Abstract
Geographic distance, different living habitats or Pleistocene climatic oscillations have frequently been found to shape population genetic structure in many species. The genetic structure of Schizothorax nukiangensis, a high altitude, valuable fish species, which is distributed throughout the Nujiang River, was investigated by mitochondrial DNA sequence analysis. The cytochrome c oxidase subunit I (COI), cytochrome b (cytb), and the mitochondrial control region (MCR) of S. nukiangensis were concatenated for examination of population structure and demographic history. The concatenated data set (2405 bp) implied a pronounced genetic population structure (overall FST = 0.149) and defined two population units. Strong differentiation was detected between the Sanjiangkou (SJK) population and other populations due to environmental heterogeneity, dispersal ability, and/or glacial cycles. Additional DNA sequencing of the nuclear RAG2 gene also examined significant differentiation between two units and between SJK and the upstream populations (U-unit). Recent expansion events suggest that S. nukiangensis may have undergone a rapid increase during warm interglacial periods. Surprisingly, S. nukiangensis appears to have undergone an obvious expansion during the last glaciations (LG) for cold hardiness and a sharp contraction from 1.5 ka to the present. However, two population units exhibited different reflections during the LG, which might be closely related to their living habitats and cold hardiness. A clear pattern of isolation by distance was detected in S. nukiangensis due to feeding habits, limited dispersal ability, and/or philopatry. It is vitally important that more attention be given to S. nukiangensis due to low genetic diversity, lack of gene flow, and recent population contraction.
Keywords: Expansion event, genetic structure, isolated by distance, Nujiang River, population bottleneck, Schizothorax nukiangensis
Introduction
The Nujiang River is an important river in China that flows from north to south. This region has retained a rich biodiversity due to a changeable climate and unique geographic features. The Nujiang River is famous for its distinct fish fauna. In fact, a total of 77 species have been reported to reside in this river in China, many of which are endemic (Chu and Chen 1989; Chen 1998, 2013; Fu et al. 2012). However, few studies have been conducted examining fish in the Nujiang River, particularly the endemic species. Four schizothoracine fishes (Schizothorax lissolabiatus, S. gongshanensis, S. yunnanensis paoshanensis, and S. nukiangensis) have been documented in the Nujiang River system (Chu and Chen 1989; Chen and Cao 2000). S. nukiangensis (Cypriniformes: Cyprinidae) (Fig.1) is a plateau fish that is economically valuable. It is widely distributed in the Nujiang River and is specialized for high elevation, exhibiting a number of unique adaptations. However, the stock of S. nukiangensis has declined dramatically in recent years due to overfishing, mining, the construction of hydropower stations in some tributaries and environmental disruption. In addition, there are plans to build 13 dam cascades along the Nujiang River mainstream, which will exacerbate the decline of S. nukiangensis (Dudgeon 2011). Consequently, it is crucial that the current stock of S. nukiangensis, and its resources, be examined and protected.
Figure 1.

The study species, Schizothorax nukiangensis in the Nujiang River.
Freshwater ecosystems all over the world have been heavily exploited and degraded by human activities, affecting both fish and fisheries; thus, it is vital that conservation efforts are undertaken quickly. Population genetic analysis is a widely used approach for assessing the genetic divergence in populations (Crandall et al. 1999) and for guiding conservation work. There is a growing body of population genetic structure studies primarily focusing on fragmented environments, while few have focused on continuous habitats. Therefore, in many species, we often know less about the patterns of genetic differentiation in continuous habitats (Cabe et al. 2007). However, understanding of patterns of genetic architecture in continuous habitats is important for determination of how these patterns are altered by fragmentation (Cabe et al. 2007). Therefore, it is critical that more population analyses are conducted in continuous environments. As S. nukiangensis is distributed throughout the Nujiang River, it is an ideal candidate for the study of population genetics in a continuous model.
Pleistocene climatic oscillations played an important role in the contemporary diversity in many species and communities (Hewitt 2000, 2004). Glacial cycles during the Quaternary Period resulted in the periodic expansions and contractions of population sizes and distribution ranges of species. In this study, the effects of climatic oscillations on the historical demography of S. nukiangensis were examined. Limited dispersal capacity can cause small-scale genetic differentiation in populations. Because of feeding habits of preying phytoplankton attached to the stone and hypognathous mouth of S. nukiangensis (Chu and Chen 1989; Chen 1998; Chen and Cao 2000), we hypothesize that S. nukiangensis has limited dispersal capacity and may fit the isolation by distance (IBD) model (Wright 1943). IBD, in the context of population genetics, is the process by which a genetic structure is generated via geographically restricted gene flow due to the fact that random genetic drift is occurring locally (Hardy and Vekemans 1999).
In this study, three mitochondrial DNA sequences (the cytochrome c oxidase submit I (COI), cytochrome b gene (cytb), and control region (MCR)) were employed as a mitochondrial concatenated data set (MCD) to evaluate genetic diversity, the population genetic structure, and population demographic history of S. nukiangensis. Furthermore, whether IBD plays a role in shaping the structure of S. nukiangensis populations was investigated and whether the height of water acts as barrier to gene flow among S. nukiangensis populations was examined. This study seeks to develop meaningful recommendations for conservation policies and the preservation of S. nukiangensis.
Materials and Methods
Samples and laboratory analyses
A total of 224 specimens of S. nukiangensis were collected from 9 localities along the Nujiang River in March and October 2012 and between May and July 2013 (Fig.2; Table1). A small piece of white muscle tissue or fin was dissected from the right body side of each specimen. All tissues used for genomic DNA extraction were preserved in 95% ethanol and deposited in the Freshwater Fish Museum at the Institute of Hydrobiology, Chinese Academy of Sciences.
Figure 2.
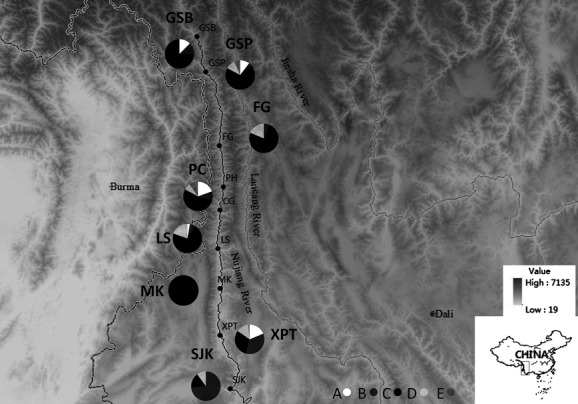
A map of the Nujiang River showing the nine sampling sites and group frequencies in each population. The information of sampling sites referred to Table1, and the five groups were defined by BAPS.
Table 1.
Descriptive statistics by sampling site for the Schizothorax nukiangensis in this study
| Collection site | PA | Coordinates | Altitude | N | NH | LSH | h (D) | π (D) |
|---|---|---|---|---|---|---|---|---|
| Bingzhongluo | GSB | 28.026/98.633 | 1536 | 8 | 7 | 2 | 0.964 (0.077) | 0.0016 (0.0004) |
| Puladi | GSP | 27.668/98.728 | 1422 | 29 | 20 | 14 | 0.966 (0.019) | 0.0025 (0.0004) |
| Fugong | FG | 26.909/98.867 | 1177 | 16 | 11 | 9 | 0.925 (0.050) | 0.0017 (0.0003) |
| Pihe | PC | 26.486/98.903 | 1036 | 37 | 20 | 10 | 0.959 (0.016) | 0.0026 (0.0004) |
| Chenggan | PC | 26.252/98.870 | 936 | 4 | 4 | 2 | ||
| Lushui | LS | 25.855/98.852 | 824 | 36 | 18 | 7 | 0.900 (0.036) | 0.0018 (0.0002) |
| Mangkuan | MK | 25.450/98.872 | 747 | 5 | 4 | 1 | 0.900 (0.161) | 0.0014 (0.0003) |
| Xiaopingtian | XPT | 24.970/98.868 | 666 | 59 | 21 | 9 | 0.926 (0.015) | 0.0021 (0.0002) |
| Sanjingkou | SJK | 24.423/98.974 | 612 | 30 | 12 | 6 | 0.772 (0.075) | 0.0014 (0.0003) |
| Nujiang River | Total | 224 | 83 | 60 | 0.965 (0.005) | 0.0024 (0.0012) |
PA, population abbreviation; N, population size; Nh, number of haplotypes; LSH, location-special haplotypes; π, nucleotide diversity; h, haplotype diversity; D, standard deviation.
Total genomic DNA was extracted from muscle tissue or fin by standard salt extraction. The mitochondrial COI was amplified using the universal barcoding primers FishF1 and FishR1 (Ward et al. 2005). The partial mitochondrial cytb was amplified using the universal primers L14724 and H15915 (Xiao et al. 2001). The mitochondrial MCR was amplified and sequenced with the primers GEDL200 and GEDH860 (Zhao et al. 2009). The PCR contained approximately 100 ng of template DNA, 1 μL of each primer (10 pmol/μL), 3 μL of 10× reaction buffer, 1.5 μL of dNTPs (2.5 mmol/L each), and 2.0 U of Taq DNA polymerase in a total volume of 30 μL. The PCR conditions for COI were as follows: initial denaturation at 95°C for 5 min, followed by 30 cycles of denaturation at 95°C for 1 min, annealing at 50°C for 45 sec, extension at 72°C for 45 sec, followed by a final extension at 72°C for 10 min. The PCR conditions were identical for the cytb gene and MCR, with an initial denaturation at 94°C for 3 min, followed by 30 cycles of denaturation at 94°C for 1 min, annealing at 58–64°C for 1 min, extension at 72°C for 1 min, followed by a final extension at 72°C for 5 min. The partial nuclear recombinase activating gene 2 (RAG2) sequences were obtained from a subset of samples consisting of 33 individuals with the primers RAG2-f2a and RAG2-R6a (Lovejoy and Collette 2001). The PCR amplification profile included an initial denaturation step at 94°C for 3 min, followed by 35 cycles of denaturation for 30 sec at 94°C, annealing for 30 sec at 55°C, extension for 90 sec at 72°C, and a final extension for 10 min at 72°C. The PCR products were purified by 1.0% low-melting agarose gel electrophoresis and sequenced with the same primer pair through an ABI PRISM 3700 sequencing system.
Data analysis
The sequences were initially edited using DNASTAR multiple package (DNASTAR Inc., Madison, WI) and aligned using the CLUSTALX 2.0 program (Thompson et al. 1997). After trimming to the same length, MCD was directly used for subsequent analyses. RAG2 sequences containing more than one ambiguous site were resolved using PHASE 2.1.1 (Stephens et al. 2001; Smith et al. 2005), for which input files were prepared using SEQPHASE (Flot 2010). In our study, RAG2 sequences were only used to estimate pairwise differentiation between populations.
Genetic diversities are reflected in the nucleotide diversity (π) and haplotype diversity (h) (Nei 1987), and the standard errors for each population were calculated using DnaSP 5.10 (Librado and Rozas 2009). A network analysis was performed with Haploviewer software to estimate gene genealogies. Haploviewer software converts trees built from traditional phylogenetic methods into haplotype genealogies (Salzburger et al. 2011). The phylogeny was estimated with PhyML 3.0 (Guindon et al. 2010) using a maximum-likelihood method, and the most appropriate model of DNA substitution (HKY + I + G model), as identified by Modeltest 3.7 (Posada and Crandall 1998).
Due to the small sample size (N = 4), the Chenggan (CG) population was analyzed with the geographically closest population, Pihe (PH). The concatenated population was named PC and the midpoint was defined as the sample site. Pairwise genetic divergences between populations were estimated using F-statistics (FST) (Weir and Cockerham 1984) with 10,000 permutations, based on the distance method. A hierarchical analysis of molecular variance (AMOVA) was used to search for significant genetic partitions among populations (Excoffier et al. 1992). Both pairwise FST comparisons and AMOVA were performed in Arlequin 3.5 (Excoffier et al. 2009). To evaluate genetic boundaries between the sampling locations studied, a spatial analysis of molecular variance (SAMOVA) was performed (Dupanloup et al. 2002). The SAMOVA algorithm was employed to search for 2 to 6 potential population units. The Bayesian method in BAPS, version 6.0 (Corander et al. 2003), was used to conduct hierarchical clustering analyses. BAPS was run using linked molecular loci with 100 replicates for each of max K set to 5, 10, 15, 20, 25, 30, and 35. Subsequent admixture was conducted using the number of clusters chosen in the mixture analysis with 100 iterations, 60 reference individuals, and 10 iterations for each reference individual.
Pairwise values of FST and FST(1 − FST)−1 (Rousset 1997) were plotted against geographic distances and the height of water among sample sites to test for IBD (Slatkin 1993). The MK population was excluded due to a small sample size and a smaller gap in the distance and water height between the LS and XPT populations. The mean of the altitude values for the PH and CG populations was used as the altitude value for the PC population. Mantel tests for IBD and water height were performed using Arlequin 3.5. Geographic distances among populations were estimated using Google Earth v.4.3.
Historical changes in effective population size were assessed using three approaches. First, Tajima's D (Tajima 1989) and Fu's Fs (Fu 1997) statistics were calculated using Arlequin 3.5 to seek evidence of demographic expansions. Second, pairwise mismatch distributions (Schneider and Excoffier 1999) were used to infer the demographic history of S. nukiangensis and were performed using Arlequin 3.5 and DnaSP 5.10. The previously established average substitution rate of 1.69% per million years (Zhao et al. 2009) was assumed and has been calibrated for MCR and the cytb gene in the schizothoracine fish. The average substitution rate was set at 1.50% for MCD due to the added COI. The expansion time was estimated from the equation τ = 2ut (Nei and Tajima 1981; Rogers and Harpending 1992), where u is the mutation rate per sequence and per generation. The value of u was calculated from u = 2μk, where μ is the mutation rate per nucleotide, and k is the number of nucleotides in the analyzed fragment. The approximate time of expansion was calculated by multiplying t by the generation time (4 years; Cao et al. 1981) of the schizothoracine fish. Finally, Bayesian skyline plots (BSP) (Drummond et al. 2005) were generated using BEAST 1.6.1 (Drummond and Rambaut 2007) for MCD with 100 million generations in order to explore demographic history. The HKY + I + G model and the strict clock, with a divergence rate of 1.50% per million years, were employed for MCD.
Results
Sequence information
All three mitochondrial fragments were successfully amplified for the 224 S. nukiangensis specimens. After alignment, the COI (675 bp) contained 13 variable sites, 7 of which were parsimony informative. The cytb (1063 bp) had 28 variable sites, 16 of which were parsimony informative. The MCR (667 bp) demonstrated 41 variable sites, 31 of which were parsimony informative. No insertions and deletions were found in any of the three gene parts. A total of 83 haplotypes were defined among the all of the MCD sequences. For RAG2, we sequenced 33 individuals from a subset of samples selected based on the MCD haplotypes. The 1126-bp fragment included five polymorphic sites, and all sequences contained five alleles. The haplotype and allele sequences were deposited in GenBank (Table S1).
Genetic diversity and genetic structure
The number of haplotypes, location-special haplotypes and the haplotype diversity (h), and nucleotide diversity (π) values within each population and in the overall population are presented in Table1. The overall haplotype and nucleotide diversity values were 0.965 (0.005) and 0.0024 (0.0012), respectively. The highest haplotype and nucleotide diversities were discovered in the GSP and PC populations, whereas the lowest both were found in the SJK population. A total of 60 private haplotypes were detected among all sites.
The haplotype genealogy, constructed with haplotypes from different sampling localities, demonstrated little association between haplotypes and geography (Fig. S1). A star-like geography genealogy, with four main haplotypes and numerous haplotypes contained in a single individual, was obtained. Phylogenetic analyses were also conducted; however, the trees could not be resolved due to low intraspecific sequence divergence (data not shown).
The nonhierarchical AMOVA revealed that most of the variation (85.14%, P < 0.001) occurred within sampling sites (Table S2). Global FST in S. nukiangensis was 0.149 and was significantly different from zero (p < 0.001). The largest mean FCT index was found for two populations units (FCT = 0.120, P < 0.05) referred to as follows: (1) XPT/SJK (D-unit); (2) the other six upstream populations (U-unit) (Table S2, Fig. S2). The estimated FST value in the hierarchical AMOVA based on the two units was 0.193, which was highly significant (P < 0.001) (Table S2). Similarly, significant genetic differentiation based on RAG2 between the two units also obtained in this study (FST = 0.067, P < 0.05) (Table S3). Clustering was performed using an admixture model in BAPS that incorporated the population of origin for each sample recovered in five groups. As shown in Figure3, most of the individuals in group B came from XPT and SJK and group C had the largest specimen number from every population. XPT contained a large number of specimens from every group, more than 70% of individuals in SJK were from group B and more than 50% of individuals in the upstream populations were derived from group C (Fig.2). Thus, Bayesian analysis showed a detectable fine-scale population structure in S. nukiangensis.
Figure 3.
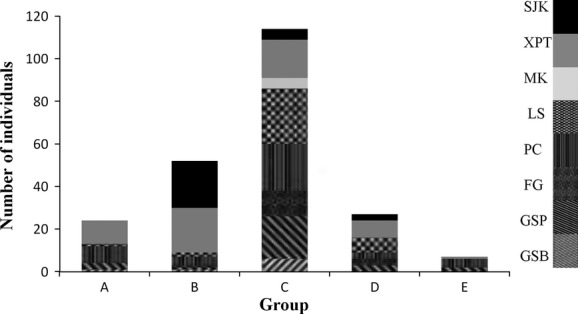
Visualization of the results of the admixture clustering that was performed in BAPS.
Pairwise comparisons of genetic differentiation (FST) ranged from −0.019 to 0.463 and are shown in Table2. Highly significant genetic differentiations were detected between the SJK population and other populations. Furthermore, significant difference between SJK and U-unit was also found based on RAG2 (Table S3). Statistically significant differences with low values were also found when comparing the FG population to the other populations. No significant differentiations, coupled with low values, were found in the other 8 of 28 pairwise comparisons of FST among the sampling areas.
Table 2.
Pairwise FST values and significance probability estimates
| GSB | GSP | FG | PC | LS | MK | XPT | SJK | |
|---|---|---|---|---|---|---|---|---|
| GSB | NS | *** | NS | * | NS | NS | *** | |
| GSP | −0.019 | *** | NS | *** | NS | *** | *** | |
| FG | 0.214 | 0.123 | *** | *** | *** | *** | *** | |
| PC | −0.018 | 0.000 | 0.158 | *** | NS | *** | *** | |
| LS | 0.053 | 0.046 | 0.112 | 0.061 | NS | *** | *** | |
| MK | 0.077 | 0.029 | 0.219 | 0.031 | 0.023 | ** | *** | |
| XPT | 0.096 | 0.100 | 0.151 | 0.109 | 0.115 | 0.175 | *** | |
| SJK | 0.386 | 0.324 | 0.355 | 0.322 | 0.345 | 0.463 | 0.119 |
NS, nonsignificant.
Above diagonal: *<0.05, **<0.005, ***<0.0005.
There was a statistically significant relationship between population differentiation and geographic distance (FST: R = 0.5064, P = 0.0232; FST(1 − FST)−1: R = 0.5388, P = 0.0195) (Fig.4), but no significant relationship between population and water height (FST: R = 0.2453, P = 0.1398; FST(1−FST)−1: R = 0.2780, P = 0.1101).
Figure 4.
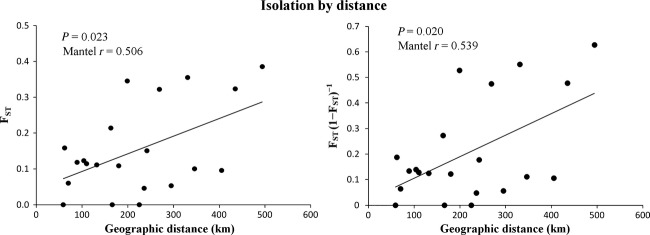
Correlation between genetic differences and geographic distances (km). p and r represent probability estimates and the correlation coefficient, respectively.
Demographic history
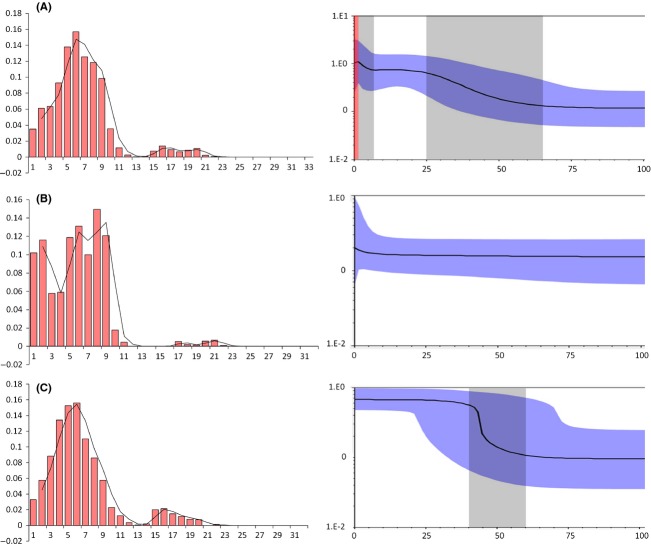
The D and FS tests implied a significant departure from neutrality for the overall population, and two population units and negative values were generated for each population by the FS test (Table3). Additionally, mismatch analysis revealed approximately unimodal distribution of pairwise differences in the overall population and U-unit, but bimodal distribution in the D-unit (Fig.5). The model of sudden demographic expansion for the overall population was not rejected by the generalized least square procedure (SSD = 0.002, P = 0.492) or by the raggedness index of the distribution (Rag = 0.010, P = 0.681) (Table3). Using the evolutionary rate calibrated in this study (1.50% per Myr), the expansion times for each population and unit were estimated to be between approximately 0.069 and 0.222 Ma (Table3). For all populations, similar results were obtained with BSP, suggesting that remarkable expansion happened from 7 to 1.5 ka and moderate expansion occurred from 65 to 25 ka (Fig.5). A sharp contraction was discovered from approximately 1.5 ka to the present. For two units, by contrast, the BSP suggested that the effective population size was relatively stable in the D-unit but sharp expansion occurred in the U-unit occurred from 60 to 40 ka.
Table 3.
Neutrality tests and mismatch distribution values for all populations
| Tajima' D (P) | Fu's Fs (P) | SSD (P) | Hri (P) | τ | T (Ma) | |
|---|---|---|---|---|---|---|
| GSB | −1.13 (0.153) | −2.53 (0.047) | 0.004 (0.959) | 0.027 (0.961) | 4.0 | 0.110 |
| GSP | −1.63 (0.030) | −8.39 (0.003) | 0.009 (0.238) | 0.024 (0.419) | 5.1 | 0.140 |
| FG | −1.52 (0.056) | −3.67 (0.030) | 0.010 (0.532) | 0.038 (0.535) | 3.9 | 0.107 |
| PC | −1.08 (0.139) | −9.04 (0.004) | 0.010 (0.324) | 0.018 (0.418) | 2.5 | 0.069 |
| LS | −1.37 (0.014) | −6.50 (0.008) | 0.013 (0.237) | 0.045 (0.159) | 5.2 | 0.143 |
| MK | 0.08 (0.586) | −0.13 (0.356) | 0.056 (0.397) | 0.150 (0.655) | 4.3 | 0.119 |
| XPT | −1.19 (0.106) | −4.74 (0.066) | 0.009 (0.374) | 0.023 (0.379) | 6.1 | 0.169 |
| SJK | −1.04 (0.154) | −2.66 (0.110) | 0.027 (0.515) | 0.040 (0.777) | 8.0 | 0.222 |
| U-unit | −1.86 (0.006) | −25.22 (0.000) | 0.002 (0.462) | 0.009 (0.795) | 4.1 | 0.114 |
| D-unit | −1.43 (0.047) | −8.34 (0.014) | 0.011 (0.424) | 0.021 (0.489) | 6.6 | 0.183 |
| Overall | −1.78 (0.005) | −24.94 (0.000) | 0.002 (0.492) | 0.010 (0.681) | 5.8 | 0.161 |
Τ, time since expansion expressed in units of mutational time; SSD, sum of squared distribution; Hri, Harpending's raggedness index; T, estimated expansion time (Ma); P, the probability value.
Figure 5.
Mismatch distribution and Bayesian skyline plots (BSP) analysis. (A) All samples. (B) D-unit. (C) U-unit. Pictures in left are results of mismatch distribution. The abscissa indicates the number of pairwise differences between compared sequences. The ordinate is the frequency for each value. Histograms are the observed frequencies of pairwise divergences among sequences and the line refers to the expectation under the model of population expansion. Pictures in right are results of BSP. The abscissa shows the time in millenniums of years ago (ka). The ordinate shows the estimated effective population size. Estimates of means are joined by a solid line while the shaded range delineates the 95% HPD limits. The gray dash areas represent timescale of the expansion events, and the red dash area represents timescale of the contraction event.
Discussion
Our study documents genetic structure and population demography within S. nukiangensis in continuous habitat. Environmental heterogeneity, limited dispersal ability and /or glacial cycles may drive genetic differentiation among populations in the fine geographic scale. Because of differential living regions and cold tolerance, S. nukiangensis shows differential responses to climatic fluctuations during the Pleistocene. Considering limited sample range and molecular markers in our study, our presented hypotheses were not very strong. Therefore, more sample size and molecular markers should be employed in the future to demonstrate our hypotheses.
Population bottleneck and fine-scale population structure
In the current study, the overall haplotype and nucleotide diversities were much lower in S. nukiangensis compared with S. prenanti, S. o'connori, Platypharodon extremus, and Gymnocypris chilianensis (He and Chen 2009; Liang et al. 2011; Zhao et al. 2011; Su et al. 2014b); however, the results were similar to other schizothoracine fishes such as Schizopygopsis pylzovi and Gymnodiptychus pachycheilus (Qi et al. 2007; Su et al., 2014a). The high level of haplotype diversity and low nucleotide diversity in S. nukiangensis are indicative of a population bottleneck, followed by rapid population growth and accumulation of mutations (Grant and Bowen 1998). Population bottlenecks appear to severely impact the extent of haplotype diversity in many fish populations (Billington and Hebert 1991). Since the Quaternary period, the Tibetan Plateau has undergone several uplifts, creating substantial changes in the climate and natural environment (Li and Fang 1999) and significantly influencing the demographic history of fish species. Neutrality tests, mismatch distribution tests, and BSP analyses confirmed that a recent expansion event likely occurred in S. nukiangensis. Furthermore, genetic bottleneck might be also suspected in S. nukiangensis due to the recent population decline in response to habitat destruction and overfishing.
In contrast to cases of absence of genetic differentiation in the schizothoracine fishes (e.g., Su et al. 2014a,b), significant but moderate population genetic structure among geographic populations was detected in continuous habitat by the AMOVA, SAMOVA, BAPS, and FST analyses. The fact that nearly 73.56% of haplotypes were only detected in single locality indicates limited gene flow among populations. The most noteworthy finding was the occurrence of significant genetic differentiation between the SJK and the other populations. The significant pairwise genetic differentiation detected between the SJK population and the other populations is consistent with Placocheilus cryptonemus in the Nujiang River (Zhang et al. 2009) and might contribute to their specific habitat and dispersal ability. First, in contrast to turbulent water between Gongshan and Lushui, the Nujiang River is comparatively peaceful in Sanjiangkou (Liu et al. 2008). Different water environments might drive distinct ecotypes, which might produce reproductive isolation. Furthermore, higher temperatures in Sanjiangkou (Fan and He 2012) might play a role in the genetic differentiation, at least in part. A study by Li et al. (2009) found that temperature is an important variable for Schizothorax species richness. Because it is a cold-water fish species, it is particularly sensitive to habitat temperature. In addition, distance might also be a key cause for population differentiation (discussed below). It is also feasible that glacial cycles may have given rise to the pattern of genetic structure. Cyclic cooling potentially restricted S. nukiangensis to a number of confined regions, which impeded gene flow among populations. The end of the glacial periods would then be followed by a population expansion event and secondary admixture; however, they might not be a panmictic population after long-term isolation.
The other interesting finding in the current study was that Xiaopingtian might act as a mixed reservoir for S. nukiangensis, as evidenced by the low genetic divergence between XPT and other populations and each BAPS group containing many individuals in XPT population; however, S. nukiangensis rarely passed through this region. The area near Lushui might form natural barriers for fish migration and dispersal, acting as a boundary line for animal fauna in the Oriental and Tibetan Plateau areas of the Nujiang River (Chen 1998). SAMOVA, BAPS, and FST analyses strongly support this hypothesis.
Complex demographic processes of S. nukiangensis
A gradual colonization process is expected to yield a decrease in genetic diversity in colonial populations (Ibrahim et al. 1996; Ramachandran et al. 2005; Krystufek et al. 2007). Fortunately, for the overall population, sudden population expansions can affect the genetic diversities of species and the relationships among the haplotypes. In a rapidly expanding population, more haplotypes and lineages are produced by mutations than are removed by genetic drift, which may increase the genetic diversity (Avise et al. 1984; Asmussen et al. 1987). A star-like haplotype genealogy, unimodal distribution of pairwise differences, BSP analyses, and significantly negative FS values all indicate that S. nukiangensis has passed through recent demographic expansions.
Climatic fluctuations during the Pleistocene play a predominant role in the patterns of genetic diversity in many animals and plants, but responses to Pleistocene glacial cycles are expected to vary among species and geographic regions, in part because of differential cold tolerance (Hewitt 1996, 2004). As the Pleistocene, four or five glaciations have occurred in the Tibetan Plateau (Zheng et al. 2002). The largest glacial (LGM) development in the Tibetan Plateau happened during the middle Pleistocene (0.5 Ma), while glacial retreat has been occurring since 0.17 Ma (Zhuo et al., 1998; Zhang et al. 2000; Zheng et al. 2002). The last glaciation (LG) in the Tibetan Plateau started at approximately 0.075 Ma and continued until 0.01 Ma (Jing et al. 2004; Yi et al. 2005), after which the plateau experienced short glacial cycles with three warmer periods (Jing et al. 2004; Yi et al. 2005).
On the basis of estimated expansion times from mismatch distribution, the typical signature of the expansion events suggests that most populations underwent a rapid increase in size after the LGM, which might be related to suitable environments within their areas during warmer periods (0.17–0.075 Ma). This sharp expansion event was also detected by BSP analyses after the LG for the overall population. However, BSP also indicated that S. nukiangensis experienced an obvious expansion in the middle of the LG (0.075–0.01 Ma) for the overall population and U-unit population. Two reasons may account for this abnormal phenomenon. First, the habitat of S. nukiangensis might not have been covered by ice during the LG. S. nukiangensis in the current study resides at low elevation (612–1536 m), much lower than the snowline (1800–3200 m) (Shi et al. 1997; Liu et al. 1999). Second, S. nukiangensis dwells in an alpine environment and endured low temperatures during the ice age. In the D-unit population, climatic vacillations had little influence on S. nukiangensis according to the BSP result, which was likely due to the lower altitude of its living habitat and its strong suffertibility against the ice age. As for dissimilar reactions to the climatic vacillations during the LGM and LG, a major factor may explain this unusual phenomenon. Temperature in the LGM was much lower than in the LG (Zheng et al. 2002), which exceeded its cold suffertibility and might give rise to bottlenecks during the LGM. Nevertheless, S. nukiangensis could pass smoothly through the LG.
Isolated by distance
Dispersal ability is severely restrained in most species (Meirmans 2012). Most animals have active locomotion, but most rarely disperse far beyond their places of birth (Greenwood 1980). The fine-scale genetic population structure seems to be a phenomenon that is generally common in poor dispersing fish species (Reusch et al. 2001; Rogers et al. 2002); thus, IBD scenarios are common phenomena in these species as well (Wang et al. 2000; Reusch et al. 2001; Taylor et al. 2001; Koskinen et al. 2002). The clear pattern of IBD in S. nukiangensis in the current study strongly supports the previous hypothesis and was predicted by the feeding habits, hypognathous mouth and limited capacity for dispersal of S. nukiangensis. Geographic distances could be a reasonable explanation for the observed geographic distribution of genetic variation between populations. S. nukiangensis mainly preys on algae attached to stones (Chen and Cao 2000), which would restrict its distribution range. Furthermore, limited dispersal capacity plays an important role in local adaption as well. Finally, philopatry is an essential factor that influences genetic differentiation. In contrast, water height does not present a significant barrier to gene flow in this species. Consequently, it appears that the continuous water habitat also represents a barrier for the dispersal of S. nukiangensis.
Unlike freshwater fish in rivers and streams with limited dispersal ability and environmental heterogeneity, pelagic fish have little opportunity for genetic diversity and isolation. The low genetic structure, on a global scale, found in Rhincodon typus due to its pelagic behavior provides a credible instance (Castro et al. 2007). Many other recent surveys also offer proof to explain this phenomenon (e.g., Liu et al. 2010; Papetti et al. 2012; Varela et al. 2012). While pelagic habits generally seem to prevent genetic divergence in allopatry, the river environment appears to facilitate isolation and differentiation in fish populations.
Implications for conservation
Schizothorax nukiangensis is a valuable fish that is distributed along the Nujiang River. However, S. nukiangensis specimens taken from the Nujiang River exhibited a miniaturized body shape, greatly reflecting human disturbance, and environmental disruption. Habitat degradation and human activity likely drive the population decline in S. nukiangensis, but genetic factors can speed up the extinction process once a population becomes very small (Westemeier et al. 1998). Because of the low genetic diversity and recent population contraction seen in this species, it is vital that protective measures be taken immediately.
We propose several recommendations based on the current research. A nature reserve should be established with strict prohibitions on overfishing and the use of poison and explosives. In addition, special attention must be paid to the XPT and SJK populations and two units should be conserved as two separated management units (MUs) (Moritz 1994). Dam construction in the mainstream will prohibit gene flow among different subpopulations, and countermeasures should be taken to manage this. Additional analyses, using larger sample sizes and more genetic markers, should be carried out to assess the extent of isolation of the putative population of the whole Nujiang River in order to develop appropriate management measures.
Acknowledgments
We are very grateful to Mao Yuntao, Yang Liandong, Lu Suxiang, Shen Yanjun, Guan Lihong, and Zhong Zaixuan for sample collection. We are thankful to Luo Si for assistance of species identification and Chen Jin for his advice about figures. This work was supported by the Key Fund and NSFC-Yunnan mutual funds of the National Natural Science Foundation of China (Grant Nos. 31130049 and U1036603).
Data Accessibility
DNA sequences have been deposited in GenBank under Accession numbers KM070564–KM070812 and KP283022–KP283026. Details regarding individual samples are available in Table1 and Table S1.
Conflict of Interest
None declared.
Supporting Information
Table S1. Schizothorax nukiangensis haplotype and allele frequency by population and GenBank accession number of each haplotype and allele.
Table S2. Results of AMOVA for two grouping options of the Schizothorax nukiangensis estimated using F–statistics (FST) based on the MCD. All P < 0.05.
Table S3. Pairwise FST values and significance probability estimates (P) based on RAG2 sequences.
Figure S1. Haplotype genealogy from Maximum Likelihood (ML) tree performed in the Haploviewer, which exhibits the relationship among haplotypes of the eight regions.
Figure S2. Summary of results of spatial analysis of molecular variance (SAMOVA) in Schizothorax nukiangensis populations.
References
- Asmussen MA, Arnold J. Avise JC. Definition and properties of disequilibrium statistics for associations between nuclear and cytoplasmic genotypes. Genetics. 1987;115:755–768. doi: 10.1093/genetics/115.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avise JC, Neigel JE. Arnold J. Demographic influences on mitochondrial DNA lineage survivorship in animal populations. J. Mol. Evol. 1984;20:99–105. doi: 10.1007/BF02257369. [DOI] [PubMed] [Google Scholar]
- Billington N. Hebert PDN. Mitochondrial DNA diversity in fishes and its implications for introductions. Can. J. Fish Aquat. Sci. 1991;48:348–355. [Google Scholar]
- Cabe PR, Page RB, Hanlon TJ, Aldrich ME, Connors L. Marsh DM. Fine-scale population differentiation and gene flow in a terrestrial salamander (Plethodon cinereus) living in continuous habitat. Heredity (Edinb) 2007;98:53–60. doi: 10.1038/sj.hdy.6800905. [DOI] [PubMed] [Google Scholar]
- Cao WX, Chen YY, Wu YF. Zhu SQ. Tibetan Expedition of the Chinese Academy of Science Studies on the period, amplitude and type of the uplift of the Qinghai–Xizang plateau. Beijing: Science Press; 1981. Origin and evolution of schizothoracine fishes in relation to the upheaval of the Xizang Plateau; pp. 118–130. [Google Scholar]
- Castro AL, Stewart BS, Wilson SG, Hueter RE, Meekan MG, Motta PJ, et al. Population genetic structure of Earth's largest fish, the whale shark (Rhincodon typus) Mol. Ecol. 2007;16:5183–5192. doi: 10.1111/j.1365-294X.2007.03597.x. [DOI] [PubMed] [Google Scholar]
- Chen YY. The Fishes of the Hengduan Mountains Region. Beijing: Science Press; 1998. [Google Scholar]
- Chen XY. Checklist of fishes of Yunnan. Zool. Res. 2013;34:281–343. doi: 10.11813/j.issn.0254-5853.2013.4.0281. [DOI] [PubMed] [Google Scholar]
- Chen YF. Cao WX. Yue P. Fauna Sinica, Osteichthyes, Cypriniformes III. Beijing: Science Press; 2000. Schizothoracinae; pp. 273–335. (in Chinese) [Google Scholar]
- Chu XL. Chen YR. The Fishes of Yunnan, China. Cyprinidae. Beijing: Sciences Press; 1989. [Google Scholar]
- Corander J, Waldmann P. Sillanpaa MJ. Bayesian analysis of genetic differentiation between populations. Genetics. 2003;163:367–374. doi: 10.1093/genetics/163.1.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crandall KA, Posada D. Vasco D. Effective population size: missing measures and missing concepts. Anim. Conserv. 1999;2:317–319. [Google Scholar]
- Drummond AJ. Rambaut A. BEAST: bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A, Shapiro B. Pybus OG. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol. Biol. Evol. 2005;22:1185–1192. doi: 10.1093/molbev/msi103. [DOI] [PubMed] [Google Scholar]
- Dudgeon D. Asian river fishes in the Anthropocene: threats and conservation challenges in an era of rapid environmental change. J. Fish Biol. 2011;79:1487–1524. doi: 10.1111/j.1095-8649.2011.03086.x. [DOI] [PubMed] [Google Scholar]
- Dupanloup I, Schneider S. Excoffier L. A simulated annealing approach to define the genetic structure of populations. Mol. Ecol. 2002;11:2571–2581. doi: 10.1046/j.1365-294x.2002.01650.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Smouse PE. Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Hofer T. Foll M. Detecting loci under selection in a hierarchically structured population. Heredity (Edinb) 2009;103:285–298. doi: 10.1038/hdy.2009.74. [DOI] [PubMed] [Google Scholar]
- Fan H. He DM. Regional climate and its change in the Nujiang River basin. Acta Geographic Sinica. 2012;67:621–630. [Google Scholar]
- Flot JF. seqphase: a web tool for interconverting phase input/output files and fasta sequence alignments. Mol. Ecol. Resour. 2010;10:162–166. doi: 10.1111/j.1755-0998.2009.02732.x. [DOI] [PubMed] [Google Scholar]
- Fu YX. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 1997;147:915–925. doi: 10.1093/genetics/147.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Sun Z, Liu S, Wang X, Yang X. Zhuang C. The ecological influences and protection measures in the Nujiang river hydropower development. J. Sichuan For. Sci. Tech. 2012;33:101–103. [Google Scholar]
- Grant WS. Bowen BW. Shallow population histories in deep evolutionar y lineages of marine fishes: Insights from sardines and anchovies and lessons for conservation. J. Hered. 1998;89:415–426. [Google Scholar]
- Greenwood PJ. Mating systems, philopatry and dispersal in birds and mammals. Anim. Behav. 1980;28:1140–1162. [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W. Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Hardy OJ. Vekemans X. Isolation by distance in a continuous population: reconciliation between spatial autocorrelation analysis and population genetics models. Heredity (Edinb) 1999;83(Pt 2):145–154. doi: 10.1046/j.1365-2540.1999.00558.x. [DOI] [PubMed] [Google Scholar]
- He D. Chen Y. Phylogeography of Schizothorax o'connori (Cyprinidae: Schizothoracinae) in the Yarlung Tsangpo River, Tibet. Hydrobiologia. 2009;635:251–262. [Google Scholar]
- Hewitt GM. Some genetic consequences of ice ages, and their role in divergence and speciation. Biol. J. Linn. Soc. 1996;58:247–276. [Google Scholar]
- Hewitt G. The genetic legacy of the Quaternary ice ages. Nature. 2000;405:907–913. doi: 10.1038/35016000. [DOI] [PubMed] [Google Scholar]
- Hewitt GM. Genetic consequences of climatic oscillations in the Quaternary. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2004;359:183–195. doi: 10.1098/rstb.2003.1388. discussion 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim KM, Nichols RA. Hewitt GM. Spatial patterns of genetic variation generated by different forms of dispersal during range expansion. Heredity (Edinb) 1996;77:282–291. [Google Scholar]
- Jing M, Yang G. Sun N. Study on the climatic changes between the last interglacial age and the last glacial age recorded by Ostracoda in eastern Qaidam Basin. J. Earth Sci. Environ. 2004;26:83–87. [Google Scholar]
- Koskinen MT, Knizhin I, Primmer CR, Schlotterer C. Weiss S. Mitochondrial and nuclear DNA phylogeography of Thymallus spp (grayling) provides evidence of ice-age mediated environmental perturbations in the world's oldest body of fresh water, Lake Baikal. Mol. Ecol. 2002;11:2599–2611. doi: 10.1046/j.1365-294x.2002.01642.x. [DOI] [PubMed] [Google Scholar]
- Krystufek B, Buzan EV, Hutchinson WF. Hanfling B. Phylogeography of the rare Balkan endemic Martino's vole, Dinaromys bogdanovi, reveals strong differentiation within the western Balkan Peninsula. Mol. Ecol. 2007;16:1221–1232. doi: 10.1111/j.1365-294X.2007.03235.x. [DOI] [PubMed] [Google Scholar]
- Li J. Fang X. Uplift of the Tibetan Plateau and environmental changes. Chin. Sci. Bull. 1999;44:2117–2124. [Google Scholar]
- Li J, He QX, Hua X, Zhou J, Xu H, Chen J, et al. Climate and history explain the species richness peak at mid-elevation for Schizothorax fishes (Cypriniformes: Cyprinidae) distributed in the Tibetan Plateau and its adjacent regions. Glob. Ecol. Biogeogr. 2009;18:264–272. [Google Scholar]
- Liang J, Liu Y, Zhang X, Yue B. Song Z. An observation of the loss of genetic variability in prenant's schizothoracin, Schizothorax prenanti, inhabiting a plateau lake. Biochem. Syst. Ecol. 2011;39:361–370. [Google Scholar]
- Librado P. Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Liu T, Zhang X, Xiong S. Qin X. Qinghai-Xizang Plateau glacial environment and global cooling. Quat. Sci. 1999;5:385–396. [Google Scholar]
- Liu D, Shen Y. Wang Z. Characteristics analysis of Nujiang river basin water resources. Yangtze River. 2008;39:64–66. [Google Scholar]
- Liu M, Lu ZC, Gao TX, Yanagimoto T. Sakurai Y. Remarkably low mtDNA control-region diversity and shallow population structure in Pacific cod Gadus macrocephalus. J. Fish Biol. 2010;77:1071–1082. doi: 10.1111/j.1095-8649.2010.02743.x. [DOI] [PubMed] [Google Scholar]
- Lovejoy NR. Collette BB. Phylogenetic relationships of new world needlefishes (Teleostei: Belonidae) and the biogeography of transitions between marine and freshwater habitats. Copeia. 2001;2001:324–338. [Google Scholar]
- Meirmans PG. The trouble with isolation by distance. Mol. Ecol. 2012;21:2839–2846. doi: 10.1111/j.1365-294X.2012.05578.x. [DOI] [PubMed] [Google Scholar]
- Moritz C. Defining ‘evolutionarily significant units’ for conservation. Trends Ecol. Evol. 1994;9:373–374. doi: 10.1016/0169-5347(94)90057-4. [DOI] [PubMed] [Google Scholar]
- Nei M. Molecular evolutionary genetics. NewYork, NY: Columbia University Press; 1987. [Google Scholar]
- Nei M. Tajima F. DNA polymorphism detectable by restriction endonucleases. Genetics. 1981;97:145–163. doi: 10.1093/genetics/97.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papetti C, Pujolar JM, Mezzavilla M, La Mesa M, Rock J, Zane L, et al. Population genetic structure and gene flow patterns between populations of the Antarctic icefish Chionodraco rastrospinosus. J. Biogeogr. 2012;39:1361–1372. [Google Scholar]
- Posada D. Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Qi DL, Guo SC, Zhao XQ, Yang J. Tang W. Genetic diversity and historical population structure of Schizopygopsis pylzovi (Teleostei: Cyprinidae) in the Qinghai-Tibetan Plateau. Freshw. Biol. 2007;52:1090–1104. [Google Scholar]
- Ramachandran S, Deshpande O, Roseman CC, Rosenberg NA, Feldman MW. Cavalli-Sforza LL. Support from the relationship of genetic and geographic distance in human populations for a serial founder effect originating in Africa. Proc. Natl Acad. Sci. USA. 2005;102:15942–15947. doi: 10.1073/pnas.0507611102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reusch TB, Wegner KM. Kalbe M. Rapid genetic divergence in postglacial populations of threespine stickleback (Gasterosteus aculeatus): the role of habitat type, drainage and geographical proximity. Mol. Ecol. 2001;10:2435–2445. doi: 10.1046/j.0962-1083.2001.01366.x. [DOI] [PubMed] [Google Scholar]
- Rogers AR. Harpending H. Population growth makes waves in the distribution of pairwise genetic differences. Mol. Biol. Evol. 1992;9:552–569. doi: 10.1093/oxfordjournals.molbev.a040727. [DOI] [PubMed] [Google Scholar]
- Rogers SM, Gagnon V. Bernatchez L. Genetically based phenotype-environment association for swimming behavior in lake whitefish ecotypes (Coregonus clupeaformis Mitchill) Evolution. 2002;56:2322–2329. doi: 10.1111/j.0014-3820.2002.tb00155.x. [DOI] [PubMed] [Google Scholar]
- Rousset F. Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics. 1997;145:1219–1228. doi: 10.1093/genetics/145.4.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzburger W, Ewing GB. Von Haeseler A. The performance of phylogenetic algorithms in estimating haplotype genealogies with migration. Mol. Ecol. 2011;20:1952–1963. doi: 10.1111/j.1365-294X.2011.05066.x. [DOI] [PubMed] [Google Scholar]
- Schneider S. Excoffier L. Estimation of past demographic parameters from the distribution of pairwise differences when the mutation rates vary among sites: application to human mitochondrial DNA. Genetics. 1999;152:1079–1089. doi: 10.1093/genetics/152.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Zheng B. Yao T. Glaciers and environments during the last glacial maximum (LGM) on the Tibetan Plateau. J. Glaciol. Geocryol. 1997;19:97–113. [Google Scholar]
- Slatkin M. Isolation by distance in equilibrium and non-equilibrium populations. Evolution. 1993;47:264–279. doi: 10.1111/j.1558-5646.1993.tb01215.x. [DOI] [PubMed] [Google Scholar]
- Smith SA, Stephens PR. Wiens JJ. Replicate patterns of species richness, historical biogeography, and phylogeny in Holarctic treefrogs. Evolution. 2005;59:2433–2450. [PubMed] [Google Scholar]
- Stephens M, Smith NJ. Donnelly P. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su JH, Ji WH, Wei YM, Zhang Y, Gleeson DM, Lou Z, et al. Genetic structure and demographic history of the endangered and endemic schizothoracine fish Gymnodiptychus pachycheilus in Qinghai-Tibetan Plateau. Zoolog. Sci. 2014a;31:512–522. doi: 10.2108/zs130238. [DOI] [PubMed] [Google Scholar]
- Su JH, Ji WH, Zhang YP, Gleeson DM, Lou Z, Ren J, et al. Genetic diversity and demographic history of the endangered and endemic fish (Platypharodon extremus): implications for stock enhancement in Qinghai Tibetan Plateau. Environ. Biol. Fish. 2014b;2014:1–12. [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MI, Ruber L. Verheyen E. Microsatellites reveal high levels of population substructuring in the species-poor Eretmodine cichlid lineage from Lake Tanganyika. Proc. Biol. Sci. 2001;268:803–808. doi: 10.1098/rspb.2000.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F. Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela AI, Ritchie PA. Smith PJ. Low levels of global genetic differentiation and population expansion in the deep-sea teleost Hoplostethus atlanticus revealed by mitochondrial DNA sequences. Mar. Biol. 2012;159:1049–1060. [Google Scholar]
- Wang JP, Hsu KC. Chiang TY. Mitochondrial DNA phylogeography of Acrossocheilus paradoxus (Cyprinidae) in Taiwan. Mol. Ecol. 2000;9:1483–1494. doi: 10.1046/j.1365-294x.2000.01023.x. [DOI] [PubMed] [Google Scholar]
- Ward RD, Zemlak TS, Innes BH, Last PR. Hebert PD. DNA barcoding Australia's fish species. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2005;360:1847–1857. doi: 10.1098/rstb.2005.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BS. Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Westemeier RL, Brawn JD, Simpson SA, Esker TL, Jansen RW, Walk JW, et al. Tracking the long-term decline and recovery of an isolated population. Science. 1998;282:1695–1698. doi: 10.1126/science.282.5394.1695. [DOI] [PubMed] [Google Scholar]
- Wright S. Isolation by Distance. Genetics. 1943;28:114–138. doi: 10.1093/genetics/28.2.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W, Zhang Y. Liu H. Molecular systematics of Xenocyprinae (teleostei: cyprinidae): taxonomy, biogeography, and coevolution of a special group restricted in East Asia. Mol. Phylogenet. Evol. 2001;18:163–173. doi: 10.1006/mpev.2000.0879. [DOI] [PubMed] [Google Scholar]
- Yi C, Cui Z. Xiong H. Numerical periods of quaternary glaciations in China. Quat. Sci. 2005;25:609–619. [Google Scholar]
- Zhang DF, Fengquan L. Jianmin B. Eco-environmental effects of the Qinghai-Tibet plateau uplift during the Quaternary in China. Environ. Geol. 2000;39:1352–1358. [Google Scholar]
- Zhang DY, Wang DQ, Liu SP, Guo ZQ, Yue XJ, Deng HT, et al. Population genetic structure analysis of endangered species Placocheilus cryptonemus in the Nujiang River based on Cyt b sequences of mtDNA. J. Fish. Sci. China. 2009;16:477–486. [Google Scholar]
- Zhao K, Duan ZY, Peng ZG, Guo SC, Li JB, He SP, et al. The youngest split in sympatric schizothoracine fish (Cyprinidae) is shaped by ecological adaptations in a Tibetan Plateau glacier lake. Mol. Ecol. 2009;18:3616–3628. doi: 10.1111/j.1365-294X.2009.04274.x. [DOI] [PubMed] [Google Scholar]
- Zhao K, Duan Z, Peng Z, Gan X, Zhang R, He S, et al. Phylogeography of the endemic Gymnocypris chilianensis (Cyprinidae): sequential westward colonization followed by allopatric evolution in response to cyclical Pleistocene glaciations on the Tibetan Plateau. Mol. Phylogenet. Evol. 2011;59:303–310. doi: 10.1016/j.ympev.2011.02.001. [DOI] [PubMed] [Google Scholar]
- Zheng BX, Xu QQ. Shen YP. The relationship between climate change and Quaternary glacial cycles on the Qinghai-Tibetan plateau: review and speculation. Quatern. Int. 2002;97–98:93–101. [Google Scholar]
- Zhuo Z, Baoyin Y. Petit-Maire N. Paleoenvironments in China during the Last Glacial Maximum and the Holocene optimum. Episodes. 1998;21:152–158. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Schizothorax nukiangensis haplotype and allele frequency by population and GenBank accession number of each haplotype and allele.
Table S2. Results of AMOVA for two grouping options of the Schizothorax nukiangensis estimated using F–statistics (FST) based on the MCD. All P < 0.05.
Table S3. Pairwise FST values and significance probability estimates (P) based on RAG2 sequences.
Figure S1. Haplotype genealogy from Maximum Likelihood (ML) tree performed in the Haploviewer, which exhibits the relationship among haplotypes of the eight regions.
Figure S2. Summary of results of spatial analysis of molecular variance (SAMOVA) in Schizothorax nukiangensis populations.
Data Availability Statement
DNA sequences have been deposited in GenBank under Accession numbers KM070564–KM070812 and KP283022–KP283026. Details regarding individual samples are available in Table1 and Table S1.