Abstract
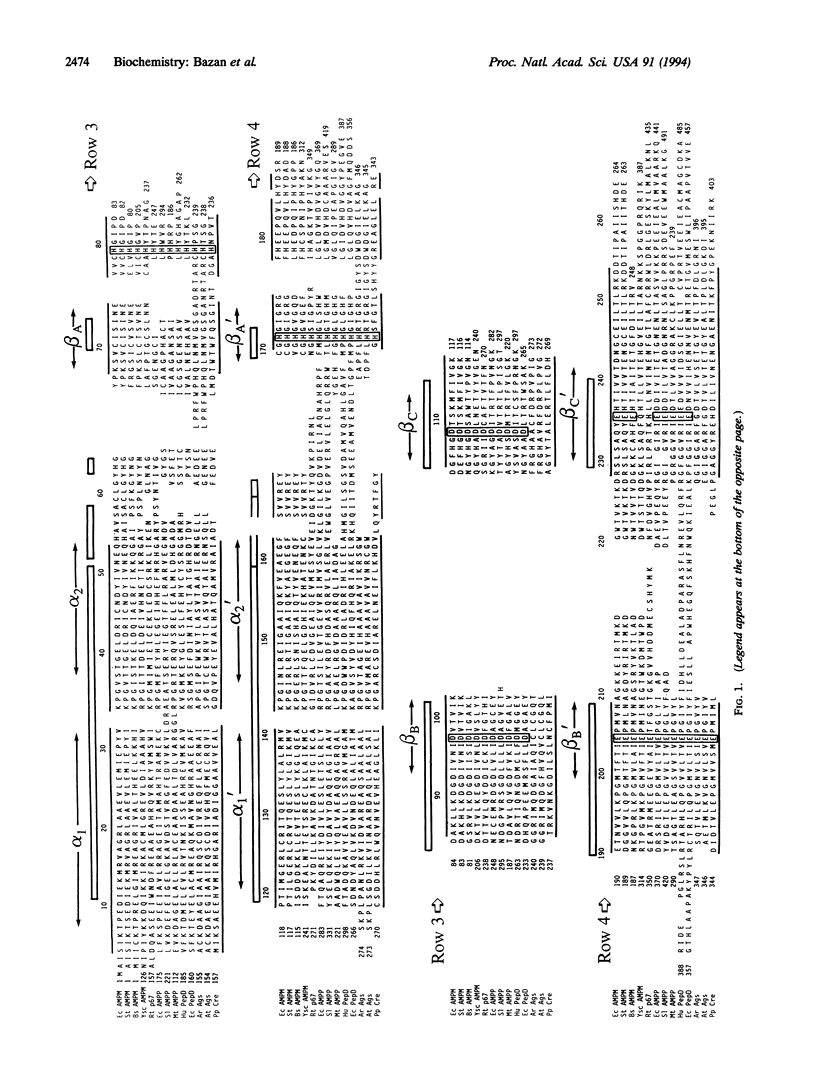
Amino acid sequence comparison suggests that the structure of Escherichia coli methionine aminopeptidase (EC 3.4.11.18) and the C-terminal domain of Pseudomonas putida creatinase (EC 3.5.3.3) are related. A detailed comparison of the three-dimensional folds of the two enzymes confirms this homology: with an approximately 260-residue chain segment, 218 C alpha atoms of the structures superimpose within 2.5 A; only 41 of these overlapping positions (i.e., 19%) feature identical amino acids in the two protein chains. Notwithstanding this striking correspondence in structure, methionine aminopeptidase binds and is stimulated by Co2+, while creatinase is not a metal-dependent enzyme. Searches of protein data banks using sequence and structure-based profiles reveal other enzymes, including aminopeptidase P (EC 3.4.11.9), prolidase (EC 3.4.13.9), and agropine synthase, that likely share the same "pita-bread" fold common to creatinase and methionine aminopeptidase.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Artymiuk P. J., Grindley H. M., Park J. E., Rice D. W., Willett P. Three-dimensional structural resemblance between leucine aminopeptidase and carboxypeptidase A revealed by graph-theoretical techniques. FEBS Lett. 1992 May 25;303(1):48–52. doi: 10.1016/0014-5793(92)80475-v. [DOI] [PubMed] [Google Scholar]
- Blundell T. L., Johnson M. S. Catching a common fold. Protein Sci. 1993 Jun;2(6):877–883. doi: 10.1002/pro.5560020602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P., Sander C., Valencia A. An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc Natl Acad Sci U S A. 1992 Aug 15;89(16):7290–7294. doi: 10.1073/pnas.89.16.7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchez D., Tourneur J. Organization of the agropine synthesis region of the T-DNA of the Ri plasmid from Agrobacterium rhizogenes. Plasmid. 1991 Jan;25(1):27–39. doi: 10.1016/0147-619x(91)90004-g. [DOI] [PubMed] [Google Scholar]
- Butler M. J., Bergeron A., Soostmeyer G., Zimny T., Malek L. T. Cloning and characterisation of an aminopeptidase P-encoding gene from Streptomyces lividans. Gene. 1993 Jan 15;123(1):115–119. doi: 10.1016/0378-1119(93)90549-i. [DOI] [PubMed] [Google Scholar]
- Chang Y. H., Teichert U., Smith J. A. Molecular cloning, sequencing, deletion, and overexpression of a methionine aminopeptidase gene from Saccharomyces cerevisiae. J Biol Chem. 1992 Apr 25;267(12):8007–8011. [PubMed] [Google Scholar]
- Chothia C., Lesk A. M. The relation between the divergence of sequence and structure in proteins. EMBO J. 1986 Apr;5(4):823–826. doi: 10.1002/j.1460-2075.1986.tb04288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chothia C. Proteins. One thousand families for the molecular biologist. Nature. 1992 Jun 18;357(6379):543–544. doi: 10.1038/357543a0. [DOI] [PubMed] [Google Scholar]
- Coll M., Knof S. H., Ohga Y., Messerschmidt A., Huber R., Moellering H., Rüssmann L., Schumacher G. Enzymatic mechanism of creatine amidinohydrolase as deduced from crystal structures. J Mol Biol. 1990 Jul 20;214(2):597–610. doi: 10.1016/0022-2836(90)90201-v. [DOI] [PubMed] [Google Scholar]
- DAVIS N. C., SMITH E. L. Purification and some properties of prolidase of swine kidney. J Biol Chem. 1957 Jan;224(1):261–275. [PubMed] [Google Scholar]
- Endo F., Tanoue A., Kitano A., Arata J., Danks D. M., Lapière C. M., Sei Y., Wadman S. K., Matsuda I. Biochemical basis of prolidase deficiency. Polypeptide and RNA phenotypes and the relation to clinical phenotypes. J Clin Invest. 1990 Jan;85(1):162–169. doi: 10.1172/JCI114407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo F., Tanoue A., Nakai H., Hata A., Indo Y., Titani K., Matsuda I. Primary structure and gene localization of human prolidase. J Biol Chem. 1989 Mar 15;264(8):4476–4481. [PubMed] [Google Scholar]
- Fleminger G., Yaron A. Soluble and immobilized clostridial aminopeptidase and aminopeptidase P as metal-requiring enzymes. Biochim Biophys Acta. 1984 Sep 25;789(3):245–256. doi: 10.1016/0167-4838(84)90180-8. [DOI] [PubMed] [Google Scholar]
- Garbe T., Servos S., Hawkins A., Dimitriadis G., Young D., Dougan G., Charles I. The Mycobacterium tuberculosis shikimate pathway genes: evolutionary relationship between biosynthetic and catabolic 3-dehydroquinases. Mol Gen Genet. 1991 Sep;228(3):385–392. doi: 10.1007/BF00260631. [DOI] [PubMed] [Google Scholar]
- Gibson T. J., Rice P. M., Thompson J. D., Heringa J. KH domains within the FMR1 sequence suggest that fragile X syndrome stems from a defect in RNA metabolism. Trends Biochem Sci. 1993 Sep;18(9):331–333. doi: 10.1016/0968-0004(93)90068-x. [DOI] [PubMed] [Google Scholar]
- Gribskov M., McLachlan A. D., Eisenberg D. Profile analysis: detection of distantly related proteins. Proc Natl Acad Sci U S A. 1987 Jul;84(13):4355–4358. doi: 10.1073/pnas.84.13.4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas E. S., Daniels C. J., Reeve J. N. Genes encoding 5S rRNA and tRNAs in the extremely thermophilic archaebacterium Methanothermus fervidus. Gene. 1989 Apr 30;77(2):253–263. doi: 10.1016/0378-1119(89)90073-5. [DOI] [PubMed] [Google Scholar]
- Henrich B., Monnerjahn U., Plapp R. Peptidase D gene (pepD) of Escherichia coli K-12: nucleotide sequence, transcript mapping, and comparison with other peptidase genes. J Bacteriol. 1990 Aug;172(8):4641–4651. doi: 10.1128/jb.172.8.4641-4651.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins D. G., Bleasby A. J., Fuchs R. CLUSTAL V: improved software for multiple sequence alignment. Comput Appl Biosci. 1992 Apr;8(2):189–191. doi: 10.1093/bioinformatics/8.2.189. [DOI] [PubMed] [Google Scholar]
- Hoeffken H. W., Knof S. H., Bartlett P. A., Huber R., Moellering H., Schumacher G. Crystal structure determination, refinement and molecular model of creatine amidinohydrolase from Pseudomonas putida. J Mol Biol. 1988 Nov 20;204(2):417–433. doi: 10.1016/0022-2836(88)90586-4. [DOI] [PubMed] [Google Scholar]
- Holm L., Sander C. Protein structure comparison by alignment of distance matrices. J Mol Biol. 1993 Sep 5;233(1):123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- Hooper N. M., Hryszko J., Turner A. J. Purification and characterization of pig kidney aminopeptidase P. A glycosyl-phosphatidylinositol-anchored ectoenzyme. Biochem J. 1990 Apr 15;267(2):509–515. doi: 10.1042/bj2670509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King G. F., Crossley M. J., Kuchel P. W. Inhibition and active-site modelling of prolidase. Eur J Biochem. 1989 Mar 15;180(2):377–384. doi: 10.1111/j.1432-1033.1989.tb14659.x. [DOI] [PubMed] [Google Scholar]
- Koyama Y., Kitao S., Yamamoto-Otake H., Suzuki M., Nakano E. Cloning and expression of the creatinase gene from Flavobacterium sp. U-188 in Escherichia coli. Agric Biol Chem. 1990 Jun;54(6):1453–1457. [PubMed] [Google Scholar]
- Matthews B. W., Rossmann M. G. Comparison of protein structures. Methods Enzymol. 1985;115:397–420. doi: 10.1016/0076-6879(85)15029-9. [DOI] [PubMed] [Google Scholar]
- Mock W. L., Green P. C. Mechanism and inhibition of prolidase. J Biol Chem. 1990 Nov 15;265(32):19606–19610. [PubMed] [Google Scholar]
- Ollis D. L., Cheah E., Cygler M., Dijkstra B., Frolow F., Franken S. M., Harel M., Remington S. J., Silman I., Schrag J. The alpha/beta hydrolase fold. Protein Eng. 1992 Apr;5(3):197–211. doi: 10.1093/protein/5.3.197. [DOI] [PubMed] [Google Scholar]
- Orengo C. A., Flores T. P., Taylor W. R., Thornton J. M. Identification and classification of protein fold families. Protein Eng. 1993 Jul;6(5):485–500. doi: 10.1093/protein/6.5.485. [DOI] [PubMed] [Google Scholar]
- Orengo C. A., Thornton J. M. Alpha plus beta folds revisited: some favoured motifs. Structure. 1993 Oct 15;1(2):105–120. doi: 10.1016/0969-2126(93)90026-d. [DOI] [PubMed] [Google Scholar]
- Ray M. K., Chakraborty A., Datta B., Chattopadhyay A., Saha D., Bose A., Kinzy T. G., Wu S., Hileman R. E., Merrick W. C. Characteristics of the eukaryotic initiation factor 2 associated 67-kDa polypeptide. Biochemistry. 1993 May 18;32(19):5151–5159. doi: 10.1021/bi00070a026. [DOI] [PubMed] [Google Scholar]
- Ray M. K., Datta B., Chakraborty A., Chattopadhyay A., Meza-Keuthen S., Gupta N. K. The eukaryotic initiation factor 2-associated 67-kDa polypeptide (p67) plays a critical role in regulation of protein synthesis initiation in animal cells. Proc Natl Acad Sci U S A. 1992 Jan 15;89(2):539–543. doi: 10.1073/pnas.89.2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roderick S. L., Matthews B. W. Structure of the cobalt-dependent methionine aminopeptidase from Escherichia coli: a new type of proteolytic enzyme. Biochemistry. 1993 Apr 20;32(15):3907–3912. doi: 10.1021/bi00066a009. [DOI] [PubMed] [Google Scholar]
- Rossmann M. G., Argos P. Exploring structural homology of proteins. J Mol Biol. 1976 Jul 25;105(1):75–95. doi: 10.1016/0022-2836(76)90195-9. [DOI] [PubMed] [Google Scholar]
- Rusu I., Yaron A. Aminopeptidase P from human leukocytes. Eur J Biochem. 1992 Nov 15;210(1):93–100. doi: 10.1111/j.1432-1033.1992.tb17395.x. [DOI] [PubMed] [Google Scholar]
- Tanoue A., Endo F., Matsuda I. Structural organization of the gene for human prolidase (peptidase D) and demonstration of a partial gene deletion in a patient with prolidase deficiency. J Biol Chem. 1990 Jul 5;265(19):11306–11311. [PubMed] [Google Scholar]
- Taylor A. Aminopeptidases: towards a mechanism of action. Trends Biochem Sci. 1993 May;18(5):167–171. [PubMed] [Google Scholar]
- Tricot C., Piérard A., Stalon V. Comparative studies on the degradation of guanidino and ureido compounds by Pseudomonas. J Gen Microbiol. 1990 Nov;136(11):2307–2317. doi: 10.1099/00221287-136-11-2307. [DOI] [PubMed] [Google Scholar]
- Wilmanns M., Hyde C. C., Davies D. R., Kirschner K., Jansonius J. N. Structural conservation in parallel beta/alpha-barrel enzymes that catalyze three sequential reactions in the pathway of tryptophan biosynthesis. Biochemistry. 1991 Sep 24;30(38):9161–9169. doi: 10.1021/bi00102a006. [DOI] [PubMed] [Google Scholar]
- Wu S., Gupta S., Chatterjee N., Hileman R. E., Kinzy T. G., Denslow N. D., Merrick W. C., Chakrabarti D., Osterman J. C., Gupta N. K. Cloning and characterization of complementary DNA encoding the eukaryotic initiation factor 2-associated 67-kDa protein (p67). J Biol Chem. 1993 May 25;268(15):10796–10801. [PubMed] [Google Scholar]
- Yaron A., Naider F. Proline-dependent structural and biological properties of peptides and proteins. Crit Rev Biochem Mol Biol. 1993;28(1):31–81. doi: 10.3109/10409239309082572. [DOI] [PubMed] [Google Scholar]
- Yoshimoto T., Tone H., Honda T., Osatomi K., Kobayashi R., Tsuru D. Sequencing and high expression of aminopeptidase P gene from Escherichia coli HB101. J Biochem. 1989 Mar;105(3):412–416. doi: 10.1093/oxfordjournals.jbchem.a122678. [DOI] [PubMed] [Google Scholar]