Abstract
Eukaryotic cells synthesize multiple classes of lipids by distinct metabolic pathways in order to generate membranes with optimal physical and chemical properties. As a result, complex regulatory networks are required in all organisms to maintain lipid and membrane homeostasis as well as to rapidly and efficiently respond to cellular stress. The unicellular nature of yeast makes it particularly vulnerable to environmental stress and yeast has evolved elaborate signaling pathways to maintain lipid homeostasis. In this article we highlight the recent advances that have been made using the budding and fission yeasts and we discuss potential roles for the unfolded protein response (UPR) and the SREBP-Scap pathways in coordinate regulation of multiple lipid classes.
Introduction
Phospholipids, sphingolipids and sterols are the primary structural components of eukaryotic cell membranes and hence, organisms have evolved complex regulatory mechanisms to control their synthesis. The plasma membrane defines the outer most boundary of the cell while the internal organelles compartmentalize the chemical reactions that constitute cellular metabolism, and not surprisingly their lipid composition is tailored to suit their individual functions. The budding and fission yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe have proven to be excellent models for the study of regulation of lipid metabolism, a result of a synergism of genetic and biochemical approaches, and decades of hard work. Much of this work laid the foundation for our understanding of the fundamental lipid metabolic pathways, and now, recent work in the yeasts is providing novel insights into the mechanisms underlying homeostatic control of lipid synthesis and in response to cellular stresses.
Regulation of phospholipid and sphingolipid synthesis
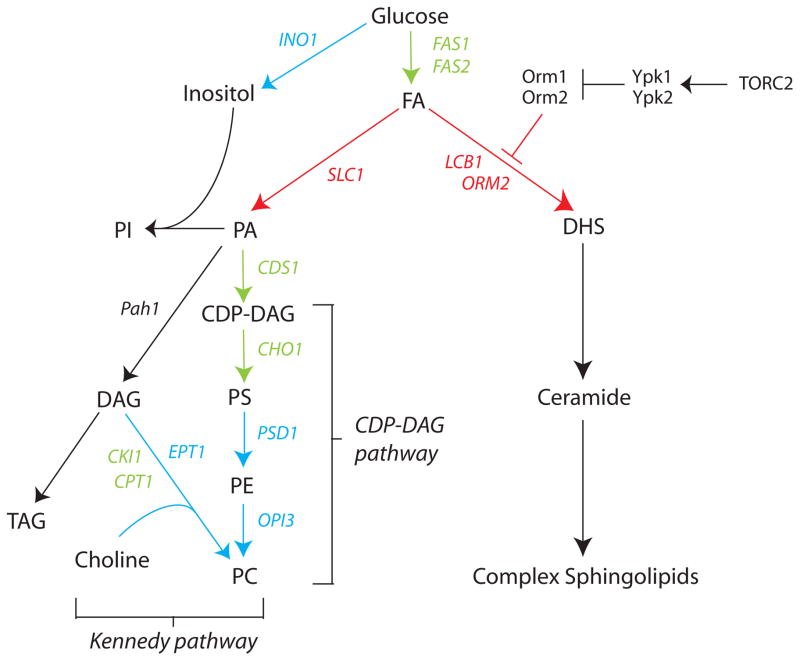
Yeast synthesize phospholipids by three pathways, a de novo CDP-DAG pathway [1], and by two salvage pathways, the Kennedy pathway [1] and the recently described exogenous lysolipid metabolism (ELM) pathway [2]. These pathways are largely conserved among all eukaryotes, with a few differences arising between fungi, plants and animals. The CDP-DAG and Kennedy pathways (Figure 1) are regulated primarily at the transcriptional level through a DNA element called UASINO, that is present in the promoters of nearly 30 of the phospholipid synthesis genes [3]. Transcriptional control is maintained by Opi1, which binds and represses the Ino2-Ino4 activator complex in the nucleus. Opi1 is repressed through its sequestration on the ER by binding the signaling lipid phosphatidic acid (PA) [1,4]. Binding of Opi1 to PA is regulated by intracellular pH, in which the lipid acts as the pH sensor, defining an unprecedented role for lipids as pH biosensors [5]. PA is a precursor in the synthesis of both phospholipids and neutral lipids and comprises only ~1% of ER phospholipids. Thus, the concentration of PA in the ER, together with intracellular pH, plays a central role in coordinate regulation of phospholipid synthesis by Opi1 [6]. In mammals, the PA hydrolases of the lipin family are emerging as critical regulators of lipid homeostasis [7••] and have been linked to metabolic diseases including type 2 diabetes [8], suggesting PA might also play a signaling role regulating lipid metabolism in humans.
Figure 1. Outline of pathways of phospholipid and sphingolipid metabolism in budding yeast.
Pathways and genes co-regulated by Opi1 and the UPR are in blue; those activated by the UPR are in red; and those repressed by Opi1 in green. Steps are not necessarily direct. Abbreviations: FA = fatty acids; PI = phosphatidylinositol; PA = phosphatidic acid; DAG = diacylglycerol; PS = phosphatidylserine; PE = phosphatidylethanolamine; TAG = triacylglycerol; PC = phosphatidylcholine; DHS = dihydro-sphingosine; TORC2 = Target of Rapamycin Complex 2, SPT = serine palmitoyltransferase.
Regulation of sphingolipid synthesis, until very recently, has been largely undefined in any organism [9]. The highly conserved Orm family of phosphoproteins, which have been linked to childhood asthma [10], have now been identified as negative regulators of serine palmitoyltransferase (SPT), which catalyses the first and rate-limiting step in the sphingolipid biosynthetic pathway [11] (Figure 1). Yeast Orm1 and Orm2 form a stable complex with SPT, and when dephosphorylated, repress SPT activity via an unknown mechanism [11]. Phosphorylation of Orm1 and Orm2 by the conserved kinase Ypk1 in response to sphingolipid deficiency relieves Orm repression of SPT, thus providing feedback control of sphingolipid production [12]. Ypk1 is in turn activated through phosphorylation by TOR kinase in response to sphingolipid levels. An exciting next step will be to determine if TOR senses lipid levels directly and which lipid acts as the signal. Interestingly, TOR kinase is a PA binding protein [13] and PA levels are altered in orm mutants [14], suggesting PA might play a role in regulation of sphingolipid metabolism.
Lipid stress sensing by the UPR
Lipid homeostasis is perturbed by different environmental stresses. But how do cells respond to these stresses? It has been recently found that deletion of several key phospholipid synthesis genes in both the CDP-DAG and Kennedy pathways results in activation of the unfolded protein response (UPR) in yeast [15]. The UPR is an ER stress response pathway that detects misfolded proteins in the ER and activates transcription of genes that facilitate proper protein folding [16]. The UPR also upregulates key phospholipid synthesis genes [17,18], thus providing a potential mechanism to respond to lipid stress (Figure 1). Tellingly, some of these target genes are the very same genes that when deleted also activate the UPR. Similarly, loss of Orm1 and Orm2 in yeast activates the UPR [14], and Orm2 is a UPR target gene [18], suggesting that dysregulated sphingolipid synthesis is another form of lipid stress detected by the UPR. Cholesterol overloading of the macrophage ER [19], and altered PC synthesis in the ER of livers from obese mice fed a high-fat diet [20], also cause ER stress and UPR activation, suggesting that lipid stress sensing by the UPR might provide coordinated feedback control of multiple lipid classes to balance membrane lipid composition.
How might the UPR detect lipid stress? The ER stress sensor Ire1 is an integral membrane protein of the ER that directly detects misfolded proteins in the ER lumen and activates the UPR [21]. Recent work now indicates that the lumenal domain of Ire1 that binds misfolded proteins is dispensable for UPR activation under conditions of lipid stress [22]. A truncated form of Ire1 containing only the transmembrane and cytoplasmic domains is sufficient to activate the UPR under starvation conditions for the lipid precursor inositol and in a CDP-DAG pathway mutant. This implies that Ire1 detects lipid stress through a mechanism distinct from protein misfolding, although it does not rule out the possibility that other domains in Ire1 also detect protein misfolding under these conditions.
An innovative new assay that is capable of monitoring levels of misfolded proteins in the ER of living cells suggests lipid stress-sensing does not occur via accumulation of unfolded proteins. Lajoie et al. measured protein misfolding under a variety of conditions that induce the UPR by monitoring the mobility of the ER chaperone Kar2 tagged with GFP [23]. Under conditions known to cause protein misfolding, Kar2-GFP mobility decreases as a result of its interaction with client proteins. However, no such decrease in Kar2-GFP mobility is observed under conditions of UPR activation by inositol starvation [23], suggesting the UPR is activated through a separate mechanism in response to lipid stress.
Is the lipid stress signal a lipid?
A clue for a possible mechanism for lipid stress sensing comes from experiments that link the UPR to synthesis of inositol. Inositol is a potent regulator of phospholipid metabolism in yeast, because its incorporation into phosphatidylinositol (PI) depletes the ER of PA [4] (Figure 1). Lowered PA releases Opi1 from the ER and its subsequent translocation to the nucleus represses phospholipid synthesis genes. INO1 is the gene most highly regulated by Opi1 and encodes the rate-limiting enzyme in inositol synthesis. INO1 is also one of the genes most highly induced by the UPR [18], which occurs via repression of Opi1 [24], suggesting that UPR activation correlates with ER PA levels. Consistent with this, addition of inositol leads to rapid inactivation of the UPR [25], which is strikingly similar to the rate at which it depletes PA in the ER [4]. However, this inactivation of the UPR is independent of Opi1 [26]. PA acts upstream of Opi1 in regulation of phospholipid synthesis genes, indicating the possibility that accumulation of PA in the ER signals lipid stress to the UPR. In support of this idea, loss of Pah1, the major ER PA hydrolase in yeast and a member of the conserved lipin family, results in both elevated PA [27] and activation of the UPR [15]. Such a signaling role for PA is not unprecedented. In plants, PA plays a central role in stress response signaling through its rapid generation and direct activation of a variety of stress effector proteins [28], although a role in UPR activation has yet to be demonstrated.
Sterol biosynthesis is under multivalent regulatory control
Sterols play essential roles in cell physiology, from membrane building blocks [29] to Hedgehog signaling during development [30], and consequently sterol synthesis is subject to regulation at multiple levels. Feedback mechanisms that control cholesterol biosynthesis and uptake through transcriptional control and regulated enzyme degradation are well-established in mammalian cells [31]. Ergosterol is the fungal analog of cholesterol, and both the budding yeast S. cerevisiae and fission yeast S. pombe have proven to be powerful models in which to study sterol biosynthesis. In this section, we discuss sterol regulatory mechanisms in these yeasts in relationship to those operating in mammalian cells.
Transcriptional control of sterol synthesis
Production of cholesterol synthesis enzymes in mammalian cells is tightly regulated at the transcriptional level through a feedback mechanism that responds to the end product cholesterol (Figure 2). The key player in this pathway is an ER membrane-bound transcription factor, SREBP (Sterol Regulatory Element Binding Protein) [31, 32]. SREBP consists of a N-terminal transcription factor domain and a C-terminal domain that forms a complex with the sterol sensing protein, SREBP cleavage activating protein (Scap) [31]. When the ER membrane is sterol-rich, Scap binds cholesterol, and the SREBP-Scap complex is retained in the ER by binding to the resident protein Insig [33]. When ER cholesterol drops, Scap undergoes a conformational change and SREBP-Scap enters COPII vesicles for transport to the Golgi [34,35•]. SREBP is cleaved sequentially in the Golgi by the Site-1 and Site-2 proteases to generate the functional transcription factor that travels to nucleus [31]. Nuclear SREBP activates transcription of cholesterol synthesis enzymes and the LDL receptor to increase sterol supply and restore homeostasis.
Figure 2. Multivalent regulation of sterol homeostasis.
Cholesterol (ergosterol in fungi) synthesis requires more than 20 enzymes and is controlled by negative feedback. The biosynthetic pathway is outlined inside the box showing key intermediates and regulated enzymes. Upon sterol depletion, transcription of sterol synthesis enzymes is activated by sterol-regulated transcription factors, SREBP in mammals, Sre1 in S. pombe, and Upc2 and Ecm22 in S. cerevisiae. In sterol-rich conditions, the end product reduces biosynthesis by inhibiting transcription factor activity. In S. pombe, low oxygen additionally activates Sre1 and sterol enzymes. In mammalian cells, two enzymes, 3-hydroxymethyl-3-methylglutaryl-coenzyme A reductase (HMGR) and squalene monooxygenase (SM) are subject to sterol-dependent degradation. HMGR catalyzes the synthesis of mevalonate from HMG-CoA and is degraded by ERAD in response to 24,25-dihydrolanosterol and an isoprenoid derived from mevalonate. SM catalyzes the first oxygenation reaction of the pathway and is regulated by cholesterol-dependent proteasomal degradation.
The SREBP pathway is functionally conserved in fungi like S. pombe [36], Cryptococcus neoformans [37,38], and Aspergillus fumigatus [39]. S. pombe SREBP, called Sre1, binds the Scap homolog, Scp1, and the mechanism of sterol regulation is conserved [36]. Interestingly, S. pombe and other ascomyceteous fungi like Aspergillus lack homologs of the Site-1 and Site-2 proteases necessitating a different processing mechanism [40]. Recently, two independent genetic screens identified a 5 subunit, membrane-bound Golgi ubiquitin E3 ligase, named the Dsc (defective for SREBP cleavage) E3 ligase, that is required for Sre1 processing [41•,42]. Bioinformatic analysis suggests that the Dsc E3 ligase does not possess protease activity, but reveals structural and organizational similarities with the Hrd1 E3 ligase complex involved in degradation of misfolded proteins by the ER-associated degradation (ERAD) pathway [43]. Genetic experiments show that Sre1 processing requires the 26S proteasome, but how these proteins mediate Sre1 proteolysis and activation requires further study.
Budding yeast S. cerevisiae lacks SREBP, and two homologous Zn(II)2Cys6 binuclear cluster transcription factors, Upc2 and Ecm22, function in ergosterol regulation [44,45]. The DNA binding domains of Upc2 and Ecm22 bind conserved sequences in promoters of ergosterol synthesis genes to activate transcription under conditions of sterol depletion [45,46]. Both Upc2 and Ecm22 contain nuclear localization signals, and Upc2 displays both nuclear and cytoplasmic localization in genome-wide localization studies [47,48]. While it has been proposed that Upc2 and Ecm22p function like SREBP [44,49], no sterol sensor has been identified and whether activation requires proteolysis is unknown.
Post-translational regulation of sterol synthesis
HMG-CoA reductase (HMGR) is a rate-limiting enzyme in sterol synthesis that catalyzes the conversion of HMG-CoA to mevalonate. In mammalian cells, HMGR is regulated at the level of transcription by SREBP, enzyme activity by AMP-activated protein kinase, and degradation by ERAD (recently reviewed in [50,51]) (Figure 2). HMGR degradation by the proteasome is accelerated by the sterol intermediate 24,25-dihydrolanosterol and the isoprenoid geranylgeranyl pyrophosphate (GGPP) through the action of the ER protein Insig and its associated ubiquitin E3 ligases, gp78 and Trc8 [52]. Recent studies demonstrate that squalene monooxygenase is also subject to regulated proteasomal degradation [53•] (Figure 2). However unlike HMGR, degradation of squalene monooxygenase is accelerated by elevated cholesterol.
Budding yeast expresses two isozymes of HMGR, Hmg1 and Hmg2, and either is sufficient for cell growth. Differential regulation of HMG1 and HMG2 transcription by oxygen and sterol intermediates has been suggested but no molecular mechanism for this regulation has been described [54]. Hmg2, like mammalian HMGR, undergoes lipid-regulated turnover mediated by the ERAD pathway [54]. Two lipid signals, the isoprenoid GGPP and an oxysterol, control the rate of Hmg2p degradation [55•]. Elevated GGPP alters Hmg2 conformation and stimulates degradation through an N-terminal sterol sensing domain and oxysterol enhances this degradation signal [56]. Unlike mammalian cells, the S. cerevisiae Insig homolog Nsg1 stabilizes Hmg2 by direct binding [57].
Fission yeast codes for one HMGR enzyme Hmg1 and the Insig homolog Ins1. In contrast to S. cerevisiae and mammalian cells, Hmg1 protein in S. pombe is neither regulated by SREBP nor degraded by Insig-mediated ERAD [58]. Rather, Ins1 regulates the catalytic activity of S. pombe Hmg1 by controlling phosphorylation of active site residues [58]. Ins1 binding to Hmg1 stimulates its phosphorylation through a stress responsive MAP kinase Sty1 inhibiting enzyme activity. Hmg1 activity is also suppressed by glucose starvation through by the inhibitory effect of Sds23 on the phosphatase Ppe1 [59].
Stress-induced regulation of sterol synthesis
Molecular oxygen is essential for cholesterol and ergosterol synthesis from squalene, requiring 11 and 12 oxygen molecules respectively [60]. Recent work on SREBP in fission yeast highlights the interplay between hypoxic stress and sterol homeostasis [40]. In addition to controlling sterol homeostasis, Sre1 functions as a hypoxic transcription factor by monitoring oxygen-dependent sterol synthesis as an indirect measure of oxygen supply [36]. Sterol synthesis decreases under low oxygen, and Sre1 is proteolytically activated to increase transcription of genes required for ergosterol synthesis and cell growth. Oxygen additionally regulates activity of the Sre1 N-terminal transcription factor by (1) blocking DNA binding and (2) accelerating degradation by the ubiquitin-proteasome system [60,61•]. Both mechanisms require Ofd1, a putative prolyl-4-hydroxylase of the 2-OG-Fe(II) dioxygenase family, and its inhibitor Nro1 [62]. The catalytic role of Ofd1 is not fully understood, but enzyme activity allows Ofd1 to function as an oxygen sensor to regulate Sre1-dependent transcription.
Anaerobic gene expression in S. cerevisiae is centrally mediated by the transcriptional repressor Rox1 [63]. Upc2, a target gene of Rox1, is derepressed under hypoxic conditions and activates a large number of anaerobically expressed genes in budding yeast, including ergosterol biosynthesis genes. In addition, Upc2 controls the expression of two sterol transporters Aus1 and Pdr11. Unlike S. pombe that cannot import exogenous sterol, S. cerevisiae takes up sterol under low oxygen when biosynthesis is compromised [36,64]. Recently, two studies identified pathways for ergosterol synthesis gene activation under extracellular stress. During hypoxic growth when heme and ergosterol levels are low, the Hog1 MAP kinase cascade is activated leading to Upc2 activation [65]. Conversely, under hyperosmotic stress Hog1 activates the repressors Rox1 and Mot3. Mot3 acts through Ecm22 to repress sterol synthesis [66].
Regulatory crosstalk?
Studies of lipid synthesis regulation have largely focused on individual classes of lipids: phospholipids, sterols and sphingolipids. But molecular interactions among these lipids seemingly necessitate coordinated control of their supply. Emerging data hints at co-regulation of these lipid classes [67]. In addition to sterols, the major phospholipids PE and PC have been shown to regulate activity of SREBP in Drosophila and mammalian cells, respectively [68••,69]. Pah1 is a PA hydrolase and regulator of phospholipid synthesis in budding yeast [6] (Figure 1). Lipin1, the mammalian Pah1 homolog, is a key regulator of phospholipid and neutral lipid balance in cells and coordinates mTORC1-dependent nutrient signaling to SREBP [7••]. Future studies will be aimed at understanding the crosstalk between these lipids and how established homeostatic mechanisms are employed to respond to diverse environmental stresses.
Acknowledgments
We extend a sincere apology to those whose work was not discussed or cited in this review owing to limitations in space and scope. This work was supported by grants from the National Institutes of Health HL077588 (PJE); and NSERC, CIHR, MSFHR, and CFI (CJRL). PJE is an Established Investigator of the American Heart Association. CJRL is a recipient of a CIHR New Investigator and a MSFHR Junior Scholar award.
Bibliography
• of special interest
•• of outstanding interest
- 1.Carman GM, Han GS. Regulation of Phospholipid Synthesis in the Yeast Saccharomyces cerevisiae. Annu Rev Biochem. 2011;80:859–883. doi: 10.1146/annurev-biochem-060409-092229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Riekhof WR, Wu J, Gijon MA, Zarini S, Murphy RC, Voelker DR. Lysophosphatidylcholine metabolism in Saccharomyces cerevisiae: the role of P-type ATPases in transport and a broad specificity acyltransferase in acylation. J Biol Chem. 2007;282:36853–36861. doi: 10.1074/jbc.M706718200. [DOI] [PubMed] [Google Scholar]
- 3.Chen M, Hancock, Lopes JM. Transcriptional regulation of yeast phospholipid biosynthetic genes. Biochim Biophys Acta. 2007;1771:310–321. doi: 10.1016/j.bbalip.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 4.Loewen CJ, Gaspar ML, Jesch SA, Delon C, Ktistakis NT, Henry SA, Levine TP. Phospholipid metabolism regulated by a transcription factor sensing phosphatidic acid. Science. 2004;304:1644–1647. doi: 10.1126/science.1096083. [DOI] [PubMed] [Google Scholar]
- 5.Young BP, Shin JJ, Orij R, Chao JT, Li SC, Guan XL, Khong A, Jan E, Wenk MR, Prinz WA, et al. Phosphatidic acid is a pH biosensor that links membrane biogenesis to metabolism. Science. 2010;329:1085–1088. doi: 10.1126/science.1191026. [DOI] [PubMed] [Google Scholar]
- 6.Loewen CJ. Lipids as conductors in the orchestra of life. F1000 Biol Reports. 2012;4:1–6. doi: 10.3410/B4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7••.Peterson TR, Sengupta SS, Harris TE, Carmack AE, Kang SA, Balderas E, Guertin DA, Madden KL, Carpenter AE, Finck BN, et al. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell. 2011;146:408–420. doi: 10.1016/j.cell.2011.06.034. This paper describes that loss of mTORC1-mediated phosphorylation of Lipin1 promotes its nuclear localization causing nuclear remodeling and downregulation of SREBP transcriptional activity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reue K. The lipin family: mutations and metabolism. Curr Opin Lipidol. 2009;20:165–170. doi: 10.1097/MOL.0b013e32832adee5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dickson RC. Roles for sphingolipids in Saccharomyces cerevisiae. Adv Exp Med Biol. 2010;688:217–231. doi: 10.1007/978-1-4419-6741-1_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S, Depner M, von Berg A, Bufe A, Rietschel E. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–473. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 11•.Breslow DK, Collins SR, Bodenmiller B, Aebersold R, Simons K, Shevchenko A, Ejsing CS, Weissman JS. Orm family proteins mediate sphingolipid homeostasis. Nature. 2010;463:1048–1053. doi: 10.1038/nature08787. This paper identifies the Orm family of proteins as key regulators of sphingolipid metabolism through formation of a complex with the enzyme serine palmitoyltransferase. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roelants FM, Breslow DK, Muir A, Weissman JS, Thorner J. Protein kinase Ypk1 phosphorylates regulatory proteins Orm1 and Orm2 to control sphingolipid homeostasis in Saccharomyces cerevisiae. PNAS. 2011;108:19222–19227. doi: 10.1073/pnas.1116948108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fang Y, Vilella-Bach M, Bachmann R, Flanigan A, Chen J. Phosphatidic acid-mediated mitogenic activation of mTOR signaling. Science. 2001;294:1942–1945. doi: 10.1126/science.1066015. [DOI] [PubMed] [Google Scholar]
- 14•.Han S, Lone MA, Schneiter R, Chang A. Orm1 and Orm2 are conserved endoplasmic reticulum membrane proteins regulating lipid homeostasis and protein quality control. PNAS. 2010;107:5851–5856. doi: 10.1073/pnas.0911617107. This study demonstrates that the Orm1 and Orm2 are important for regulating sphingolipid metabolism and that disrupted sphingolipid homeostasis leads to activation of the UPR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jonikas MC, Collins SR, Denic V, Oh E, Quan EM, Schmid V, Weibezahn J, Schwappach B, Walter P, Weissman JS, et al. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science. 2009;323:1693–1697. doi: 10.1126/science.1167983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 17.Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- 18.Kimata Y, Ishiwata-Kimata Y, Yamada S, Kohno K. Yeast unfolded protein response pathway regulates expression of genes for anti-oxidative stress and for cell surface proteins. Genes Cells. 2006;11:59–69. doi: 10.1111/j.1365-2443.2005.00921.x. [DOI] [PubMed] [Google Scholar]
- 19•.Feng B, Yao PM, Li Y, Devlin CM, Zhang D, Harding HP, Sweeney M, Rong JX, Kuriakose G, Fisher EA, Marks AR, et al. The endoplasmic reticulum is the site of cholesterol-induced cytotoxicity in macrophages. Nat Cell Biol. 2003;5:781–792. doi: 10.1038/ncb1035. This paper describes how TOR signalling regulates sphingolipid metabolism through phosphorylation of the Ypk1/2 kinases which in turn phosphorylate the Orm proteins. [DOI] [PubMed] [Google Scholar]
- 20•.Fu S, Yang L, Li P, Hofmann O, Dicker L, Hide W, Lin X, Watkins SM, Ivanov AR, Hotamisligil GS. Aberrant lipid metabolism disrupts calcium homeostasis causing liver endoplasmic reticulum stress in obesity. Nature. 2011;473:528–531. doi: 10.1038/nature09968. In this study, the authors show that abnormal lipid metabolism and altered calcium homeostasis are associated with hepatic ER stress in the obese mouse. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gardner BM, Walter P. Unfolded proteins are Ire1-activating ligands that directly induce the unfolded protein response. Science. 2011 doi: 10.1126/science.1209126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishiwata-Kimata Y, Promlek T, Shido M, Sakuramoto M, Kohno K, Kimata Y. Membrane aberrancy and unfolded proteins activate the endoplasmic reticulum-stress sensor Ire1 by different manners. Mol Biol Cell. 2011 doi: 10.1091/mbc.E11-04-0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23••.Lajoie P, Moir RD, Willis IM, Snapp EL. Kar2p availability defines distinct forms of endoplasmic reticulum stress in living cells. Mol Biol Cell. 2012 doi: 10.1091/mbc.E11-12-0995. By studying the mobility of the chaperone Kar2, the authors demonstrate that lipid stress and protein misfolding activate the UPR by distinct mechanisms. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cox JS, Chapman RE, Walter P. The unfolded protein response coordinates the production of endoplasmic reticulum protein and endoplasmic reticulum membrane. Mol Biol Cell. 1997;8:1805–1814. doi: 10.1091/mbc.8.9.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jesch SA, Liu P, Zhao X, Wells MT, Henry SA. Multiple endoplasmic reticulum-to-nucleus signaling pathways coordinate phospholipid metabolism with gene expression by distinct mechanisms. J Biol Chem. 2006;281:24070–24083. doi: 10.1074/jbc.M604541200. [DOI] [PubMed] [Google Scholar]
- 26.Jesch SA, Zhao X, Wells MT, Henry SA. Genome-wide analysis reveals inositol, not choline, as the major effector of Ino2p-Ino4p and unfolded protein response target gene expression in yeast. J Biol Chem. 2005;280:9106–9118. doi: 10.1074/jbc.M411770200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han GS, Siniossoglou S, Carman GM. The cellular functions of the yeast lipin homolog Pah1p are dependent on its phosphatidate phosphatase activity. J Biol Chem. 2007;282:37026–37035. doi: 10.1074/jbc.M705777200. [DOI] [PubMed] [Google Scholar]
- 28.Testerink C, Munnik T. Molecular, cellular, and physiological responses to phosphatidic acid formation in plants. J Exp Bot. 2011;62:2349–2361. doi: 10.1093/jxb/err079. [DOI] [PubMed] [Google Scholar]
- 29.Maxfield FR, van Meer G. Cholesterol, the central lipid of mammalian cells. Curr Opin Cell Biol. 2010;22:422–429. doi: 10.1016/j.ceb.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bidet M, Joubert O, Lacombe B, Ciantar M, Nehme R, Mollat P, Bretillon L, Faure H, Bittman R, Ruat M, et al. The hedgehog receptor patched is involved in cholesterol transport. PLoS One. 2011;6:e23834. doi: 10.1371/journal.pone.0023834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goldstein JL, DeBose-Boyd RA, Brown MS. Protein sensors for membrane sterols. Cell. 2006;124:35–46. doi: 10.1016/j.cell.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 32.Espenshade PJ, Hughes AL. Regulation of sterol synthesis in eukaryotes. Annu Rev Genet. 2007;41:401–427. doi: 10.1146/annurev.genet.41.110306.130315. [DOI] [PubMed] [Google Scholar]
- 33.Motamed M, Zhang Y, Wang ML, Seemann J, Kwon HJ, Goldstein JL, Brown MS. Identification of luminal Loop 1 of Scap protein as the sterol sensor that maintains cholesterol homeostasis. J Biol Chem. 2011;286:18002–18012. doi: 10.1074/jbc.M111.238311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun LP, Seemann J, Goldstein JL, Brown MS. Sterol-regulated transport of SREBPs from endoplasmic reticulum to Golgi: Insig renders sorting signal in Scap inaccessible to COPII proteins. Proc Natl Acad Sci U S A. 2007;104:6519–6526. doi: 10.1073/pnas.0700907104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35•.Radhakrishnan A, Goldstein JL, McDonald JG, Brown MS. Switch-like control of SREBP-2 transport triggered by small changes in ER cholesterol: a delicate balance. Cell Metab. 2008;8:512–521. doi: 10.1016/j.cmet.2008.10.008. This paper determined the levels of ER cholesterol that control SREBP-Scap transport from the ER by maintaining cooperative interactions between cholesterol, SCAP and Insig. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hughes AL, Todd BL, Espenshade PJ. SREBP pathway responds to sterols and functions as an oxygen sensor in fission yeast. Cell. 2005;120:831–842. doi: 10.1016/j.cell.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 37.Chun CD, Liu OW, Madhani HD. A link between virulence and homeostatic responses to hypoxia during infection by the human fungal pathogen Cryptococcus neoformans. PLoS Pathog. 2007;3:e22. doi: 10.1371/journal.ppat.0030022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang YC, Ingavale SS, Bien C, Espenshade P, Kwon-Chung KJ. Conservation of the sterol regulatory element-binding protein pathway and its pathobiological importance in Cryptococcus neoformans. Eukaryot Cell. 2009;8:1770–1779. doi: 10.1128/EC.00207-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Willger SD, Puttikamonkul S, Kim KH, Burritt JB, Grahl N, Metzler LJ, Barbuch R, Bard M, Lawrence CB, Cramer RA., Jr A sterol-regulatory element binding protein is required for cell polarity, hypoxia adaptation, azole drug resistance, and virulence in Aspergillus fumigatus. PLoS Pathog. 2008;4:e1000200. doi: 10.1371/journal.ppat.1000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bien CM, Espenshade PJ. Sterol regulatory element binding proteins in fungi: hypoxic transcription factors linked to pathogenesis. Eukaryot Cell. 2010;9:352–359. doi: 10.1128/EC.00358-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41•.Stewart EV, Nwosu CC, Tong Z, Roguev A, Cummins TD, Kim DU, Hayles J, Park HO, Hoe KL, Powell DW, et al. Yeast SREBP cleavage activation requires the Golgi Dsc E3 ligase complex. Mol Cell. 2011;42:160–171. doi: 10.1016/j.molcel.2011.02.035. The authors identify the Golgi Dsc E3 ligase, which is similar to the Hrd1 E3 ligase in ERAD, as required for fission yeast SREBP processing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stewart EV, Lloyd SJ, Burg JS, Nwosu CC, Lintner RE, Daza R, Russ C, Ponchner K, Nusbaum C, Espenshade PJ. Yeast Sterol Regulatory Element-Binding Protein (SREBP) cleavage requires Cdc48 and Dsc5, a ubiquitin regulatory X domain-containing subunit of the Golgi Dsc E3 ligase. J Biol Chem. 2012;287:672–681. doi: 10.1074/jbc.M111.317370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xie W, Ng DT. ERAD substrate recognition in budding yeast. Semin Cell Dev Biol. 2010;21:533–539. doi: 10.1016/j.semcdb.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 44.Davies BS, Wang HS, Rine J. Dual activators of the sterol biosynthetic pathway of Saccharomyces cerevisiae: similar activation/regulatory domains but different response mechanisms. Mol Cell Biol. 2005;25:7375–7385. doi: 10.1128/MCB.25.16.7375-7385.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vik A, Rine J. Upc2p and Ecm22p, dual regulators of sterol biosynthesis in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21:6395–6405. doi: 10.1128/MCB.21.19.6395-6405.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Germann M, Gallo C, Donahue T, Shirzadi R, Stukey J, Lang S, Ruckenstuhl C, Oliaro-Bosso S, McDonough V, Turnowsky F, et al. Characterizing sterol defect suppressors uncovers a novel transcriptional signaling pathway regulating zymosterol biosynthesis. J Biol Chem. 2005;280:35904–35913. doi: 10.1074/jbc.M504978200. [DOI] [PubMed] [Google Scholar]
- 47.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O’Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 48.Habeler G, Natter K, Thallinger GG, Crawford ME, Kohlwein SD, Trajanoski Z. YPL. db: the Yeast Protein Localization database. Nucleic Acids Res. 2002;30:80–83. doi: 10.1093/nar/30.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marie C, Leyde S, White TC. Cytoplasmic localization of sterol transcription factors Upc2p and Ecm22p in S. cerevisiae. Fungal Genet Biol. 2008;45:1430–1438. doi: 10.1016/j.fgb.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Burg JS, Espenshade PJ. Regulation of HMG-CoA reductase in mammals and yeast. Prog Lipid Res. 2011;50:403–410. doi: 10.1016/j.plipres.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jo Y, Debose-Boyd RA. Control of cholesterol synthesis through regulated ER-associated degradation of HMG CoA reductase. Crit Rev Biochem Mol Biol. 2010;45:185–198. doi: 10.3109/10409238.2010.485605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jo Y, Lee PCW, Sguigna PV, DeBose-Boyd RA. Sterol-induced degradation of HMG CoA reductase depends on interplay of two Insigs and two ubiquitin ligases, gp78 and Trc8. PNAS. 2011;108:20503–20508. doi: 10.1073/pnas.1112831108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53•.Gill S, Stevenson J, Kristiana I, Brown AJ. Cholesterol-dependent degradation of squalene monooxygenase, a control point in cholesterol synthesis beyond HMG-CoA reductase. Cell Metab. 2011;13:260–273. doi: 10.1016/j.cmet.2011.01.015. This paper describes an additional regulatory mechanism to control flux through the sterol biogenesis pathway beyond mevalonate. [DOI] [PubMed] [Google Scholar]
- 54.Hampton R, Dimster-Denk D, Rine J. The biology of HMG-CoA reductase: the pros of contra-regulation. Trends Biochem Sci. 1996;21:140–145. [PubMed] [Google Scholar]
- 55•.Garza RM, Tran PN, Hampton RY. Geranylgeranyl pyrophosphate is a potent regulator of HRD-dependent 3-Hydroxy-3-methylglutaryl-CoA reductase degradation in yeast. J Biol Chem. 2009;284:35368–35380. doi: 10.1074/jbc.M109.023994. This paper demonstrates isoprenoid regulation of HMGR is conserved in both yeast and mammalian cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Theesfeld CL, Pourmand D, Davis T, Garza RM, Hampton RY. The sterol-sensing domain (SSD) directly mediates signal-regulated endoplasmic reticulum-associated degradation (ERAD) of 3-hydroxy-3-methylglutaryl (HMG)-CoA reductase isozyme Hmg2. J Biol Chem. 2011;286:26298–26307. doi: 10.1074/jbc.M111.244798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Flury I, Garza R, Shearer A, Rosen J, Cronin S, Hampton RY. INSIG: a broadly conserved transmembrane chaperone for sterol-sensing domain proteins. EMBO J. 2005;24:3917–3926. doi: 10.1038/sj.emboj.7600855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Burg JS, Powell DW, Chai R, Hughes AL, Link AJ, Espenshade PJ. Insig regulates HMG-CoA reductase by controlling enzyme phosphorylation in fission yeast. Cell Metab. 2008;8:522–531. doi: 10.1016/j.cmet.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Burg JS, Espenshade PJ. Glucose controls phosphoregulation of hydroxymethylglutaryl coenzyme A reductase through the protein phosphatase 2A-related phosphatase protein, Ppe1, and Insig in fission yeast. J Biol Chem. 2011;286:27139–27146. doi: 10.1074/jbc.M111.233452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hughes BT, Espenshade PJ. Oxygen-regulated degradation of fission yeast SREBP by Ofd1, a prolyl hydroxylase family member. EMBO J. 2008;27:1491–1501. doi: 10.1038/emboj.2008.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61•.Lee CY, Yeh TL, Hughes BT, Espenshade PJ. Regulation of the Sre1 hypoxic transcription factor by oxygen-dependent control of DNA binding. Mol Cell. 2011;44:225–234. doi: 10.1016/j.molcel.2011.08.031. Paper describes a new mechanism for control of oxygen-dependent gene expression through regulation of SREBP DNA binding in fission yeast. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee CY, Stewart EV, Hughes BT, Espenshade PJ. Oxygen-dependent binding of Nro1 to the prolyl hydroxylase Ofd1 regulates SREBP degradation in yeast. EMBO J. 2009;28:135–143. doi: 10.1038/emboj.2008.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kwast KE, Lai LC, Menda N, James DT, 3rd, Aref S, Burke PV. Genomic analyses of anaerobically induced genes in Saccharomyces cerevisiae: functional roles of Rox1 and other factors in mediating the anoxic response. J Bacteriol. 2002;184:250–265. doi: 10.1128/JB.184.1.250-265.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wilcox LJ, Balderes DA, Wharton B, Tinkelenberg AH, Rao G, Sturley SL. Transcriptional profiling identifies two members of the ATP-binding cassette transporter superfamily required for sterol uptake in yeast. J Biol Chem. 2002;277:32466–32472. doi: 10.1074/jbc.M204707200. [DOI] [PubMed] [Google Scholar]
- 65.Hickman MJ, Spatt D, Winston F. The Hog1 mitogen-activated protein kinase mediates a hypoxic response in Saccharomyces cerevisiae. Genetics. 2011;188:325–338. doi: 10.1534/genetics.111.128322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Montanes FM, Pascual-Ahuir A, Proft M. Repression of ergosterol biosynthesis is essential for stress resistance and is mediated by the Hog1 MAP kinase and the Mot3 and Rox1 transcription factors. Mol Microbiol. 2011;79:1008–1023. doi: 10.1111/j.1365-2958.2010.07502.x. [DOI] [PubMed] [Google Scholar]
- 67.Gulati S, Liu Y, Munkacsi AB, Wilcox L, Sturley SL. Sterols and sphingolipids: dynamic duo or partners in crime? Prog Lipid Res. 2010;49:353–365. doi: 10.1016/j.plipres.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68••.Walker AK, Jacobs RL, Watts JL, Rottiers V, Jiang K, Finnegan DM, Shioda T, Hansen M, Yang F, Niebergall LJ, et al. A conserved SREBP-1/phosphatidylcholine feedback circuit regulates lipogenesis in metazoans. Cell. 2011;147:840–852. doi: 10.1016/j.cell.2011.09.045. This paper describes that limiting amount of phosphatidylcholine or S-adenosylmethionine can activate SREBP-1 by a new mechanism. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dobrosotskaya IY, Seegmiller AC, Brown MS, Goldstein JL, Rawson RB. Regulation of SREBP processing and membrane lipid production by phospholipids in Drosophila. Science. 2002;296:879–883. doi: 10.1126/science.1071124. [DOI] [PubMed] [Google Scholar]