Abstract
The nuclear exosome targeting complex (NEXT) directs a major 3′–5′ exonuclease, the RNA exosome, for degradation of nuclear noncoding (nc) RNAs. We identified the RNA-binding component of the NEXT complex, RBM7, as a substrate of p38MAPK/MK2-mediated phosphorylation at residue S136. As a result of this phosphorylation, RBM7 displays a strongly decreased RNA-binding capacity, while inhibition of p38MAPK or mutation of S136A in RBM7 increases its RNA association. Interestingly, promoter-upstream transcripts (PROMPTs), such as proRBM39, proEXT1, proDNAJB4, accumulated upon stress stimulation in a p38MAPK/MK2-dependent manner, a process inhibited by overexpression of RBM7S136A. While there are no stress-dependent changes in RNA-polymerase II (RNAPII) occupation of PROMPT regions representing unchanged transcription, stability of PROMPTs is increased. Hence, we propose that phosphorylation of RBM7 by the p38MAPK/MK2 axis increases nuclear ncRNA stability by blocking their RBM7-binding and subsequent RNA exosome targeting to allow stress-dependent modulations of the noncoding transcriptome.
Keywords: ncRNAs, RNA stability, RNA binding, RBM7, phosphorylation, p38MAPK/MK2 signaling
INTRODUCTION
The well-characterized p38 mitogen-activated protein kinase (MAPK)/MAP kinase-activated protein (MAPKAP) kinase 2 (p38MAPK/MK2) signaling module is typically linked to the stress response, which is involved in the transcription of immediate early genes (IEGs) and the post-transcriptional control of proinflammatory cytokine mRNAs (summarized in Gaestel 2006; Cargnello and Roux 2011; Tiedje et al. 2014). By phosphorylating a variety of substrates, diverse functions ranging from cell-cycle control to keratin-regulation can be performed by this signaling module (Manke et al. 2005; Menon et al. 2010). Interestingly, in innate immunity p38MAPK/MK2 regulates the transient LPS-driven cytokine response of macrophages by regulating a feedback loop which confers alterations in mRNA stability and translation of tumor necrosis factor (TNF) and the TNF–mRNA-binding protein tristetraprolin (TTP) (Tiedje et al. 2012). The p38MAPK/MK2 axis is also involved in regulation of miRNA levels and their Ago2-mediated function (Zeng et al. 2008; Cannell et al. 2010; Horman et al. 2013). It remains unclear whether the p38MAPK/MK2 axis can also regulate other RNAs.
The human RNA exosome complex is the major nuclear ribonuclease endowed with 3′–5′ exo- and endoribonucleolytic activities. The RNA exosome is evolutionary conserved from Archaea and yeast to human and is distributed throughout cellular compartments. The multisubunit nature of the RNA exosome and its associated cofactors links the complex to key cellular RNA processing and decay reactions (summarized in Chlebowski et al. 2013). In Saccharomyces cerevisiae, the nuclear exosome interacts with the DEVH-box RNA helicase Mtr4p (de la Cruz et al. 1998) that itself organizes a trimeric and exosome-regulating Trf4/5p-Air1/2p-Mtr4p polyadenylation (TRAMP) complex (Houseley and Tollervey 2009). In human cells, hMTR4 forms at least two distinct complexes with different subunit compositions and subcellular localization (Lubas et al. 2011). In the nucleoplasm, hMTR4 and the two putative RNA-binding proteins RBM7 and ZCCHC8 comprise the nuclear exosome targeting (NEXT) complex, whereas in the nucleolus, hMTR4 forms a TRAMP-like complex (hTRAMP) with hTRF4-2 and ZCCHC7. In addition, the exosome was recently linked to the cap-binding complex (CBC) via ARS2 and ZC3H18 interactions with the NEXT complex, which altogether assemble the CBC-NEXT (CBCN) complex (Andersen et al. 2013).
Evidence demonstrates a role of the nuclear exosome and NEXT complexes in the decay of long noncoding RNA (lncRNAs), such as promoter-upstream transcripts (PROMPTs), enhancer RNAs (eRNAs), and extended U snRNAs (Preker et al. 2008; Lubas et al. 2011; Andersen et al. 2013; Andersson et al. 2014). PROMPTs comprise an >100 nt and 5′ capped species of lncRNAs, which arise in antisense direction from upstream of active transcription start sites (TSSs) (Ntini et al. 2013). Depletion of RNA exosome activity results in the robust stabilization of PROMPTs, which are normally subjected to rapid nucleolysis (Preker et al. 2008). Although, the exact mechanism of PROMPT decay remains unresolved, it has been shown that PROMPT regions, unlike their promoter-downstream counterparts, harbor early termination (polyadenylation-like) signals, which make them susceptible to rapid decay (Ntini et al. 2013). In another study, a potential binding of the NEXT components RBM7 and ZCCHC8 to PROMPTs was postulated (Lubas et al. 2011), which was later supported by RNA-immunoprecipitation (RIP) analysis (Andersen et al. 2013) and narrowed down to RBM7 using an individual nucleotide resolution cross-linking IP (iCLIP) approach (M Lubas, PR Andersen, A Schein, A Dziembowski, G Kudla, and TH Jensen, in prep.). Collectively, these studies suggest a role of exosome cofactors in the control of the degradation of PROMPTs.
Both RBM7 and ZCCHC8 undergo phosphorylation events. ZCCHC8 is a substrate of the glycogen synthase kinase-3 (GSK-3) (Gustafson et al. 2005) and RBM7 is phosphorylated in vitro by the p38-activated kinase MK2 (Rousseau et al. 2002). This indicates a role of post-translational modification as a regulatory layer of nuclear RNA decay and has been the subject of this study. Here, we used phospho-proteomics to identify a MK2-mediated phosphorylation of RBM7 at position S136. Cellular levels of unstable ncRNAs like PROMPTs and U snRNA metabolites were strongly dependent on the RBM7 phosphorylation status and CLIP experiments showed that this is due to altered RBM7–RNA-binding capacity which likely impacts subsequent substrate delivery to the nuclear exosome. As a consequence, PROMPTs accumulate during MK2-activating stress conditions. These effects could be reversed by using pharmacological inhibitors toward the p38MAPK/MK2 signaling axis and by point mutation of S136 in RBM7, respectively. Thus, a stress-induced post-transcriptional regulation of nuclear RNA decay through the p38MAPK/MK2 signaling axis connects the cellular stress response to nuclear ncRNA metabolism.
RESULTS
Stress-induced phosphorylation of RBM7S136 by MK2
In a phospho-proteomic approach (PhosphoScan) carried out with Cell Signaling technology comparing phospho-PKD-motif-directed protein phosphorylation (anti-LXRXXpS/T) of MK2/3-deficient and MK2-rescued mouse cells (Menon et al. 2013), we identified the RNA-binding protein Rbm7 as an in vivo substrate of MK2 with strongly increased phosphorylation in LPS-stimulated bone marrow-derived macrophages. Using mass spectrometry, the phosphorylated peptide and residue was determined as SFpS136SPEDYQR in mouse Rbm7. This was in agreement with a previous finding that Rbm7 is an in vitro substrate of MK2 (Rousseau et al. 2002). MK2 directly phosphorylates GST-Rbm7 at S136 in vitro, as determined by detecting radioactivity in Edman degradation of in vitro phosphorylated protein using [γ-32P]ATP labeling (cf. Materials and Methods). In human RBM7, S136 is located C-terminal to the RRM domain in a perfect MK2 phosphorylation site motif, ΦXRXXS (where Φ is a hydrophobic residue such as L, I, F, or V), which is specific for RBM7 of higher vertebrates, especially for human, chimp, mouse, and frog (Fig. 1A).
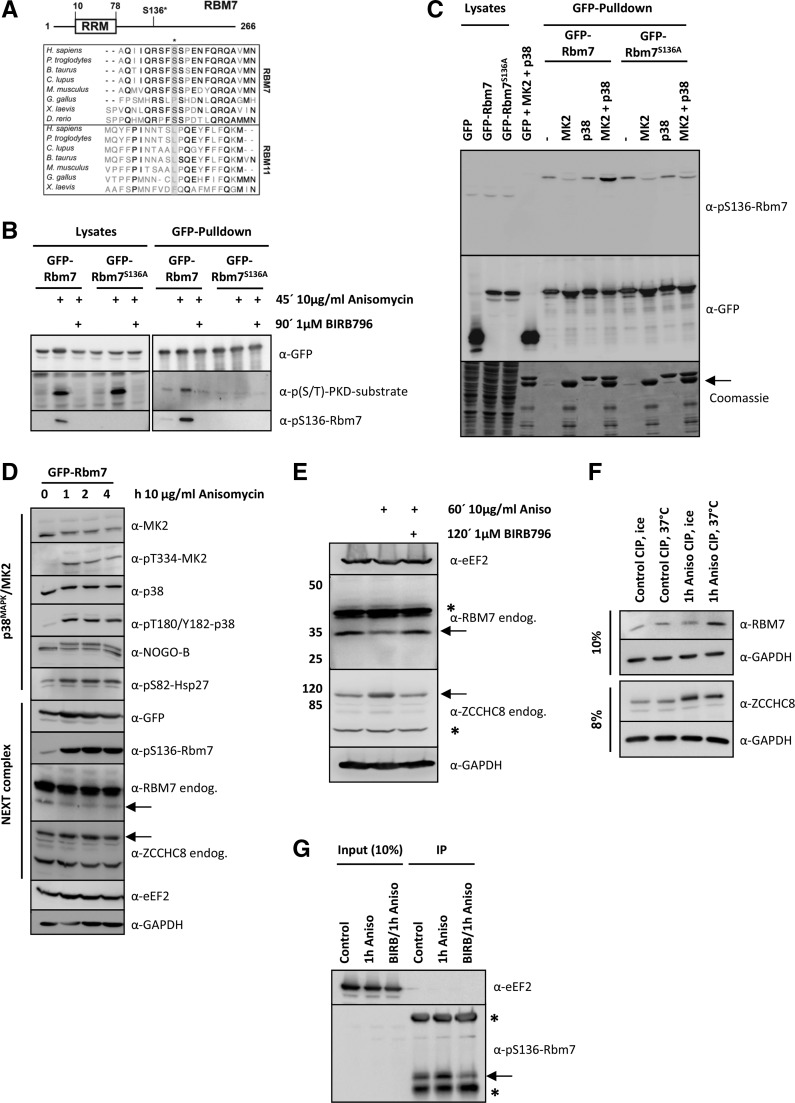
FIGURE 1.
RBM7 is phosphorylated by MK2 in vivo and in vitro. (A) Multiple sequence alignment of vertebrate RBM7 and its closest homolog RBM11. Displayed amino acid sequences span the human RBM7 S136 MK2 phosphorylation site (indicated by a star). The MK2-site is absent in RBM11. (B) Murine GFP-Rbm7 and GFP-Rbm7S136A were overexpressed in HeLa cells and treated as indicated. Upon precipitation with the help of a GFP-Nanotrap lysates and precipitates were developed with p(S/T)-PKD-substrate and pS136-Rbm7 antibodies to analyze Rbm7 phosphorylation and a GFP-antibody as loading control. Only wild-type Rbm7 gave rise to a specific phosphorylation on S136 upon anisomycin stimulation, whereas BIRB796-treatment and S136 to A mutation resulted in a loss of the phospho-signal. (C) Murine GFP-Rbm7 and GFP-Rbm7S136A were again overexpressed in HeLa cells and precipitated with a GFP-Nanotrap upon stimulation as indicated. The precipitates were then incubated with recombinant MK2 and p38 proteins for 30 min at 30°C to trigger in vitro phosphorylation. Afterward, samples were resolved by SDS-PAGE and either transferred to a nitrocellulose membrane and developed with the specific pS136-Rbm7- or GFP-antibody to monitor equal precipitation or stained by Coomassie. As demonstrated with the in vivo experiments (2B and 2C) only active MK2 (MK2 + p38) could phosphorylate Rbm7 in vitro. Precipitated GFP-Rbm7 and GFP-Rbm7S136A was detected at the same molecular weight as recombinant MK2. Its positions are indicated by an arrow. (D) Murine GFP-Rbm7 was expressed in HeLa cells and stimulated with anisomycin for 0, 1, 2, and 4 h to gain information on the phosphorylation kinetics of RBM7 and the p38MAPK-signaling pathway components and targets. The anti-pS136-Rbm7 antibody only detected the phosphorylated exogenous GFP-tagged Rbm7 protein. The positions of endogenous RBM7 and ZCCHC8 are indicated by arrows. Anisomycin stimulation altered the band intensities of RBM7 and ZCCHC8. (E) To analyze the potential phosphorylation of endogenous RBM7 and ZCCHC8 in more detail, extracts of HeLa cells treated as indicated were resolved on 8% and 12% SDS gels, respectively. Still, the endogenous RBM7-signal is weaker when cells are treated with anisomycin and the ZCCHC8 signal shifts slightly to a higher molecular weight. Both effects could point to a phosphorylation of the endogenous proteins. Unspecific bands coming up with antibodies detecting endogenous RBM7 and ZCCHC8 are marked with a star. (F) To verify endogenous RBM7 and ZCCHC8 phosphorylation, cell lysates that were treated as indicated and incubated with phosphatase (CIP, see Materials and Methods) were analyzed by Western blotting upon running 8% or 10% gels, respectively. (G) Endogenous RBM7 from HeLa cells that were treated as in F was immunoprecipitated using the anti-pS136-Rbm7 antibody (see Materials and Methods for details). Samples were analyzed for the presence of pS136-RBM7 using the same antibody and eEF2 for equal loading. The arrow indicates endogenous phosphorylated RBM7 and the stars indicate for the heavy and the light of the antibody used for IP.
To further validate this phosphorylation, mouse wild-type (WT) GFP-tagged Rbm7 was ectopically expressed in HeLa cells and enriched by a GFP-Pulldown (Rothbauer et al. 2008). Precipitated proteins were probed with either anti-p-PKD-substrate-motif (LXRXXpS/T) antibody or an anti-pS136-Rbm7 antibody, both of which gave rise to a phospho-specific signal when HeLa cells were stimulated with anisomycin (Fig. 1B). This indicated the involvement of p38MAPK in the phosphorylation event, as anisomycin is known to be a strong activator of this pathway (Hazzalin et al. 1996). Importantly, inhibition of p38 using BIRB796 (Kuma et al. 2005) led to a complete loss of the phospho-signal, further indicating a role of the p38MAPK/MK2 signaling module in the Rbm7 phosphorylation (Fig. 1B). This result was confirmed for human FLAG-tagged RBM7 in HeLa cells (Supplemental Fig. S1A). The putatively nonphosphorylatable mutant of murine Rbm7, Rbm7S136A, was generated and similar pulldown experiments were conducted and showed no phosphorylation of Rbm7S136A in response to anisomycin (Fig. 1B). Finally, the direct phosphorylation of Rbm7 by MK2 was analyzed by an in vitro phosphorylation assay (Fig. 1C). Recombinant MK2 and/or p38 were mixed with GFP-Rbm7 or GFP-Rbm7S136A and incubated at 30°C in the presence of ATP. Subsequently, samples were resolved by SDS-PAGE and analyzed by Western blotting using the pS136-Rbm7-specific antibody. Only the p38-activated MK2 was able to phosphorylate WT Rbm7 in vitro and gave rise to a robust phosphorylation-specific signal, whereas Rbm7S136A phosphorylation was unaffected by the same treatment (Fig. 1C). Alterations in the appearance of nonphosphorylated GFP-fusion proteins were due to the fact that they migrated at the same position as recombinant MK2 (Fig. 1C, lower panel). The phosphorylation result was confirmed by a radioactive kinase assay using [γ-33P]ATP and showing that only activated MK2, and not p38 alone, was able to phosphorylate Rbm7 (Supplemental Fig. S1B). Notably, mutant Rbm7S136A was still phosphorylated by active MK2 to a certain degree (see Discussion). Taken together, our results indicate that RBM7S136 is a major MK2-phosphorylation site that is conserved in most vertebrates (Fig. 1A).
Next, we measured the kinetics of anisomycin-induced phosphorylation of murine Rbm7 in HeLa cells. Rbm7 phosphorylation at S136 as well as activity of p38 and MK2 (as measured by the activating phosphorylations of pT180/pY182 and pT334, respectively) could be clearly detected after 1 h of anisomycin treatment and were sustained during the full 4 h observation time (Fig. 1D). Notably, only GFP-tagged Rbm7 protein was detected by the anti-pS136-Rbm7 antibody. Simultaneously, the subcellular localizations of the overexpressed GFP-Rbm7 and GFP-Rbm7S136A were monitored (Supplemental Fig. S2A). As reported for human RBM7 (Lubas et al. 2011; Hett and West 2014), these proteins were exclusively localized to the nuclear compartment and did not display notable changes in localization upon anisomycin-treatment. Established “bona fide” substrates of MK2, such as Hsp27 (Stokoe et al. 1992) and NOGO-B (Rousseau et al. 2005), displayed similar kinetics of phosphorylation (Fig. 1D). We also detected endogenous NEXT complex proteins RBM7 and ZCCHC8 (Fig. 1D). Anisomycin stimulation resulted in a reduction of endogenous RBM7, which correlated well with the MK2-mediated phosphorylation of murine Rbm7 and could be used to monitor the in vivo phosphorylation of endogenous RBM7 (Fig. 1D–F). The disappearance of endogenous RBM7 signal was similar to the weaker signal of total MK2 upon anisomycin stimulation that is due to weaker detection of the phospho-protein by the antibody (see MK2 panel in Fig. 1D and Tiedje et al. 2012). The effect on RBM7 detection was reversed by pretreatment with the p38MAPK-inhibitor BIRB796 (Fig. 1E) and could also indicate phosphorylation or protein stabilization. To exclude a role for stress-induced proteasomal degradation of RBM7, anisomycin stimulation was combined with proteasome inhibitor treatment. In the presence of the proteasome inhibitor MG-132, stress stimulation also altered the RBM7 signals (Supplemental Fig. S2B), arguing against the involvement of proteasomal degradation of RBM7 and substantiating the potential monitoring of endogenous RBM7 phosphorylation by immunoblotting signals. To confirm the in vivo phosphorylation of endogenous RBM7, lysates of HeLa cells were treated with calf intestine phosphatase (CIP) before analysis (Fig. 1F). Indeed, the endogenous RBM7 signal was turned into a distinct intense band upon this treatment compared with the endogenous RBM7-signal of anisomycin-treated cells not treated (ice) (Fig. 1F). Interestingly, the ZCCHC8 signal was increased and shifted to lower mobility upon anisomycin treatment and also collapsed to a sharper band upon CIP treatment (Fig. 1F). Further evidence for an in vivo phosphorylation of endogenous RBM7 was provided by precipitating RBM7 using the phospho-specific anti-pS136-Rbm7 antibody. Clearly, in anisomycin-treated cells, the immunoprecipitate is enriched for the phosphorylated RBM7 protein (Fig. 1G). Importantly, this effect was reversed by the use of the p38MAPK-inhibitor BIRB796. In sum, we further confirmed that RBM7 undergoes MK2-dependent phosphorylation in vivo and in vitro, which is sustained following activation kinetics of the p38MAPK/MK2 pathway.
Phosphorylation of RBM7 does not alter interaction with ZCCHC8 but decreases its RNA-binding capacity
RBM7 associates with ZCCHC8 and hMTR4 forming the trimeric NEXT complex (Lubas et al. 2011), which interacts with both the nuclear RNA exosome and the nuclear CBC (Andersen et al. 2013). To assess any functional consequences of RBM7 phosphorylation, we first monitored a possible phosphorylation-dependent interaction of murine N-terminally tagged GFP-Rbm7 with FLAG-tagged human ZCCHC8. Importantly for the pull down, we found that GFP-Rbm7 and ZCCHC8-FLAG can form a complex in vivo (Fig. 2A). However, neither stimulation of cells by anisomycin nor the S136A mutation of Rbm7 altered its pulldown efficiency of FLAG-ZCCHC8 (Fig. 2A). Thus, RBM7 phosphorylation does not influence the composition of the RBM7/ZCCHC8 dimer. Consistently, coprecipitation of GFP-tagged Rbm7 by FLAG-ZCCHC8 was also not affected by the same manipulations (Supplemental Fig. S3A). To study the interaction of tagged RBM7 or ZCCHC8 with its endogenous interacting proteins, the experiment was repeated using cells overexpressing only one of the proteins. Under these conditions, GFP-Rbm7 and GFP-ZCCHC8 did interact with endogenous ZCCHC8 and RBM7, respectively, independently of mutations and the activation status of the cells (Supplemental Fig. S3B,C). GFP-Rbm7 and its mutant were additionally analyzed for their ability to pull down CBP80 protein (Supplemental Fig. S3C). This revealed a loss of interaction of both proteins upon stimulation independent of p38MAPK inhibition. Notably, overexpressed GFP-ZCCHC8 protein was unstable and difficult to pull down (Supplemental Fig. S3B). This notion was supported by the finding that tagged and endogenous ZCCHC8 protein levels were stabilized when Rbm7 was coexpressed (Fig. 2A; Supplemental Fig. S3A).
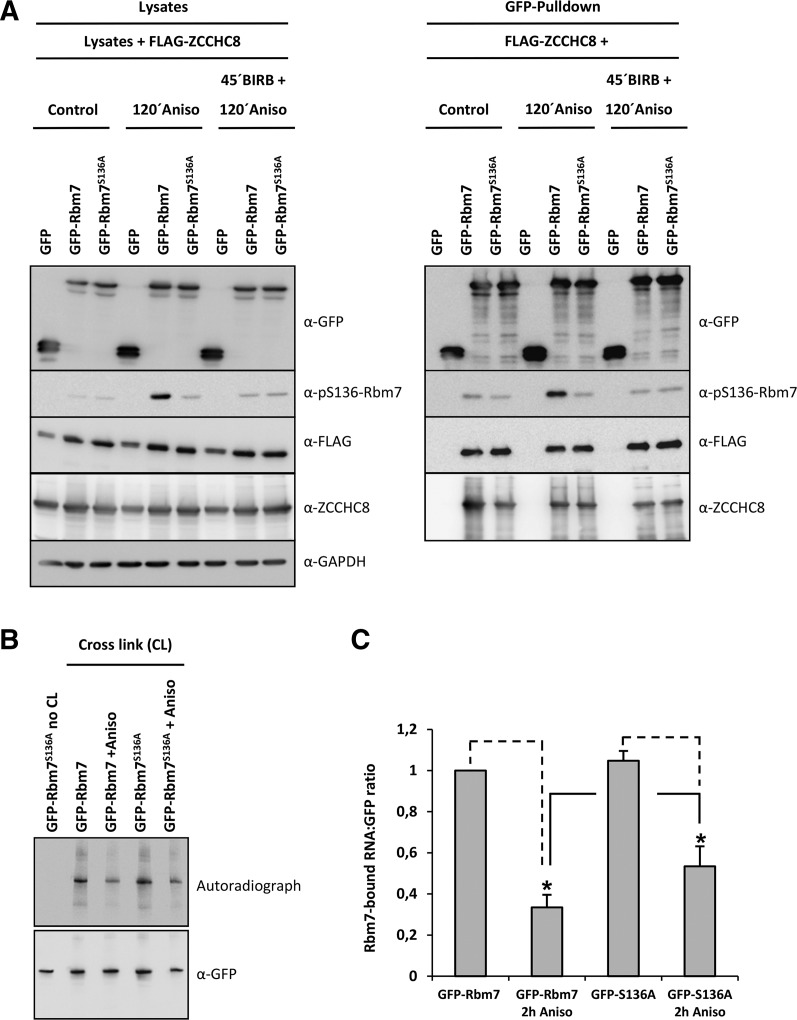
FIGURE 2.
Consequences of MK2-mediated phosphorylation of RBM7 on its protein–protein interactions and its RNA-binding capacity. (A) N-terminally GFP-tagged Rbm7 and Rbm7S136A were coexpressed with FLAG-tagged ZCCHC8 in HeLa cells. Their interaction was analyzed in nonstimulated cells, in cells stimulated with anisomycin for 2 h or in cells stimulated with anisomycin after BIRB796-mediated p38MAPK-inhibtion. GFP-fusion proteins were precipitated as mentioned in the Materials and Methods. Equal expression of GFP- and FLAG-tagged fusion proteins and GAPDH (loading control) in the lysate was monitored by Western blotting. In addition, the stimulation with anisomycin and the inhibition by BIRB796 was checked with the above-mentioned pS136-Rbm7-specific antibody. It became clear that the interaction of exogenous Rbm7 and ZCCHC8 independent of the activation status of the cells and the phosphorylation status of the proteins was not altered. (B) HeLa cells overexpressing GFP-Rbm7 or GFP-Rbm7S136A were cross-linked with 300 mJ/cm2 UV-C light prior to cell lysis to covalently connect Rbm7 to the RNAs to which it is bound. As indicated, different conditions were analyzed and the cross-linked RNAs were radioactively labeled, visualized by autoradiography and developed afterward with an anti-GFP-antibody by Western blotting. As a negative control GFP-Rbm7S136A expressing cells were not cross-linked but otherwise treated like the other samples. Consistently, the precipitate did not produce signals of copurifying RNAs. (C) Rbm7-bound RNA signals of the autoradiograph were normalized to anti-GFP Western blot signals derived from the same membrane (see Materials and Methods). The star indicates P < 0.01. The replicate for parts of this experiment is shown in Supplemental Figure S4B.
Next we analyzed a potential phosphorylation-dependent change in the RNA-binding capacity of RBM7. To this end, GFP-Rbm7 or GFP-Rbm7S136A expressed in HeLa cells were cross-linked (CL) to cellular RNA using UV-C light prior to precipitation (CLIP). Notably, RNaseI treatment was not sufficient to fully digest RNAs cross-linked to Rbm7 even at high concentrations and resulted in a smear of various RNA-Rbm7 cross-linking products (Supplemental Fig. S4A). However, a mixture of RNaseA and RNaseT1 was able to digest copurifying RNAs to the minimal sizes protected by the cross-linked Rbm7 protein (Fig. 2B). As controls, expressing GFP alone (Supplemental Fig. S4A) or omitting the UV treatment (Fig. 2B, “no CL”) resulted in no RNA copurification. Importantly, anisomycin-treatment reduced the RNA levels immunoprecipitated by Rbm7 (to ∼30%), a reduction which was partially rescued by the S136A mutation (to ∼50%) (Fig. 2B,C; Supplemental Fig. S4B). The effect of p38MAPK/MK2-activation on the Rbm7–RNA-binding capacity was confirmed by treating GFP-Rbm7-expressing HeLa cells with BIRB796 prior to anisomycin-treatment (Supplemental Fig. S4C). In addition, RNA mobility shift assays and AU-rich element firefly reporter systems were utilized to determine any potential AU-rich element (ARE) binding capacity of Rbm7 (Rousseau et al. 2002). Using both assays, no ARE-binding activity of Rbm7 could be detected (Supplemental Fig. S5A,B). Together, our findings suggest that the p38MAPK/MK2 axis lowers the target RNA recruitment by the NEXT complex component RBM7 in an ARE-independent manner.
Stress-induced and p38-dependent accumulation of nuclear ncRNAs
To assay the influence of phosphorylation of RBM7 on cellular RNA, we analyzed the expression of the exosome-sensitive PROMPTs in anisomycin or UV-C treated HeLa cells. Anisomycin-treatment induced a strong and sustained increase in the p38MAPK/MK2 activity (Figs. 1D, 3A) whereas the effect of UV-C was rather moderate (Fig. 3A). The levels of four prominent PROMPTs (proRBM39, proIFNAR1, proEXT1, and proDNAJB4) were investigated by qRT-PCR after 1, 2, and 4 h of anisomycin treatment of HeLa cells. The proEXT1, proRBM39, and proDNAJB4 transcripts were strongly accumulating with maxima after 1 and 2 h, respectively, while proIFNAR1 showed a more moderate accumulation peaking at ∼2 h (Fig. 3B). The kinetics of PROMPTs induction followed the kinetics of p38 activation (Fig. 1D) and pretreatment of cells with BIRB796 fully inhibited their stress-induced accumulation, suggesting the involvement of the p38MAPK/MK2 signaling axis. This idea was further supported by our findings that addition of the structurally unrelated p38MAPK-inhibitors SB203580 and SB202190 lead to a similar dampening of PROMPTs levels (Supplemental Fig. S6A). Only SB203580 failed to inhibit proIFNAR1 increase and could indicate its lower efficiency to block p38MAPK-activity (Menon et al. 2011). Moreover, the previously described GSK-3-mediated phosphorylation of ZCCHC8 (Gustafson et al. 2005) could also contribute. However, when assaying PROMPT levels after inhibition of GSK-3 with LiCl treatment we found no clear changes of proRBM39 and proEXT1 RNAs and only a minor alteration of proDNAJB4 levels (Supplemental Fig. S6B). The diminished levels of endogenous RBM7 protein were also not reversed by LiCl treatment (Supplemental Fig. S6B). To test whether the observed effects were specific for PROMPTs, we compared anisomycin-stimulated expression levels of proRBM39 with its neighboring RBM39 mRNA. While proRBM39 was strongly increased peaking after 2 h (Fig. 3B), the RBM39 mRNA showed only a weak linear accumulation which is only slightly inhibited by BIRB796 (Fig. 3C). Thus, p38MAPK-activity appears to specifically affect the RBM39 PROMPT. To verify that the observed effects were not due to an anisomycin-triggered off-target effect, we also analyzed PROMPT expressions triggered by UV-C-light treatment. The UV-C dose chosen stimulated p38MAPK-activity (Fig. 3A) and also led to a BIRB796-sensitive accumulation of proEXT1, proRBM39, proDNAJB4, and proIFNAR1 (Fig. 3D). Thus, increased PROMPTs levels are a general stress response for different stimuli-activating p38MAPK.
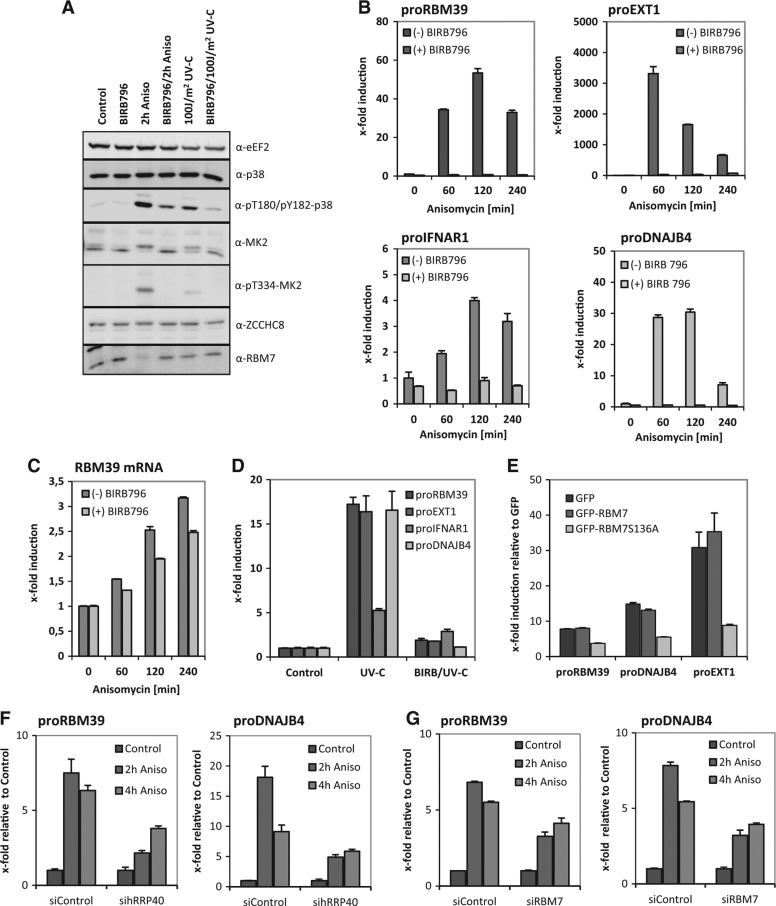
FIGURE 3.
PROMPTs are accumulating by anisomycin- and UV-C light-induced p38MAPK-activation in HeLa cells. (A) Treatment of HeLa cells with 10 µg/mL anisomycin for 2 h or irradiation with 100 J/m2 UV-C light results in the strong activation of p38MAPK as shown by Western blot for phospho-T180 and phospho-Y182 in p38 and the activation of its downstream kinase MK2 as monitored by T334 phosphorylation. Inhibition of p38MAPK-activity by the inhibitor BIRB796 (BIRB) prevents p38- and MK2-phosphorylation. Total p38 and MK2 protein remained unchanged throughout the experiments. The translation elongation factor eEF2 served as a loading control and RBM7 and ZCCHC8 showed similar band patterns in response to stress like in previous experiments (see Figs. 1E–G, 4A,C). (B) Using the anisomycin stimulation experimental setup, expression of four well-characterized PROMPTs (proRBM39, proEXT1, proIFNAR1, and proDNAJB4) was analyzed at different time points (0, 60, 120, and 240 min anisomycin stimulation) by qRT-PCR. (C) As a control the anisomycin-induced downstream expression of RBM39 was analyzed. (D) Like the anisomycin-induced up-regulation of PROMPTs the UV-C light-mediated accumulation can be abolished by p38MAPK-inhibition using BIRB796. All results are shown as x-fold induction relative to the nonstimulated expression that was set to 1. All graphs are displayed with standard deviations. All experiments were performed independently a minimum of two times and showed similar effects. (E) Overexpression of GFP-RBM7S136A leads to diminished PROMPTs accumulation upon anisomycin stimulation indicating that stress targets the exosomal degradation of PROMPTs via RBM7. (F,G) HeLa cells were treated with either a Control siRNA or hRRP40- and RBM7-specific siRNAs and 48 h post-transfection treated as indicated. Levels of proRBM39 and proDNAJB4 were determined by qRT-PCR and accumulation was calculated relative to untreated cells for each siRNA knockdown. Knockdown efficiency of hRRP40 and RBM7 was assayed by Western blot (Supplemental Fig. S7B).
As different stress inductions led to the accumulation of PROMPTs, we speculated that the MK2-mediated and stress-induced phosphorylation of RBM7 is connected to this effect. Therefore, we overexpressed the S136A mutant of RBM7 in HeLa cells and reanalyzed the PROMPTs accumulation. Indeed, the S136A mutant prevented PROMPTs accumulation to >50%, whereas overexpression of the wild-type protein did not (Fig. 3E). Equal expression of the GFP-fusion proteins was monitored by Western blotting (Supplemental Fig. S7A). As RBM7 participates in the RNA exosome-mediated decay of PROMPTs, we further tested whether anisomycin could work through this pathway. The RNA exosome core subunit, hRRP40, and RBM7 were siRNA-depleted in HeLa cells (Supplemental Fig. S7B) and subjected to qRT-PCR analysis. Accumulation of proDNAJB4 and proRBM39 was calculated relative to untreated cells for each siRNA knockdown (Fig. 3F,G). Depletion of either hRRP40 or RBM7 obviously lowered the observed anisomycin induction of PROMPTs. We also noted that the decrease in PROMPTs accumulation was stronger for the more efficient hRRP40 depletion (cf. Supplemental Fig. S7B and Fig. 3F,G). Having observed that partial block in the exosome/RBM7 decay pathway lowered anisomycin-induced accumulation of PROMPTs, we conclude that p38MAPK/MK2-mediated phosphorylation of RBM7 affects nuclear exosome-mediated decay of PROMPTs.
Besides PROMPTs, extended U2 snRNA has been described as a target of the NEXT complex (Andersen et al. 2013) and U6atac levels are affected by anisomycin-induced p38MAPK activation (Younis et al. 2013). Hence, we asked whether U snRNAs could also be affected by anisomycin. Using PCR primers, that do not discriminate between premature, or mature U snRNAs, we determined the levels of snRNAs U1, U2, U4, U11, U12, and U6atac in HeLa cells before and after anisomycin treatment. Interestingly, U1, U11, and U6atac snRNA metabolites, which end up as parts of the major and minor spliceosomes, respectively, displayed a strong anisomycin-dependent accumulation, which was inhibited by BIRB796 (Supplemental Fig. S7C).
PROMPTs accumulation is a cell type-independent stress-induced response
We then asked whether the observed stress-induced p38MAPK/MK2-dependent PROMPT accumulation is specific for HeLa cells. Hence, we analyzed PROMPTs expression in the monocytic line THP-1, in which specific-induction of p38MAPK activity could also proceed by LPS addition via TLR4-mediated signaling (Han et al. 1994; Lee et al. 1994). As seen in Figure 4A, anisomycin treatment led to the same phosphorylation-dependent change in the RBM7 band as observed in HeLa cells (Fig. 1D,E). Consistently, anisomycin stimulation resulted in high PROMPTs levels (Fig. 4B), demonstrating that PROMPTs-accumulation is not cell-type specific. In contrast, LPS treatment that induced TNFα expression by the monocytes (Fig. 4C) led only to a weakly increased proIFNAR abundance but unaltered levels of proRBM39, proEXT1, and proDNAJB4 (Fig. 4D). This suggests that PROMPTs-induction might depend on the quality of the stress stimulus. Similarly, IL-1α-stimulated HeLa cells showed a light p38MAPK-dependent up-regulation of only proRBM39 after 60 min of stimulation (Supplemental Fig. S8A). These findings and the observation that osmotic stress only weakly and selectively stimulates PROMPTs level in HeLa cells (Supplemental Fig. S8B) imply that activation of the p38MAPK/MK2 axis is necessary but not sufficient for the response observed.
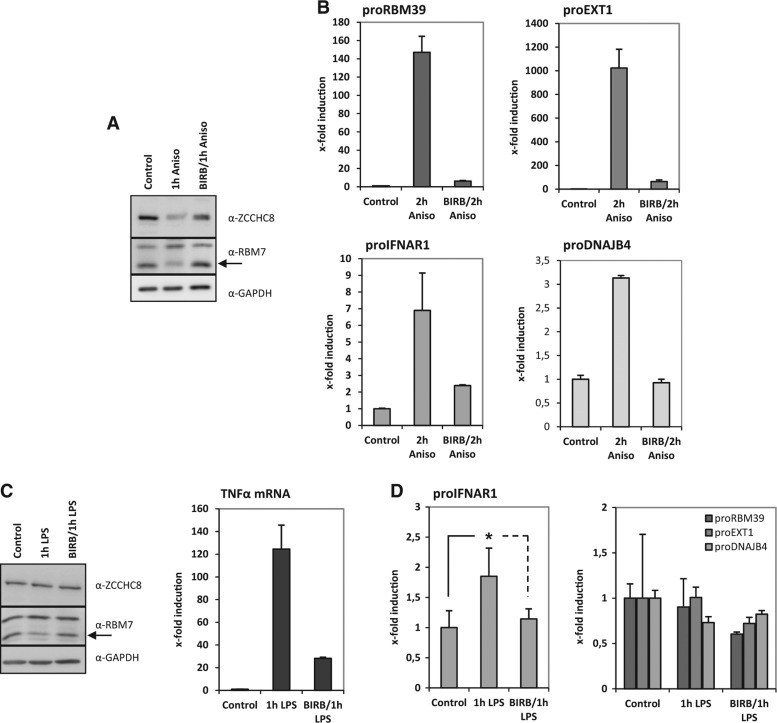
FIGURE 4.
PROMPTs are accumulating by anisomycin and partially by LPS stimulations in monocytic THP-1 cells. (A) THP-1 cells were stimulated for 1 h with 10 µg/mL anisomycin in the presence or absence of the p38-inhibitor BIRB796 and the indicated proteins were assayed by Western blot. (B) THP-1 cells were stimulated for 2 h with 10 µg/mL anisomycin in the presence or absence of the p38-inhibitor BIRB796. Consequently, PROMPTs RNA levels were determined by qRT-PCR and shown as x-fold induction relative to the nonstimulated expression levels. (C) LPS stimulation (1 µg/mL) for 1 h of THP-1 cells resulted in a strong induction of TNFα mRNA in THP-1 cells (right panel). Induction of TNFα was sensitive to inhibition of p38MAPK activity. Blotting for endogenous proteins showed similar loss of RBM7 band intensity as in Figure 1E–G or A. (D) The same cDNAs as in C were analyzed for PROMPTs RNA levels. While proRBM39, proEXT1, and proDNJAB4 levels were not changed, proIFNAR1 levels were twofold up-regulated by LPS stimulation in a p38-dependent manner. All graphs are displayed with standard deviations. The star indicates P < 0.04 (as determined by two-sided Student's t-test) for the LPS-induced up-regulation of proIFNAR1.
Pharmacological inhibition and shRNA-mediated knockdown of MK2 reduce stress-dependent PROMPTs accumulation
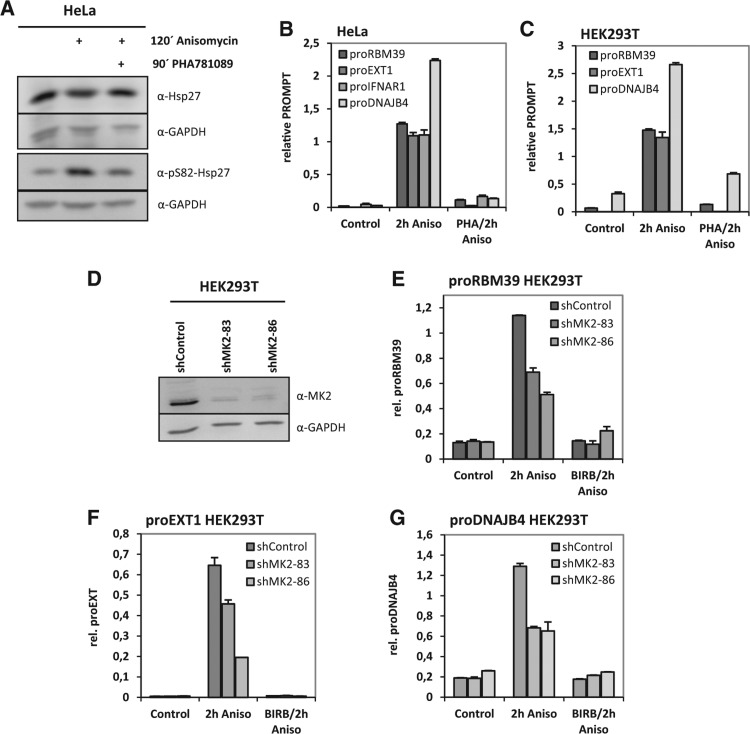
To investigate whether the protein kinase downstream to p38MAPK, MAPK-activated protein kinase 2 (MK2), is involved in the observed effects on PROMPTs, we treated HeLa cells with the MK2-inhibitor PHA781089 (Anderson et al. 2007) before anisomycin stimulation and analyzed MK2 activity toward its bona fide substrate Hsp27 as well as PROMPTs. MK2 activity was induced by anisomycin and inhibited by PHA781089 as seen by the diagnostic changes of Hsp27 phosphorylation (Fig. 5A). Anisomycin-induced up-regulation of PROMPTs was clearly inhibited by PHA781089 while the expression of the RBM39 mRNA was only weakly affected (Fig. 5B; Supplemental Fig. S9A). Of the PROMPTs investigated, proRBM39, proEXT1, and proDNAJB4 are also expressed in HEK293T cells and their expression was strongly inhibited by PHA781089 (Fig. 5C). The level of proIFNAR1 in HEK293T is very low and its accumulation by anisomycin is not as efficient as in HeLa cells (Supplemental Fig. S9B).
FIGURE 5.
PROMPTs accumulation is sensitive to inhibition of the p38MAPK-activted kinase MK2 in HeLa and HEK293T cells. (A) Prior to 2 h anisomycin-stimulation HeLa cells were preincubated for 90 min with MK2-inhibitor PHA781089 (15 µM). The phosphorylation of MK2's bona fide substrate Hsp27 was monitored through Western blot for pS82-Hsp27. Blots for total Hsp27 and GAPDH served as loading controls. (B) Under the same conditions as in A. PROMPTs RNA levels were determined and are shown as x-fold inductions. MK2-inhibition led to a strong down-regulation of PROMPTs levels under these conditions. (C) PROMPTs were detected in HEK293T cells. Although up-regulation of proIFNAR1 was not detectable in these cells, the anisomycin-induction of proRM39 and proEXT1 was highly sensitive to the p38MAPK-activated kinase MK2 as shown by qRT-PCR. (D) Three different stable HEK293T cell lines expressing two distinct MK2-specific shRNAs and a sh-control-RNA were generated. MK2-knockdown efficacy was controlled through blotting for MK2. GAPDH served as a loading control. MK2 levels were strongly reduced in the two MK2-specific knockdown cell lines when compared with the control cell line. (E–G) The anisomycin-induced levels of proRBM39 (E), proEXT1 (F), and proDNAJB4 (G) were assayed in the control and knockdown cell lines. Both knockdown cell lines showed reduced PROMPTs levels upon anisomycin stimulation compared with the control cell line. Notably, PROMPTs levels were further reduced by pretreatment of the cells with the p38-inhibitor BIRB796. All graphs are displayed with standard deviations. All experiments were performed independently a minimum of two times and showed similar effects.
As small molecule inhibitors are rarely specific to only one protein kinase, we also performed MK2-knockdown experiments in HEK293T cells that were stably expressing two different MK2-specific shRNAs and one control shRNA (Fig. 5D). Both MK2-knockdown cell lines displayed reduced anisomycin-stimulated levels of proRBM39, proEXT1, and proDNAJB4 (Fig. 5E–G). Probably due to residual MK2 levels in these cell lines, there was a further reduction of PROMPTs levels in the presence of BIRB796 (Fig. 5E–G). Taken together, these data support MK2 in p38-mediated PROMPTs regulation.
Anisomycin stimulation does not affect RNAPII occupancy at the proRBM39 locus but increases PROMPTs stability
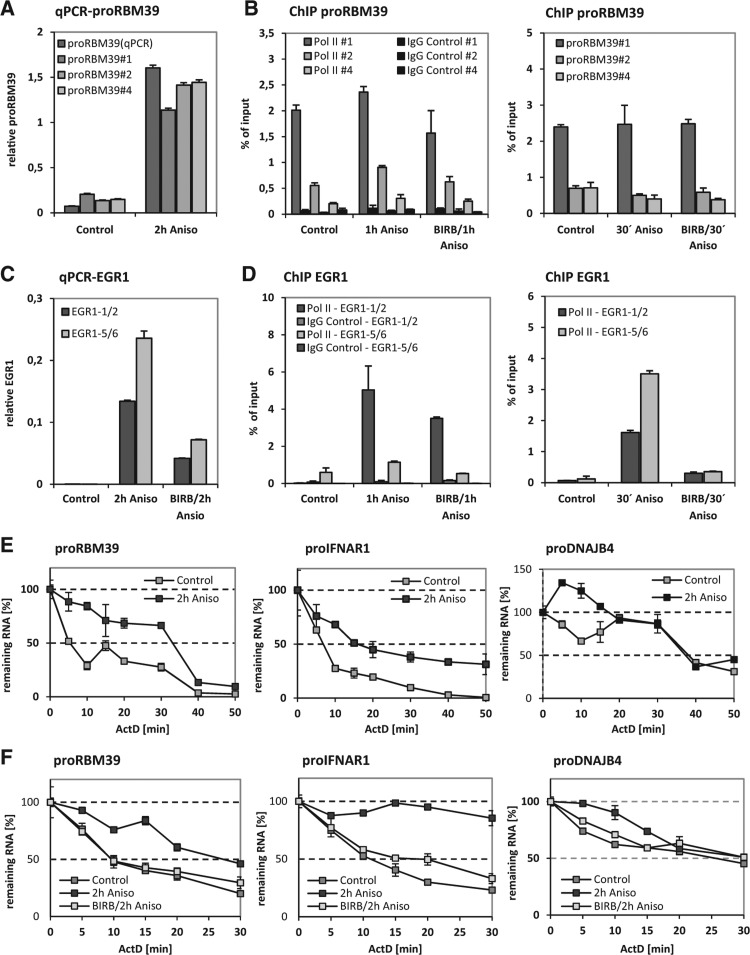
To address a possible influence of the p38MAPK/MK2 axis on the transcription of PROMPTs, we performed RNAPII chromatin-IP (ChIP) assays of the proRBM39 region. Different published amplicons along the proRBM39 transcribed region were analyzed (Andersen et al. 2013). As a control, qRT-PCR analysis of cDNA samples conducted with the ChIP primers showed anisomycin-dependent accumulation of RNA at all selected sites (Fig. 6A). However, no obvious change in RNAPII occupancy was detected by ChIP after 30 and 60 min of anisomycin treatment (Fig. 6B). Furthermore, there was no effect of BIRB796 coadditon on RNAPII occupancy at the proRBM39 locus (Fig. 6B). As a positive control, RNAPII ChIP showed increased anisomycin-stimulated occupancy at the stress-activated EGR1 gene at both time points using amplicons spanning the 5′ UTR and early coding sequence (CDS) of EGR1 (primers 1 and 2) or a CDS downstream region (primers 5 and 6) (Fig. 6D,E). In parallel, EGR1 transcript levels were strongly induced upon anisomycin stimulation (Fig. 6C) resulting in high occupancy levels in the gene body of EGR1 (Fig. 6D). Together, these data argue against a role of transcription in the p38MAPK/MK2-induced accumulation of proRBM39.
FIGURE 6.
PROMPTs are regulated at the level of stability and not by RNAPII-mediated changes in transcription regulation. (A) The ability of three different proRBM39 amplicons along the transcript (#1, #2, and #4) (Andersen et al. 2013) to assess the anisomycin-induced up-regulation of proRBM39 in comparison to the primer pair used in all other qRT-PCR experiments was proven. All primers were similarly effective in detecting the strong induction. We note that the ChIP primer showed higher background amplification than the common qRT-PCR primers. (B) RNAPII ChIP analyses were performed after treating HeLa cells for 30 and for 60 min with anisomycin (10 µg/mL). The DNA-fragments cross-linked to RNAPII and to the control IgG were analyzed by qPCR. Only minor differences in RNAPII occupancy of the proRBM39 transcript region were detected. Notably, the occupancy along the transcript region decreases strongly toward upstream. (C) The anisomycin-induced up-regulation shows a pattern similar to PROMPTs, but the RNAPII occupancy pattern differs strikingly from PROMPTs. (D) Regardless of the duration of anisomycin stimulation the occupancy along the transcript is increased under stimulatory conditions. (E) The stability of PROMPTs was assayed in nonstimulated HeLa cells or in cells after 2 h of anisomycin stimulation by inhibiting RNA-Polymerase II transcription by the addition of 10 µg/mL actinomycin D. Every 5 min the PROMPTs RNA levels were determined by qRT-PCR and are shown as the remaining RNA amount (% of time point 0, start of actinomycin D addition). proRBM39, proIFNAR1, and proDNAJB4 transcripts were stabilized upon stimulation (increase of their half-life from 5–10 min to 20–35 min). The half-life of proEXT1 during anisomycin stimulation was ∼20 min and in the same range of the other PROMPTs that were analyzed (Supplemental Fig. S10A). Under nonstimulated conditions only the first time point before adding actinomycin D was accessible for qRT-PCR measurement as levels after actinomycin treatment were equal to background detection. (F) BIRB796-mediated p38MAPK-inhibtion reverses the gain in RNA stability of PROMPTs. The stability of proRBM39, proIFNAR1, and proDNJAB4 from derived HeLa cells treated as indicated was determined. BIRB796-treament led to an intermediate half-life of PROMPTs RNAs. All graphs are displayed with standard deviations. All experiments were performed independently a minimum of two times and showed similar effects.
As transcription seemed not to be the reason for regulating the abundance of PROMPTs, we determined a possible role of the p38MAPK/MK2 axis in the stability of PROMPTs and blocked transcription by actinomycin D to perform chase experiments in HeLa cells. Importantly, due to the very unstable character of these RNAs, a high-resolution analysis was performed determining remaining transcript levels every 5–10 min. In untreated cells, proRBM39, proIFNAR1, and proDNAJB4 transcripts were highly unstable exhibiting half-lives between 5 and 10 min (Fig. 6E). Owing to even lower stability, proper proEXT1 RNA decay kinetics could not be detected in nonstimulated cells. Critically, anisomycin treatment led to increased stability of proRBM39 and proIFNAR1 raising their half-lives to ∼30 and 20 min, respectively, and a superinduction of proDNAJB4 (Fig. 6E). In addition, proEXT1 mRNA now became detectable with a half-life of ∼20 min (Supplemental Fig. S10A). For proDNAJB4 effects of hyperstabilization were observed at early time points (Fig. 6E). We speculate that it might have been due to the kinetics of actinomycin D action or to the phenomenon of superinduction. Parallel treatment with anisomycin and BIRB796 partially reversed the increased stabilization of proRBM39, proIFNAR1, and proDNAJB4 (Fig. 6F). We also determined stability of U1 and U11 snRNAs metabolites (including pre-, mature-, and extended U snRNA transcripts) in the presence and absence of anisomycin and could determine an increased stability upon anisomycin treatment that was reversed by the p38MAPK-inhibitor BIRB796 (Supplemental Fig. S10B). These anisomycin-induced increases in PROMPTs and U snRNA metabolite stability detected here could explain the increased abundance of these ncRNAs under stress.
DISCUSSION
A variety of proteins bind to specific mRNA-elements, often located in 5′ or 3′ UTRs, and post-transcriptionally regulate transcript stability or translation mainly in the cytoplasm (for review, see Anderson 2008; Ray et al. 2013). Well-characterized proteins like HuR and TTP that undergo signal-dependent phosphorylation by p38MAPK/MK2 signaling are prominent examples of this class of proteins (for review, see Tiedje et al. 2014). Here we report a nucleus-specific stress-induced mechanism of ncRNA regulation functioning through an activated p38MAPK/MK2 signaling axis modulating the general RNA-binding function of RBM7.
We show that MK2, a protein kinase activated in the nucleus (Engel et al. 1998), phosphorylates the human and murine NEXT complex subunit RBM7 at serine 136 in a stress-dependent manner and by this regulates the turnover of nuclear ncRNAs like PROMPTs. As interactions of NEXT complex partners remain unchanged, functional alterations of the NEXT were analyzed by RNA-CLIP assays and revealed a decreased RNA-binding capacity of RBM7 during stress. As a consequence of RBM7 phosphorylation ncRNAs become stabilized and are no more delivered effectively to the RNA exosome as detected by their accumulation. The mechanism can be triggered in HeLa, HEK293T, and THP-1 cells (summarized in Fig. 7) and can be partially reversed by either BIRB796-mediated p38MAPK-inhibition or the use of the S136A Rbm7 mutant. Four well-described PROMPTs, proRBM39, proIFNAR1, proEXT1, and proDNAJB4 (Lubas et al. 2011; Andersen et al. 2013; Ntini et al. 2013) were investigated upon anisomycin-induced stress. Under more physiological conditions proIFNAR1 is also up-regulated by LPS stimulation in the human monocytic THP-1 cell line. More work is needed to decipher whether LPS stimulation results in the altered levels of other PROMPTs.
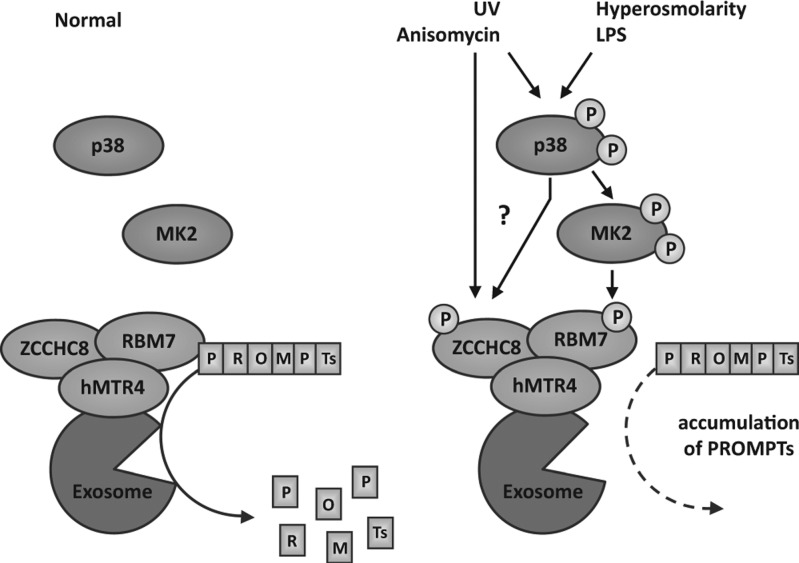
FIGURE 7.
Proposed model for the regulation of PROMPTs RNAs by the p38MAPK/MK2 signaling module upon stress. Upon stress, induced by anisomycin- or UV-C light-stimulation, p38MAPK becomes activated through phosphorylation. In turn, activated p38 is able to phosphorylate MK2, its downstream kinase. MK2 phosphorylates RBM7 at S136. It is possible that p38 or other stress-activated kinases are able to phosphorylate ZCCHC8 in parallel. Phosphorylated RBM7, as part of the trimeric NEXT complex (composed of RBM7, ZCCHC8, and hMTR4) is less effective in binding and in properly guiding PROMPTs to nuclear exosomal degradation. This is supported by a massive gain in PROMPTs RNA transcript stability and easy detection in qRT-PCR analyses because of their accumulation. Notably, p38MAPK/MK2-activation seems to be necessary but not sufficient for complete activation of this mechanism as LPS-, Il-1α stimulation and high osmolarity only lead to moderate and selective PROMPTs accumulation (Fig. 4D; Supplemental Fig. S8A,B).
A more complex regulation could also be envisioned, as the interaction between RBM7 and ZCCHC8 leads to their mutual stabilization (see Fig. 2A) and as ZCCHC8 could also be directly modified in a p38-dependent manner (see Fig. 1E,F). By inhibiting GSK-3 using LiCl, we can exclude the possibility that anisomycin stimulation activates the documented GSK-3-mediated phosphorylation of ZCCHC8 (Gustafson et al. 2005), that then potentially contributes to ncRNA regulation. Perhaps other NEXT-associated proteins can be involved in RBM7 recruitment to RNA. Indeed, in recent studies it was shown that NEXT can associate with the cap-binding complex (CBC) via ARS2 and ZC3H18 forming CBCN (Andersen et al. 2013; Hallais et al. 2013). The consequences of RBM7 phosphorylation and potentially ZCCHC8 could involve modulated interactions with other proteins and thereby influencing RBM7–RNA interactions. This could add another layer of regulation to the PROMPTs accumulation. This is indicated by the partial loss of RBM7–CBP80 interaction that is p38MAPK-independent (Supplemental Fig. S3C). Phosphorylation of proteins often serves as a proteasomal degradation signal due to their recognition by ubiquitin-ligases (i.e., Chen et al. 1995; Leboucher et al. 2012). No effect of proteasomal inhibition using MG-132 on RBM7 protein levels argues against such a mechanism (Supplemental Fig. S2B). RBPs that undergo signal-dependent phosphorylation and target the 3′ UTRs of mRNAs can show prominent affinity changes toward its targets. This is exemplified by MK2-dependent loss of TTP affinity toward the AU-rich element in TNF mRNA (Tiedje et al. 2012) or by HuR binding with higher or lower affinity to targets depending on the signaling cascade involved (summarized in Eberhardt et al. 2012). Similarly, phosphorylation of RBM7 on S136 could lead to changes in its RNA-binding capacity toward PROMPTs and U snRNAs and could therefore explain the accumulation of these ncRNAs during stress.
Moreover, phosphorylation of RNA-binding proteins can promote their sequestration by scaffold proteins like it was again demonstrated for the AU-rich RNA-binding protein TTP in the nucleus (Johnson et al. 2002; Stoecklin et al. 2004). Indeed, in a very recent publication, the phosphorylation of RBM7 by MK2 upon UV-C light exposure on S136 and S204 was demonstrated and constitutes a 14-3-3 protein binding site preventing rapid PROMPTs degradation by the nuclear RNA exosome (Blasius et al. 2014). These findings clearly complement the mechanism elaborated in this manuscript and could explain the remaining phosphorylation signal seen for Rbm7S136A in the radioactive kinase assay (Supplemental Fig. S1B). In the same study, RBM7 and RBM7S136A,S204A were used to specifically precipitate PROMPTs. It was shown that RBM7 loses its PROMPTs-binding capacity during UV-stress and that this loss is completely abrogated using the RBM7S136A,S204A double mutant (Blasius et al. 2014). These observations are in line with our CLIP-results although the effects for the Rbm7S136A single mutant and the p38MAPK-inhibitor BIRB796 (Fig. 2B,C; Supplemental Fig. S4B,C) are less pronounced. This could indicate that PROMPTs only describe a minimal portion of all RBM7-bound RNAs that are visualized by the CLIP experiments. Both studies share the observation of stress-induced PROMPTs accumulation in dependence of MK2 that is due to misregulated guidance of PROMPTs to exosomal degradation by RBM7/NEXT. Therefore, the MK2-dependent binding of RBM7 to 14-3-3 proteins can explain the increased PROMPTs RNA stability shown in this study (Fig. 6E,F).
Apart from PROMPTs, a noteworthy up-regulation of U snRNA metabolites is observed in the current study. Nevertheless, this class of transcripts is of low complexity and harbors multiple pseudogenes and results have to be handled with care (e.g., U1 snRNA; O'Reilly et al. 2013). Of these, a pseudogene-free U6atac, component of the minor splicing complex, is known to be stimulated and in part stabilized by anisomycin treatment in a p38-dependent manner (Younis et al. 2013). As a consequence, Pten is differentially expressed and TNFα expression is modulated. Like in the recent work (Younis et al. 2013), we find a 2.5-fold up-regulation of U6atac, indicating a potential involvement of RBM7 in this regulation. The U1/U11 induction could influence post-transcriptional mRNA processing at the level of splicing. The strong induction of single components of the major (U1) and minor (U11) spliceosome probably disturbs the stoichiometry of these complexes and could exert an inhibitory effect on the splice reaction. Indeed, under stress conditions an increased expression of intron-free mRNAs and an inhibition of splicing have been known for a long time (Yost and Lindquist 1986; Biamonti and Caceres 2009; Shalgi et al. 2014).
Could stress-induced lncRNA accumulation have a physiological meaning? In addition to PROMPTs, enhancer (e)RNAs are also targeted for rapid nuclear decay by the RNA exosome (Andersson et al. 2014) and thus potentially regulated via RBM7 phosphorylation upon stress. Some eRNAs function as regulators of cell type-specific gene expression events (Kim et al. 2010; Ilott et al. 2014). Furthermore, several examples have assigned specific functions to lncRNAs during the classical stress response, e.g., the lncRNA HSR1 acts as an RNA stress sensor, which contributes to HSF1 trimerization (Shamovsky et al. 2006), and the lncRNA 7SK functions as repressor of the P-TEFb complex blocking RNAPII CTD-phosphorylation and elongation of transcription (Nguyen et al. 2001).
The provided data of this study raises the question whether a global regulation of nuclear ncRNA decay is important for the stress response. Therefore, it is tempting to speculate that altered PROMPTs levels upon stress can influence the transcription of stress-induced mRNAs by guiding the transcription machinery in a spatial manner to sites of opened chromatin. It will be a major future task to elucidate detailed molecular mechanisms of PROMPTs action under stress.
MATERIALS AND METHODS
Cell culture of HeLa, HEK293T, THP-1 cells, and cell treatments
Cells were grown in DMEM (HeLa and HEK293T) on plates or in RPMI1640 (THP-1) as a suspension containing 2 mM l-glutamine, 10% (v/v) FCS, 100 units penicillin G/mL, and 100 mg/mL streptomycin (all purchased from Life Technologies) with 5% CO2 in a humidified atmosphere at 37°C. The HeLa cells were kindly provided by H. Holtmann (Hannover, DE). All cells were treated as indicated with 10 µg/mL Anisomycin (dissolved in DMSO, Sigma), 1 µg/mL lipopolysaccharide (LPS, dissolved in water, Escherichia coli 0127:B8, Sigma), or pretreated for a minimum of 45 min with BIRB796, SB203580, and SB202190 (all dissolved in DMSO, Axon Medchem) at concentrations of 1 µM (BIRB796) and 5 µM (SB compounds), respectively. For inhibition of the proteasome, cells were incubated with 20 µM MG-132 (dissolved in DMSO, Peptide Institute). The MK2-inhibitor PHA78108 was a kind gift from Robert J. Mourey (Pfizer).
Transfection of HeLa and HEK293T cells
Cells were seeded the day before transfection to reach 70%–80% density the next day. The medium was then changed to penicillin/streptomycin-free. DNA was mixed in Opti-MEM (Life Technologies) and complexed with polyethylenimine (PEI, pH 7.0, Serva). The mix was incubated for a minimum of 10 min at room temperature and then added drop wise to the cells. Medium was changed to full medium 6 h and cells were treated 24 h post-transfection.
siRNA transfection of HeLa cells
HeLa cells were transfected according to the manufacturer's instruction using HiPerFect (Qiagen). Specific RBM7 or hRRP40 siRNAs and a control siRNA (Allstars negative Control siRNA, Qiagen # 1027281) were transfected at final concentrations of 2 nM. After 24 h medium was exchanged and 48 h post-transfection the cells were stimulated. The following siRNAs were used (Andersen et al. 2013): GGAUAAAGGCAUUGCUUAAdTdT (siRBM7) and CACGCACAGUACUAGGUCAdTdT (sihRRP40).
Generation of stable shMK2-knockdown HEK293T cells
MK2-knockdown HEK293T cells were generated using “SHVRS MISSION shRNA Lentiviral Particles” (Sigma) as previously described for HT29 cells (Menon et al. 2010). Two different lentiviral particles were used (catalog #TRCN22-83 and -86) together with the control lentiviral particles (MISSION Non-Target shRNA Control). The cells were transduced in 96-well plates at an MOI of 2. After transduction, cells were selected with puromycin (2 µg/mL) for 2 wk and MK2 expression was analyzed by Western blot.
Plasmids and mutagenesis
Plasmids coding N- and C-terminally GFP- and FLAG-tagged hRBM7 and hZCCHC8 were described before (Lubas et al. 2011). The murine Rbm7 coding sequence was PCR-amplified from total MEF cDNA (fwd: CACCATGGGGGCGGCGGCCGCAGA and rev: CAGTGTCCTTGGAATGCTTG) and cloned into pENTR-D-TOPO (Life Technologies) following the manufacturer's instructions. Destination vectors with GFP-tags were generated by LR-mediated recombination reactions (Life Technologies). Site-directed mutagenesis was performed using the Quickchange kit (Agilent).
Identification of Ser136 as the amino acid residue on Rbm7 phosphorylated by MK2
Rbm7 was expressed as a glutathione-S-transferase fusion protein in E. coli, purified by affinity chromatography on glutathione-sepharose and phosphorylated by incubation for 60 min at 30°C with MK2 and Mg-[γ-32P]ATP (106 cpm/nmol) to a stoichiometry approaching 1.0 mol phosphate per mol protein. Following digestion with trypsin and chromatography on a C18 column the major phosphopeptide with a molecular mass corresponding to the monophosphorylated form of the peptide comprising amino acid residues 134–143 of Rbm7 was pooled and analyzed by solid phase sequencing. These experiments showed that 32P-radioactivity was released from the peptide after the third cycle of Edman degradation corresponding to Ser136.
Phospho-specific antibody that recognizes Rbm7 phosphorylated at Ser136
An antibody was raised in sheep against the synthetic phosphopeptide CVQRSFS*SPEDY (where S* is phosphoserine in the murine Rbm7 sequence) corresponding to amino acid residues 131–141 of Rbm7 plus an extra cysteine residue at the N-terminus. The peptide was coupled via the cysteine residue to both keyhole limpet hemocyanin (Calbiochem) and bovine serum albumin (Sigma) using m-maleimidobenzoyl-N-hydroxysuccinimide ester (Sigma). The antigen–protein conjugates were mixed and 0.3 mg injected into a sheep (S041B) every 4 wk. Serum was removed 1 wk after the second injection (first bleed) and every 4 wk thereafter. The anti-sera were purified by affinity chromatography against the phosphopeptide immunogene that had been coupled covalently to vinylsulfone-activated-sepharose. Antibody purified from the first bleed was used in the present study.
GFP- and FLAG-Pulldown
A GFP-Nanotrap (Rothbauer et al. 2008) was used for pull downs of GFP-tagged proteins. For this, the lama-derived GFP-binding protein was expressed in E. coli and FPLC-purified using a HisTrap Ni-Sepharose column on an ÄKTA system (GE Healthcare) as described in Rothbauer et al. (2008). To allow protein-purification with low background under high-salt conditions the GFP-Nanotrap was then covalently coupled to magnetic Epoxy Dynabeads (M-270, Life Technologies) in line with the manufacturer's instructions and as described in Domanski et al. (2012). Pull downs for protein–protein interactions were performed under native conditions as also described in Rothbauer et al. (2008). Cell lysates were prepared in buffer containing 20 mM Tris/HCl, pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 0.5% [v/v] NP-40, and protease inhibitors. Lysates were then adjusted to equal protein contents and diluted with lysis buffer containing no detergent to a final concentration of NP-40 under 0.2% to prevent unspecific binding. Before binding, lysates were sonicated with two strokes of each 20s and 20% power output and treated on ice for 10 min with 2 µL of Turbo DNase I (2 units/µL, Ambion) and 5 µL of RNaseA (10 mg/mL, Thermo). A measure of 10 µL slurry of preequilibrated magnetic GFP-Nanotrap (in lysis buffer) was used in each pull down. Pull downs tumbled for 1 h at 4°C in the cold room and were then washed twice with dilution buffer (20 mM Tris/HCl, pH 7.5, 150 mM NaCl, 0.5 mM EDTA) and another two times with high-salt wash buffer (20 mM Tris/HCl, pH 7.5, 300 mM NaCl, 0.5 mM EDTA) using a magnetic separator. Finally, the beads were re-suspended in 40 µL 2× SDS-PAGE buffer and boiled for 5 min at 95°C. For FLAG-Pulldowns the same conditions were chosen. The M2-FLAG agarose beads were purchased from Sigma.
Immunoprecipitation of endogenous RBM7
HeLa cells (5 × 106), treated as indicated, were lysed in 500 µL cell lysis buffer (20 mM Tris [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 1 mM β-glycerophosphate, 1 mM Na3VO4, 1 μg/mL Leupeptin containing a 1:100 dilution of phosphatase inhibitor cocktail 3 [Sigma] mix). Lysates were then sonicated three times for 5 sec at 5% power output and cleared by centrifugation. A measure of 200 µL of each lysate was precleared before IP by incubating with Protein G Sepharose beads (GE Healthcare) tumbling at 4°C for 1 h. The precleared lysates were incubated overnight (4°C, tumbling) with 2.5 µg of the sheep-derived anti-pS136-Rbm7 antibody. Twenty-five milliliters of Protein G Sepharose slurry was added afterward for another hour tumbling at 4°C. IPs were finally washed three times using cell lysis buffer and eluted with 40 µL 2× SDS-PAGE loading buffer at 95°C for 5 min.
Dephosphorylation of lysates
HeLa cells (5 × 106), treated as indicated, were lysed in 750 µL lysis buffer (containing 20 mM Tris/HCl, pH 8.0, 50 mM KCl, 2 mM MgCl2, [v/v] 0.2% NP-40) containing protease inhibitors; 40 µL lysate were incubated with CutSmart Buffer (1× final, New England Biolabs) and 15 units calf intestine phosphatase (CIP, 10 units/µL, New England Biolabs) for 45 min at 37°C. Reactions were stopped by adding 2× SDS-PAGE loading buffer and denaturation for 5 min at 95°C.
SDS-PAGE Western blotting
Protein extracts were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) on 7.5%–16% gradient gels and transferred by semi-dry blotting to Hybond ECL nitrocellulose membranes (GE Healthcare). The following primary antibodies were used to develop the blots: anti-MK2, anti-p38, anti-pT180/Y182-p38, anti-pT334-MK2, anti-p(S/T)-PKD-substrate, anti-Hsp27, anti-pS82-Hsp27 (all purchased from Cell Signaling), anti-GAPDH (Millipore), anti-eEF2, anti-GFP, anti-hRRP40 (all from Santa Cruz Biotechnology), anti-RBM7, anti-ZCCHC8 (both from Atlas antibodies), anti-pS136-Rbm7 (see above); anti-NOGO-B (Rousseau et al. 2005), and anti-CBP80 (Andersen et al. 2013) antibody were described before. The anti-FLAG (M2) antibody was purchased from Agilent. ECL detection was carried out on a chemo-luminescence detector (LAS 3000 FujiFilm).
In vitro kinase assay
In vitro kinase assays were carried out as described previously (Menon et al. 2010; Tiedje et al. 2012).
RNA isolation, cDNA synthesis, and qRT-PCR
Total RNAs were isolated using TRIzol (Life Technologies) according to the manufacturer's instructions. cDNA from 500 to 100 ng total RNA was synthesized using the first strand cDNA synthesis kit (Fermentas/Thermo) in combination with random hexamer primers. qRT-PCRs were run on a Rotor-Gene-Q (Qiagen) device using the 2× SYBR-Green mix (no ROX, Bioline) according to the instructions. The following commercial primers were used for the detection of human β-actin and TNFα: QuantiTect Primer Assays (Qiagen): Hs_TNF_3_SG and Hs_ACTB_2_SG. For the detection of PROMPTs, the following primers for proRBM39 (Lubas et al. 2011), proEXT1 and proIFNAR1 (Andersen et al. 2013), and proDNAJB4 (M Lubas, PR Andersen, A Schein, A Dziembowski, G Kudla, and TH Jensen, in prep.) were chosen: proRBM39-fwd (GGAAATAGTGGAGAAAAGCA), proRB39-rev (CATTTTTGAAGGAACGGTAG), proEXT1-fwd (TCTAATGGCTGCAGGGAAAC), proEXT1-rev (TAGCTGGGACAGTTGGCAAT), proIFNAR1-fwd (TTGCTAATTAATTGCTTGGTTGTT), proIFNAR1-rev (GCATTGTGAAAATATGCGAAATA), proDNAJB4-fwd (TTTCTGGCGTTTCTGATTGA), proDNAJB4-rev (ACCAAAACGCAGGTTGTTTA). RBM39-fwd (GTGCTGTGGCAGAATTCTCTT) and RBM39-rev (TTGTGTTGCAAGTGGCTGA) primers to detect the RBM39 coding gene were designed with the help of the Primer3 online tool (Primer3, http://www-genome.wi.mit.edu/genome_software/other/primer3.html). Similarly, primers to detect U snRNAs metabolites were designed: U1-fwd: ATACTTACCTGGCAGGGGAG, U1-rev: CAGGGGAAAGCGCGAACGCA, U2-fwd: ATCGCTTCTCGGCCTTTTGG, U2-rev: GGGTGCACCGTTCCTGGAGG, U4-fwd: AGCTTTGCGCAGTGGCAGTA, U4-rev: TCCGTAGAGACTGTCAAAAA, U11- fwd: AGGGCTTCTGTCGTGAGTGG, U11-rev: AAAGGGCGCCGGGACCAACG, U12-fwd: TGCCTTAAACTTATGAGTAA, U12-rev: CGGGCAGATCGCAACTCCCA, U6atac-fwd: GGTTAGCACTCCCCTTGACA, U6atac- rev: ACGATGGTTAGATGCCACGA.
The threshold cycle (CT) of each individual PCR-product for relative quantifications was determined with the help of the machine's software. PROMPTs RNA levels were normalized to β-Actin mRNA and shown as x-fold induction relative to the basal (nonstimulated) condition. U snRNAs were normalized to 18S rRNA levels that were determined with the primers 18S-fwd: TTGTTGGTTTTCGGAACTGAG and 18S-rev: GCAAATGCTTCGGCTCTGGTC.
RNA-stability measurements
For RNA-stability measurements, cells were treated as indicated above before stopping RNA-Polymerase II transcription with 10 µg/mL actinomycin D (dissolved in DMSO, Sigma). RNA-samples were taken every 5–10 min using TRIzol (Life Technologies). RNA isolation, cDNA synthesis, and qRT-PCRs were carried out as described before.
RNA-Polymerase II (RNAPII) chromatin-immunoprecipitation (ChIP)
RNAPII ChIP experiments were performed using the ZymoSpin ChIP Kit (Zymo Research) according to instructions. A ChIP-grade anti-RNA-Polymerase II antibody (H-224, sc-9001, Santa Cruz Biotechnology) and as a control normal rabbit IgG (sc-2027, Santa Cruz Biotechnology) were used at concentrations of 2.5 µg/ChIP experiment. The sequences of the primer pairs for the detection of proRBM39 transcript occupancy were described before (Andersen et al. 2013) (proRBM39#1: fwd: GGGTGGAGAGGAAGTGGAAAAGTG, rev: CCCCTCTCCCCCTACCCCTAAAGA proRBM39#2: fwd: TGTGCCCCGAATTTAATCAGC, rev: TCAGGAAGGGGAACATTTTTGAA and proRBM39#4: fwd: AACAGCTAACTTCTGTTTCTGCT, rev: ACCTGAGAGGCAAAACCACT). Control primers detecting the RNA-Polymerase II occupancy of EGR1 were designed using the Primer3 online tool mentioned above: EGR1-1(fwd): CCGACACCAGCTCTCCAG, EGR1-2(rev): TCAGCAGCATCATCTCCTCC, EGR1-5(fwd): CAGTGGCCTAGTGAGCATGA, EGR1-6(rev): TTGTGGCTCAGGGAAAATGT. The combination EGR1-1/2 amplifies base pairs 239–394 and EGR1-5/6 the nucleotides 744–933 of human EGR1 (NM_001964.2).
Cross-linking immunoprecipitation (CLIP)
For CLIP experiments under urea–salt conditions 4 × 106 HeLa cells transfected with GFP-RBM7 constructs, were cross-linked with 300 mJ/cm2 UV-C light in a Stratalinker (Stratagene/Agilent). Cell pellets were lysed in 1 mL of lysis buffer (50 mM Tris/HCl, pH 8.0, 150 mM NaCl, 0.5% [v/v] Triton X-100, 1 mM EDTA, protease, and phosphatase inhibitors [Roche and Sigma]). Samples were sonicated three times using a Branson sonicator (3 × 20 sec at 20% power). Lysates were then clarified by full-speed centrifugation at 4°C in a table-top centrifuge for 15 min. Cleared lysates were treated for 5 min at 37°C in an Eppendorf-shaker (1100 rpm) with 2 µL Turbo DNase I (Ambion) and 10 µL of a 1:10 dilution of an RNaseA/T1 (Thermo/Roche) mixture in lysis buffer. Afterward, together with 5 µL of RiboLock (Thermo), urea was added to a final concentration of 0.5 M and lysates were incubated on ice for 5 min; 15 µL magnetic GFP-Nanotrap-beads were equilibrated with lysis buffer for each pulldown and incubated with the DNase-/RNase-treated lysates for 1 h at 4°C in the cold room. The pull-down samples were then washed once with lysis buffer containing 0.5 M urea and three times with low salt buffer (50 mM Tris/HCl, pH 8.0, 150 mM NaCl, 0.1% [v/v] Triton X-100, and 0.5 M urea). Then, the beads were briefly washed twice with high-salt buffer (50 mM Tris/HCl, pH 8.0, 1000 mM NaCl, 0.5% [v/v] Triton X-100, and 2 M urea) and washed once for 15 min on a rotator in the cold room at 4°C with the same buffer. Finally, beads were washed three times with PNK buffer (20 mM Tris/HCl, pH 7.4, 10 mM MgCl2, and 0.2% [v/v] Tween 20); 50% of the beads were used for labeling of the associated RNA fragments. For this, the beads were dissolved in 4 µL of labeling mix (3 µL H2O, 0.2 µL PNK [NEB], 0.4 µL 10X PNK buffer [NEB], and 0.4 µL [γ-32P]ATP [Hartmann Analytic]), and incubated for 5 min at 37°C. The supernatant was removed and complexes were eluted with 15 µL 1× NuPAGE loading buffer (Life Technologies) for 10 min at 70°C. Samples were resolved on a 4%–12% Novex NuPAGE Bis–Tris gel (Life Technologies), transferred to a nitrocellulose membrane (GE Healthcare) and then exposed to a FujiFilm Imager Plate for 30 min to overnight. The plate was read with a Fuji FLA-5000 system. The membrane was finally developed with a GFP antibody to monitor equal precipitation. For quantification of the autoradiography signals a minimum of two different exposures of the autoradiograph were normalized to at least three different exposures of the GFP Western blots that were derived from the radioactive membrane used for autoradiography. Western blot signals were obtained using a quantitative chemo-luminescence detector (LAS 3000 FujiFilm). All band intensities (blots and autoradiographs) were determined using the Multi Gauge V3.2 software (FujiFilm). For Figure 2C, the signal of untreated GFP-Rbm7 was set to 1 and the other relative signals were calculated in relation to this signal. For Supplemental Figure S4B,C, the signal of anisomycin-treated GFP-Rbm7 was set to 1 for each replicate to evaluate the effect of S136A mutation or the use the BIRB796 inhibitor. Significant changes in relative signals were determined calculating P-values using two-sided Student's t-tests.
RNA-EMSA
Shift assays with AU-rich and Random-RNAs (Kratochvill et al. 2011) were performed basically as described in Tiedje et al. (2012). To improve the shifts using 10 µg of HEK293T lysates overexpressing the indicated proteins, 20 µg of yeast tRNA in combination with 10 µg of salmon sperm DNA were preincubated with the lysates for 30 min prior to the addition of the DY681 and DY781-5′ labeled RNAs.
Luciferase measurements
For luciferase-reporter measurements GFP- and FLAG-tagged RBM7 was overexpressed in HEK293T cells. Firefly coding reporter plasmids (pGL3, Promega) bearing no additional cis-element, the complete 3′ UTR of murine TNFα or human IL-6 or a mutated 3′ UTR of murine TNFα with no AU-rich element just downstream from the firefly coding sequence were coexpressed and served as a readout for changes in transcript stability and translation in response to RBM7 overexpression. To normalize firefly values, a renilla-luciferase-coding plasmid (phRL-TK, Promega) was cotransfected and measured separately on a GloMax (Promega) device using buffers and substrates purchased from Promega.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank David Campbell (MRC Protein Phosphorylation Unit [MRC-PPU] Dundee) for identifying Ser136 as the major site on RBM7 phosphorylated by MK2 in vitro, James Hastie (MRC-PPU, Dundee) for making and purifying the phospho-specific antibody recognizing Ser136 of RBM7, Dr. Robert J. Mourey (Pfizer) for providing PHA781089, and Dr. Andrzej Dziembowski (Warsaw) for the GFP-Nanotrap-coding plasmid. Dr. Maya Sethi and Dr. Hans Bakker (both Hannover) are acknowledged for help with the FPLC-purification of the GFP-Nanotrap, Dr. Helmut Holtmann (Hannover) for providing HeLa cells and the firefly reporter plasmids, and Kathrin Laaß for expert technical assistance. This work was supported by Deutsche Forschungsgemeinschaft (DFG) grant Ga453/13-1 (to C.T. and M.G.) and the “Zytokin-Preis” to C.T. funded by “GdF der MHH e.V.”
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.048090.114.
REFERENCES
- Andersen PR, Domanski M, Kristiansen MS, Storvall H, Ntini E, Verheggen C, Schein A, Bunkenborg J, Poser I, Hallais M, et al. 2013. The human cap-binding complex is functionally connected to the nuclear RNA exosome. Nat Struct Mol Biol 20:1367–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P 2008. Post-transcriptional control of cytokine production. Nat Immunol 9:353–359. [DOI] [PubMed] [Google Scholar]
- Anderson DR, Meyers MJ, Vernier WF, Mahoney MW, Kurumbail RG, Caspers N, Poda GI, Schindler JF, Reitz DB, Mourey RJ 2007. Pyrrolopyridine inhibitors of mitogen-activated protein kinase-activated protein kinase 2 (MK-2). J Med Chem 50:2647–2654. [DOI] [PubMed] [Google Scholar]
- Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, Chen Y, Zhao X, Schmidl C, Suzuki T, et al. 2014. An atlas of active enhancers across human cell types and tissues. Nature 507:455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biamonti G, Caceres JF 2009. Cellular stress and RNA splicing. Trends Biochem Sci 34:146–153. [DOI] [PubMed] [Google Scholar]
- Blasius M, Wagner SA, Choudhary C, Bartek J, Jackson SP 2014. A quantitative 14-3-3 interaction screen connects the nuclear exosome targeting complex to the DNA damage response. Genes Dev 28:1977–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannell IG, Kong YW, Johnston SJ, Chen ML, Collins HM, Dobbyn HC, Elia A, Kress TR, Dickens M, Clemens MJ, et al. 2010. p38 MAPK/MK2-mediated induction of miR-34c following DNA damage prevents Myc-dependent DNA replication. Proc Natl Acad Sci 107:5375–5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cargnello M, Roux PP 2011. Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol Mol Biol Rev 75:50–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Hagler J, Palombella VJ, Melandri F, Scherer D, Ballard D, Maniatis T 1995. Signal-induced site-specific phosphorylation targets I κ B α to the ubiquitin-proteasome pathway. Genes Dev 9:1586–1597. [DOI] [PubMed] [Google Scholar]
- Chlebowski A, Lubas M, Jensen TH, Dziembowski A 2013. RNA decay machines: the exosome. Biochim Biophys Acta 1829:552–560. [DOI] [PubMed] [Google Scholar]
- de la Cruz J, Kressler D, Tollervey D, Linder P 1998. Dob1p (Mtr4p) is a putative ATP-dependent RNA helicase required for the 3′ end formation of 5.8S rRNA in Saccharomyces cerevisiae. EMBO J 17:1128–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domanski M, Molloy K, Jiang H, Chait BT, Rout MP, Jensen TH, LaCava J 2012. Improved methodology for the affinity isolation of human protein complexes expressed at near endogenous levels. Biotechniques 0:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhardt W, Doller A, Pfeilschifter J 2012. Regulation of the mRNA-binding protein HuR by posttranslational modification: spotlight on phosphorylation. Curr Protein Pept Sci 13:380–390. [DOI] [PubMed] [Google Scholar]
- Engel K, Kotlyarov A, Gaestel M 1998. Leptomycin B-sensitive nuclear export of MAPKAP kinase 2 is regulated by phosphorylation. EMBO J 17:3363–3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaestel M 2006. MAPKAP kinases—MKs—two's company, three's a crowd. Nat Rev Mol Cell Biol 7:120–130. [DOI] [PubMed] [Google Scholar]
- Gustafson MP, Welcker M, Hwang HC, Clurman BE 2005. Zcchc8 is a glycogen synthase kinase-3 substrate that interacts with RNA-binding proteins. Biochem Biophys Res Commun 338:1359–1367. [DOI] [PubMed] [Google Scholar]
- Hallais M, Pontvianne F, Andersen PR, Clerici M, Lener D, Benbahouche Nel H, Gostan T, Vandermoere F, Robert MC, Cusack S, et al. 2013. CBC-ARS2 stimulates 3′-end maturation of multiple RNA families and favors cap-proximal processing. Nat Struct Mol Biol 20:1358–1366. [DOI] [PubMed] [Google Scholar]
- Han J, Lee JD, Bibbs L, Ulevitch RJ 1994. A MAP kinase targeted by endotoxin and hyperosmolarity in mammalian cells. Science 265:808–811. [DOI] [PubMed] [Google Scholar]
- Hazzalin CA, Cano E, Cuenda A, Barratt MJ, Cohen P, Mahadevan LC 1996. p38/RK is essential for stress-induced nuclear responses: JNK/SAPKs and c-Jun/ATF-2 phosphorylation are insufficient. Curr Biol 6:1028–1031. [DOI] [PubMed] [Google Scholar]
- Hett A, West S 2014. Inhibition of U4 snRNA in human cells causes the stable retention of polyadenylated pre-mRNA in the nucleus. PLoS One 9:e96174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horman SR, Janas MM, Litterst C, Wang B, MacRae IJ, Sever MJ, Morrissey DV, Graves P, Luo B, Umesalma S, et al. 2013. Akt-mediated phosphorylation of argonaute 2 downregulates cleavage and upregulates translational repression of microRNA targets. Mol Cell 50:356–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J, Tollervey D 2009. The many pathways of RNA degradation. Cell 136:763–776. [DOI] [PubMed] [Google Scholar]
- Ilott NE, Heward JA, Roux B, Tsitsiou E, Fenwick PS, Lenzi L, Goodhead I, Hertz-Fowler C, Heger A, Hall N, et al. 2014. Long non-coding RNAs and enhancer RNAs regulate the lipopolysaccharide-induced inflammatory response in human monocytes. Nat Commun 5:3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BA, Stehn JR, Yaffe MB, Blackwell TK 2002. Cytoplasmic localization of tristetraprolin involves 14-3-3-dependent and -independent mechanisms. J Biol Chem 277:18029–18036. [DOI] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. 2010. Widespread transcription at neuronal activity-regulated enhancers. Nature 465:182–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratochvill F, Machacek C, Vogl C, Ebner F, Sedlyarov V, Gruber AR, Hartweger H, Vielnascher R, Karaghiosoff M, Rülicke T, et al. 2011. Tristetraprolin-driven regulatory circuit controls quality and timing of mRNA decay in inflammation. Mol Syst Biol 7:560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuma Y, Sabio G, Bain J, Shpiro N, Márquez R, Cuenda A 2005. BIRB796 inhibits all p38 MAPK isoforms in vitro and in vivo. J Biol Chem 280:19472–19479. [DOI] [PubMed] [Google Scholar]
- Leboucher GP, Tsai YC, Yang M, Shaw KC, Zhou M, Veenstra TD, Glickman MH, Weissman AM 2012. Stress-induced phosphorylation and proteasomal degradation of mitofusin 2 facilitates mitochondrial fragmentation and apoptosis. Mol Cell 47:547–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JC, Laydon JT, McDonnell PC, Gallagher TF, Kumar S, Green D, McNulty D, Blumenthal MJ, Heys JR, Landvatter SW, et al. 1994. A protein kinase involved in the regulation of inflammatory cytokine biosynthesis. Nature 372:739–746. [DOI] [PubMed] [Google Scholar]
- Lubas M, Christensen MS, Kristiansen MS, Domanski M, Falkenby LG, Lykke-Andersen S, Andersen JS, Dziembowski A, Jensen TH 2011. Interaction profiling identifies the human nuclear exosome targeting complex. Mol Cell 43:624–637. [DOI] [PubMed] [Google Scholar]
- Manke IA, Nguyen A, Lim D, Stewart MQ, Elia AE, Yaffe MB 2005. MAPKAP kinase-2 is a cell cycle checkpoint kinase that regulates the G2/M transition and S phase progression in response to UV irradiation. Mol Cell 17:37–48. [DOI] [PubMed] [Google Scholar]
- Menon MB, Schwermann J, Singh AK, Franz-Wachtel M, Pabst O, Seidler U, Omary MB, Kotlyarov A, Gaestel M 2010. p38 MAP kinase and MAPKAP kinases MK2/3 cooperatively phosphorylate epithelial keratins. J Biol Chem 285:33242–33251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon MB, Kotlyarov A, Gaestel M 2011. SB202190-induced cell type-specific vacuole formation and defective autophagy do not depend on p38 MAP kinase inhibition. PLoS One 6:e23054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon MB, Tiedje C, Lafera J, Ronkina N, Konen T, Kotlyarov A, Gaestel M 2013. Endoplasmic reticulum-associated ubiquitin-conjugating enzyme Ube2j1 is a novel substrate of MK2 (MAPKAP kinase-2) involved in MK2-mediated TNFα production. Biochem J 456:163–172. [DOI] [PubMed] [Google Scholar]
- Nguyen VT, Kiss T, Michels AA, Bensaude O 2001. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature 414:322–325. [DOI] [PubMed] [Google Scholar]
- Ntini E, Järvelin AI, Bornholdt J, Chen Y, Boyd M, Jørgensen M, Andersson R, Hoof I, Schein A, Andersen PR, et al. 2013. Polyadenylation site-induced decay of upstream transcripts enforces promoter directionality. Nat Struct Mol Biol 20:923–928. [DOI] [PubMed] [Google Scholar]
- O'Reilly D, Dienstbier M, Cowley SA, Vazquez P, Drozdz M, Taylor S, James WS, Murphy S 2013. Differentially expressed, variant U1 snRNAs regulate gene expression in human cells. Genome Res 23:281–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preker P, Nielsen J, Kammler S, Lykke-Andersen S, Christensen MS, Mapendano CK, Schierup MH, Jensen TH 2008. RNA exosome depletion reveals transcription upstream of active human promoters. Science 322:1851–1854. [DOI] [PubMed] [Google Scholar]
- Ray D, Kazan H, Cook KB, Weirauch MT, Najafabadi HS, Li X, Gueroussov S, Albu M, Zheng H, Yang A, et al. 2013. A compendium of RNA-binding motifs for decoding gene regulation. Nature 499:172–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbauer U, Zolghadr K, Muyldermans S, Schepers A, Cardoso MC, Leonhardt H 2008. A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol Cell Proteomics 7:282–289. [DOI] [PubMed] [Google Scholar]
- Rousseau S, Morrice N, Peggie M, Campbell DG, Gaestel M, Cohen P 2002. Inhibition of SAPK2a/p38 prevents hnRNP A0 phosphorylation by MAPKAP-K2 and its interaction with cytokine mRNAs. EMBO J 21:6505–6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau S, Peggie M, Campbell DG, Nebreda AR, Cohen P 2005. Nogo-B is a new physiological substrate for MAPKAP-K2. Biochem J 391:433–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalgi R, Hurt JA, Lindquist S, Burge CB 2014. Widespread inhibition of posttranscriptional splicing shapes the cellular transcriptome following heat shock. Cell Rep 7:1362–1370. [DOI] [PubMed] [Google Scholar]
- Shamovsky I, Ivannikov M, Kandel ES, Gershon D, Nudler E 2006. RNA-mediated response to heat shock in mammalian cells. Nature 440:556–560. [DOI] [PubMed] [Google Scholar]
- Stoecklin G, Stubbs T, Kedersha N, Wax S, Rigby WF, Blackwell TK, Anderson P 2004. MK2-induced tristetraprolin:14-3-3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J 23:1313–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokoe D, Engel K, Campbell DG, Cohen P, Gaestel M 1992. Identification of MAPKAP kinase 2 as a major enzyme responsible for the phosphorylation of the small mammalian heat shock proteins. FEBS Lett 313:307–313. [DOI] [PubMed] [Google Scholar]
- Tiedje C, Ronkina N, Tehrani M, Dhamija S, Laass K, Holtmann H, Kotlyarov A, Gaestel M 2012. The p38/MK2-driven exchange between tristetraprolin and HuR regulates AU-rich element-dependent translation. PLoS Genet 8:e1002977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiedje C, Holtmann H, Gaestel M 2014. The role of mammalian MAPK signaling in regulation of cytokine mRNA stability and translation. J Interferon Cytokine Res 34:220–232. [DOI] [PubMed] [Google Scholar]
- Yost HJ, Lindquist S 1986. RNA splicing is interrupted by heat shock and is rescued by heat shock protein synthesis. Cell 45:185–193. [DOI] [PubMed] [Google Scholar]
- Younis I, Dittmar K, Wang W, Foley SW, Berg MG, Hu KY, Wei Z, Wan L, Dreyfuss G 2013. Minor introns are embedded molecular switches regulated by highly unstable U6atac snRNA. eLife 2:e00780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Sankala H, Zhang X, Graves PR 2008. Phosphorylation of Argonaute 2 at serine-387 facilitates its localization to processing bodies. Biochem J 413:429–436. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.