Abstract
Epigenetic reprogramming of parental genomes following fertilisation is important to ensure compatibility for totipotency and development thereafter. New studies by Jiang et al. (2013) and Potok et al. (2013) now demonstrate how the parental DNA methylomes are reset in zebrafish, and reveal striking differences from events in mammals.
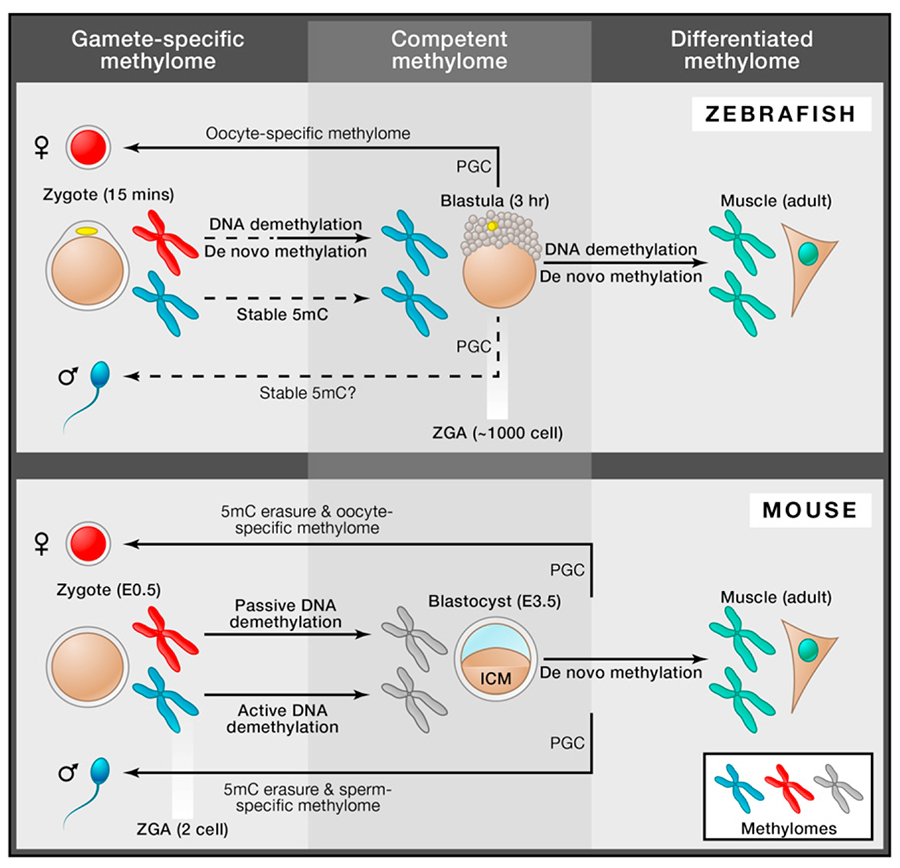
Sperm and oocytes are highly distinct and specialised cell types, yet together they generate the totipotent state following fertilisation. Significantly, while they make an equivalent genetic contribution to the zygote, their epigenetic states are highly asymmetric due to their diverse origins, and are therefore reset soon after fertilisation. In vertebrates, this involves global remodelling of the parental DNA methylation (5mC) patterns, which is thought to generate an epigenetic state competent for totipotency (Surani et al. 2007). At the same time, however, the extent to which the inherited parental-epigenomes are themselves important for development is unclear. Indeed, resetting of parental epigenomes occurs in the overall context of development, which differs markedly amongst vertebrates, and which may therefore influence the balance between reprogramming and inheritance. In this issue of Cell, reports by Jiang et al. (2013) and Potok et al. (2013) now demonstrate how genome-wide DNA methylation transitions of parental genomes occur during zebrafish development. Notably, while the maternal methylome undergoes striking remodelling during early development, the paternal methylome is stably inherited in a remarkably unchanged state. The strategy for reprogramming parental epigenomes is thus fundamentally different between vertebrates (see Fig 1)(Smith et al. 2012).
Fig 1.
Zebrafish development proceeds through synchronous cleavage divisions every ~15mins until the mid-blastula transition (MBT), when major zygotic gene activation (ZGA) commences (~1000 cells). (Tadros and Lipshitz 2009). To track DNA methylation transitions during this period, both groups generate whole-genome bisulfite sequencing maps from gametes and early developmental time points flanking the ZGA. Zebrafish oocytes are hypomethylated (75-80% CpG methylation) relative to sperm (91-95%), similarly to mice. However, upon fertilisation the paternally-derived methylome is stably inherited without significant changes throughout early zebrafish development. In parallel, the maternal methylome is initially stable but subsequently undergoes extensive remodelling that resets its epigenetic state to that of the paternal genome. This occurs through simultaneous DNA demethylation of oocyte-specific hypermethylated regions, and de novo methylation of oocyte-specific hypomethylated regions. Thus, by the time of ZGA, the parental genomes reach epigenomic equivalence through selective resetting of the maternal methylome to resemble the stable paternal methylome. At this time, the methylome acquires competence for further development, including primordial germ cell (PGC) specification through the inheritance of preformed germ cell determinants (Fig. 1).
The reprogramming strategy in zebrafish contrasts markedly with mice, where both parental genomes undergo extensive DNA demethylation via active (paternal) and passive (maternal) mechanisms, leading to a shared hypomethylated state, that is distinct from both gametic methylomes (Gu et al., 2011; Inoue and Zhang, 2011; Smith et al. 2012; Wossidlo et al. 2011). The different strategies may reflect of the underlying developmental programs of mammals and fish; mice activate transcription of the zygotic genome (2 cell) and undergo the first lineage-restricted commitment (~32 cell) relatively early during development, whereas zebrafish rely on maternal factors for ~10 divisions until their ZGA. Thus, mammalian development is under pressure to rapidly generate a methylome that is competent for the switch from a germ cell to a totipotent gene expression program, for example by demethylation of paternal Nanog (Farthing et al. 2008). In contrast, because early development in zebrafish is regulated by maternally inherited factors, the emphasis on rapid epigenomic competence for totipotency may be reduced. Indeed, the greater reliance on maternally inherited determinants may underpin the observed zebrafish oocyte-specific methylation of germline (e.g. Dazl, Piwil1) and early developmental (e.g. Hox, Pax) genes, which are presumably methylated to prevent their precocious accumulation as maternal factors in oocytes, which might otherwise skew lineage priming prior to ZGA. The paternal methylome lacking such constraints is apparently already primed for early development at the time of fertilisation. It is unclear how DNA demethylation (or de novo methylation) is precisely targeted to specific regions of the maternal genome to progressively reprogram it to the paternal pattern. However, the process appears to be passive, and apparently occurs independently of conversion to 5-hydroxymethylcytosine, and of the involvement of AID/GADD45 activity, which cannot be detected during the time of demethylation (Rai et al. 2008).
The inheritance of the sperm methylome without significant changes until ZGA is a striking observation, and raises several questions: Is the inherited sperm methylome important for embryogenesis? How is it recognised and maintained during extensive remodelling of the maternal methylome? Can it be inherited over multiple generations? To evaluate the significance of paternal epigenetic inheritance, Jiang and colleagues find that enucleated oocytes could only initiate development following transfer of a sperm nucleus, but not an oocyte nucleus, implying a fundamental epigenetic asymmetry that is consistent with the sperm methylome being in a competent state (Jiang et al. 2013). However, Potok and colleagues find that gynogenetic embryos fertilised with UV-exposed sperm (that carry non-replicating DNA) develop apparently normally with appropriate remodelling of the maternal methylomes (Potok et al. 2013). This argues that stable inheritance of the sperm methylome per se does not have a key early developmental role or act as a ‘template’ for maternal reprogramming, but rather that sperm may contribute other factors, perhaps including small RNAs. Further studies are required to reach definitive conclusions concerning the functional role of parentally contributed epigenetic states.
How the paternal methylome is protected from remodelling during development is unclear, but could be related to its chromatin state, as unlike mice, zebrafish sperm are not associated with protamines (Wu et al. 2011). Alternatively, the de novo mechanism that establishes the paternal DNA methylation pattern may also maintain it during early development, while also promoting a progressive resetting of the maternal methylome. In any case, the striking similarity between the ZGA-stage methylome and sperm methylome raises the additional intriguing possibility that the paternal DNA methylation pattern may avoid reprogramming throughout the entire zebrafish life-cycle. That is, after the paternal methylome is stably maintained until the ZGA-stage, when PGC specification occurs, it could subsequently be inherited through germ cell development to mature sperm, as the sperm methylome is near-identical to ZGA-stage cells (Fig 1). If so, this suggests a potential route for transgenerational epigenetic inheritance through the paternal germline in zebrafish. However, it remains to be established that germline fated cells formed through the inheritance of ‘germplasm’ have a comparable methylome to their somatic fated neighbours at ZGA, and that it remains stable through germ cell development.
Overall the recent studies on zebrafish reveal a distinct strategy of vertebrate epigenetic reprogramming, which does not rely on comprehensive genome-wide DNA demethylation to generate a methylome that is competent to commit to all lineages. This may inform on the functional significance of the process in other vertebrates, where genome-wide demethylation may be a necessary requirement for establishing a permissive epigenetic state at just a few key genes. These studies illustrate that the regulation of epigenetic changes should be considered in the context of the diversity of development.
References
- Farthing CR, Ficz G, et al. Global mapping of DNA methylation in mouse promoters reveals epigenetic reprogramming of pluripotency genes. PLoS Genet. 2008;4(6):e1000116. doi: 10.1371/journal.pgen.1000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, Xie ZG, Shi L, He X, Jin SG, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- Inoue A, Zhang Y. Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science. 2011;334:194. doi: 10.1126/science.1212483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Zhang J, Wang JJ, Wang L, Zhang L, Li G, Yang X, Ma X, Sun X, Cai J, et al. Sperm, but not oocyte, DNA methylome is inherited by zebrafish early embryos. Cell. 2013;153:773–784. doi: 10.1016/j.cell.2013.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potok ME, Nix DA, Parnell TJ, Cairns BR. Reprogramming the maternal zebrafish genome after fertilization to match the paternal methylation pattern. Cell. 2013;153:759–772. doi: 10.1016/j.cell.2013.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai K, Huggins IJ, et al. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135(7):1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, Chan MM, et al. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature. 2012;484(7394):339–344. doi: 10.1038/nature10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surani MA, Hayashi K, et al. Genetic and Epigenetic Regulators of Pluripotency. Cell. 2007;128(4):747–762. doi: 10.1016/j.cell.2007.02.010. [DOI] [PubMed] [Google Scholar]
- Tadros W, Lipshitz HD. The maternal-to-zygotic transition: a play in two acts. Development. 2009;136(18):3033–3042. doi: 10.1242/dev.033183. [DOI] [PubMed] [Google Scholar]
- Wossidlo M, Nakamura T, et al. 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat Commun. 2011;2:241. doi: 10.1038/ncomms1240. [DOI] [PubMed] [Google Scholar]
- Wu SF, Zhang H, et al. Genes for embryo development are packaged in blocks of multivalent chromatin in zebrafish sperm. Genome Res. 2011;21(4):578–589. doi: 10.1101/gr.113167.110. [DOI] [PMC free article] [PubMed] [Google Scholar]