Abstract
Clustered regularly interspaced short palindromic repeats (CRISPR) are polymorphic elements found in the genome of some or all strains of particular bacterial species, providing them with a system of acquired immunity against invading bacteriophages and plasmids. Two CRISPR-Cas systems have been identified in Acinetobacter baumannii, an opportunistic pathogen with a remarkable capacity for clonal dissemination. In this study, we investigated the mode of evolution and diversity of spacers of the CRISPR-cas subtype I-Fb locus in a global collection of 76 isolates of A. baumannii obtained from 14 countries and 4 continents. The locus has basically evolved from a common ancestor following two main lineages and several pathways of vertical descent. However, this vertical passage has been interrupted by occasional events of horizontal transfer of the whole locus between distinct isolates. The isolates were assigned into 40 CRISPR-based sequence types (CST). CST1 and CST23-24 comprised 18 and 9 isolates, representing two main sub-clones of international clones CC1 and CC25, respectively. Epidemiological data showed that some of the CST1 isolates were acquired or imported from Iraq, where it has probably been endemic for more than one decade and occasionally been able to spread to USA, Canada, and Europe. CST23-24 has shown a remarkable ability to cause national outbreaks of infections in Sweden, Argentina, UAE, and USA. The three isolates of CST19 were independently imported from Thailand to Sweden and Norway, raising a concern about the prevalence of CST19 in Thailand. Our study highlights the dynamic nature of the CRISPR-cas subtype I-Fb locus in A. baumannii, and demonstrates the possibility of using a CRISPR-based approach for subtyping a significant part of the global population of A. baumannii.
Introduction
Clustered regularly interspaced short palindromic repeats (CRISPR) are DNA elements present in the genome of 1176 out of 2612 fully sequenced bacterial strains (http://crispr.u-psud.fr/crispr/, last accessed December 2014). These elements together with neighboring genes, called cas for “CRISPR-associated”, provide bacteria with an adaptive immunity system against invading genetic elements, such as bacteriophages and plasmids [1]. The CRISPR-Cas immunity system functions in three steps: adaptation, expression, and interference [2]. Upon the entry of an invading element, the Cas machinery takes up a short sequence(s), a proto-spacer, from the invasive DNA and integrates it into the CRISPR array, where the adjacent direct repeat (DR) is duplicated and the integrated sequence becomes a new spacer. Next, the CRISPR array is transcribed and the produced RNA is cleaved and processed into small mature CRISPR RNAs (crRNA). Finally, the crRNAs guide the Cas nucleases for a sequence-specific cleavage of the invader. Flanking one side of the proto-spacers, a conserved short sequence, called PAM for Proto-spacer Adjacent Motif, has been identified [1]. PAMs, representing the recognition sites for the CRISPR-Cas machinery, play an important role in the adaptation and interference steps [3].
CRISPR—Cas systems are classified into three major types and various subtypes, based on the phylogenies of the conserved cas genes and the gene composition and architecture of the cas operons [4]. In addition, the CRISPR DRs are divided into at least 12 clusters, based on their sequence similarity and ability to form stable secondary structures [5]. Some of the DR clusters clearly correspond to particular CRISPR-Cas subtypes [4, 5]. Importantly, a linkage between the PAM sequences, typically 2–5 nucleotides long, and the main DR clusters has also been reported [3].
Generally, each CRISPR-cas locus includes a strain-specific array of spacers that has expanded and diversified over time [1, 6]. Due to their dynamic nature, comparative analysis of the arrays of spacers has successfully been used for subtyping isolates from several Gram-positive and-negative bacteria, including Mycobacterium tuberculosis, Yersinia pestis and the plant pathogen Erwinia amylovora (reviewed in [6]). Arrays of spacers were found to be highly polymorphic in Salmonella and a strong correlation was detected between polymorphisms in the arrays and the serotypes [7, 8]. In fact, analyzing only newly incorporated spacers gave results that were highly consistent with traditional serotyping of Salmonella isolates [8]. Furthermore, all the S. enterica Typhi and Paratyphi A isolates carried serotype-specific spacers that were exploited in the development of PCR assays able to identify these serotypes [7]. Other studies have focused on the evolutionary history of the CRISPR-Cas systems [9, 10]. For instance, frequent non-vertical transmission events have occurred throughout the evolution of the CRISPR-Cas system in S. enterica ssp. enterica [10].
Acinetobacter baumannii is an important opportunistic pathogen responsible for a wide range of hospital-acquired infections, including ventilator-associated pneumonia and catheter-related bloodstream infections [11]. Multilocus sequence typing (MLST), based on comparative sequence analyses of the loci of seven house-keeping genes, has demonstrated the frequent occurrence of A. baumannii isolates sharing similar allelic profiles although obtained independently from different countries [12, 13]. Using the MLST terminology, a “sequence type” (ST) refers to a particular allelic profile and “clonal complex” (CC) refers to a group of related STs sharing the same alleles at 5/7 or 6/7 of the loci (http://eburst.mlst.net). On the other hand, a “clone” is a general term that has been used to describe a group of phenotypically and genotypically related but epidemiologically unrelated isolates, which are believed to be a progeny of a common ancestor [14, 15]. Therefore, an ST or CC represents a “clone” only when it includes epidemiologically unrelated isolates, otherwise the terms are not interchangeable [14]. Two MLST schemes are currently available for Acinetobacter (http://pubmlst.org/abaumannii/). CC2, according to the Pasteur’s MLST scheme, is currently the largest and most widely distributed clone in the global population of A. baumannii [12, 13]. Nonetheless, several other clones have co-dominated or recently emerged as important international actors. For instance, CC1 ranks as the second largest clone of A. baumannii, with a broad international distribution in more than 30 countries from all continents [14]. Isolates from this clone have commonly showed a multidrug-resistance phenotype and frequently carried AbaR3-like resistance islands [16]. In parallel, a growing occurrence of CC25 has recently been reported from different countries in Europe, South and North America, Africa, and Asia [13, 14, 17, 18]. In addition to their extensive resistance to antibiotics, the CC25 isolates have shown the ability to resist desiccation, form biofilms on abiotic surfaces, and adhere to human alveolar epithelial cells [19].
Two CRISPR-Cas systems have recently been found in the genome of particular A. baumannii strains [20, 21]. The CRISPR-cas locus in strain AYE belongs to subtype I-Fb, herein denoted as CRISPR-cas subtype I-Fb throughout the manuscript [4, 22]. Genomic islands carrying this locus in strains 4190, AB0057 and AYE were found to be closely related, indicating potential inter-strain horizontal transfer [20]. Comparative analysis of partial sequences of the CRISPR-cas subtype I-Fb locus was useful in detecting the occurrence of an intra-clonal diversity among clinical isolates of international clone CC1 [21]. The aim of this study was to investigate the evolutionary history of CRISPR-cas subtype I-Fb in A. baumannii and to determine the genetic relatedness among a collection of CRISPR-positive clinical isolates of A. baumannii, based on comparative sequence analysis of the arrays of spacers located in their CRISPR-cas subtype I-Fb locus.
Material and Methods
A. baumannii isolates
The study included 74 isolates of A. baumannii carrying the CRISPR-Cas subtype I-Fb system (Table 1). The isolates were collected from the United States of America (USA; n = 29), Sweden (n = 12), Norway (n = 10), Iraq (n = 5), Netherlands (n = 3), Czech Republic (n = 3), Germany (n = 3), Canada (n = 2), Greece (n = 1), France (n = 1), Italy (n = 1), Argentina (n = 1), Colombia (n = 1), and United Arab Emirates (UAE; n = 1). The country of isolation was unknown for one isolate. Twenty-five isolates were part of three ongoing projects involving whole-genome sequencing of carbapenem-resistant A. baumannii isolates obtained in Norway between 2010 and 2013 (project I), Sweden between 2012 and 2013 (project II), or representatives of an international collection of A. baumannii isolates belonging to CC25 (project III). Forty-four isolates with sequenced genomes were selected from the records of the International Nucleotide Sequence Database Collaboration (http://www.insdc.org/). Occurrence of CRISPR-cas subtype I-Fb in these isolates was detected by Nucleotide BLAST algorithm (http://blast.ncbi.nlm.nih.gov/Blast.cgi), against both the “Nucleotide collection (nr/nt)” and “Whole-genome shotgun contigs (wgs)” databases. The remaining 5 isolates belonged to CC1 or CC25 and were part of previously published studies [23, 24]. The online multilocus sequence typing (MLST) service hosted by the Center for Genomic Epidemiology (http://www.genomicepidemiology.org/) in Denmark was used to determine the ST of all the isolates for which a full genome sequence was available [25]. The assignment was performed according to the Institute Pasteur’s MLST scheme [13] (http://pubmlst.org/abaumannii/). In order to group the isolates into CCs, a minimum spanning tree was generated from all the allelic profiles in the database using PhyloWeb and the MSTree application assimilated in the Institute Pasteur’s MLST web site (http://www.pasteur.fr/mlst).
Table 1. Epidemiological data on the 74 Acinetobacter baumannii isolates included in this study.
| Isolate | Date and place of isolation a | Source and type of sample a | Import and other epidemiological data a | ST / CST a | GenBank accession no., [reference] |
|---|---|---|---|---|---|
| K48–42 | Feb 2008, Unilabs Telelab, Skien, Norway | Tracheal aspirate | Import from India | ST1 / CST6 | KM998765, [23] |
| K55–61 | Mar 2009, Vestfold Hospital, Vestfold, Norway | Abdominal cavity | Import from India | ST94 / CST13 | KM998766, [23] |
| K57–06 | Mar 2009, Oslo University Hospital (Ullevål), Oslo, Norway | Trachael aspirate | Import from India, same PFGE pattern as K55–61 | ST94 / CST13 | [23] |
| AO-471 | 2005, Karolinska University Hospital, Stockholm, Sweden | Wound | Import from Thailand, tsunami-related | ST25 / CST30 | KM998767, [24] |
| AO-21841 | 2006, Karolinska University Hospital, Stockholm, Sweden | Intra-abdominal | No history of import | ST25 / CST31 | KM998768, [24] |
| K63–58 | Mar 2010, Oslo University Hospital (Ullevål), Oslo, Norway | Cerebrospinal fluid | Import from Iraq | ST94 / CST12 | KM998769 |
| 50509585 | May 2011, Oslo University Hospital (Aker), Oslo, Norway | Trachael aspirate | Import from Greece | ST1 / CST8 | KM998770 |
| 50525357 | Jul 2011, Haukeland University Hospital, Bergen, Norway | Drain tube | Import from Romania | ST1 / CST9 | KM998771 |
| 50535631 | Sep 2012, Vestfold Hospital, Vestfold, Norway | Perineum | Import from Thailand | ST25 / CST20 | KM998772 |
| 50678066 | Dec 2012, Innlandet Hospital, Levanger, Norway | Tracheal | Import from Thailand | ST25 / CST19 | KM998773 |
| 50691529 | Feb 2013, Oslo University Hospital (Ullevål), Oslo, Norway | Urine | Import from Thailand | ST25 / CST19 | Our unpublished data |
| 50695882 | Feb 2013, Sørlandet Hospital, Kristiansand, Norway | Urine | No history of import | ST1 / CST8 | Our unpublished data |
| A068 | Apr 2012, Blekinge Hospital, Blekinge, Sweden | - | - | ST25 / CST23 | KM998774 |
| A069 | May, 2012, Halmstad Hospital, Halland, Sweden | Feces | Import from Thailand | ST25 / CST19 | Our unpublished data |
| A076 | Jan 2013, Skåne University Hospital, Skåne, Sweden | Rectum | - | ST1 / CST1 | KM998775 |
| A082 | Mar 2013, Linköping University Hospital, Östergötland, Sweden | Wound | - | ST1 / CST4 | KM998776 |
| A092 | Mar 2013, Linköping University Hospital, Östergötland, Sweden | Blood | - | ST25 / CST23 | Our unpublished data |
| A093 | Mar 2013, Linköping University Hospital, Östergötland, Sweden | - | - | ST25 / CST23 | Our unpublished data |
| A094 | Mar 2013, Linköping University Hospital, Östergötland, Sweden | Feces | - | ST25 / CST23 | Our unpublished data |
| A096 | Mar 2013, Linköping University Hospital, Östergötland, Sweden | Thorax | - | ST25 / CST23 | Our unpublished data |
| A097 | Mar 2013, Linköping University Hospital, Östergötland, Sweden | Nose | - | ST25 / CST23 | Our unpublished data |
| A100 | Oct 2013, Skåne University Hospital, Skåne, Sweden | - | - | ST1 / CST5 | KM998777 |
| RUH1486 | 1985, Rotterdam, Netherlands | Umbilicus | - | ST25 / CST29 | KM998778, [12] |
| LUH7841 | 2002, Leiden, Netherlands | Intra-venous catheter tip | - | ST402 / CST15 | KM998779 |
| LUH6220 | 2000, Leiden, Netherlands | Sputum | - | ST25 / CST16 | KM998780 |
| 4390 | 2003, Hippokration, Athens, Greece | Bronchial | Representing 3 isolates with the same PFGE pattern | ST25 / CST16 | [40] |
| 161/07 | 2007, Frankfurt University Hospital, Frankfurt, Germany | Respiratory tract | Import from Serbia | ST25 / CST28 | KM998781, [41] |
| NM3 | 2008, Abu Dhabi, UAE | Sputum | Representing 4 isolates with the same PFGE pattern | ST25 / CST23 | [39] |
| 741019 | 2011, H7, Buenos Aires, Argentina | Pleural Fluid | Representing 7 isolates with the same PFGE pattern collected from 3 hospitals | ST25 / CST24 | KM998782, [38] |
| 4190 | 2009, Monaldi Hospital, Naples, Italy | Blood | Representing 3 isolates with the same PFGE pattern | ST25 / CST32 | KM998783, [17, 20] |
| AYE | 2001, Kremlin-Bicetre, France | Patient with pneumonia and urinary tract infection | Epidemic in 54 healthcare facilities in eight French administrative regions | ST1 / CST7 | CU459141, [42] |
| AB0057 | 2004, WRAMC, Washington DC, USA | Blood | WRAMC was the major USA site receiving casualties from the conflict in Iraq/Kuwait and Afghanistan | ST1 / CST1 | CP001182, [33] |
| Canada BC1 | 2007, a civilian hospital, Canada | - | Due to nosocomial spread of a war-related isolate introduced by a soldier evacuated via Landstuhl Regional Medical Center (Germany) | ST1 / CST1 | AMSZ00000000 |
| Canada BC-5 | 2007, a civilian hospital, Canada | - | Due to nosocomial spread of a war-related isolate introduced by a soldier evacuated via Landstuhl Regional Medical Center (Germany) | ST1 / CST1 | AFDN00000000 |
| IS-58 | Feb 2008, Ibn Sina, Iraq | Respiratory tract | - | ST1 / CST1 | AMGH00000000 |
| IS-235 | Aug 2008, Ibn Sina, Iraq | Blood | - | ST1 / CST1 | AMEI00000000 |
| IS-251 | Sep 2008, Ibn Sina, Iraq | Respiratory tract | - | ST1 / CST1 | AMEJ00000000 |
| AB5075 | 2009, WRAMC, Maryland, USA | Bone infection | - | ST1 / CST1 | AHAH00000000, [34] |
| AB_908–13 | 2007, Centers for Disease Control, USA | Urine | - | ST1 / CST1 | AMHW00000000, [43] |
| AB_909–02–7 | 2007, Centers for Disease Control, USA | Sputum | - | ST1 / CST1 | AMHZ00000000, [43] |
| TG20277 | Jun 2006, Landstuhl Regional Medical Center, Landstuhl, Germany | Sputum | Isolated from a Canadian soldier injured in Afghanistan | ST1 / CST1 | ASFH00000000 |
| TG22112 | 2011, Arizona State Labs, USA | Trachael aspirate | - | ST1 / CST1 | ASFK00000000 |
| TG22148 | 2011, Arizona State Labs, USA | Trachael aspirate | - | ST1 / CST1 | ASFN00000000 |
| TG22190 | 2011, Arizona State Labs, USA | Trachael aspirate | - | ST1 / CST1 | ASFP00000000 |
| TG22194 | 2011, Arizona State Labs, USA | Trachael aspirate | - | ST1 / CST1 | ASFR00000000 |
| TG22196 | 2011, Arizona State Labs, USA | Blood | - | ST1 / CST1 | ASFS00000000 |
| TG22214 | 2011, Arizona State Labs, USA | Sputum | - | ST1 / CST1 | ASFX00000000 |
| 1605 | J. Craig Venter Institute, USA | - | - | ST1 / CST1 | AUWL00000000 |
| Naval-83 | Oct 2006, National Naval Medical Center, Bethesda, Maryland, USA | Wound | - | ST20 / CST3 | AMFK00000000 |
| ABNIH19 | 2009, National Institutes of Health Clinical Center, Bethesda, Maryland, USA | Trachael aspirate | - | ST1 / CST2 | APBH00000000, [44] |
| MRSN58 | Jun 2010, WRAMC, USA | Wound | - | ST1 / CST2 | JABU00000000 |
| NIPH 290 | Oct 1994, Příbram, Czech Republic | ICU | - | ST1 / CST2 | APRD00000000, [37] |
| AB307–0294 | 1994, Buffalo, New York, USA | Blood | - | ST1 / CST10 | CP001172, [33] |
| TG19582 | American Type Culture Collection | - | - | ST1 / CST11 | AMIV00000000, [43] |
| OIFC074 | Jun 2003, Landstahl Regional Medical Center, Landstahl, Germany | - | - | ST19 / CST13 | AMDE00000000 |
| MRSN 3405 | Mar 2011, WRAMC, USA | Wound | - | ST94 / CST13 | JNOU00000000 |
| NIPH 201 | Jul 1992, Liberec, Czech Republic | ICU | - | ST38 / CST35 | APQV00000000, [37] |
| ab233846 | USA | Sputum | - | ST126 / CST38 | JMOG00000000 |
| ab299505 | USA | Perirectal | - | ST508 / CST36 | JEWY00000000 |
| NIPH 615 | Jan 1994, Praha, Czech Republic | Burns unit | - | ST12 / CST39 | APOV00000000, [37] |
| TG27391 | Arizona State Labs, USA | - | - | ST427 / CST40 | ASGK00000000 |
| ab532279 | USA | Perirectal | - | ST519 / CST37 | JEYH00000000 |
| ab1106579 | USA | Perirectal | - | ST505 / CST33 | JEXN00000000 |
| Naval-82 | Oct 2006, National Naval Medical Center, Bethesda, Maryland, USA | Blood | - | ST428 / CST34 | AMSW00000000 |
| OIFC143 | Jul 2003, WRAMC, USA | - | - | ST25 / CST25 | AFDL00000000 |
| abC179 | Mar 2005, Iraq | Respiratory tract | From military personnel, combat-related | ST25 / CST26 | AVOD00000000, [45] |
| abCI86 | Feb 2005, Iraq | Superficial wound | From military personnel, combat-related | ST25 / CST26 | AVOB00000000, [45] |
| ab984213 | USA | Perirectal | - | ST25 / CST17 | JEVX00000000 |
| Naval-18 | Jun 2006, National Naval Medical Center, Bethesda, Maryland, USA | - | - | ST25 / CST27 | AFDA00000000 |
| AB-1650–8 | 2006, Arizona State Labs, USA | Bone (from hip) | - | ST113 / CST14 | AMHG00000000, [43] |
| ab1429530 | USA | Perirectal | - | ST25 / CST21 | JEWM00000000 |
| 107m | Colombia | - | - | ST25 / CST22 | CBSG00000000 |
| AB_2008–15–69 | 2008, Centers for Disease Control, USA | - | - | ST25 / CST18 | AMHN00000000, [43] |
| AB5256 | 2009, WRAMC, Maryland, USA | Blood | Representing a clonal group | ST25 / CST23 | AHAI00000000, [34] |
a ST, Sequence type; CST, CRISPR-based sequence type; PFGE, Pulsed-field gel electrophoresis; UAE, United Arab Emirates; WRAMC, Walter Reed Army Medical Center; USA, United States of America; ICU, Intensive care unit.
Phylogenetic analysis
Phylogenetic trees were generated based on nucleotide sequences alignments of (i) a conserved segment of 101 bp located downstream of the array of spacers, (ii) 920 bp of the cas1 gene, (iii) 1251 bp of the csy1 (Cas system-associated) gene, (iv) 615 bp of the csy4 gene, and (v) 2976 bp of the concatenated MLST sequences. Only isolates with sequenced genomes were included in the phylogenetic analyses. The online available package of programs (MUSCLE, Gblocks, PhyML, and TreeDyn) was used for nucleotide alignment, tree construction, and tree rendering [26]. One hundred bootstraps were used for bootstrap analysis.
CRISPR-based subtyping
DNA sequences of the CRISPR arrays of spacers were either retrieved from the GenBank nucleotide database or amplified and sequenced using the BigDye 3.1 technology (Applied Biosystems). Amplification of the full length of the arrays was performed using a pair of external primers, Ab-CRIS-F and Ab-CRIS-R, targeting conserved flanking regions (S1 Table). PCR products were purified using ExoProStar (GE Healthcare Bio-Sciences). The purified PCR amplicons were subsequently sequenced using the two external and several internal primers designed in tandem as required. Arrays of spacers were identified using CRISPRFinder [27]. A dictionary of annotated spacers was created using CRISPRtionary (http://crispr.u-psud.fr/CRISPRcompar/Dict/Dict.php) and revised manually. CRISPRtionary was also used to create a binary file of presence (1) or absence (0) of spacers. Each spacer with a newly defined sequence was assigned a new consecutive number, and each array with a newly defined assortment of spacers represented a new CRISPR-based sequence type (CST), as previously described [28].
GenBank Accession Numbers
Nucleotide sequences of the CRISPR arrays in isolates K48–42, K55–61, AO-471, AO-21841, K63–58, 50509585, 50525357, 50535631, 50678066, A068, A076, A082, A100, RUH1486, LUH7841, LUH6220, 161/07, 741019, and 4190 were deposited in the GenBank nucleotide database under accession numbers KM998765 to KM998783, respectively.
Results and Discussion
Description and distribution of the CRISPR-cas subtype I-Fb locus
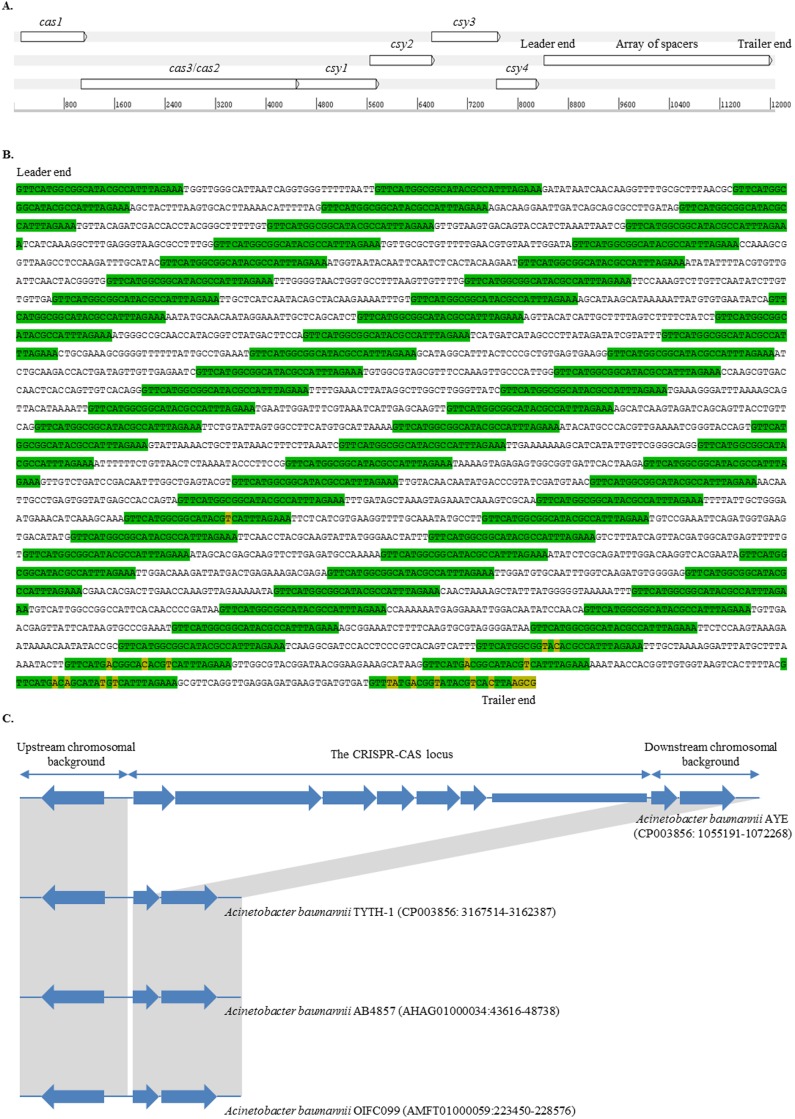
The CRISPR-cas subtype I-Fb locus, located at position 1,057,691 to 1,069,768 of the genome of A. baumannii strain AYE (GenBank accession number: CU459141), consisted of six genes consecutively encoding the Cas1 endonuclease, Cas3/cas2 helicase/RNAse, and four Csy proteins (Fig. 1A and B). csy4 (cas6f) was followed by a short non-coding TA-rich leader sequence. Then, the locus enclosed an array of spacers, where each spacer was flanked by two DRs. The spacers had variable sequences and a common length of 32 bp, whereas the DRs had the 5′-GTTCATGGCGGCATACGCCATTTAGAAA-3′ consensus sequence and were 28 bp long. Similarly to the CRISPR-Cas subtype I-F systems in other genera, the consensus sequence belonged to the DR cluster 4, showing a nucleotide similarity of 65–75% with those of Escherichia coli, Pseudomonas aeruginosa, Yersinia pestis, Shewanella spp., and Pectobacterium atrosepticum [3, 5, 29, 30]. Overall, 876 distinct A. baumannii-spacers (Ab-1 to-876) were identified (S2 Table). The size of the arrays was remarkably different among the isolates, ranging from 148 bp (2 spacers) up to 7354 bp (122 spacers).
Fig 1. Genetic structure of the CRISPR-cas subtype I-Fb locus in Acinetobacter baumannii.
A) The CRISPR-cas subtype I-Fb locus in A. baumannii strain AYE (GenBank accession number: CU459141, located at position 1,057,691 to 1,069,768). The locus consisted of two CRISPR–associated genes (cas1 and cas3/cas2), four Cas system-associated genes (csy1, csy2, csy3, and csy4), and an array of spacers. The map was created using Artemis (http://www.sanger.ac.uk/resources/software/artemis/). B) Nucleotide sequence of the array of spacers in A. baumannii strain AYE. The array included 59 spacers surrounded by 60 direct repeats (marked in green). Some repeats, mainly at the trailer end of the array, included degenerated nucleotides (marked in yellow). C) Comparative analysis of the genetic surroundings. The comparison was performed between the locus-positive strain AYE which belonged to sequence type (ST1) and the locus-negative strains TYTH-1, AB4857, and OIFC099 which belonged to ST2, ST3, and ST32, respectively. Homologous sequences shared by all the isolates were indicated by gray zones.
A complete CRISPR-cas subtype I-Fb locus was present in all isolates belonging to CC1 and CC25. The locus was also present in isolates from ST113, ST12, ST38, ST126, ST427, ST505, ST508 and ST519. An internal deletion was detected in the locus of only one isolate, Naval-82 that belonged to ST428, resulting in the absence of cas3/cas2, csy1, csy2 and part of csy3 (GenBank accession number: AMSW01000159). On the other hand, the locus was replaced by a short sequence of 128 bp in the genome of A. baumannii strain TYTH-1 (GenBank accession number: CP003856), which was assigned to ST2 (Fig. 1C). The locus was also not present in the genome of other isolates from CC2 or isolates AB4857 and OIFC099 from the epidemic clones CC3 and CC32, respectively (Fig. 1C and data not shown). Notably, the locus was present in isolates from other Acinetobacter species such as Acinetobacter haemolyticus TG19602 and Acinetobacter gyllenbergii NIPH 230 (GenBank accession numbers: AMJB01000210 and AYEQ01000180, respectively). In addition, a common occurrence of CRISPR-cas subtype I-Fb in Acinetobacter parvus has recently been reported [22].
A Blastn search failed to detect proto-spacers with homologous sequences to 42/106 of the spacers present in A. baumannii CC1 isolates (S2 Table). This could be due to the limited number of A. baumannii phages that have been sequenced and deposited in the GenBank databases [22]. Nonetheless, phage- and plasmid-related DNA elements were the source of 50/106 and 12/106 of the spacers, respectively. The remaining 2/106 spacers originated from A. baumannii DNA that was most likely not related to phages or plasmids. These results were comparable to those of previous studies on the CRISPR arrays in Streptococcus thermophilus and Y. pestis [9, 31]. For example, only 500 out of 952 unique spacers in S. thermophilus showed similarity to viral (n = 384), plasmid (n = 80), or chromosomal (n = 33) sequences [9]. A prophage of 42778 bp, located on the genome of A. baumannii NIPH 527 (APQW01000004: 179581–222358), represented the main foreign DNA encountered by our isolates, being the source of 16/106 of the spacers (S2 Table and S1 Fig.). The proto-spacers in this prophage were carried on a variety of genes, such as the replicative DNA helicase, glycosyl hydrolase, tail tape measure, integrase, terminase, and GDSL-family lipase genes, or located in inter-gene regions. The proto-spacers were found either on the sense or antisense strand (S1 Fig.), as previously described [3].
Alignment of sequences surrounding the proto-spacers identified the dinucleotide “CC”, or “GG” on the complementary strand, to be the PAM for the CRISPR-Cas subtype I-Fb machinery of A. baumanni (S2 Table and S2 Fig.). The PAM motif was located on the 5′ end of the proto-spacers, as previously reported for other CRISPR-Cas type I systems [2]. Our results were also consistent with earlier studies showing that GG has been the signature PAM for the DR cluster 4 and CRISPR-Cas subtype I-F systems [3]. A set of 4 spacers (Ab-62 to Ab-65) was most likely acquired by one of our isolates, assigned to CST8, after a single contact with a plasmid from A. baumannii IS-58 assigned to CST1 (S2 Table). Similarly, Ab-77 to Ab-80, Ab-92 to Ab-94 (S1 Fig.), and Ab-97 to Ab-99 could also be acquired after single interactions with particular plasmid or phage DNA molecules.
Evolution of the CRISPR-cas subtype I-Fb locus
Arrays of spacers have mainly evolved by adding new spacers in response to contacts with foreign genetic elements [9]. Since the addition takes place in a one-way direction, spacers present at one end (the trailer end) of the arrays had been integrated earlier than those present in the other end (the leader end), and the order of spacers generally provides a chronological narration of former exposures to invading phages and plasmids [31]. Studies have reported that trailer end spacers are generally conserved among different isolates and can be used to anchor clusters and detect common ancestors of the arrays and probably of the isolates themselves [9]. In contrast, the leader end spacers are usually polymorphic, reflecting the existence of distinctive phage/plasmid pools at a particular era in different geographic locations [8]. Ab-1 and Ab-107 were the first spacers acquired by the vast majority of our isolates (S2 Table). Spacer Ab-1 was present in the locus of all the isolates from CC1, ST38, ST428, ST505, and ST519, and also isolate 4190 from CC25. In contrast, spacer Ab-107 was shared by all the CC25 isolates, except isolate 4190, and the ST113, ST126, ST12, and ST427 isolates. The conservation of Ab-1 and Ab-107 at the trailer end of the arrays putatively assembled our arrays into two major groups with a different first step of descending from a common ancestor empty of spacers. In agreement with previous studies, the DRs at the trailer end of our isolates were degenerated (Fig. 1B). However, the number and sequence of the degenerated nucleotides were following two patterns corresponding to the Ab-1 and Ab-107 grouping of the isolates, which confirmed the occurrence of two main lineages in the early evolutionary history of the locus (S3 Fig.). Furthermore, comparative sequence analysis of a conserved segment of 101-bp located adjacent to the trailer end of the arrays showed a precisely matching assembly of the isolates (S4 Fig.).
The CRISPR arrays have frequently been reshaped by internal deletions and duplications, of individual spacers or sets of consecutive spacers, leading to further diversification of the arrays [32]. Interestingly, Ab-5 was located first at the trailer end of the array of spacers in isolate ab299505 (ST508). This was probably due to an internal deletion of 240 bp, erasing Ab-1 to Ab-4, caused by a recombination event between the two DRs surrounding the deleted region (Fig. 2). The last DR in this array had a unique sequence that was most likely derived from the two recombined DRs, consistent with the occurrence of such a recombination event. Accordingly, the locus in ab299505 also belonged to the Ab-1 lineage (S4 Fig.).
Fig 2. Schematic comparison of the trailer end of the arrays of spacers in Acinetobacter baumannii AYE and A. baumannii ab299505.
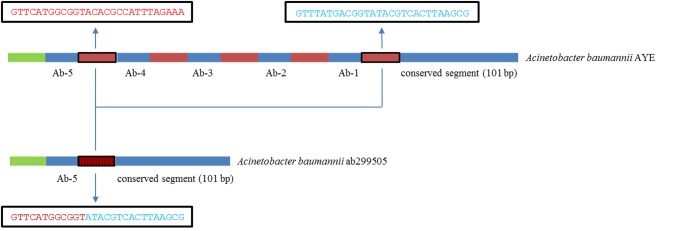
The region carrying spacers Ab-1 to Ab-4 in A. baumannii AYE was missing in A. baumannii ab299505, most likely due to an internal deletion caused by a recombination event between the two direct repeats (highlighted by a black frame) surrounding the deleted region. This created a unique direct repeat (highlighted by a black frame and vertical lines) characterized by a novel mosaic sequence derived from the recombined direct repeats. Sequence of the direct repeats involved in the recombination was presented in adjacent black boxes.
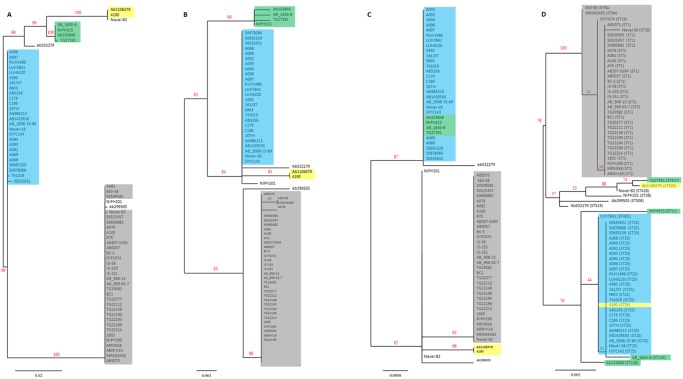
Phylogenetic trees of cas1, csy1, and csy4 identified seven distinct pathways of evolution for the CRISPR-cas subtype I-Fb locus in A. baumannii (Fig. 3A, B, and C). Pathways 1, 4, 5, 6, and 7 branched from the Ab-1 lineage whereas pathways 2 and 3 descended from the Ab-92 lineage. Pathway 1 ended with a cluster including all the alleles of the locus present in isolates from CC1. Similarly, pathway 2 created a cluster of all the alleles of isolates from CC25, except for strain 4190. Pathway 3 was shared by the alleles of isolates AB_1650–8 (ST113), NIPH615 (ST12), ab233846 (ST126), and TG23791 (ST427), whereas the two alleles of isolates 4190 (CC25) and ab1106579 (ST505) evolved following pathway 4. The alleles of isolates NIPH201 (ST38), ab299505 (ST508), and ab532279 (ST519) showed individual pathways of evolution. However, the latter three alleles had mosaic sequences proposing the occurrence of penetrations mediated by recombination events with the locus of other strains. Due to the internal deletion, the pathway of evolution could not be determined for the locus in Naval-82 (ST428). Comparing the topology of the isolates in these phylogenetic trees with the one based on the concatenated MLST sequences showed a congruent positioning of the isolates of pathways 1 and 2, suggesting a vertical spread of the locus in these two clusters (Fig. 3D). On the other hand, the incongruent standing of the isolates of pathways 3 and 4 proposed the occurrence of recent horizontal acquisitions of the whole locus between isolates of each cluster [10].
Fig 3. Phylogenetic trees of cas1, csy1, csy4 and the concatenated MLST sequences.
The phylogenetic trees were based on aligned nucleotide sequences of (A) 920 bp of cas1 from 69 isolates of Acinetobacter baumannii, (B) 1251 bp of csy-1 from 68 isolates of A. baumannii since csy1 was lacking in isolate Naval-82, (C) 615 bp of csy4 from 69 isolates of A. baumannii, and (D) 2976 bp of concatenated MLST sequences of 69 isolates of A. baumannii. MUSCLE, Gblocks, PhyML, and TreeDyn were used for nucleotide alignment and tree construction. One hundred bootstraps were used for bootstrap analysis. Branch support values were displayed in %. Isolates of the CRISPR-cas subtype I-Fb pathways of evolution 1, 2, 3, and 4 were highlighted in gray, blue, green, and yellow, respectively. The corresponding sequence type (ST) was presented next to the name of each isolate in (D).
CRISPR-based subtyping
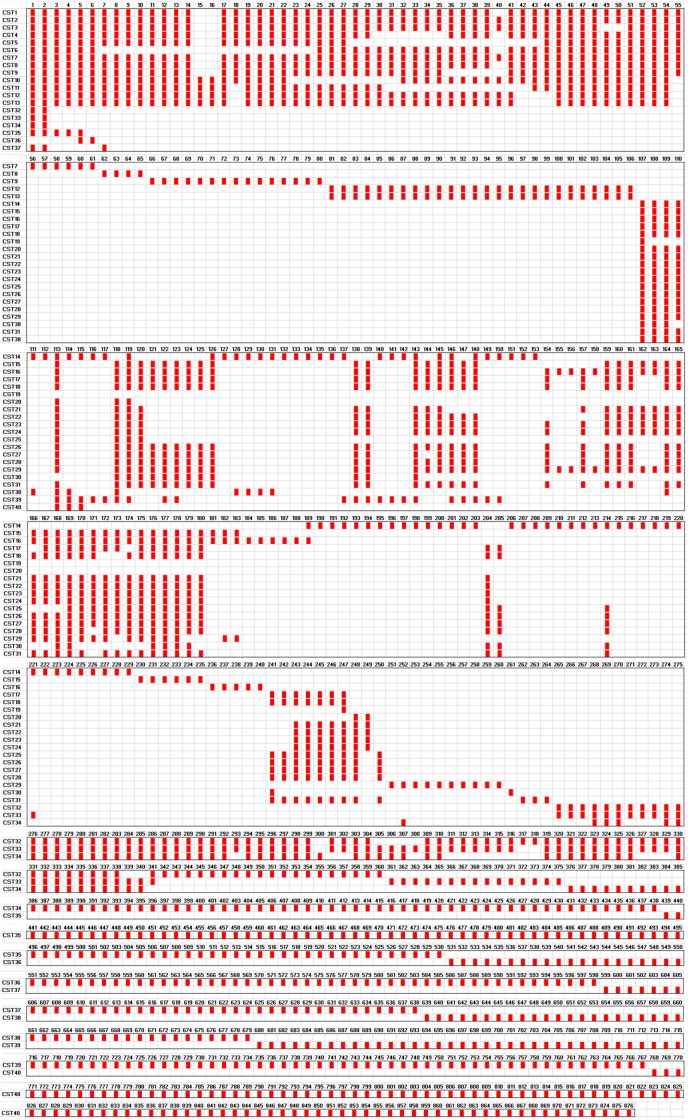
Different assortments of the spacers divided the isolates into 40 CSTs (Fig. 4 and S3 Table). Isolates from CC1 (n = 36) belonged to 13 CSTs, with some CSTs being different from each other only by a duplication or deletion of 1 spacer. CST1 included 18 isolates recovered between 2004 and 2013 from USA (n = 11), Iraq (n = 3), Canada (n = 2), Germany (n = 1), and Sweden (n = 1). Tracking the epidemiological data showed that eight of the isolates were obtained during the military operations in Iraq and Afghanistan [33, 34]. CST1 could be an Iraq-endemic sub-clone of CC1 that was able to spread to USA, Canada, and Europe. A previous study comparing the DNA profiles of A. baumannii isolates from USA and the United Kingdom that were associated with casualties returning from the Iraq conflict has also demonstrated the import of at least one strain responsible for outbreaks of infections in the two countries [35]. Adaptation of CST1 to the pool of phages and plasmids present in a particular geographical site resulted in the acquisition of specific spacers which might be used as a genomic signature of this sub-clone and a biological marker of this particular geographic ecosystem [10, 36]. On the other hand, CST2 included 3 isolates obtained from Czech Republic in 1994 and USA in 2009 and 2010. However, the Czech isolate was not reported to be epidemiologically linked with the two American isolates [37].
Fig 4. Graphic representation of the arrays of spacers in the CRISPR-cas subtype I-Fb locus of Acinetobacter baumannii.
The figure demonstrated the assortment of 74 A. baumannii isolates into 40 CRISPR sequence types (CST) based on the spacer content of their CRISPR arrays. Spacers were represented by red rectangles. Each unique spacer was assigned a number (1–876). Spacers were sequentially aligned in order to facilitate comparison among the CSTs.
CST8 included two isolates obtained in 2011 and 2013 by two different diagnostic laboratories in Norway. Since no other epidemiological linkage was detected, inter-hospital transfer of patients or medical staff could be responsible for the spread of CST8. However, the long gap in the time of acquisition obviously excluded the occurrence of an outbreak. CST12 was different from CST13 only by having a duplication of spacer Ab-49. Five isolates belonged to CST12–13. Three of these isolates, obtained from Germany, Norway, and USA between 2003 and 2011, shared a history of import from Iraq (Table 1). The other two isolates were obtained from different laboratories in Norway in 2009 but were both imported from India [23]. Interestingly, none of these isolates belong to ST1, the founder of CC1. In contrast, the isolates mainly belong to ST94, indicating a considerable sub-clonal demarcation of ST94, able to overcome the origination of the isolates from two geographically disconnected countries. Three isolates, obtained from patients in one military hospital in France in 2009, were found to carry arrays of spacers with a high similarity to each other [21]. The three isolates were linked to CST12–13 according to our subtyping scheme. However, the comparison was incomplete since only the leader end of the CRISPR arrays was amplified and partially sequenced in the French isolates. Of note, the three French isolates were recovered from skin samples while our isolates came from various sources [21].
The CC25 isolates (n = 30) were divided into 19 CSTs (Table 1 and Fig. 4). Three isolates, obtained from Sweden in 2012 and Norway in 2012 and 2013, belonged to CST19. Interestingly, these isolates were recovered from patients previously hospitalized in Thailand. The molecular similarity between the isolates suggested the existence of a Thai strain and supported the history of import of these isolates, pointing once more to the necessity of having a screening program for patients after hospitalization abroad [23]. CST23 was identical to CST24 apart from having a duplication of three spacers. CST23–24 included 6 isolates obtained from Sweden and 3 isolates obtained from Argentina, UAE, and USA. The first Swedish isolate was collected in Blekinge in April 2012 while the other 5 isolates came from 5 different patients hospitalized at the same medical center in Östergötland in August 2013. The Swedish isolates, particularly the latter five, could represent a single strain responsible for a small-sized outbreak taking place in Östergötland. The Argentinean strain represents 7 isolates sharing the same pulsed-field gel electrophoresis (PFGE) pattern [38]. These isolates were recovered between 2009 and 2012, during an endemic setting in three hospitals located in two different cities in Argentina. Similarly, the UAE and USA strains are representatives of two groups of isolates sharing same PFGE patterns [34, 39]. The ability of CST23–24 to be endemic and to cause outbreaks of infections highlights the need to precisely distinguish such highly-successful sub-clones, which requires the application of more strict infection control procedures. Isolates from singleton STs (n = 8) belonged to 8 distinct CSTs. These isolates carried >80% of the unique isolate-specific spacers. The broad polymorphism detected among the spacers reflects the complicity and extensive diversity of phage and plasmid populations, facilitating the occurrence of several events of independent interactions and leading to separated pathways of evolution of the CRISPR arrays.
Conclusion
Vertical transmission of the CRISPR-cas subtype I-Fb locus in our global collection of A. baumannii clinical isolates took place following two main lineages and several pathways of descent from a common ancestor. Occasional events of horizontal transfer have increased the diversification and facilitated further dissemination of the locus. Using the CRISPR-based subtyping approach, we were able to detect a sub-clone of A. baumannii CC1, probably originating in Iraq and spreading internationally to the USA and Europe. The study also detected a sub-clone of A. baumannii CC25 with a remarkable ability to cause outbreaks of infections. The unambiguous data generated by this approach can readily be exchanged in silico, used by other groups, and expanded by forthcoming projects. Overall, CRISPR-based subtyping supplements MLST and can be used to track the source and dissemination routes of particular strains.
Supporting Information
The prophage, 42778-bp long, was located on the genome of A. baumannii NIPH 527 (APQW01000004: 179581–222358). The prophage was shown as a white box on the graph and described as a “mobile_element” in the feature list. Genes and open reading frames were shown as blue arrows, with the arrowheads indicating the direction of transcription. Proto-spacers were presented as labeled green arrows, with the arrowheads indicating the direction of their integration as spacers in the CRISPR arrays. The prophage was surrounded by two identical 20-bp repeat regions, for which the sequences were indicated on the graph. The map was created using Artemis (http://www.sanger.ac.uk/resources/software/artemis/).
(TIF)
The sequence logo was created based on an alignment of 10-bp sequences adjacent to 63 proto-spacers targeted by the CRISPR-Cas subtype I-Fb system of A. baumannii. The alignment and the logo were created using WebLogo (http://weblogo.berkeley.edu/logo.cgi). The logo consisted of stacks of letters, with a maximum height of 2 bits. The height of letters within each stack reflects the relative frequency of the corresponding nucleotide at that position. The alignment defined CC as the proto-spacer adjacent motif (PAM) for the CRISPR-Cas subtype I-Fb machinery of A. baumannii.
(TIF)
Pattern 1 was detected in all isolates from CC1, ST38, ST428, ST505, and ST519, and isolate 4190 from CC25. Pattern 2 was detected in all isolates from CC25, except for strain 4190, and in the isolates from ST113, ST126, ST12, and ST427. Preserved and degenerated nucleotides of the direct repeats were marked in green and yellow, respectively.
(TIF)
The tree was based on aligned nucleotide sequences of 101 bp from 69 isolates of Acinetobacter baumannii. MUSCLE, Gblocks, PhyML, and TreeDyn were used for nucleotide alignment and tree construction. One hundred bootstraps were used for bootstrap analysis. Branch support values were displayed in %. Isolates of linages Ab-1 and Ab-107 were indicated by braces.
(TIF)
(XLS)
(XLS)
(XLS)
Acknowledgments
The Authors thank all colleagues who generously provided strains for this study: Mariana Catalana, Lenie Dijkshoorn, Tibor Pàl, Laurent Poirel, Spyros Pournaras, and the Public Health Agency of Sweden represented by Barbro Olsson Liljequist.
Data Availability
Nucleotide sequences are available on 01.01.2015 in the GenBank nucleotide database under accession numbers KM998765 to KM998783. All other relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by project grants from the Swedish Research Council (VR-MH 2010-3031, VR-NT 2012-4638, and VR-MH 2013-3878). The work was performed as part of the Umeå Centre for Microbial Research (UCMR) Linnaeus Program supported by Umeå University and the Swedish Research Council (349-2007-8673). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Horvath P, Barrangou R (2010) CRISPR/Cas, the Immune System of Bacteria and Archaea. Science 327: 167–170a. 10.1126/science.1179555 [DOI] [PubMed] [Google Scholar]
- 2. Barrangou R, Marraffini LA (2014) CRISPR-Cas systems: Prokaryotes upgrade to adaptive immunity. Mol Cell 54: 234–244. 10.1016/j.molcel.2014.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mojica FJ, Díez-Villasenor C, García-Martínez J, Almendros C (2009) Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 155: 733–740. 10.1099/mic.0.023960-0 [DOI] [PubMed] [Google Scholar]
- 4. Makarova KS, Haft DH, Barrangou R, Brouns SJJ, Charpentier E, et al. (2011) Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol 9:467–477. 10.1038/nrmicro2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kunin V, Sorek R, Hugenholtz P (2007). Evolutionary conservation of sequence and secondary structures in CRISPR repeats. Genome Biol 8:R61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shariat N, Dudley EG (2014) CRISPRs: Molecular Signatures Used for Pathogen Subtyping. Appl Environ Microbiol 80: 430–439. 10.1128/AEM.02790-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fabre L, Zhang J, Guigon G, Le Hello S, Guibert V, et al. (2012) CRISPR typing and subtyping for improved laboratory surveillance of Salmonella infections. PLoS One 7: e36995 10.1371/journal.pone.0036995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li H, Li P, Xie J, Yi S, Yang C, et al. (2014) New Clustered Regularly Interspaced short palindromic repeat Locus spacer pair typing method based on the newly incorporated spacer for Salmonella enterica . J Clin Microbiol 52: 2955–2962. 10.1128/JCM.00696-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Horvath P, Romero DA, Coûté-Monvoisin A-C, Richards M, Deveau H, et al. (2008) Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus . J Bacteriol 190: 1401–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pettengill JB, Timme RE, Barrangou R, Toro M, Allard MW, et al. (2014) The evolutionary history and diagnostic utility of the CRISPR-Cas system within Salmonella enterica ssp. enterica . PeerJ 2: e340 10.7717/peerj.340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dijkshoorn L, Nemec A, Seifert H (2007) An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii . Nat Rev Microbiol 5: 939–951. [DOI] [PubMed] [Google Scholar]
- 12. Diancourt L, Passet V, Nemec A, Dijkshoorn L, Brisse S (2010) The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS One 5: e10034 10.1371/journal.pone.0010034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zarrilli R, Pournaras S, Giannouli M, Tsakris A (2013) Global evolution of multidrug-resistant Acinetobacter baumannii clonal lineages. Int J Antimicrob Agents 41: 11–19. 10.1016/j.ijantimicag.2012.09.008 [DOI] [PubMed] [Google Scholar]
- 14. Karah N, Sundsfjord A, Towner K, Samuelsen O (2012) Insights into the global molecular epidemiology of carbapenem non-susceptible clones of Acinetobacter baumannii . Drug Resist Updat 15: 237–247. 10.1016/j.drup.2012.06.001 [DOI] [PubMed] [Google Scholar]
- 15. Orskov F, Orskov I (1983) From the national institutes of health. Summary of a workshop on the clone concept in the epidemiology, taxonomy, and evolution of the Enterobacteriaceae and other bacteria. J Infect Dis 148: 346–357. [DOI] [PubMed] [Google Scholar]
- 16. Krizova L, Dijkshoorn L, Nemec A (2011) Diversity and evolution of AbaR genomic resistance islands in Acinetobacter baumannii strains of European clone I. Antimicrobial Agents and Chemotherapy 55: 3201–3206. 10.1128/AAC.00221-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Di Popolo A, Giannouli M, Triassi M, Brisse S, Zarrilli R (2011) Molecular epidemiological investigation of multidrug-resistant Acinetobacter baumannii strains in four Mediterranean countries with a multilocus sequence typing scheme. Clin Microbiol Infect 17: 197–201. 10.1111/j.1469-0691.2010.03254.x [DOI] [PubMed] [Google Scholar]
- 18. Revathi G, Siu LK, Lu PL, Huang LY (2013) First report of NDM-1-producing Acinetobacter baumannii in East Africa. Int J Infect Dis 17: e1255–e1258. 10.1016/j.ijid.2013.07.016 [DOI] [PubMed] [Google Scholar]
- 19. Giannouli M, Antunes LCS, Marchetti V, Triassi M, Visca P, et al. (2013) Virulence-related traits of epidemic Acinetobacter baumannii strains belonging to the international clonal lineages I-III and to the emerging genotypes ST25 and ST78. BMC Infect Dis 13: 282 10.1186/1471-2334-13-282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Di Nocera PP, Rocco F, Giannouli M, Triassi M, Zarrilli R (2011) Genome organization of epidemic Acinetobacter baumannii strains. BMC Microbiol 11: 224 10.1186/1471-2180-11-224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hauck Y, Soler C, Jault P, Merens A, Gerome P, et al. (2012) Diversity of Acinetobacter baumannii in four French military hospitals, as assessed by multiple locus variable number of tandem repeats analysis. PLoS ONE 7: e44597 10.1371/journal.pone.0044597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Touchon M, Cury J, Yoon EJ, Krizova L, Cerqueira GC, et al. (2014) The genomic diversification of the whole Acinetobacter genus: Origins, mechanisms, and consequences. Genome Biol Evol 6(10):2866–2882. 10.1093/gbe/evu225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Karah N, Haldorsen B, Hermansen NO, Tveten Y, Ragnhildstveit E, et al. (2011) Emergence of OXA-carbapenemase- and 16S rRNA methylase-producing international clones of Acinetobacter baumannii in Norway. J Med Microbiol 60: 515–521. 10.1099/jmm.0.028340-0 [DOI] [PubMed] [Google Scholar]
- 24. Karah N, Giske CG, Sundsfjord A, Samuelsen O (2011) A diversity of OXA-carbapenemases and class 1 integrons among carbapenem-resistant Acinetobacter baumannii clinical isolates from Sweden belonging to different international clonal lineages. Microb Drug Resist 17: 545–549. 10.1089/mdr.2011.0089 [DOI] [PubMed] [Google Scholar]
- 25. Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, et al. (2012) Multilocus sequence typing of total genome sequenced bacteria. J Clin Microbiol 50: 1355–1361. 10.1128/JCM.06094-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, et al. (2008) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36: W465–469. 10.1093/nar/gkn180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Grissa I, Vergnaud G, Pourcel C (2007) CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res 35: W52–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schouls LM, Reulen S, Duim B, Wagenaar JA, Willems RJL, et al. (2003) Comparative genotyping of Campylobacter jejuni by amplified fragment length polymorphism, multilocus sequence typing, and short repeat sequencing: strain diversity, host range, and recombination. J Clin Microbiol 41: 15–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Almendros C, Guzmán NM, Díez-Villaseñor C, García-Martínez J, Mojica FJM (2012) Target motifs affecting natural immunity by a constitutive CRISPR-Cas system in Escherichia coli . PLoS ONE 7(11): e50797 10.1371/journal.pone.0050797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Richter C, Fineran PC (2013) The subtype I-F CRISPR—Cas system influences pathogenicity island retention in Pectobacterium atrosepticum via crRNA generation and Csy complex formation. Biochem Soc Trans. 41(6): 1468–1474. 10.1042/BST20130151 [DOI] [PubMed] [Google Scholar]
- 31. Cui Y, Li Y, Gorge O, Platonov ME, Yan Y, et al. (2008) Insight into microevolution of Yersinia pestis by clustered regularly interspaced short palindromic repeats. PLoS One 3: e2652 10.1371/journal.pone.0002652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. DiMarzio MJ, Shariat N, Kariyawasam S, Barrangou R, Dudley EG (2013) Antibiotic resistance in Salmonella enterica serovar Typhimurium associates with CRISPR sequence type. Antimicrob Agents Chemother 57: 4282–4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Adams MD, Goglin K, Molyneaux N, Hujer KM, Lavender H, et al. (2008) Comparative genome sequence analysis of multidrug-resistant Acinetobacter baumannii . J Bacteriol 190: 8053–8064. 10.1128/JB.00834-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zurawski DV, Thompson MG, McQueary CN, Matalka MN, Sahl JW, et al. (2012) Genome sequences of four divergent multidrug-resistant Acinetobacter baumannii strains isolated from patients with sepsis or osteomyelitis. J Bacteriol 194: 1619–1620. 10.1128/JB.06749-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Turton JF, Kaufmann ME, Gill MJ, Pike R, Scott PT, et al. (2006) Comparison of Acinetobacter baumannii isolates from the United Kingdom and the United States that were associated with repatriated casualties of the Iraq conflict. J Clin Microbiol 44: 2630–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kunin V, He S, Warnecke F, Peterson SB, Martin HG, et al. (2008) A bacterial metapopulation adapts locally to phage predation despite global dispersal. Genome Res 18: 293–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nemec A, Janda L, Melter O, Dijkshoorn L (1999) Genotypic and phenotypic similarity of multiresistant Acinetobacter baumannii isolates in the Czech Republic. J Med Microbiol 48: 287–296. [DOI] [PubMed] [Google Scholar]
- 38. Stietz MS, Ramírez MS, Vilacoba E, Merkier AK, Limansky AS, et al. (2013) Acinetobacter baumannii extensively drug resistant lineages in Buenos Aires hospitals differ from the international clones I–III. Infect Genet Evol 14: 294–301. 10.1016/j.meegid.2012.12.020 [DOI] [PubMed] [Google Scholar]
- 39. Sonnevend A, Ghazawi A, Al Munthari N, Pitout M, Hamadeh MB, et al. (2013) Characteristics of epidemic and sporadic strains of Acinetobacter baumannii isolated in Abu Dhabi hospitals. J Med Microbiol 62(4): 582–590. 10.1099/jmm.0.055681-0 [DOI] [PubMed] [Google Scholar]
- 40. Gogou V, Pournaras S, Giannouli M, Voulgari E, Piperaki ET, et al. (2011) Evolution of multidrug-resistant Acinetobacter baumannii clonal lineages: a 10 year study in Greece (2000–2009). J Antimicrob Chemother 66: 2767–2772. 10.1093/jac/dkr390 [DOI] [PubMed] [Google Scholar]
- 41. Bonnin RA, Poirel L, Naas T, Pirs M, Seme K, et al. (2012) Dissemination of New Delhi metallo-β-lactamase-1-producing Acinetobacter baumannii in Europe. Clin Microbiol Infect 18(9): E362–365. 10.1111/j.1469-0691.2012.03928.x [DOI] [PubMed] [Google Scholar]
- 42. Fournier PE, Vallenet D, Barbe V, Audic S, Ogata H, et al. (2006) Comparative genomics of multidrug resistance in Acinetobacter baumannii . PLoS Genetics 2: e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sahl JW, Gillece JD, Schupp JM, Waddell VG, Driebe EM, et al. (2013). Evolution of a pathogen: a comparative genomics analysis identifies a genetic pathway to pathogenesis in Acinetobacter . PLoS ONE 8: e54287 10.1371/journal.pone.0054287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Snitkin ES, Zelazny AM, Gupta J, NISC Comparative Sequencing Program, Palmore TN, et al. (2013) Genomic insights into the fate of colistin resistance and Acinetobacter baumannii during patient treatment. Genome Res 23: 1155–1162. 10.1101/gr.154328.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ketter P, Guentzel MN, Chambers JP, Jorgensen J, Murray CK, et al. (2014) Genome sequences of four Acinetobacter baumannii-A. calcoaceticus complex isolates from combat-related infections sustained in the Middle East. Genome Announc 2(1): e00026–14. 10.1128/genomeA.00026-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The prophage, 42778-bp long, was located on the genome of A. baumannii NIPH 527 (APQW01000004: 179581–222358). The prophage was shown as a white box on the graph and described as a “mobile_element” in the feature list. Genes and open reading frames were shown as blue arrows, with the arrowheads indicating the direction of transcription. Proto-spacers were presented as labeled green arrows, with the arrowheads indicating the direction of their integration as spacers in the CRISPR arrays. The prophage was surrounded by two identical 20-bp repeat regions, for which the sequences were indicated on the graph. The map was created using Artemis (http://www.sanger.ac.uk/resources/software/artemis/).
(TIF)
The sequence logo was created based on an alignment of 10-bp sequences adjacent to 63 proto-spacers targeted by the CRISPR-Cas subtype I-Fb system of A. baumannii. The alignment and the logo were created using WebLogo (http://weblogo.berkeley.edu/logo.cgi). The logo consisted of stacks of letters, with a maximum height of 2 bits. The height of letters within each stack reflects the relative frequency of the corresponding nucleotide at that position. The alignment defined CC as the proto-spacer adjacent motif (PAM) for the CRISPR-Cas subtype I-Fb machinery of A. baumannii.
(TIF)
Pattern 1 was detected in all isolates from CC1, ST38, ST428, ST505, and ST519, and isolate 4190 from CC25. Pattern 2 was detected in all isolates from CC25, except for strain 4190, and in the isolates from ST113, ST126, ST12, and ST427. Preserved and degenerated nucleotides of the direct repeats were marked in green and yellow, respectively.
(TIF)
The tree was based on aligned nucleotide sequences of 101 bp from 69 isolates of Acinetobacter baumannii. MUSCLE, Gblocks, PhyML, and TreeDyn were used for nucleotide alignment and tree construction. One hundred bootstraps were used for bootstrap analysis. Branch support values were displayed in %. Isolates of linages Ab-1 and Ab-107 were indicated by braces.
(TIF)
(XLS)
(XLS)
(XLS)
Data Availability Statement
Nucleotide sequences are available on 01.01.2015 in the GenBank nucleotide database under accession numbers KM998765 to KM998783. All other relevant data are within the paper and its Supporting Information files.