (FIG. 3). Over the past decade, aberrant choline metabolism is one common pathway that has been consistently revealed by bench-to-bedside MRS studies evaluating tCho levels as a biomarker for detection and monitoring response to treatment (FIG. 1). Elevated levels of PCho and tCho have been observed in almost every cancer type studied. Although substantial advances have been made in understanding the molecular mechanisms that drive these differences, the challenges for the future are to comprehensively understand the mechanisms underlying this elevation, interactions between cancer cells and stromal cells, and the transcription factors that regulate enzymes and transporters in the choline pathway. New methodologies such as mass spectrometric imaging172 in combination with molecular biology techniques such as PCR carried out on tissue sections should allow a genomic- to proteomic-scale characterization of deregulated choline metabolism. Although 31P MRS studies have identified increased PEtn levels in tumours13, the role of the CDP-ethanolamine pathway in malignant transformation and progression is almost completely unexplored and merits further investigation. The apparent positive feedback that is mediated by enzymes in the choline cycle, as well as the interaction and compensatory mechanisms between enzymes in the choline cycle and the ethanolamine cycle that may allow cancer cells to adapt and survive the downregulation of individual enzymes, are other areas that require focus in the future. As the connectivity, networks and feedback mechanisms in choline metabolism become more evident, a systems biology approach will be necessary to identify the crucial nodes that should be targeted in this pathway. Other challenges are the sequencing of all genes in the choline cycle to allow a better understanding of deregulated choline metabolism in cancers. Targeting the enzymes involved in choline metabolism may prove to be highly effective against cancer cells, and MRSI would lend itself most easily to clinical applications to detect the effect of targeting choline metabolism non-invasively in vivo for image-guided therapy. The development of image-guided siRNA delivery to target enzymes173 could allow the downregulation of multiple enzymes in the choline cycle, and the clinical translation of these technologies provides hope for the future.
At a glance.
Some of the levels of metabolic intermediates that are generated in choline phospholipid metabolism, particularly phosphocholine (PCho) and total choline (tCho), are elevated in cancer, and can be used for non-invasive detection in cancer diagnosis and staging by using magnetic resonance spectroscopy (MRS) or positron emission tomography (PET).
Several enzymes in choline metabolism, such as choline transporter-like protein 1 (CTL1), choline kinase-α (CHKα), CTP:phosphocholine cytidylyltransferase (CCT), phosphatidylcholine-specific phospholipase D (PC-PLD) and PC-PLC, are overexpressed and/or activated in cancer and can potentially be used as prognostic markers.
Overexpression and activation of choline cycle enzymes are mediated by oncogenic signalling via pathways such as the RAS and PI3K–AKT pathways, and by transcription factors associated with oncogenesis such as hypoxia-inducible factor 1 (HIF1). Recent studies point to a reciprocal interaction between choline cycle enzymes and oncogenic signalling, where modulation of the enzymes can contribute to enhancing oncogenic signalling.
The therapeutic response of tumours can be monitored non-invasively by MRS of the tCho signal because treatment with conventional chemotherapeutic agents results in a decrease of tCho levels in responding, but not in non-responding, tumours.
Inhibition of oncogenic signalling pathways with targeted anticancer drugs results in altered choline-containing metabolite levels. Therefore, these metabolites could provide downstream metabolic readouts of the effective inhibition of these pathways.
New molecularly targeted therapeutic opportunities arise from targeting choline cycle enzymes such as CHKα, which is currently being tested in Phase I clinical trials.
Acknowledgements
The authors gratefully acknowledge useful discussions with F. Podo, R. Gillies, H. Degani and J. Delikatny. The authors apologize for only including a limited number of studies owing to journal restrictions for the number of citations. Support from P50 CA103175, P30 CA006973, R01 CA73850, R01 CA82337, R01 CA136576, R01 CA138515, R01 CA138264, R0 1 CA134695, R01 CA154725, R21 CA120010, RO1 CA130819 and P50 CA097257 is gratefully acknowledged.
Glossary
- Total choline-containing compounds
(tCho). The sum of choline (Cho),phosphocholine (PCho) and glycerophosphocholine (GPC),this term was coined because these three metabolite signals appear as one overlapping signal in non-invasive in vivo 1H magnetic resonance spectra owing to limited spectral resolution
- Magnetic resonance spectroscopy
(MRS). A technique that measures the nuclear magnetic resonance of 1H,31P and other magnetic resonance-active nuclei to determine their physical and chemical properties either non-invasively in live organisms or at high spectral resolution in ex vivo samples
- Phosphomonoesters
(PMEs). Phospholipid metabolism intermediates, such as phosphocholine (PCho) and phosphoethanolamine (PEtn), which contain one ester bond between the phosphate group and the specific phospholipid head group alcohol
- Phosphodiesters
(PDEs). Phospholipid metabolism intermediates, such as glycerophosphocholine (GPC) and glycerophosphoethanolamine (GPE),which contain two ester bonds at the phosphate group,one to the specific phospholipid head group alcohol and a second one to glycerol
- Magic angle spinning
(MAS). A specialized magnetic resonance spectroscopy technique that avoids tissue extraction to detect high-resolution spectra by spinning solid tissue samples at the angle of 54.74°with respect to the magnetic field. These tissue samples can be used for subsequent histological,biochemical and genetic analyses
- Phorbol esters
A class of compounds originally derived from esterification of the plant compound phorbol,which promote tumorigenesis through the activation of protein kinase C
- MRS imaging
(MRSI). Applies magnetic resonance spectroscopy (MRS) in a spatially resolved manner, thereby providing metabolite concentrations in tissue in two or three spatial dimensions and allowing for the display of metabolic maps throughout the tissue
Footnotes
Competing interests statement
The authors declare no competing financial interests.
FURTHER INFORMATION
Kristine Glunde’s homepage: http://www.hopkinsradiology.org/Radiology%20Faculty/Research%20Faculty%20Bios/Glunde
Zaver M. Bhujwalla’s homepage: http://icmic.rad.jhmi.edu/
Sabrina M. Ronen’s homepage: http://www.radiology.ucsf.edu/research/labs/ronen
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
Contributor Information
Kristine Glunde, Email: kglunde@mri.jhu.edu.
Zaver M. Bhujwalla, Email: zaver@mri.jhu.edu.
Sabrina M. Ronen, Email: Sabrina.Ronen@ucsf.edu.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2010;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Griffiths JR, Stevens AN, Iles RA, Gordon RE, Shaw D. 31P–NMR investigation of solid tumours in the living rat. Biosci. Rep. 1981;1:319–325. doi: 10.1007/BF01114871. [DOI] [PubMed] [Google Scholar]
- 3. Griffiths JR, et al. 31P–NMR studies of a human tumour in situ . Lancet. 1983;1:1435–1436. doi: 10.1016/s0140-6736(83)92375-9. 31P MRS spectrum of rhabdomyosarcoma was recorded and showed elevated levels of PMEs when compared with normal tissue.
- 4.Daly PF, Lyon RC, Faustino PJ, Cohen JS. Phospholipid metabolism in cancer cells monitored by 31P NMR spectroscopy. J. Biol. Chem. 1987;262:14875–14878. [PubMed] [Google Scholar]
- 5.Aboagye EO, Bhujwalla ZM. Malignant transformation alters membrane choline phospholipid metabolism of human mammary epithelial cells. Cancer Res. 1999;59:80–84. [PubMed] [Google Scholar]
- 6.Gillies RJ, Raghunand N, Karczmar GS, Bhujwalla ZM. MRI of the tumor microenvironment. J. Magn. Reson. Imaging. 2002;16:430–450. doi: 10.1002/jmri.10181. [DOI] [PubMed] [Google Scholar]
- 7.Galons JP, Job C, Gillies RJ. Increase of GPC levels in cultured mammalian cells during acidosis. A 31P MR spectroscopy study using a continuous bioreactor system. Magn. Reson. Med. 1995;33:422–426. doi: 10.1002/mrm.1910330317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Glunde K, et al. Hypoxia regulates choline kinase expression through hypoxia-inducible factor-1 α signaling in a human prostate cancer model. Cancer Res. 2008;68:172–180. doi: 10.1158/0008-5472.CAN-07-2678. First proof that hypoxia regulates CHKα expression through HIF1 binding to hypoxia response elements in the CHKA promoter of a human prostate cancer model, thereby increasing PCho levels in hypoxic tumour regions.
- 9.Mori N, Natarajan K, Chacko VP, Artemov D, Bhujwalla ZM. Choline phospholipid metabolites of human vascular endothelial cells altered by cyclooxygenase inhibition, growth factor depletion, and paracrine factors secreted by cancer cells. Mol. Imaging. 2003;2:124–130. doi: 10.1162/15353500200303127. [DOI] [PubMed] [Google Scholar]
- 10.Podo F, et al. MR evaluation of response to targeted treatment in cancer cells. NMR Biomed. 2011;24:648–672. doi: 10.1002/nbm.1658. [DOI] [PubMed] [Google Scholar]
- 11.Lean CL, et al. Assessment of human colorectal biopsies by 1H MRS: correlation with histopathology. Magn. Reson. Med. 1993;30:525–533. doi: 10.1002/mrm.1910300502. [DOI] [PubMed] [Google Scholar]
- 12.Cheng LL, Chang IW, Smith BL, Gonzalez RG. Evaluating human breast ductal carcinomas with high-resolution magic-angle spinning proton magnetic resonance spectroscopy. J. Magn. Reson. 1998;135:194–202. doi: 10.1006/jmre.1998.1578. [DOI] [PubMed] [Google Scholar]
- 13.Negendank W. Studies of human tumors by MRS: a review. NMR Biomed. 1992;5:303–324. doi: 10.1002/nbm.1940050518. [DOI] [PubMed] [Google Scholar]
- 14.Mountford CE, Wright LC. Organization of lipids in the plasma membranes of malignant and stimulated cells: a new model. Trends Biochem. Sci. 1988;13:172–177. doi: 10.1016/0968-0004(88)90145-4. [DOI] [PubMed] [Google Scholar]
- 15.Kennedy EP, Weiss SB. The function of cytidine coenzymes in the biosynthesis of phospholipides. J. Biol. Chem. 1956;222:193–214. [PubMed] [Google Scholar]
- 16.Cuadrado A, Carnero A, Dolfi F, Jimenez B, Lacal JC. Phosphorylcholine: a novel second messenger essential for mitogenic activity of growth factors. Oncogene. 1993;8:2959–2968. [PubMed] [Google Scholar]
- 17.Price BD, Morris JD, Marshall CJ, Hall A. Stimulation of phosphatidylcholine hydrolysis, diacylglycerol release, and arachidonic acid production by oncogenic ras is a consequence of protein kinase C activation. J. Biol. Chem. 1989;264:16638–16643. [PubMed] [Google Scholar]
- 18.Besterman JM, Duronio V, Cuatrecasas P. Rapid formation of diacylglycerol from phosphatidylcholine: a pathway for generation of a second messenger. Proc. Natl Acad. Sci. USA. 1986;83:6785–6789. doi: 10.1073/pnas.83.18.6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bogin L, Papa MZ, Polak-Charcon S, Degani H. TNF-induced modulations of phospholipid metabolism in human breast cancer cells. Biochim. Biophys. Acta. 1998;1392:217–232. doi: 10.1016/s0005-2760(98)00035-6. [DOI] [PubMed] [Google Scholar]
- 20.Katz-Brull R, Degani H. Kinetics of choline transport and phosphorylation in human breast cancer cells; NMR application of the zero trans method. Anticancer Res. 1996;16:1375–1380. [PubMed] [Google Scholar]
- 21.Okuda T, et al. Identification and characterization of the high-affinity choline transporter. Nature Neurosci. 2000;3:120–125. doi: 10.1038/72059. [DOI] [PubMed] [Google Scholar]
- 22.O’Regan S, et al. An electric lobe suppressor for a yeast choline transport mutation belongs to a new family of transporter-like proteins. Proc. Natl Acad. Sci. USA. 2000;97:1835–1840. doi: 10.1073/pnas.030339697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Traiffort E, Ruat M, O’Regan S, Meunier FM. Molecular characterization of the family of choline transporter-like proteins and their splice variants. J. Neurochem. 2005;92:1116–1125. doi: 10.1111/j.1471-4159.2004.02962.x. [DOI] [PubMed] [Google Scholar]
- 24.Yuan Z, Tie A, Tarnopolsky M, Bakovic M. Genomic organization, promoter activity, and expression of the human choline transporter-like protein 1. Physiol. Genomics. 2006;26:76–90. doi: 10.1152/physiolgenomics.00107.2005. [DOI] [PubMed] [Google Scholar]
- 25.Koepsell H, Schmitt BM, Gorboulev V. Organic cation transporters. Rev. Physiol. Biochem. Pharmacol. 2003;150:36–90. doi: 10.1007/s10254-003-0017-x. [DOI] [PubMed] [Google Scholar]
- 26.Wang T, et al. Choline transporters in human lung adenocarcinoma: expression and functional implications. Acta Biochim. Biophys. Sin (Shanghai) 2007;39:668–674. doi: 10.1111/j.1745-7270.2007.00323.x. [DOI] [PubMed] [Google Scholar]
- 27.Kouji H, et al. Molecular and functional characterization of choline transporter in human colon carcinoma HT-29 cells. Arch. Biochem. Biophys. 2009;483:90–98. doi: 10.1016/j.abb.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 28.Eliyahu G, Kreizman T, Degani H. Phosphocholine as a biomarker of breast cancer: molecular and biochemical studies. Int. J. Cancer. 2007;120:1721–1730. doi: 10.1002/ijc.22293. [DOI] [PubMed] [Google Scholar]
- 29.Hara T, Bansal A, DeGrado TR. Choline transporter as a novel target for molecular imaging of cancer. Mol. Imaging. 2006;5:498–509. [PubMed] [Google Scholar]
- 30.Wittenberg J, Kornberg A. Choline phosphokinase. J. Biol. Chem. 1953;202:431–444. [PubMed] [Google Scholar]
- 31.Hosaka K, Tanaka S, Nikawa J, Yamashita S. Cloning of a human choline kinase cDNA by complementation of the yeast cki mutation. FEBS Lett. 1992;304:229–232. doi: 10.1016/0014-5793(92)80625-q. [DOI] [PubMed] [Google Scholar]
- 32.Aoyama C, Liao H, Ishidate K. Structure and function of choline kinase isoforms in mammalian cells. Prog. Lipid Res. 2004;43:266–281. doi: 10.1016/j.plipres.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 33.Aoyama C, Ohtani A, Ishidate K. Expression and characterization of the active molecular forms of choline/ethanolamine kinase-α and -β in mouse tissues, including carbon tetrachloride-induced liver. Biochem. J. 2002;363:777–784. doi: 10.1042/0264-6021:3630777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gallego-Ortega D, et al. Differential role of human choline kinase α and β enzymes in lipid metabolism: implications in cancer onset and treatment. PLoS ONE. 2009;4:e7819. doi: 10.1371/journal.pone.0007819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sher RB, et al. A rostrocaudal muscular dystrophy caused by a defect in choline kinase β, the first enzyme in phosphatidylcholine biosynthesis. J. Biol. Chem. 2006;281:4938–4948. doi: 10.1074/jbc.M512578200. [DOI] [PubMed] [Google Scholar]
- 36.Wu G, Aoyama C, Young SG, Vance DE. Early embryonic lethality caused by disruption of the gene for choline kinase α, the first enzyme in phosphatidylcholine biosynthesis. J. Biol. Chem. 2008;283:1456–1462. doi: 10.1074/jbc.M708766200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wieprecht M, Wieder T, Geilen CC. N-[2-bromocin namyl(amino)ethyl]-5-isoquinolinesulphonamide (H-89) inhibits incorporation of choline into phosphatidylcholine via inhibition of choline kinase and has no effect on the phosphorylation of CTP:phosphocholine cytidylyltransferase. Biochem. J. 1994;297(Pt 1):241–247. doi: 10.1042/bj2970241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramirez de Molina A, et al. Overexpression of choline kinase is a frequent feature in human tumor-derived cell lines and in lung, prostate, and colorectal human cancers. Biochem. Biophys. Res. Commun. 2002;296:580–583. doi: 10.1016/s0006-291x(02)00920-8. [DOI] [PubMed] [Google Scholar]
- 39.Ramirez de Molina A, et al. Expression of choline kinase α to predict outcome in patients with early-stage non-small-cell lung cancer: a retrospective study. Lancet Oncol. 2007;8:889–897. doi: 10.1016/S1470-2045(07)70279-6. [DOI] [PubMed] [Google Scholar]
- 40.Nakagami K, et al. Increased choline kinase activity and elevated phosphocholine levels in human colon cancer. Jpn J. Cancer Res. 1999;90:419–424. doi: 10.1111/j.1349-7006.1999.tb00764.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramirez de Molina A, et al. Increased choline kinase activity in human breast carcinomas: clinical evidence for a potential novel antitumor strategy. Oncogene. 2002;21:4317–4322. doi: 10.1038/sj.onc.1205556. [DOI] [PubMed] [Google Scholar]
- 42.Gribbestad IS, Sitter B, Lundgren S, Krane J, Axelson D. Metabolite composition in breast tumors examined by proton nuclear magnetic resonance spectroscopy. Anticancer Res. 1999;19:1737–1746. [PubMed] [Google Scholar]
- 43.Borkenhagen LF, Kennedy EP. The enzymatic synthesis of cytidine diphosphate choline. J. Biol. Chem. 1957;227:951–962. [PubMed] [Google Scholar]
- 44.Kent C. CTP:phosphocholine cytidylyltransferase. Biochim. Biophys. Acta. 1997;1348:79–90. doi: 10.1016/s0005-2760(97)00112-4. [DOI] [PubMed] [Google Scholar]
- 45.Cornell RB, Northwood IC. Regulation of CTP:phosphocholine cytidylyltransferase by amphitropism and relocalization. Trends Biochem. Sci. 2000;25:441–447. doi: 10.1016/s0968-0004(00)01625-x. [DOI] [PubMed] [Google Scholar]
- 46.Karim M, Jackson P, Jackowski S. Gene structure, expression and identification of a new CTP:phosphocholine cytidylyltransferase β isoform. Biochim. Biophys. Acta. 2003;1633:1–12. doi: 10.1016/s1388-1981(03)00067-2. [DOI] [PubMed] [Google Scholar]
- 47.Jackowski S, Fagone P. CTP: Phosphocholine cytidylyltransferase: paving the way from gene to membrane. J. Biol. Chem. 2005;280:853–856. doi: 10.1074/jbc.R400031200. [DOI] [PubMed] [Google Scholar]
- 48.Tessitore L, Dianzani I, Cui Z, Vance DE. Diminished expression of phosphatidylethanolamine N-methyltransferase 2 during hepatocarcinogenesis. Biochem. J. 1999;337(Pt 1):23–27. [PMC free article] [PubMed] [Google Scholar]
- 49.Bell JD, Bhakoo KK. Metabolic changes underlying 31P MR spectral alterations in human hepatic tumours. NMR Biomed. 1998;11:354–359. doi: 10.1002/(sici)1099-1492(1998110)11:7<354::aid-nbm515>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 50.Dueck DA, et al. The modulation of choline phosphoglyceride metabolism in human colon cancer. Mol. Cell Biochem. 1996;162:97–103. doi: 10.1007/BF00227535. [DOI] [PubMed] [Google Scholar]
- 51.Hanahan DJ, Chaikoff IL. On the nature of the phosphorus-containing lipides of cabbage leaves and their relation to a phospholipide-splitting enzyme contained in these leaves. J. Biol. Chem. 1948;172:191–198. [PubMed] [Google Scholar]
- 52.Exton JH. Regulation of phospholipase D. FEBS Lett. 2002;531:58–61. doi: 10.1016/s0014-5793(02)03405-1. [DOI] [PubMed] [Google Scholar]
- 53.Hammond SM, et al. Characterization of two alternately spliced forms of phospholipase D1. Activation of the purified enzymes by phosphatidylinositol 4, 5-bisphosphate, ADP-ribosylation factor, and Rho family monomeric GTP-binding proteins and protein kinase C-α. J. Biol. Chem. 1997;272:3860–3868. doi: 10.1074/jbc.272.6.3860. [DOI] [PubMed] [Google Scholar]
- 54.Steed PM, Clark KL, Boyar WC, Lasala DJ. Characterization of human PLD2 and the analysis of PLD isoform splice variants. FASEB J. 1998;12:1309–1317. doi: 10.1096/fasebj.12.13.1309. [DOI] [PubMed] [Google Scholar]
- 55.Freyberg Z, Siddhanta A, Shields D. “Slip, sliding away”: phospholipase D and the Golgi apparatus. Trends Cell Biol. 2003;13:540–546. doi: 10.1016/j.tcb.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 56.Liscovitch M, Czarny M, Fiucci G, Lavie Y, Tang X. Localization and possible functions of phospholipase D isozymes. Biochim. Biophys. Acta. 1999;1439:245–263. doi: 10.1016/s1388-1981(99)00098-0. [DOI] [PubMed] [Google Scholar]
- 57. Buchanan FG, et al. Requirement of phospholipase D1 activity in H-RasV12-induced transformation. Proc. Natl Acad. Sci. USA. 2005;102:1638–1642. doi: 10.1073/pnas.0406698102. Rat-2 fibroblasts transfected with HRAS-G12V formed colonies in agar and tumours in mice. However, HRAS-G12V–transfected Rat-2 cells with reduced PLD1 activity did not, thus showing the requirement for normal PLD1 activity during transformation.
- 58.Uchida N, Okamura S, Kuwano H. Phospholipase D activity in human gastric carcinoma. Anticancer Res. 1999;19:671–675. [PubMed] [Google Scholar]
- 59. Iorio E, et al. Activation of phosphatidylcholine cycle enzymes in human epithelial ovarian cancer cells. Cancer Res. 2010;70:2126–2135. doi: 10.1158/0008-5472.CAN-09-3833. Epithelial ovarian cancer cells were compared with normal and non-tumour immortalized counterparts, demonstrating elevated PCho levels in tumour cells owing to upregulation or activation of CHKα and PC-PLC.
- 60.Uchida N, Okamura S, Nagamachi Y, Yamashita S. Increased phospholipase D activity in human breast cancer. J. Cancer Res. Clin. Oncol. 1997;123:280–285. doi: 10.1007/BF01208639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Noh DY, et al. Overexpression of phospholipase D1 in human breast cancer tissues. Cancer Lett. 2000;161:207–214. doi: 10.1016/s0304-3835(00)00612-1. [DOI] [PubMed] [Google Scholar]
- 62.Zhong M, et al. Phospholipase D prevents apoptosis in v-Src-transformed rat fibroblasts and MDA-MB-231 breast cancer cells. Biochem. Biophys. Res. Commun. 2003;302:615–619. doi: 10.1016/s0006-291x(03)00229-8. [DOI] [PubMed] [Google Scholar]
- 63.Oka M, et al. Protein kinase C α associates with phospholipase D1 and enhances basal phospholipase D activity in a protein phosphorylation-independent manner in human melanoma cells. J. Invest. Dermatol. 2003;121:69–76. doi: 10.1046/j.1523-1747.2003.12300.x. [DOI] [PubMed] [Google Scholar]
- 64.Oshimoto H, Okamura S, Yoshida M, Mori M. Increased activity and expression of phospholipase D2 in human colorectal cancer. Oncol. Res. 2003;14:31–37. doi: 10.3727/000000003108748586. [DOI] [PubMed] [Google Scholar]
- 65.Saito M, et al. Expression of phospholipase D2 in human colorectal carcinoma. Oncol. Rep. 2007;18:1329–1334. [PubMed] [Google Scholar]
- 66.Zhao Y, et al. Increased activity and intranuclear expression of phospholipase D2 in human renal cancer. Biochem. Biophys. Res. Commun. 2000;278:140–143. doi: 10.1006/bbrc.2000.3719. [DOI] [PubMed] [Google Scholar]
- 67.Yamada Y, et al. Association of a polymorphism of the phospholipase D2 gene with the prevalence of colorectal cancer. J. Mol. Med. 2003;81:126–131. doi: 10.1007/s00109-002-0411-x. [DOI] [PubMed] [Google Scholar]
- 68.Joseph T, et al. Transformation of cells overexpressing a tyrosine kinase by phospholipase D1 and D2. Biochem. Biophys. Res. Commun. 2001;289:1019–1024. doi: 10.1006/bbrc.2001.6118. [DOI] [PubMed] [Google Scholar]
- 69.Min DS, et al. Neoplastic transformation and tumorigenesis associated with overexpression of phospholipase D isozymes in cultured murine fibroblasts. Carcinogenesis. 2001;22:1641–1647. doi: 10.1093/carcin/22.10.1641. [DOI] [PubMed] [Google Scholar]
- 70.Fiucci G, et al. Changes in phospholipase D isoform activity and expression in multidrug-resistant human cancer cells. Int. J. Cancer. 2000;85:882–888. doi: 10.1002/(sici)1097-0215(20000315)85:6<882::aid-ijc24>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 71.Foster DA, Xu L. Phospholipase D in cell proliferation and cancer. Mol. Cancer Res. 2003;1:789–800. [PubMed] [Google Scholar]
- 72.Imamura F, et al. Induction of in vitro tumor cell invasion of cellular monolayers by lysophosphatidic acid or phospholipase D. Biochem. Biophys. Res. Commun. 1993;193:497–503. doi: 10.1006/bbrc.1993.1651. [DOI] [PubMed] [Google Scholar]
- 73.Pai JK, Frank EA, Blood C, Chu M. Novel ketoepoxides block phospholipase D activation and tumor cell invasion. Anticancer Drug Des. 1994;9:363–372. [PubMed] [Google Scholar]
- 74.Williger BT, Ho WT, Exton JH. Phospholipase D mediates matrix metalloproteinase-9 secretion in phorbol ester-stimulated human fibrosarcoma cells. J. Biol. Chem. 1999;274:735–738. doi: 10.1074/jbc.274.2.735. [DOI] [PubMed] [Google Scholar]
- 75.Aguirre-Ghiso JA, et al. RalA requirement for v-Src-and v-Ras-induced tumorigenicity and overproduction of urokinase-type plasminogen activator: involvement of metalloproteases. Oncogene. 1999;18:4718–4725. doi: 10.1038/sj.onc.1202850. [DOI] [PubMed] [Google Scholar]
- 76.Exton JH. Phosphatidylcholine breakdown and signal transduction. Biochim. Biophys. Acta. 1994;1212:26–42. doi: 10.1016/0005-2760(94)90186-4. [DOI] [PubMed] [Google Scholar]
- 77.Ramoni C, Spadaro F, Barletta B, Dupuis ML, Podo F. Phosphatidylcholine-specific phospholipase C in mitogen-stimulated fibroblasts. Exp. Cell Res. 2004;299:370–382. doi: 10.1016/j.yexcr.2004.05.037. [DOI] [PubMed] [Google Scholar]
- 78.Wu X, Lu H, Zhou L, Huang Y, Chen H. Changes of phosphatidylcholine-specific phospholipase C in hepatocarcinogenesis and in the proliferation and differentiation of rat liver cancer cells. Cell Biol. Int. 1997;21:375–381. doi: 10.1006/cbir.1997.0148. [DOI] [PubMed] [Google Scholar]
- 79.Bjorkoy G, Overvatn A, Diaz-Meco MT, Moscat J, Johansen T. Evidence for a bifurcation of the mitogenic signaling pathway activated by Ras and phosphatidylcholine-hydrolyzing phospholipase C. J. Biol. Chem. 1995;270:21299–21306. doi: 10.1074/jbc.270.36.21299. [DOI] [PubMed] [Google Scholar]
- 80.Spadaro F, et al. Phosphatidylcholine-specific phospholipase C activation in epithelial ovarian cancer cells. Cancer Res. 2008;68:6541–6549. doi: 10.1158/0008-5472.CAN-07-6763. [DOI] [PubMed] [Google Scholar]
- 81.Paris L, et al. Inhibition of phosphatidylcholine-specific phospholipase C downregulates HER2 overexpression on plasma membrane of breast cancer cells. Breast Cancer Res. 2010;12:R27. doi: 10.1186/bcr2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Glunde K, Jie C, Bhujwalla ZM. Molecular causes of the aberrant choline phospholipid metabolism in breast cancer. Cancer Res. 2004;64:4270–4276. doi: 10.1158/0008-5472.CAN-03-3829. [DOI] [PubMed] [Google Scholar]
- 83.Moestue SA, et al. Distinct choline metabolic profiles are associated with differences in gene expression for basal-like and luminal-like breast cancer xenograft models. BMC Cancer. 2010;10:433. doi: 10.1186/1471-2407-10-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wu BX, Clarke CJ, Hannun YA. Mammalian neutral sphingomyelinases: regulation and roles in cell signaling responses. Neuromolecular Med. 2010;12:320–330. doi: 10.1007/s12017-010-8120-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rodriguez-Gonzalez A, Ramirez de Molina A, Fernandez F, Lacal JC. Choline kinase inhibition induces the increase in ceramides resulting in a highly specific and selective cytotoxic antitumoral strategy as a potential mechanism of action. Oncogene. 2004;23:8247–8259. doi: 10.1038/sj.onc.1208045. [DOI] [PubMed] [Google Scholar]
- 86.Warden CH, Friedkin M, Geiger PJ. Acid-soluble precursors and derivatives of phospholipids increase after stimulation of quiescent Swiss 3T3 mouse fibroblasts with serum. Biochem. Biophys. Res. Commun. 1980;94:690–696. doi: 10.1016/0006-291x(80)91287-5. [DOI] [PubMed] [Google Scholar]
- 87.Warden CH, Friedkin M. Regulation of choline kinase activity and phosphatidylcholine biosynthesis by mitogenic growth factors in 3T3 fibroblasts. J. Biol. Chem. 1985;260:6006–6011. [PubMed] [Google Scholar]
- 88.Macara IG. Elevated phosphocholine concentration in ras-transformed NIH 3T3 cells arises from increased choline kinase activity, not from phosphatidylcholine breakdown. Mol. Cell Biol. 1989;9:325–328. doi: 10.1128/mcb.9.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lacal JC, Moscat J, Aaronson SA. Novel source of 1, 2-diacylglycerol elevated in cells transformed by Ha-ras oncogene. Nature. 1987;330:269–272. doi: 10.1038/330269a0. [DOI] [PubMed] [Google Scholar]
- 90. Ramirez de Molina A, Penalva V, Lucas L, Lacal JC. Regulation of choline kinase activity by Ras proteins involves Ral-GDS and PI3K. Oncogene. 2002;21:937–946. doi: 10.1038/sj.onc.1205144. NIH3T3 cells that overexpress RAS-G12V mutants that specifically interact either solely with RALGDS or solely with PI3K were used to show that CHK activation is mediated by signalling via RALGDS and PI3K.
- 91.Ramirez de Molina A, et al. Choline kinase is a novel oncogene that potentiates RhoA-induced carcinogenesis. Cancer Res. 2005;65:5647–5653. doi: 10.1158/0008-5472.CAN-04-4416. [DOI] [PubMed] [Google Scholar]
- 92.Bakovic M, Waite K, Vance DE. Oncogenic Ha-Ras transformation modulates the transcription of the CTP:phosphocholine cytidylyltransferase α gene via p42/44MAPK and transcription factor Sp3. J. Biol. Chem. 2003;278:14753–14761. doi: 10.1074/jbc.M300162200. [DOI] [PubMed] [Google Scholar]
- 93.Sugimoto H, Banchio C, Vance DE. Transcriptional regulation of phosphatidylcholine biosynthesis. Prog. Lipid Res. 2008;47:204–220. doi: 10.1016/j.plipres.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 94.Wieprecht M, Wieder T, Paul C, Geilen CC, Orfanos CE. Evidence for phosphorylation of CTP:phosphocholine cytidylyltransferase by multiple proline-directed protein kinases. J. Biol. Chem. 1996;271:9955–9961. doi: 10.1074/jbc.271.17.9955. [DOI] [PubMed] [Google Scholar]
- 95.Ryan AJ, Andrews M, Zhou J, Mallampalli RK. c-Jun N-terminal kinase regulates CTP:phosphocholine cytidylyltransferase. Arch. Biochem. Biophys. 2006;447:23–33. doi: 10.1016/j.abb.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 96.Teegarden D, Taparowsky EJ, Kent C. Altered phosphatidylcholine metabolism in C3H10T1/2 cells transfected with the Harvey-ras oncogene. J. Biol. Chem. 1990;265:6042–6047. [PubMed] [Google Scholar]
- 97.Carnero A, Cuadrado A, del Peso L, Lacal JC. Activation of type D phospholipase by serum stimulation and ras-induced transformation in NIH3T3 cells. Oncogene. 1994;9:1387–1395. [PubMed] [Google Scholar]
- 98.Lucas L, Penalva V, Ramirez de Molina A, Del Peso L, Lacal JC. Modulation of phospholipase D by Ras proteins mediated by its effectors Ral-GDS, PI3K and Raf-1. Int. J. Oncol. 2002;21:477–485. [PubMed] [Google Scholar]
- 99.Zhao C, Du G, Skowronek K, Frohman MA, Bar-Sagi D. Phospholipase D2-generated phosphatidic acid couples EGFR stimulation to Ras activation by Sos. Nature Cell Biol. 2007;9:706–712. doi: 10.1038/ncb1594. [DOI] [PubMed] [Google Scholar]
- 100.Kang DW, et al. Phospholipase D1 drives a positive feedback loop to reinforce the Wnt/β-catenin/TCF signaling axis. Cancer Res. 2010;70:4233–4242. doi: 10.1158/0008-5472.CAN-09-3470. [DOI] [PubMed] [Google Scholar]
- 101.Larrodera P, et al. Phospholipase C-mediated hydrolysis of phosphatidylcholine is an important step in PDGF-stimulated DNA synthesis. Cell. 1990;61:1113–1120. doi: 10.1016/0092-8674(90)90074-o. [DOI] [PubMed] [Google Scholar]
- 102.Plevin R, Cook SJ, Palmer S, Wakelam MJ. Multiple sources of sn-1, 2-diacylglycerol in platelet-derived-growth-factor-stimulated Swiss 3T3 fibroblasts. Evidence for activation of phosphoinositidase C and phosphatidylcholine-specific phospholipase D. Biochem. J. 1991;279(Pt 2):559–565. doi: 10.1042/bj2790559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Van Putten V, et al. Induction of cytosolic phospholipase A2 by oncogenic Ras is mediated through the JNK and ERK pathways in rat epithelial cells. J. Biol. Chem. 2001;276:1226–1232. doi: 10.1074/jbc.M003581200. [DOI] [PubMed] [Google Scholar]
- 104.van Rossum GS, Klooster R, van den Bosch H, Verkleij AJ, Boonstra J. Phosphorylation of p42/44(MAPK) by various signal transduction pathways activates cytosolic phospholipase A(2) to variable degrees. J. Biol. Chem. 2001;276:28976–28983. doi: 10.1074/jbc.M101361200. [DOI] [PubMed] [Google Scholar]
- 105.Yoo MH, et al. Role of the cytosolic phospholipase A2-linked cascade in signaling by an oncogenic, constitutively active Ha-Ras isoform. J. Biol. Chem. 2001;276:24645–24653. doi: 10.1074/jbc.M101975200. [DOI] [PubMed] [Google Scholar]
- 106.Ramirez de Molina A, et al. Choline kinase as a link connecting phospholipid metabolism and cell cycle regulation: implications in cancer therapy. Int. J. Biochem. Cell Biol. 2008;40:1753–1763. doi: 10.1016/j.biocel.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 107. Yalcin A, et al. Selective inhibition of choline kinase simultaneously attenuates MAPK and PI3K/AKT signaling. Oncogene. 2009;29:139–149. doi: 10.1038/onc.2009.317. CHK expression was silenced by siRNA in transformed HeLa cells. This reduced PCho levels, as well as signalling through the MAPK and PI3K–AKT pathways, which was associated with a decrease in the growth of cells in soft agar and in athymic mice.
- 108.Chua BT, et al. Regulation of Akt(ser473) phosphorylation by choline kinase in breast carcinoma cells. Mol. Cancer. 2009;8:131. doi: 10.1186/1476-4598-8-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Miyake T, Parsons SJ. Functional interactions between choline kinase α, epidermal growth factor receptor (EGFR), and c-Src in breast cancer cell proliferation. Oncogene. 2011 Aug;8 doi: 10.1038/onc.2011.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Aoyama C, Ishidate K, Sugimoto H, Vance DE. Induction of choline kinase α by carbon tetrachloride (CCl4) occurs via increased binding of c-jun to an AP-1 element. Biochim. Biophys. Acta. 2007;1771:1148–1155. doi: 10.1016/j.bbalip.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 111.Morrish F, Isern N, Sadilek M, Jeffrey M, Hockenbery DM. c-Myc activates multiple metabolic networks to generate substrates for cell-cycle entry. Oncogene. 2009;28:2485–2491. doi: 10.1038/onc.2009.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Banchio C, Lingrell S, Vance DE. Sp-1 binds promoter elements that are regulated by retinoblastoma and regulate CTP:phosphocholine cytidylyltransferase-α transcription. J. Biol. Chem. 2007;282:14827–14835. doi: 10.1074/jbc.M700527200. [DOI] [PubMed] [Google Scholar]
- 113.Gao S, et al. Mutated ras induced PLD1 gene expression through increased Sp1 transcription factor. Nagoya J. Med. Sci. 2009;71:127–136. [PMC free article] [PubMed] [Google Scholar]
- 114.Katz-Brull R, Lavin PT, Lenkinski RE. Clinical utility of proton magnetic resonance spectroscopy in characterizing breast lesions. J. Natl Cancer Inst. 2002;94:1197–1203. doi: 10.1093/jnci/94.16.1197. [DOI] [PubMed] [Google Scholar]
- 115.Horska A, Barker PB. Imaging of brain tumors: MR spectroscopy and metabolic imaging. Neuroimaging Clin. N. Am. 2010;20:293–310. doi: 10.1016/j.nic.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kurhanewicz J, Swanson MG, Nelson SJ, Vigneron DB. Combined magnetic resonance imaging and spectroscopic imaging approach to molecular imaging of prostate cancer. J. Magn. Reson. Imaging. 2002;16:451–463. doi: 10.1002/jmri.10172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Scheenen TW, et al. Discriminating cancer from noncancer tissue in the prostate by 3-dimensional proton magnetic resonance spectroscopic imaging: a prospective multicenter validation study. Invest. Radiol. 2011;46:25–33. doi: 10.1097/rli.0b013e3181f54081. [DOI] [PubMed] [Google Scholar]
- 118.Chen JH, et al. Clinical characteristics and biomarkers of breast cancer associated with choline concentration measured by (1) H MRS. NMR Biomed. 2011;24:316–324. doi: 10.1002/nbm.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.McKnight TR, et al. Correlation of magnetic resonance spectroscopic and growth characteristics within Grades II and III gliomas. J. Neurosurg. 2007;106:660–666. doi: 10.3171/jns.2007.106.4.660. [DOI] [PubMed] [Google Scholar]
- 120.Zeng Q, Liu H, Zhang K, Li C, Zhou G. Noninvasive evaluation of cerebral glioma grade by using multivoxel 3D proton MR spectroscopy. Magn. Reson. Imaging. 2011;29:25–31. doi: 10.1016/j.mri.2010.07.017. [DOI] [PubMed] [Google Scholar]
- 121.Chuang CF, et al. Potential value of MR spectroscopic imaging for the radiosurgical management of patients with recurrent high-grade gliomas. Technol. Cancer Res. Treat. 2007;6:375–382. doi: 10.1177/153303460700600502. [DOI] [PubMed] [Google Scholar]
- 122.Srinivasan R, et al. Ex vivo MR spectroscopic measure differentiates tumor from treatment effects in GBM. Neuro Oncol. 2010;12:1152–1161. doi: 10.1093/neuonc/noq075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lin AP, Tran TT, Ross BD. Impact of evidence-based medicine on magnetic resonance spectroscopy. NMR Biomed. 2006;19:476–483. doi: 10.1002/nbm.1058. [DOI] [PubMed] [Google Scholar]
- 124.Gabellieri C, et al. Therapeutic target metabolism observed using hyperpolarized 15N choline. J. Am. Chem. Soc. 2008;130:4598–4599. doi: 10.1021/ja8001293. [DOI] [PubMed] [Google Scholar]
- 125.Cudalbu C, et al. Feasibility of in vivo 15N MRS detection of hyperpolarized 15N labeled choline in rats. Phys. Chem. Chem. Phys. 2010;12:5818–5823. doi: 10.1039/c002309b. [DOI] [PubMed] [Google Scholar]
- 126.Allouche-Arnon H, et al. A hyperpolarized choline molecular probe for monitoring acetylcholine synthesis. Contrast Media Mol. Imaging. 2010;6:139–147. doi: 10.1002/cmmi.418. [DOI] [PubMed] [Google Scholar]
- 127.Rice SL, Roney CA, Daumar P, Lewis J. The next generation of positron emission tomography radiopharmaceuticals in oncology. Seminars in Nuclear Medicine. 2011;41:265–282. doi: 10.1053/j.semnuclmed.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Morse DL, et al. Characterization of breast cancers and therapy response by MRS and quantitative gene expression profiling in the choline pathway. NMR Biomed. 2009;22:114–127. doi: 10.1002/nbm.1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Baek HM, et al. Predicting pathologic response to neoadjuvant chemotherapy in breast cancer by using MR imaging and quantitative 1H MR spectroscopy. Radiology. 2009;251:653–662. doi: 10.1148/radiol.2512080553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Danishad KK, et al. Assessment of therapeutic response of locally advanced breast cancer (LABC) patients undergoing neoadjuvant chemotherapy (NACT) monitored using sequential magnetic resonance spectroscopic imaging (MRSI) NMR Biomed. 2010;23:233–241. doi: 10.1002/nbm.1436. [DOI] [PubMed] [Google Scholar]
- 131.Ronen SM, Jackson LE, Beloueche M, Leach MO. Magnetic resonance detects changes in phosphocholine associated with Ras activation and inhibition in NIH 3T3 cells. Br. J. Cancer. 2001;84:691–696. doi: 10.1054/bjoc.2000.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Al-Saffar NM, et al. The phosphoinositide 3-kinase inhibitor PI-103 downregulates choline kinaseα leading to phosphocholine and total choline decrease detected by magnetic resonance spectroscopy. Cancer Res. 2010;70:5507–5517. doi: 10.1158/0008-5472.CAN-09-4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Beloueche-Babari M, et al. Identification of magnetic resonance detectable metabolic changes associated with inhibition of phosphoinositide 3-kinase signaling in human breast cancer cells. Mol. Cancer Ther. 2006;5:187–196. doi: 10.1158/1535-7163.MCT-03-0220. [DOI] [PubMed] [Google Scholar]
- 134.Beloueche-Babari M, et al. Magnetic resonance spectroscopy monitoring of mitogen-activated protein kinase signaling inhibition. Cancer Res. 2005;65:3356–3363. doi: 10.1158/10.1158/0008-5472.CAN-03-2981. [DOI] [PubMed] [Google Scholar]
- 135.Koul D, et al. Cellular and in vivo activity of a novel PI3K inhibitor, PX-866, against human glioblastoma. Neuro Oncol. 2010;12:559–569. doi: 10.1093/neuonc/nop058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Buganim Y, et al. p53 Regulates the Ras circuit to inhibit the expression of a cancer-related gene signature by various molecular pathways. Cancer Res. 2010;70:2274–2284. doi: 10.1158/0008-5472.CAN-09-2661. [DOI] [PubMed] [Google Scholar]
- 137.Mori N, et al. Loss of p53 function in colon cancer cells results in increased phosphocholine and total choline. Mol. Imaging. 2004;3:319–323. doi: 10.1162/15353500200404121. [DOI] [PubMed] [Google Scholar]
- 138.Jordan BF, et al. Metabolite changes in HT-29 xenograft tumors following HIF-1α inhibition with PX-478 as studied by MR spectroscopy in vivo and ex vivo . NMR Biomed. 2005;18:430–439. doi: 10.1002/nbm.977. [DOI] [PubMed] [Google Scholar]
- 139.Griffiths JR, et al. Metabolic changes detected by in vivo magnetic resonance studies of HEPA-1 wild-type tumors and tumors deficient in hypoxia-inducible factor-1β (HIF-1β): evidence of an anabolic role for the HIF-1 pathway. Cancer Res. 2002;62:688–695. [PubMed] [Google Scholar]
- 140. Chung YL, et al. Magnetic resonance spectroscopic pharmacodynamic markers of the heat shock protein 90 inhibitor 17-allylamino, 17-demethoxygeldanamycin (17AAG) in human colon cancer models. J. Natl Cancer Inst. 2003;95:1624–1633. doi: 10.1093/jnci/djg084. Treatment of colorectal cancer cells and xenografts with the HSP90 inhibitor 17-AAG resulted in an increase in PCho demonstrating the complexity of factors affecting choline metabolism and the value of MRS for monitoring response to targeted therapies.
- 141.Sankaranarayanapillai M, et al. Detection of histone deacetylase inhibition by noninvasive magnetic resonance spectroscopy. Mol. Cancer Ther. 2006;5:1325–1334. doi: 10.1158/1535-7163.MCT-05-0494. [DOI] [PubMed] [Google Scholar]
- 142.Monazzam A, et al. Evaluation of the Hsp90 inhibitor NVP-AUY922 in multicellular tumour spheroids with respect to effects on growth and PET tracer uptake. Nucl. Med. Biol. 2009;36:335–342. doi: 10.1016/j.nucmedbio.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 143.Brandes AH, Ward CS, Ronen SM. 17-allyamino-17-demethoxygeldanamycin treatment results in a magnetic resonance spectroscopy-detectable elevation in choline-containing metabolites associated with increased expression of choline transporter SLC44A1 and phospholipase A2. Breast Cancer Res. 2010;12:R84. doi: 10.1186/bcr2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Beloueche-Babari M, et al. Modulation of melanoma cell phospholipid metabolism in response to heat shock protein 90 inhibition. Oncotarget. 2010;1:185–197. doi: 10.18632/oncotarget.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ross J, et al. Fatty acid synthase inhibition results in a magnetic resonance-detectable drop in phosphocholine. Mol. Cancer Ther. 2008;7:2556–2565. doi: 10.1158/1535-7163.MCT-08-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Glunde K, Jie C, Bhujwalla ZM. Mechanisms of indomethacin-induced alterations in the choline phospholipid metabolism of breast cancer cells. Neoplasia. 2006;8:758–771. doi: 10.1593/neo.06187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Milkevitch M, Beardsley NJ, Delikatny EJ. Phenylbutyrate induces apoptosis and lipid accumulations via a peroxisome proliferator-activated receptor γ-dependent pathway. NMR Biomed. 2010;23:473–479. doi: 10.1002/nbm.1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Glunde K, Raman V, Mori N, Bhujwalla ZM. RNA interference-mediated choline kinase suppression in breast cancer cells induces differentiation and reduces proliferation. Cancer Res. 2005;65:11034–11043. doi: 10.1158/0008-5472.CAN-05-1807. [DOI] [PubMed] [Google Scholar]
- 149.Al-Saffar NM, et al. Noninvasive magnetic resonance spectroscopic pharmacodynamic markers of the choline kinase inhibitor MN58b in human carcinoma models. Cancer Res. 2006;66:427–434. doi: 10.1158/0008-5472.CAN-05-1338. [DOI] [PubMed] [Google Scholar]
- 150.Krishnamachary B, et al. Noninvasive detection of lentiviral-mediated choline kinase targeting in a human breast cancer xenograft. Cancer Res. 2009;69:3464–3471. doi: 10.1158/0008-5472.CAN-08-4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lacal JC. Choline kinase: a novel target for antitumor drugs. IDrugs. 2001;4:419–426. [PubMed] [Google Scholar]
- 152.Hernandez-Alcoceba R, et al. Choline kinase inhibitors as a novel approach for antiproliferative drug design. Oncogene. 1997;15:2289–2301. doi: 10.1038/sj.onc.1201414. [DOI] [PubMed] [Google Scholar]
- 153.Rodriguez-Gonzalez A, et al. Inhibition of choline kinase as a specific cytotoxic strategy in oncogene-transformed cells. Oncogene. 2003;22:8803–8812. doi: 10.1038/sj.onc.1207062. [DOI] [PubMed] [Google Scholar]
- 154.Mori N, Glunde K, Takagi T, Raman V, Bhujwalla ZM. Choline kinase down-regulation increases the effect of 5-fluorouracil in breast cancer cells. Cancer Res. 2007;67:11284–11290. doi: 10.1158/0008-5472.CAN-07-2728. [DOI] [PubMed] [Google Scholar]
- 155.Nimmagadda S, Glunde K, Pomper MG, Bhujwalla ZM. Pharmacodynamic markers for choline kinase down-regulation in breast cancer cells. Neoplasia. 2009;11:477–484. doi: 10.1593/neo.81430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lockman PR, Allen DD. The transport of choline. Drug Dev. Ind. Pharm. 2002;28:749–771. doi: 10.1081/ddc-120005622. [DOI] [PubMed] [Google Scholar]
- 157.Clive S, Gardiner J, Leonard RC. Miltefosine as a topical treatment for cutaneous metastases in breast carcinoma. Cancer Chemother. Pharmacol. 1999;44(Suppl):S29–S30. doi: 10.1007/s002800051114. [DOI] [PubMed] [Google Scholar]
- 158.Geilen CC, Wieder T, Reutter W. Hexadecylphosphocholine inhibits translocation of CTP:choline-phosphate cytidylyltransferase in Madin-Darby canine kidney cells. J. Biol. Chem. 1992;267:6719–6724. [PubMed] [Google Scholar]
- 159.Jimenez-Lopez JM, Carrasco MP, Segovia JL, Marco C. Hexadecylphosphocholine inhibits phosphatidylcholine biosynthesis and the proliferation of HepG2 cells. Eur. J. Biochem. 2002;269:4649–4655. doi: 10.1046/j.1432-1033.2002.03169.x. [DOI] [PubMed] [Google Scholar]
- 160.Lucas L, Hernandez-Alcoceba R, Penalva V, Lacal JC. Modulation of phospholipase D by hexadecylphosphorylcholine: a putative novel mechanism for its antitumoral activity. Oncogene. 2001;20:1110–1117. doi: 10.1038/sj.onc.1204216. [DOI] [PubMed] [Google Scholar]
- 161.Eisen SF, Brown HA. Selective estrogen receptor (ER) modulators differentially regulate phospholipase D catalytic activity in ER-negative breast cancer cells. Mol. Pharmacol. 2002;62:911–920. doi: 10.1124/mol.62.4.911. [DOI] [PubMed] [Google Scholar]
- 162.Chen Y, Zheng Y, Foster DA. Phospholipase D confers rapamycin resistance in human breast cancer cells. Oncogene. 2003;22:3937–3942. doi: 10.1038/sj.onc.1206565. [DOI] [PubMed] [Google Scholar]
- 163.Lerner LJ, Jordan VC. Development of antiestrogens and their use in breast cancer: eighth Cain memorial award lecture. Cancer Res. 1990;50:4177–4189. [PubMed] [Google Scholar]
- 164.Couldwell WT, et al. Clinical and radiographic response in a minority of patients with recurrent malignant gliomas treated with high-dose tamoxifen. Neurosurgery. 1993;32:485–489. doi: 10.1227/00006123-199303000-00034. discussion 489–490. [DOI] [PubMed] [Google Scholar]
- 165.Del Prete SA, Maurer LH, O’Donnell J, Forcier RJ, LeMarbre P. Combination chemotherapy with cisplatin, carmustine, dacarbazine, and tamoxifen in metastatic melanoma. Cancer Treat. Rep. 1984;68:1403–1405. [PubMed] [Google Scholar]
- 166.Cabot MC, et al. Tamoxifen activates cellular phospholipase C and D and elicits protein kinase C translocation. Int. J. Cancer. 1997;70:567–574. doi: 10.1002/(sici)1097-0215(19970304)70:5<567::aid-ijc13>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 167.Amtmann E. The antiviral, antitumoural xanthate D609 is a competitive inhibitor of phosphatidylcholine-specific phospholipase C. Drugs Exp. Clin. Res. 1996;22:287–294. [PubMed] [Google Scholar]
- 168.Meng A, et al. Sphingomyelin synthase as a potential target for D609-induced apoptosis in U937 human monocytic leukemia cells. Exp. Cell Res. 2004;292:385–392. doi: 10.1016/j.yexcr.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 169.Amtmann E, Sauer G. Selective killing of tumor cells by xanthates. Cancer Lett. 1987;35:237–244. doi: 10.1016/0304-3835(87)90125-x. [DOI] [PubMed] [Google Scholar]
- 170.Schick HD, et al. Antitumoral activity of a xanthate compound. II. Therapeutic studies in murine leukemia and tumor models in vivo . Cancer Lett. 1989;46:149–152. doi: 10.1016/0304-3835(89)90023-2. [DOI] [PubMed] [Google Scholar]
- 171.Vogt A, Pestell KE, Day BW, Lazo JS, Wipf P. The antisignaling agent SC-α α δ 9, 4-(benzyl-(2-[(2, 5-diphenyloxazole-4-carbonyl)amino]ethyl)carbamoyl)-2-decanoylaminobutyric acid, is a structurally unique phospholipid analogue with phospholipase C inhibitory activity. Mol. Cancer Ther. 2002;1:885–892. [PubMed] [Google Scholar]
- 172.Amstalden van Hove ER, et al. Multimodal mass spectrometric imaging of small molecules reveals distinct spatio-molecular signatures in differentially metastatic breast tumor models. Cancer Res. 2010;70:9012–9021. doi: 10.1158/0008-5472.CAN-10-0360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Li C, et al. Nanoplex delivery of siRNA and prodrug enzyme for multimodality image-guided molecular pathway targeted cancer therapy. ACS Nano. 2010;4:6707–6716. doi: 10.1021/nn102187v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Katz-Brull R, Margalit R, Bendel P, Degani H. Choline metabolism in breast cancer; 2H-, 13C- and 31P–NMR studies of cells and tumors. MAGMA. 1998;6:44–52. doi: 10.1007/BF02662511. [DOI] [PubMed] [Google Scholar]
- 175.Glunde K, Bhujwalla ZM. Metabolic tumor imaging using magnetic resonance spectroscopy. Semin Oncol. 2011;38:26–41. doi: 10.1053/j.seminoncol.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Hernando E, et al. A critical role for choline kinase-α in the aggressiveness of bladder carcinomas. Oncogene. 2009;28:2425–2435. doi: 10.1038/onc.2009.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Usenius JP, Vainio P, Hernesniemi J, Kauppinen RA. Choline-containing compounds in human astrocytomas studied by 1H NMR spectroscopy in vivo and in vitro . J. Neurochem. 1994;63:1538–1543. doi: 10.1046/j.1471-4159.1994.63041538.x. [DOI] [PubMed] [Google Scholar]
- 178.Kinoshita Y, Yokota A. Absolute concentrations of metabolites in human brain tumors using in vitro proton magnetic resonance spectroscopy. NMR Biomed. 1997;10:2–12. doi: 10.1002/(sici)1099-1492(199701)10:1<2::aid-nbm442>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 179.Kuang Y, et al. Transport and metabolism of radiolabeled choline in hepatocellular carcinoma. Mol. Pharm. 2010;7:2077–2092. doi: 10.1021/mp1001922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Onodera K, et al. 31P nuclear magnetic resonance analysis of lung cancer: the perchloric acid extract spectrum. Jpn J. Cancer Res. 1986;77:1201–1206. [PubMed] [Google Scholar]
- 181.Rocha CM, et al. Metabolic profiling of human lung cancer tissue by 1H high resolution magic angle spinning (HRMAS) NMR spectroscopy. J. Proteome Res. 2010;9:319–332. doi: 10.1021/pr9006574. [DOI] [PubMed] [Google Scholar]
- 182.Swanson MG, et al. Quantification of choline- and ethanolamine-containing metabolites in human prostate tissues using 1H HR-MAS total correlation spectroscopy. Magn. Reson. Med. 2008;60:33–40. doi: 10.1002/mrm.21647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Navon G, Ogawa S, Shulman RG, Yamane T. 31P nuclear magnetic resonance studies of Ehrlich ascites tumor cells. Proc. Natl Acad. Sci. USA. 1977;74:87–91. doi: 10.1073/pnas.74.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Evanochko WT, et al. NMR study of in vivo RIF-1 tumors. Analysis of perchloric acid extracts and identification of 1H, 31P and 13C resonances. Biochim. Biophys. Acta. 1984;805:104–116. doi: 10.1016/0167-4889(84)90042-9. Extracts of RIF-1 tumours were analysed by multinuclear MRS and chromatographic methods, and it was shown that levels of PMEs that are elevated in cancer are comprised of PCho and PEtn.
- 185. Bruhn H, et al. Noninvasive differentiation of tumors with use of localized H-1 MR spectroscopy in vivo: initial experience in patients with cerebral tumors. Radiology. 1989;172:541–548. doi: 10.1148/radiology.172.2.2748837. 1H MRS was used to monitor brain tumours in vivo, demonstrating an elevated tCho signal and a drop in N -acetyl-aspartate compared with normal tissue.
- 186. Ramirez de Molina A, et al. Choline kinase activation is a critical requirement for the proliferation of primary human mammary epithelial cells and breast tumor progression. Cancer Res. 2004;64:6732–6739. doi: 10.1158/0008-5472.CAN-04-0489. Primary human mammary epithelial and breast tumour-derived cells were investigated, and it was shown that CHK activation is a crucial requirement for proliferation.