Abstract
The microbial community of spiders is little known, with previous studies focussing primarily on the medical importance of spiders as vectors of pathogenic bacteria and on the screening of known cytoplasmic endosymbiont bacteria. These screening studies have been performed by means of specific primers that only amplify a selective set of endosymbionts, hampering the detection of unreported species in spiders. In order to have a more complete overview of the bacterial species that can be present in spiders, we applied a combination of a cloning assay, DGGE profiling and high-throughput sequencing on multiple individuals of the dwarf spider Oedothorax gibbosus. This revealed a co-infection of at least three known (Wolbachia, Rickettsia and Cardinium) and the detection of a previously unreported endosymbiont bacterium (Rhabdochlamydia) in spiders. 16S rRNA gene sequences of Rhabdochlamydia matched closely with those of Candidatus R. porcellionis, which is currently only reported as a pathogen from a woodlouse and with Candidatus R. crassificans reported from a cockroach. Remarkably, this bacterium appears to present in very high proportions in one of the two populations only, with all investigated females being infected. We also recovered Acinetobacter in high abundance in one individual. In total, more than 99% of approximately 4.5M high-throughput sequencing reads were restricted to these five bacterial species. In contrast to previously reported screening studies of terrestrial arthropods, our results suggest that the bacterial communities in this spider species are dominated by, or even restricted to endosymbiont bacteria. Given the high prevalence of endosymbiont species in spiders, this bacterial community pattern could be widespread in the Araneae order.
Introduction
It has become clear that many arthropods harbour a wealth of symbiotic bacteria exerting a strong effect on host adaptation and, hence, host evolution [1–6]. Though recent advances in molecular methods have allowed for a comprehensive quantification of microbial communities in a diverse set of species (e.g. [7–9]), the majority of arthropod groups still remain largely unexplored. This is particularly the case for spiders, for which only little research on their microbial community has currently been performed.
The few studies on microbial assemblages associated with spiders concentrated on the medical importance of spiders as vectors of potentially human pathogenic bacteria. Here, emphasis has been put on the general internal and external bacterial community of the spider [10,11], human serum and blood after presumed spider bites [12] and the presence of pathogenic bacteria associated with venom and fangs [13,14].
On the other hand, microbial investigations in spiders focussed on infection by endosymbiont bacteria. These maternally inherited bacteria that reside obligatorily in the cells or intercellular lumen of their host may profoundly alter host reproduction by killing male offspring, feminizing genetic males and inducing parthenogenesis and cytoplasmic incompatibility [15–17]. One of the earliest accounts of endosymbiont infections in spiders were made by microscope studies that report Rickettsia and Chlamydia like bacteria [18–22]. However, electron microscopy is limited in the observation of bacterial species as the positive identification is based on phenotypic features. A more systematic approach through PCR based screening of endosymbionts, with particular emphasis on the endosymbionts Wolbachia, Rickettsia, Spiroplasma and Cardinium, showed that spiders may show a remarkable diversity and high prevalence of cytoplasmic bacteria [23–27]. Currently, up to six different bacterial taxa that potentially affect host reproduction have been detected so far (i.e. Wolbachia, Rickettsia, Spiroplasma ixodetis, Spiroplasma poulsonii, Cardinium and Arsenophonus [24,28]). In few cases, these endosymbionts appeared to have a pronounced effect on their spider hosts’ biology by manipulating host reproduction [28–30] as well through affecting dispersal behaviour [31].
Quantification of bacterial species in these studies are based on directed searches that target only a predefined set of putative reproductive manipulators. These methods are therefore less suitable to provide a more comprehensive view on the complete endosymbiont community present in many arthropod groups. However, with the advent of high-throughput sequencing techniques this can readily be achieved by employing a metagenomic approach.
In this paper we aim to determine the bacterial diversity in the dwarf spider Oedothorax gibbosus. Previous work showed that two populations of this spider are infected by at least three endosymbiont species, i.e. Wolbachia (belonging to clade G), Rickettsia and Cardinium. Of these, only Wolbachia was found to affect reproduction causing a female biased sex ratio by killing male embryos [28]. Remarkably, some females produced highly distorted female biased sex ratios in the absence of Wolbachia (unpublished results), suggesting that an additional factor influences sex ratio in this species. To fully understand the different agents potentially affecting sex ratio there is a clear need of a comprehensive identification of the microbial community.
We tackle this problem by first characterizing and identifying the bacterial community of this species using a combination of high-throughput sequencing, cloning and DGGE profiling of 16S rDNA amplicons. Second, we test the prevalence of newly identified bacteria in this species in multiple individuals of both sexes by means of species specific PCR screenings.
Material and Methods
Sample origin and DNA extraction
Individuals of Oedothorax gibbosus used in the molecular analysis were sampled by hand in two different populations in Belgium i.e. Damvallei (DAM: 51°03’25.60”N, 3°49’51.13”E)) and Walenbos (WAL: 50°55’32.49”N, 4°51’48.91”E, permissions for field collections: Walenbos: Belgian Nature and Forest Agency, Damvallei: Natuurpunt). Oedothorax gibbosus is not an endangered or protected species.
Although mostly field captured specimens were used for molecular analysis, we also included specimens that were bred from wild caught females from the WAL population for six consecutive generations (F6 generation). One of these lines, further referred to as Wol+, was previously shown to be infected with Wolbachia, Rickettsia and Cardinium and previously used to investigate the effect of Wolbachia infection on sex ratio (see [28]) (average sex ratio: 0.25 ± 0.04 (n = 797 offspring)). The second maternal line, further referred to as Wol-, was even more female biased and infected with Rickettsia and Cardinium, but not with Wolbachia (average sex ratio: 0.05 ± 0.02 (n = 611 offspring)).
DNA of whole individuals was extracted using the NucleoSpin Tissue DNA extraction kit (Macherey-Nagel) following the manufacturers recommended protocol.
Identification and characterization of the bacterial community
We first attempted to obtain a complete picture of the dominant bacterial species that can be found in Oedothorax gibbosus. To achieve this, we applied the following molecular methods on either pooled or individual samples.
Cloning of bacterial 16S rRNA
Two individual females of the Wol- maternal line and two individual females of the Wol+ maternal line were used for the cloning assay (Table 1). 16S rRNA gene sequences were amplified using the universal primers F45 and R1242 [32]. PCR conditions were as following: initial denaturation at 95°C for 2 min, followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, extension at 72°C for 90 s and a final extension at 72°C during 5 min. PCR amplicons were ligated into the pCR II-TOPO vector (Invitrogen) and transferred into TOP10 chemically competent cells (Invitrogen) according to the manufacturers recommended protocol. Cloned sequences were reamplified using the M13F and M13R primers (Invitrogen) and presence of the insert was checked by gel electrophoresis. Inserts with bands of expected size were sequenced using BigDye v.1.1 Terminator Sequencing mix and run on an ABI 3130 automated sequencer. Identification of the bacteria was based on BLAST searches against both the NCBI nucleotide collection and the NCBI 16S rRNA database (bacteria and archaea).
Table 1. Overview of the different individuals (D: Damvallei, W: Walenbos; Wol individuals originate from two Walenbos matrilines) used in the cloning assay (Cloning), V3 high-throughput sequencing (HTS-V3), V4 high-throughput sequencing (HTS-V4) and Denaturating Gradient Gel Electrophoresis (DGGE) and the different bacteria found in each assay (Ac = Acinetobacter, Ca = Cardinium, Ri = Rickettsia, Rh = Rhabdochlamydia, Wo = Wolbachia).
| Individual | Origin | Cloning | HTS-V3 | HTS-V4 | DGGE |
|---|---|---|---|---|---|
| Wol-.01 | lab | Ri, Ca, Rh | Ri, Ca, Rh | Ri | |
| Wol-.02 | lab | Ri, Rh | Ri | ||
| Wol-.03 | lab | Ri, Rh | |||
| Wol+.01 | lab | Wo, Ri, Ca, Rh | Wo, Ri, Ca, Rh | Wo, Ri | |
| Wol+.02 | lab | Wo, Ri | |||
| Wol+.03 | lab | Wo, Ri | Wo, Ri | ||
| Wol+.04 | lab | Wo, Ri | Wo, Ri | ||
| D160 | wild | Wo, Ri, Ca, Ac | Ri | ||
| D023 | wild | Wo, Ri | |||
| D043 | wild | Wo, Ri | |||
| D026 | wild | Ac | |||
| D100 | wild | Ri | |||
| D121 | wild | ||||
| D306 | wild | ||||
| D054 | wild | ||||
| D057 | wild | ||||
| D327 | wild | ||||
| W121 | wild | Wo, Ri, Ca, Rh | Ri | ||
| W102 | wild | Ri | |||
| W202 | wild | Wo, Ri | |||
| W220 | wild | Ri | |||
| W011 | wild | Ri | |||
| W162 | wild | ||||
| W216 | wild | ||||
| W312 | wild | ||||
| W186 | wild | ||||
| W201 | wild | ||||
| W208 | wild | Wo |
High-throughput sequencing
Two individual females, one of the Wol+ and one of the Wol- maternal line, and two pooled samples, one from WAL (n = 10 females) and one from DAM (n = 10 females) were used for high-throughput sequencing (Table 1). For the pooled samples, DNA of individual extracts was measured with a fluorometric method (Qubit, Invitrogen) and equimolar amounts of DNA were combined from each of the 10 individuals into a single sample. We targeted the 16S rRNA V4 region for all four samples using the primers F563 and R802 (RDP website: http://pyro.cme.msu.edu/pyro/help.jsp) and the V3 region with the primers F338 and R534 (V3; [33]) for the two individual samples only. Including both the V3 and V4 regions accounts for differences between samples based on differential primer specificity. Primers were indexed with a sample specific barcode and amplicons were paired-end sequenced for 100 cycles at 1/8 of a single Illumina HiSeq2000 lane (Baseclear NV, The Netherlands).
Raw reads were subjected to an initial screening and only high quality reads with correct barcode, correct primer sequence and no ambiguous base pairs were retained for further downstream analysis (1.898.581 and 3.773.370 paired-end reads for V3 and V4 respectively).
We first combined both 100bp paired-end reads into a single sequence. For the V3 region, which spans <200bp, overlap is expected between the complementary read ends and we merged the overlapping reads with FLASH [34]. For the V4 region, which spans a region of approximately 238bp, both reads were merged while inserting a series of 38N’s between both read ends by means of a home-made Python script “ConcatenateNonoverlaps.py” (S1 Information). An initial screening of the V3 data showed considerable contamination with the 18S rRNA of the spider. We identified and removed these contaminant sequences with the standalone version of DeconSeq [35] by retaining sequences with hits to Bacterial (2,206 unique genomes and 73,337 whole genome sequences) and Archaeal (155 unique genomes) genomes and removing reads with hits to the 18S rRNA sequence of Oedothorax gibbosus. A total of 994,207 V3 sequences were retained.
We used MOTHUR v.1.26.0 [36] for further downstream analysis, data clean-up and data reduction. First, replicate sequences were eliminated to reduce the entire dataset. Remaining sequences were then aligned to the SILVA bacterial reference alignment (downloaded from http://www.mothur.org/wiki/Silva_reference_files, accessed 29/06/2013) using the Needleman alignment method and a k-mer size = 8. Sequences that did not align within the expected range were removed. The remaining sequences were preclustered into groups of sequences with maximal 2bp differences. Next, we identified chimeric sequences with ChimeraSlayer as well as UChime [37] as implemented in MOTHUR, using both the SILVA reference alignment as well as the original sequences (taking into account their frequency) as a template. Chimeric sequences constituted 132 (0.8%; V3) and 36,252 (27.2%; V4) of the preclustered sequences. The reduced dataset consisted of 18,863 (V3) and 96,789 (V4) unique sequences, representing 887,125 (V3) and 3,645,675 (V4) of the original sequences.
After data preprocessing, a phylotype analysis was conducted wherein sequences were first assigned a taxonomy and subsequently clustered according to their taxonomic identity. Taxonomic assignment was based on the Wang method [38] using a k-mer size = 8 and taxonomic classification was based on the Greengenes taxonomy as this database contained most detailed taxonomic classification of the endosymbiont species obtained from cloning analysis. We further performed taxonomic assignment of each cluster by comparing the most abundant sequence from each cluster against both the type and non-type and cultured and uncultured strain bacterial database using the Ribosomal Database Project (RDP; http://rdp.cme.msu.edu/, accessed 02/07/2013)[39] (See S2 and S3 Informations for the V3 region, and S4 and S5 Informations for the V4 region). A detailed description of the bioinformatics steps and MOTHUR commands to perform the analyses are given in S6 Information. Assembled reads were submitted to GenBank and accessible as BioSamples SAMN02903876 (Wol-V3); SAMN02903877 (Wol+_V3); SAMN02903878 (Wol-V4); SAMN02903879 (Wol+_V4); SAMN02903880 (WAL) and SAMN02903881 (DAM).
Low number of reads were observed corresponding to Wolbachia in the Wol- matriline and Rhabdochlamydia in the DAM pooled samples for the V4 region. These most likely constitute contamination due to the very low number of reads (less than 0.00002% for Wolbachia and 0.00006% for Rhabdochlamydia) and the absence of a PCR product for these endosymbionts when performed on these samples.
Denaturing gel gradient electrophoresis (DGGE)
Six and five females originating from the WAL and DAM population respectively were included in the DGGE assay (Table 1). With the exception of one female from the WAL population, these individuals were also included in the pooled samples of the high-throughput sequencing assay. Besides these wild-caught females, two females of the Wol- and four females of the Wol+ matriline were also included in the DGGE analysis. Six DNA amplicons (gene library) from the Wol- and Wol+ matriline were included as a reference.
16S rRNA gene sequences were obtained using the universal primers F45 and R1242 [32]. Next, a nested PCR was performed with use of the primers F357 (5′-CCTACGGGAGGCAGCAG-3′) and R518 (5′-ATTACCGCGGCTGCTGG-3′) which amplify the highly variable V3 region of the 16S rRNA gene [40]. A GC-clamp was added to the forward primer to ensure clear DGGE separation [41,42]. PCR conditions were as follows: initial denaturation at 95°C for 1 min, followed by 30 cycles of denaturation at 95°C for 30 s, annealing (55°C for 45 s), extension (72°C, for 60 s) and a final extension at 72°C during 7 min. DGGE analysis was carried out on PCR products using a DCode Universal Mutation Detection System device (Bio-Rad) [as described in 40,43]. Electrophoresis was performed using 35%-70% denaturating gradient polyacrylamide gels at 70 V in 1x TAE buffer at 60°C for 16.5 hours. This allows discrimination of the different amplified 16S rRNA sequences based on nucleotide composition. Gels were placed in the staining solution SYBR gold (Molecular Probes, Invitrogen) for 30 min after which the gel was visualized and photographed with the Molecular Imager Gel Doc XR System. Selected bands were excised by inserting a pipette tip (1 μl) into the gel and incubated overnight at 4°C in TE buffer. A PCR with the primers F357 and R518 [32] was performed on the DNA solution and PCR amplicons were sequenced as described above. A BLAST search was performed to obtain the best match in the nucleotide collection database.
PCR screening and phylogeny of endosymbionts
As the high throughput sequencing revealed the presence of Rhabdochlamydia, the prevalence of this endosymbiont was further investigated using PCR screening of 19 females and 38 males from the WAL population and 14 females and 14 males from the DAM population. We also included Arsenophonus in the PCR screening as this is the only reproductive endosymbiont that was not included in the previous study [28] (n = 14 and n = 27 for Damvallei and Walenbos females respectively). Based on the 16S rRNA sequence obtained from the cloning assay, we designed the primers Rhab16S-F1 5’-CGA GCC TGG GTA AGG TTC TTC-3’and Rhab16S-R1 5’-CTA TCA AAG TGG GGG CCC TTG-3’ to selectively amplify a part of the 16S rRNA gene of Rhabdochlamydia. PCR conditions were as follows: initial denaturation at 95°C for 2 min, followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 52°C for 30 s, extension at 72°C for 90 s and a final extension at 72°C during 5 min.
We also tested for the presence of Arsenophonus using the primers and protocols reported in [24]. Electrophoresis was performed on a 1,5% agarose gel. Gels were stained in a solution of GELRED for approximately 15 min and bands were visualized by UV-fluorescence. Ten PCR products were sequenced using BigDye v.1.1 Terminator Sequencing mix and run on an ABI 3130 automated sequencer to check for primer specificity. To investigate the presence of multiple strains, sequences were aligned using the Muscle algorithm [44] implemented in MEGA5 [45] and checked for the presence of single nucleotide polymorphisms.
Phylogenetic position of the Rhabdochlamydia 16S gene sequence was compared to other Chlamydiae sequences [reported in 46]. A p-distance based Neighbour joining tree was constructed as implemented in MEGA 6 [47]. Bootstrap percentage support was calculated for the nodes by generating 10000 bootstrap values.
Results
Cloning of 16S rRNA
A total of 82 clones were isolated from the two different females of the Wol- matriline and consisted of two groups of identical sequences. Sequences of the largest group (77 sequences) showed a close match with Rickettsia limoniae [Genbank:KF720712] (Table 2). Sequences obtained from the five remaining clones showed a very high similarity with Rhabdochlamydia porcellionis [KF720713] (Table 2). Sequences of the 19 clones of the two females of the Wol+ matriline could equally be grouped into two groups of identical sequences (Table 2). 13 appeared identical with our previously obtained 16S rRNA gene sequence of Wolbachia in Oedothorax gibbosus [GenBank:HQ286291], while sequences of the remaining six clones were identical with the Rickettsia sequence obtained from the Wol- matriline (Table 2).
Table 2. Top four BLAST hits against NCBI nucleotide collection and NCBI 16S rRNA databases for the three distinct sequences obtained from cloning of 16S rRNA from Oedothorax gibbosus.
| N Clones | NCBI nucleotide collection | NCBI 16S ribosomal RNA | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sequence | Wol- | Wol+ | Species [Accession number] | Score | E-value | Ident | Species [Accession number] | Score | E-value | Ident |
| KF720713 | 5 | 0 | Candidatus Rhabdochlamydia porcellionis [HF933203] | 1452 | 0 | 0,98 | Parachlamydia acanthamoebae [NR_026357] | 917 | 0 | 0,87 |
| Candidatus Rhabdochlamydia porcellionis [AY223862] | 1450 | 0 | 0,98 | Parachlamydia acanthamoebae [NR_074972] | 915 | 0 | 0,87 | |||
| Candidatus Rhabdochlamydia sp. [JF513056] | 1399 | 0 | 0,97 | Simkania negevensis [NR_074932] | 887 | 0 | 0,86 | |||
| Rhabdochlamydia crassificans [AY928092] | 1382 | 0 | 0,96 | Candidatus Protochlamydia amoebophila [NR_074271] | 878 | 0 | 0,88 | |||
| KF720712 | 77 | 6 | Rickettsia limoniae [AF322443] | 1480 | 0 | 0,99 | Rickettsia slovaca [NR_074474] | 1323 | 0 | 0,96 |
| Rickettsia limoniae [AF322442] | 1480 | 0 | 0,99 | Rickettsia slovaca [NR_074462] | 1323 | 0 | 0,96 | |||
| Rickettsia endosymbiont of Macrolophus sp. [HE583203] | 1474 | 0 | 0,99 | Rickettsia australis [NR_074496] | 1317 | 0 | 0,96 | |||
| Rickettsia endosymbiont of Asobara tabida [FJ603467] | 1474 | 0 | 0,99 | Rickettsia peacockii [NR_074488] | 1317 | 0 | 0,96 | |||
| HQ286291 | 0 | 13 | Wolbachia endosymbiont of Tetragnatha montana [EU333940] | 1450 | 0 | 1 | Wolbachia sp. wRi [NR_074437] | 1439 | 0 | 0,99 |
| Wolbachia endosymbiont of Oedemeronia lucidicollis [GU236934] | 1445 | 0 | 0,99 | Wolbachia endosymbiont of Brugia malayi [NR_074571] | 1387 | 0 | 0,98 | |||
| Wolbachia sp. Pin [GQ167635] | 1445 | 0 | 0,99 | Wolbachia endosymbiont of Culex quinquefasciatus [NR_074127] | 1378 | 0 | 0,98 | |||
| Wolbachia sp. Psq [GQ167634] | 1445 | 0 | 0,99 | Ehrlichia ruminatium [NR_074155] | 1038 | 0 | 0,9 | |||
High-throughput sequencing of the 16SrRNA V3 and V4 region
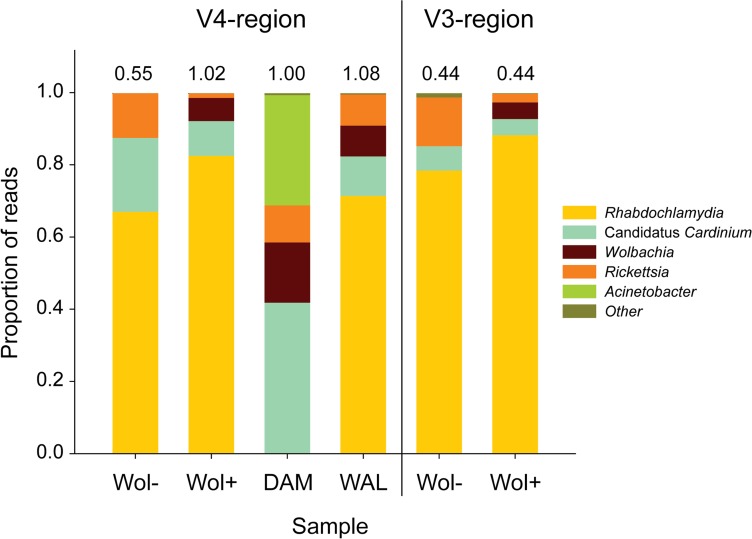
Phylotype analysis revealed that sequences matched to 210 (V3) and 327 (V4) different bacterial taxa. The distribution of the sequences among these different taxa was highly uneven and, with the exception of the pooled DAM sample, > 99% of the sequences were assigned to four bacterial taxa only. RDP based taxonomic classification of the most abundant sequences within each phylotype showed that 107 (V3) and 74 (V4) of these sequences showed similarity scores >80%. For all samples, the most frequent sequence was classified as Candidatus Rhabdochlamydia porcellionis (Fig. 1) and its sequence was identical to the 16S rRNA gene sequence obtained from cloning analysis. The other three bacteria that showed a high number of reads were classified as Wolbachia, Rickettsia and Candidatus Cardinium hertigii (Fig. 1). Also for Wolbachia and Rickettsia, representative sequences were identical to the sequences obtained from cloning. Only for the pooled DAM sample, an additional bacterium was found that represented 30% of the reads of this sample and closely matched with Acinetobacter.
Fig 1. Proportion of paired 100bp Illumina reads from the 16S rRNA-V4 and 16S rRNA-V3 region assigned to the different bacterial taxa in the dwarf spider Oedothorax gibbosus.
Wol+ and Wol- are individual females from maternal lines infected with and without Wolbachia respectively. DAM and WAL represent samples consisting of ten pooled wild caught females from population Damvallei and Walenbos respectively. Numbers above bars represent the number of reads in millions.
The remaining taxa that could be classified with relative high similarity scores were only found in very low frequencies and did not yield consistent results between the V3 and V4 region. For the V3 region, these were an unclassified Bacteriodetes (0.2%) and a Ruminococcus species (0.05%) and for the V4 region an unclassified member of the Enterobacteriaceae (0.09%) and an unclassified member of the order Saprospirales (0.017%). Despite the relatively low number of bacterial species associated with O. gibbosus, species composition was not similar between the two individuals and the two pooled population samples. For the two individual samples amplification of the V3 and V4 region produced consistent results (Fig. 1). As expected, high-throughput sequencing did not reveal any match with Wolbachia in the individual originating from the Wol- maternal line, confirming that Wolbachia is not the causative agent for the production of highly female biased clutches in this maternal line. This is in contrast with the Wol+ matriline where in concordance with previous results, Wolbachia was positively identified. Both lines showed infection with Rhabdochlamydia, Cardinium and Rickettsia. However, the Wol- individual showed a much higher proportion of Rickettsia relative to Cardinium and Rhabdochlamydia compared to the Wol+ line. The relative ratio of Rickettsia/Rhabdochlamydia and the ratio Rickettsia/Cardinium being 6.3 (V3) to 12.2 (V4) and 3.7 (V3) to 4.6 (V4) times higher in the Wol- compared to the Wol+ individual. As this increase was consistently observed in both datasets, this likely reflects a higher Rickettsia infection rate in the Wol- line.
Bacterial species composition was furthermore not similar between the two pooled population samples. While Rhabdochlamydia appears to be the predominant bacterial species present in the WAL individuals, none of the sequences in the DAM population were assigned to this or a related bacterial species. Conversely, Acinetobacter appeared to be only present in the pooled DAM sample. Cardinium, Wolbachia and Rickettsia were found to infect both populations.
Denaturing Gel Gradient Electrophoresis (DGGE)
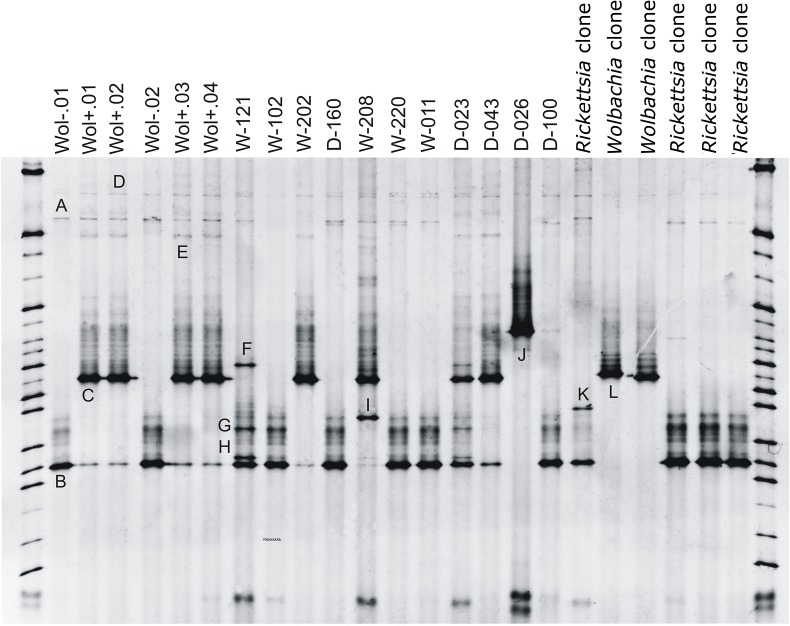
A total of 12 bands (A—L) were selected for sequencing (Fig. 2; Table 3) and revealed that these correspond to identical sequences of Wolbachia (C and L), Rickettsia (B and D) and Acinetobacter (J). Sequencing of bands A, E, F, G, H, I, and K were ambiguous. Several DNA amplicons from the cloning study were included in the DGGE as a reference. This resulted in one clear band in the DGGE profiles corresponding to Rickettsia and Wolbachia of the Wol- and Wol+ matrilines respectively.
Fig 2. DGGE profiles for the 16S rRNA amplicons of females originating from the Wol- and Wol+ matriline and females of the Damvallei (D) and Walenbos (W) population.
Clones indicate the use of DNA amplicons resulting from the cloning study. Bands B and D: Rickettsia endosymbiont; C and L: Wolbachia endosymbiont; J: Acinetobacter sp.; sequences from other bands were ambiguous.
Table 3. Taxonomic affiliation of the sequenced bands obtained by DGGE of Walenbos and Damvallei females.
PCR screening and phylogeny of endosymbionts
PCR screening revealed that Rhabdochlamydia detection was not restricted to the females used in the pooled sample for high-throughput sequencing, but that all females of the WAL population tested positive and 26% of the males (n = 19 and n = 38 for females and males respectively). Moreover, Rhabdochlamydia was not detected in the DAM population (n = 14 for both females and males). Primer specificity was subsequently confirmed by sequencing the obtained amplicons and yielded no interspecific variation.
PCR screening for Arsenophonus did not reveal any positives, indicating that this endosymbiont is absent in the two investigated populations (n = 14 and n = 27 for Damvallei and Walenbos females respectively).
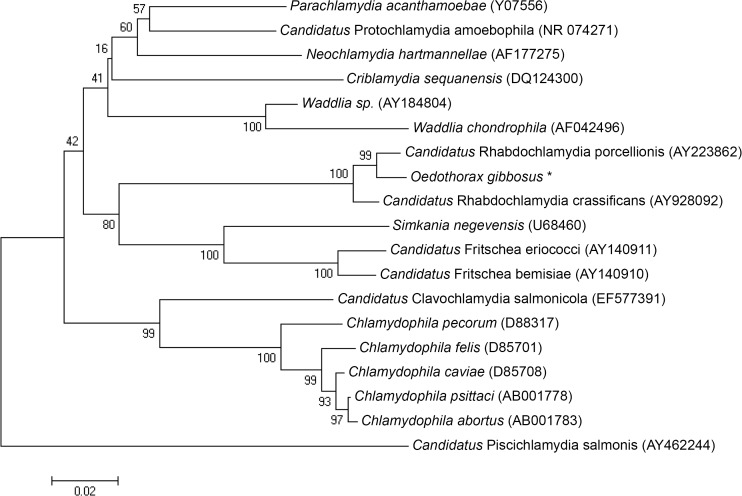
The Rhabdochlamydia 16S rRNA gene sequences showed high similarity with existing Rhabdochlamydia 16S rRNA gene sequences and were most closely related to Candidatus Rhabdochlamydia porcellionis (Genbank accession number: AY223862), followed by Candidatus Rhabdochlamydia crassificans (Genbank accession number: AY928092) (Fig. 3).
Fig 3. Phylogenetic position of the 16S rRNA gene sequence of Rhabdochlamydia of Oedothorax gibbosus.
A p-distance based Neighbour Joining tree was constructed as implemented in MEGA 6 [47] on a subset of Chlamydiae sequences available at Genbank. Percentage bootstrap support was generated for the nodes. Accession numbers are given between brackets, Rhabdochlamydia found in Oedothorax gibbosus is marked with an asterisk.
Discussion
Bacterial symbionts can have a vast diversity of relationships with their hosts and consequently have a high potential in affecting host ecology and evolution [1,3,48,49]. In Oedothorax gibbosus, where at least one endosymbiont (Wolbachia) causes a distorted sex ratio, we investigated the bacterial community in two different populations. This showed that these communities are dominated by four endosymbionts: Rhabdochlamydia, Cardinium, Wolbachia and Rickettsia. In all but one of the samples these four genera make up > 99% of the reads. Only for the DAM population, Acinetobacter sp. is found in approx. 30% of the reads with DGGE profiles showing that this is most likely due to detection of this bacterial species in a single individual. Screening of females showed that Rhabdochlamydia detection is limited to the WAL populations as all investigated females tested positive compared to none of the females in the DAM population.
Bacteria of the Chlamydiales are considered obligate intracellular parasites and are known to infect arthropods [50–52]. In scorpions, pathogenic effects are observed caused by a Porochlamydia infection in the hepatopancreas [53]. Although Chlamydia-like micro-organisms have been observed in spiders [22], this is the first report of Rhabdochlamydia as the genus has only recently been described [54,55]. Candidatus Rhabdochlamydia crassificans [55] infects cockroaches and causes pathogenic abdominal swelling of the host. The effects of Candidatus Rhabdochlamydia porcellionis [54] in the terrestrial isopod Porcellio scaber are currently not known. To date, no effects on host reproduction are documented. However, in the cockroach Blatta orientalis, Rhabdochlamydia was isolated from both fat body and ovary tissue [55]. The association with reproductive tissue could be a first indicator of potential reproductive effects. This is further suggested from our data by the difference in infection frequency between the sexes observed in the WAL population where the prevalence appeared to be much higher in females compared to males.
The NGS analysis of the pooled sample of the DAM population and a single DGGE profile revealed the presence of Acinetobacter sp. This bacterial genus is commonly found in environmental samples [56] and has been found to infect a variety of arthropods both internally as on the cuticle [57–61]. As whole spiders were used it is currently not known if Acinetobacter is located externally or internally.
Beside the Rhabdochlamydia detection pattern, both populations are similar in the frequencies of the other endosymbionts. Rickettsia and Cardinium are fixed in the two, both in males and females, while Wolbachia is found in approximately half of the male and female individuals [28, and this study] while Arsenophonus is absent in both populations. The cloning assay, DGGE profiles and high-throughput sequencing each support the absence of Wolbachia in the Wol- matriline. Still, this matriline is characterised by a highly distorted female biased sex ratio (5% males), suggesting that a second sex ratio distorting element, besides Wolbachia, is present in this species. No bacterial taxon was uniquely found in the Wol- matriline rendering it unlikely that a yet unidentified bacterial endosymbiont is responsible for the sex ratio trait. Given that this sex ratio trait is mainly inherited through daughters (Vanthournout & Hendrickx, unpublished), it is most likely that one of the endosymbionts (i.e. Rhabdochlamydia, Rickettsia or Cardinium) are responsible for the strong female bias. Of these, a higher ratio of Rickettsia was found in the Wol- matriline compared to the Wol+ matriline. This increase could reflect the absence of competition with Wolbachia and explain the manifestation of a sex ratio distortion if the penetrance of the endosymbiont is density dependent [62,63]. Alternatively, the presence of different strains of these endosymbionts in the Wol- matriline could account for a female bias in the absence of Wolbachia. As we targeted the conservative 16S rRNA gene, subtle genetic differences between strains that cause differences in the ability to manipulate sex ratios could be overlooked. Future investigations should explore the densities of the different endosymbionts in this matriline through real-time PCR and verify the presence of different strains through multi locus sequencing and 16S rRNA libraries. At last, it still remains possible that a non-bacterial species is responsible for this sex-ratio bias. Microsporidia, for example, show a high degree of vertical transmission and sex ratio manipulation of the host by these endosymbionts has been frequently reported, in particular for crustaceans such as amphipods [64]. Although both high-throughput sequencing, the cloning assay and DGGE profiles demonstrate the absence of Wolbachia in the Wol- matriline, striking differences in their detection of the other endosymbionts were observed. Illumina sequencing and the use of endosymbiont specific primers indicate that both matrilines are infected with Rickettsia, Cardinium and Rhabdochlamydia. The cloning assay failed to detect Cardinium in both maternal lines and Rhabdochlamydia in the Wol+ matriline. Similarly, DGGE profiles did not show infection with Cardinium and Rhabdochlamydia in any of the samples. This represents an underestimation of the bacterial diversity in the cloning assay and DGGE approach and illustrates the merit of high-throughput sequencing. One the one hand, our use of pooled samples demonstrates that also rare infections can be detected as was the case for Acinetobacter. However, this also illustrates that care should be taken with interpreting read frequencies of pooled samples as single infections may apparently result in a significant proportion of the reads (30%) from this species, suggesting a high prevalence of Acinetobacter in this population if not combined with individual approaches.
Notwithstanding these differences, all methods support the same conclusion: the bacterial community of this spider species appears to be almost completely restricted to endosymbiont bacteria. This contrasts with recent studies of arthropod bacterial communities where typically a large number of taxa is found of which the majority is not endosymbiotic. [7–9,65–68]. Given that whole specimens were used that did not receive any treatment for the removal of bacteria from the integument, this suggests that Oedothorax gibbosus is characterized by a unique bacterial community that consist almost solely of intracellular endosymbionts and an absence of gut microbiota. This is in line with the results of endosymbiont screening studies showing that a high number of spider species are infected [23–27]. However, in the medical studies, fangs and venom of spiders of the Loxosceles genus were surveyed for pathogenic members of Clostridium [13,14], while in Tegenaria agrestis [11] and in several species of human associated spiders [10] both the internal and external bacterial community was characterised. None of these studies detected an infection with endosymbiont bacteria. These results reflect most likely the limitations of the studies’ methodology as either a specific bacterium was targeted or a medium based rearing of bacteria was employed which excludes the possibility of finding non culturable endosymbiont bacteria.
It remains therefore most interesting to verify in multiple spider species to what extent their bacterial communities are dominated by endosymbiont bacteria.
Conclusions
In order to obtain a complete overview of the bacterial community in a dwarf spider species a next generation sequencing approach in combination with a cloning assay and DGGE profiles was used. We found that individuals can be co-infected with up to four endosymbiont bacteria, i.e. Wolbachia, Rickettsia, Cardinium and Rhabdochlamydia. In one sample a high abundance of Acinetobacter was detected. Remarkably, virtually no other bacterial species were detected in this spider species, which suggest that its bacterial community is dominated by endosymbiont bacteria. Given the high prevalence of endosymbiont infection in spiders, future screening studies need to determine the dominance of endosymbiont bacteria in spider bacterial assemblages.
Supporting Information
(DOCX)
OTU’s were determined by phylotype analysis wherein sequences were clustered according to their match with sequences in the SILVA database (as implemented in MOTHUR v 1.29.0) using the Greengenes taxonomic classification.
(PDF)
Taxonomic classification was based on comparison of the most abundant sequence from each cluster against both the type and non-type and cultured and uncultured strain bacterial database using the Ribosomal Database Project (RDP; http://rdp.cme.msu.edu/, accessed 02/07/2013)(Cole et al. 2009). Taxonomy is based on the NCBI nomenclature. OTU’s were determined by phylotype analysis wherein sequences were clustered according to their match with sequences in the SILVA database (as implemented in MOTHUR v 1.29.0) using the Greengenes taxonomic classification.
(PDF)
OTU’s were determined by phylotype analysis wherein sequences were clustered according to their match with sequences in the SILVA database (as implemented in MOTHUR v 1.29.0) using the Greengenes taxonomic classification.
(PDF)
Taxonomic classification was based on comparison of the most abundant sequence from each cluster against both the type and non-type and cultured and uncultured strain bacterial database using the Ribosomal Database Project (RDP; http://rdp.cme.msu.edu/, accessed 02/07/2013)(Cole et al. 2009). Taxonomy is based on the NCBI nomenclature. OTU’s were determined by phylotype analysis wherein sequences were clustered according to their match with sequences in the SILVA database (as implemented in MOTHUR v 1.29.0) using the Greengenes taxonomic classification.
(PDF)
(DOCX)
Acknowledgments
Illumina sequencing was performed at Baseclear NV, The Netherlands. Viki Vandomme assisted kindly in DNA extractions, PCR screening and cloning analysis. We would like to thank Joke Hollants for her most appreciated help in DGGE analysis.
Data Availability
Assembled reads were submitted to GenBank and accessible as BioSamples SAMN02903876 (Wol-V3); SAMN02903877 (Wol+_V3); SAMN02903878 (Wol-V4); SAMN02903879 (Wol+_V4); SAMN02903880 (WAL) and SAMN02903881 (DAM).
Funding Statement
Bram Vanthournout is holder of a PhD scholarship grant from the agency for Innovation by Science and Technology (IWT grant n° 73344). Additional financial support was received from the Belgian Science Policy (BELSPO, research project MO/36/O25). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Engelstädter J, Hurst GDD (2009) The ecology and evolution of microbes that manipulate host reproduction. Annu Rev Ecol Evol Syst 40: 127–149. [Google Scholar]
- 2. Moran NA, McCutcheon JP, Nakabachi A (2008) Genomics and Evolution of Heritable Bacterial Symbionts. Annual Review of Genetics 42: 165–190. 10.1146/annurev.genet.41.110306.130119 [DOI] [PubMed] [Google Scholar]
- 3. Charlat S, Hurst GDD, Merçot H (2003) Evolutionary consequences of Wolbachia infections. Trends in Genetics 19: 217–222. [DOI] [PubMed] [Google Scholar]
- 4. White JA (2011) Caught in the act: Rapid, symbiont-driven evolution. BioEssays 33: 823–829. 10.1002/bies.201100095 [DOI] [PubMed] [Google Scholar]
- 5. Saridaki A, Bourtzis K (2010) Wolbachia: more than just a bug in insects genitals. Current Opinion in Microbiology 13: 67–72. 10.1016/j.mib.2009.11.005 [DOI] [PubMed] [Google Scholar]
- 6. Serbus LR, Casper-Lindley C, Landmann F, Sullivan W (2008) The Genetics and Cell Biology of Wolbachia-Host Interactions. Annual Review of Genetics 42: 683–707. 10.1146/annurev.genet.41.110306.130354 [DOI] [PubMed] [Google Scholar]
- 7. Andreotti R, de Leon AAP, Dowd SE, Guerrero FD, Bendele KG, et al. (2011) Assessment of bacterial diversity in the cattle tick Rhipicephalus (Boophilus) microplus through tag-encoded pyrosequencing. BMC Microbiology 11:6 10.1186/1471-2180-11-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Palavesam A, Guerrero FD, Heekin AM, Wang J, Dowd SE, et al. (2012) Pyrosequencing-Based Analysis of the Microbiome Associated with the Horn Fly, Haematobia irritans . PLoS One 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kautz S, Rubin BER, Russell JA, Moreau CS (2013) Surveying the Microbiome of Ants: Comparing 454 Pyrosequencing with Traditional Methods To Uncover Bacterial Diversity. Applied and Environmental Microbiology 79: 525–534. 10.1128/AEM.03107-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Baxtrom C, Mongkolpradit T, Kasimos JN, Braune LM, Wise RD, et al. (2006) Common house spiders are not likely vectors of community-acquired methicillin-resistant Staphylococcus aureus infections. Journal of Medical Entomology 43: 962–965. [DOI] [PubMed] [Google Scholar]
- 11. Gaver-Wainwright MM, Zack RS, Foradori MJ, Lavine LC (2011) Misdiagnosis of Spider Bites: Bacterial Associates, Mechanical Pathogen Transfer, and Hemolytic Potential of Venom From the Hobo Spider, Tegenaria agrestis (Araneae: Agelenidae). Journal of Medical Entomology 48: 382–388. [DOI] [PubMed] [Google Scholar]
- 12. Mascarelli PE, Maggi RG, Hopkins S, Mozayeni BR, L. trull C, et al. (2013) Bartonella henselae infection in a family experiencing neurological and neurocognitive abnormalities after woodlouse hunter spider bites. Parasites and Vectors 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Silvestre FG, de Castro CS, de Moura JF, Giusta MS, De Maria M, et al. (2005) Characterization of the venom from the Brazilian Brown Spider Loxosceles similis Moenkhaus, 1898 (Araneae, Sicariidae). Toxicon 46: 927–936. [DOI] [PubMed] [Google Scholar]
- 14. Monteiro CLB, Rubel R, Cogo LL, Mangili OC, Gremski W, et al. (2002) Isolation and identification of Clostridium perfringens in the venom and fangs of Loxosceles intermedia (brown spider): enhancement of the dermonecrotic lesion in loxoscelism. Toxicon 40: 409–418. [DOI] [PubMed] [Google Scholar]
- 15. Engelstadter J, Hurst GDD (2007) The Impact of Male-Killing Bacteria on Host Evolutionary Processes. Genetics 175: 245–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hurst GDD, Jiggins FM (2000) Male-Killing Bacteria in Insects: Mechanisms, Incidence, and Implications. Emerging Infectious Diseases 6: 329–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stouthamer R, Breeuwer JAJ, Hurst DD (1999) Wolbachia pipientis: microbial manipulator of arthropod reproduction. Annu Rev Micribiol 53: 71–102. [DOI] [PubMed] [Google Scholar]
- 18. Cowdry EV (1923) The distribution of Rickettsia in the tissues of insects and arachnids. Journal of Experimental Medicine 37: 431–U178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Morel G (1978) 2 Chlamydiales (Rickettsias) in an arachnida—spider Pisaura mirabilis- CL. Experientia 34: 344–346. [Google Scholar]
- 20. Morel G (1977) Study of a Rickettsiella (Rickettsia) pathogen of spider Pisaura mirabilis Annales De Microbiologie A 128: 49–60. [PubMed] [Google Scholar]
- 21.Haupt J (2002) Fungal and rickettsial infections of some East Asian trapdoor spiders; Toft S, Scharff N, editors. 45–49 p.
- 22. Osaki H (1973) Electron microscopic observations of Chlamydia-like microorganism in hepatopancreas cells of the spider, Coras luctusosus . Acta Arachnol 25: 23–36. [Google Scholar]
- 23. Duron O, Hurst GDD, Hornett EA, Josling JA, Engelstädter J (2008) High incidence of the maternally inherited bacterium Cardinium in spiders. Molecular Ecology: 1–11. 10.1111/j.1365-294X.2007.03607.x [DOI] [PubMed] [Google Scholar]
- 24. Duron O, Bouchon D, Boutin S, Bellamy L, Zhou LQ, et al. (2008) The Diversity of Reproductive Parasites Among Arthropods: Wolbachia Do Not Walk Alone. BMC Biology 6:27 10.1186/1741-7007-6-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Goodacre SL, Martin OY, Thomas CFG, Hewitt GM (2006) Wolbachia and Other Endosymbiont Infections in Spiders. Molecular Ecology 15: 517–527. [DOI] [PubMed] [Google Scholar]
- 26. Martin OY, Goodacre SL (2009) Widespread Infections by the Bacterial Endosymbiont Cardinium in Arachnids. Journal of Arachnology 37: 106–108. [Google Scholar]
- 27. Rowley SM, Raven RJ, Mcgraw EA (2004) Wolbachia Pipientis in Australian Spiders. Current Microbiology 49: 208–214. [DOI] [PubMed] [Google Scholar]
- 28. Vanthournout B, Swaegers J, Hendrickx F (2011) Spiders do not escape reproductive manipulations by Wolbachia . BMC Evolutionary Biology 11:15 10.1186/1471-2148-11-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gunnarsson B, Goodacre SL, Hewitt GM (2009) Sex Ratio, Mating Behaviour and Wolbachia Infections in a Sheetweb Spider. Biological Journal of the Linnean Society 98: 181–186. [Google Scholar]
- 30. Vanthournout B, Vandomme V, Hendrickx F (2014) Sex ratio bias caused by endosymbiont infection in the dwarf spider Oedothorax retusus . Journal of Arachnology 42: 24–33. [Google Scholar]
- 31. Goodacre SL, Martin OY, Bonte D, Hutchings L, Woolley C, et al. (2009) Microbial modification of host long-distance dispersal capacity. BMC Biology 7:32 10.1186/1741-7007-7-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. O'neill SL, Giordano R, Colbert AME, Karr TL, Robertson HM (1992) 16s Ribosomal-Rna Phylogenetic Analysis of the Bacterial Endosymbionts Associated With Cytoplasmic Incompatibility in Insects. Proceedings of the National Academy of Sciences of the United States of America 89: 2699–2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dethlefsen L, Huse S, Sogin ML, Relman DA (2008) The Pervasive Effects of an Antibiotic on the Human Gut Microbiota, as Revealed by Deep 16S rRNA Sequencing. PLOS Biology 6: 2383–2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Magoc T, Salzberg SL (2011) FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27: 2957–2963. 10.1093/bioinformatics/btr507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schmieder R, Edwards R (2011) Fast Identification and Removal of Sequence Contamination from Genomic and Metagenomic Datasets. PLoS One 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, et al. (2009) Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Applied and Environmental Microbiology 75: 7537–7541. 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27: 2194–2200. 10.1093/bioinformatics/btr381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naïve Bayesian Classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Env Microbiol 73: 5261–5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cole JR, Wang QO, Cardenas E, Fish J, Chai B, et al. (2009) The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Research 37: 141–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Temmerman R, Scheirlinck I, Huys G, Swings J (2003) Culture-independent analysis of probiotic products by denaturing gradient gel electrophoresis. Applied and Environmental Microbiology 69: 220–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Myers RM, Fischer SG, Maniatis T, Lerman LS (1985) Modification of the melting properties of duplex DNA by attachment of a GC-rich DNA sequence as determined by denaturing gradient gel-electrophoresis. Nucleic Acids Research 13: 3111–3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sheffield VC, Cox DR, Lerman LS, Myers RM (1989) Attachment of a 40 base pair G+C-rich sequence (GC-clamp) to genomic DNA fragments by the polymerase chain reaction results in improved detection of single-base changes. Proceedings of the National Academy of Sciences of the United States of America 86: 232–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hollants J, Leliaert F, De Clerck O, Willems A (2010) How endo- is endo-? Surface sterilization of delicate samples: a Bryopsis (Bryopsidales, Chlorophyta) case study. Symbiosis 51: 131–138. [Google Scholar]
- 44. Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Molecular Biology and Evolution 28: 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Horn M (2008) Chlamydiae as Symbionts in Eukaryotes Annual Review of Microbiology. Palo Alto: Annual Reviews; pp. 113–131. 10.1146/annurev.micro.62.081307.162818 [DOI] [PubMed] [Google Scholar]
- 47. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Molecular Biology and Evolution 30: 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Dale C, Moran NA (2006) Molecular interactions between bacterial symbionts and their hosts. Cell 126: 453–465. [DOI] [PubMed] [Google Scholar]
- 49. Engel P, Moran NA (2013) The gut microbiota of insects—diversity in structure and function. Fems Microbiology Reviews 37: 699–735. 10.1111/1574-6976.12025 [DOI] [PubMed] [Google Scholar]
- 50. Corsaro D, Greub G (2006) Pathogenic potential of novel chlamydiae and diagnostic approaches to infections due to these obligate intracellular bacteria. Clinical Microbiology Reviews 19: 283–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Everett KDE, Thao ML, Horn M, Dyszynski GE, Baumann P (2005) Novel chlamydiae in whiteflies and scale insects: endosymbionts 'Candidatus Fritschea bemisiae' strain Falk and 'Candidatus Fritschea eriococci' strain Elm. International Journal of Systematic and Evolutionary Microbiology 55: 1581–1587. [DOI] [PubMed] [Google Scholar]
- 52. Thao ML, Baumann L, Hess JM, Falk BW, Ng JCK, et al. (2003) Phylogenetic evidence for two new insect-associated chlamydia of the family Simkaniaceae. Current Microbiology 47: 46–50. [DOI] [PubMed] [Google Scholar]
- 53. Morel G (1976) Studies on Porochlamydia-buthi g n, sp-n, an intracellular pathogen of scorpion Buthus occitanus . Journal of Invertebrate Pathology 28: 167–175. [Google Scholar]
- 54. Kostanjsek R, Strus J, Drobne D, Avgustin G (2004) 'Candidatus Rhabdochlamydia porcellionis', an intracellular bacterium from the hepatopancreas of the terrestrial isopod Porcellio scaber (Crustacea: Isopoda). International Journal of Systematic and Evolutionary Microbiology 54: 543–549. [DOI] [PubMed] [Google Scholar]
- 55. Corsaro D, Thomas V, Goy G, Venditti D, Radek R, et al. (2007) 'Candidatus Rhabdochlamydia crassificans', an intracellular bacterial pathogen of the cockroach Blatta orientalis (Insecta: Blattodea). Systematic and Applied Microbiology 30: 221–228. [DOI] [PubMed] [Google Scholar]
- 56.Towner KJ, Bergogne-Bérézin E, Fewson CA (1991) Acinetobacter: portrait of a genus. In: Towner KJ, Bergogne-Bérézin E, Fewson CA, editors. The Biology of Acinetobacter: Fems symposium no.57.
- 57. Murrell A, Dobson SJ, Yang XY, Lacey E, Barker SC (2003) A survey of bacterial diversity in ticks, lice and fleas from Australia. Parasitology Research 89: 326–334. [DOI] [PubMed] [Google Scholar]
- 58. Zouache K, Voronin D, Tran-Van V, Mousson L, Failloux AB, et al. (2009) Persistent Wolbachia and Cultivable Bacteria Infection in the Reproductive and Somatic Tissues of the Mosquito Vector Aedes albopictus . Plos One 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Montllor Curley C, Brodie EL, Lechner MG, Purcell AH (2007) Exploration for facultative endosymbionts of glassy-winged sharpshooter (Hemiptera: Cicadellidae). Annals of the Entomological Society of America 100: 345–349. [Google Scholar]
- 60. Akhoundi M, Bakhtiari R, Guillard T, Baghaei A, Tolouei R, et al. (2012) Diversity of the Bacterial and Fungal Microflora from the Midgut and Cuticle of Phlebotomine Sand Flies Collected in North-Western Iran. Plos One 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Geiger A, Fardeau ML, Grebaut P, Vatunga G, Josenando T, et al. (2009) First isolation of Enterobacter, Enterococcus, and Acinetobacter spp. as inhabitants of the tsetse fly (Glossina palpalis palpalis) midgut. Infection Genetics and Evolution 9: 1364–1370. 10.1016/j.meegid.2009.09.013 [DOI] [PubMed] [Google Scholar]
- 62. Hurst GDD, Johnson AP, von der Schulenburg JHG, Fuyama Y (2000) Male-killing Wolbachia in Drosophila: A temperature-sensitive trait with a threshold bacterial density. Genetics 156: 699–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Unckless RL, Boelio LM, Herren JK, Jaenike J (2009) Wolbachia as populations within individual insects: causes and consequences of density variation in natural populations. Proceedings of the Royal Society B-Biological Sciences 276: 2805–2811. 10.1098/rspb.2009.0287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Smith JE (2009) The ecology and evolution of microsporidian parasites. Parasitology 136: 1901–1914. 10.1017/S0031182009991818 [DOI] [PubMed] [Google Scholar]
- 65. Reid NM, Addison SL, Macdonald LJ, Lloyd-Jones G (2011) Biodiversity of Active and Inactive Bacteria in the Gut Flora of Wood-Feeding Huhu Beetle Larvae (Prionoplus reticularis). Applied and Environmental Microbiology 77: 7000–7006. 10.1128/AEM.05609-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hirsch J, Strohmeier S, Pfannkuchen M, Reineke A (2012) Assessment of bacterial endosymbiont diversity in Otiorhynchus spp. (Coleoptera: Curculionidae) larvae using a multitag 454 pyrosequencing approach. Bmc Microbiology 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Taylor M, Mediannikov O, Raoult D, Greub G (2012) Endosymbiotic bacteria associated with nematodes, ticks and amoebae. Fems Immunology and Medical Microbiology 64: 21–31. 10.1111/j.1574-695X.2011.00916.x [DOI] [PubMed] [Google Scholar]
- 68. Zheng LY, Crippen TL, Singh B, Tarone AM, Dowd S, et al. (2013) A Survey of Bacterial Diversity From Successive Life Stages of Black Soldier Fly (Diptera: Stratiomyidae) by Using 16S rDNA Pyrosequencing. Journal of Medical Entomology 50: 647–658. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
OTU’s were determined by phylotype analysis wherein sequences were clustered according to their match with sequences in the SILVA database (as implemented in MOTHUR v 1.29.0) using the Greengenes taxonomic classification.
(PDF)
Taxonomic classification was based on comparison of the most abundant sequence from each cluster against both the type and non-type and cultured and uncultured strain bacterial database using the Ribosomal Database Project (RDP; http://rdp.cme.msu.edu/, accessed 02/07/2013)(Cole et al. 2009). Taxonomy is based on the NCBI nomenclature. OTU’s were determined by phylotype analysis wherein sequences were clustered according to their match with sequences in the SILVA database (as implemented in MOTHUR v 1.29.0) using the Greengenes taxonomic classification.
(PDF)
OTU’s were determined by phylotype analysis wherein sequences were clustered according to their match with sequences in the SILVA database (as implemented in MOTHUR v 1.29.0) using the Greengenes taxonomic classification.
(PDF)
Taxonomic classification was based on comparison of the most abundant sequence from each cluster against both the type and non-type and cultured and uncultured strain bacterial database using the Ribosomal Database Project (RDP; http://rdp.cme.msu.edu/, accessed 02/07/2013)(Cole et al. 2009). Taxonomy is based on the NCBI nomenclature. OTU’s were determined by phylotype analysis wherein sequences were clustered according to their match with sequences in the SILVA database (as implemented in MOTHUR v 1.29.0) using the Greengenes taxonomic classification.
(PDF)
(DOCX)
Data Availability Statement
Assembled reads were submitted to GenBank and accessible as BioSamples SAMN02903876 (Wol-V3); SAMN02903877 (Wol+_V3); SAMN02903878 (Wol-V4); SAMN02903879 (Wol+_V4); SAMN02903880 (WAL) and SAMN02903881 (DAM).