Abstract
DNA methylation undergoes dynamic changes during development and cell differentiation. Recent genome-wide studies discovered that tissue-specific differentially methylated regions (DMRs) often overlap tissue-specific distal cis-regulatory elements. However, developmental DNA methylation dynamics of the majority of the genomic CpGs outside gene promoters and CpG islands has not been extensively characterized. Here we generate and compare comprehensive DNA methylome maps of zebrafish developing embryos. From these maps we identify thousands of developmental stage-specific DMRs (dsDMR) across zebrafish developmental stages. The dsDMRs contain evolutionarily conserved sequences, are associated with developmental genes, and are marked with active enhancer histone post-translational modifications. Their methylation pattern correlates much stronger than promoter methylation with expression of putative target genes. When tested in vivo using a transgenic zebrafish assay, 20 out of 20 selected candidate dsDMRs exhibit functional enhancer activities. Our data suggest that developmental enhancers are a major target of DNA methylation changes during embryogenesis.
Enhancers are DNA sequences that can influence transcription of nearby genes in an orientation- and position-independent manner. Enhancers can activate transcription of their target genes from long distances1, and play essential roles in driving spatial and temporal patterns of gene expression during development2. Since the location of enhancers relative to their target genes is highly variable and the activity of enhancers can be restricted to a specific cell type or a specific developmental stage, the identification of enhancers has been challenging3. Comparative genomic studies found that non-coding sequences that are highly conserved between different species were enriched for enhancers4,5, but only a small fraction was validated as functional enhancers in transgenic mouse reporter assays6. Furthermore, many functional enhancers showed modest or no sequence conservation between species7. Chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq) of specific epigenomic marks such as histone H3 mono-methylated lysine 4 (H3K4me1)8 or p3009 has also been used to identify enhancers. Recent efforts in annotating genome-wide enhancers in a variety of cell types, including those of the ENCODE project, identified hundreds of thousands putative enhancers in human and mouse10,11. However, the majority of these predicted enhancers are still waiting to be tested by in vivo enhancer assays; a substantial portion of tested ones failed to be validated as active enhancers in vivo12. Thus, current enhancer prediction methods have both high false negative rate (missed enhancers) and high false positive rate (validated as non-enhancers in other assays)13. This implies that alternative methods to predict active and functional enhancers in a certain cell type or at a certain developing stage need to be considered to complement and enhance the efficacy of the existing methods.
DNA methylation is one of the best-studied epigenetic modifications. It plays crucial roles in diverse biological processes including transcription, tissue-specific gene expression, and normal development14. Methylated DNA sequences in the genome differ between different cell types and developmental stages15,16. DNA methylation is known to define and stabilize cellular identity and developmental state17. Until recently, many studies on DNA methylation focused on CpG-rich regions such as CpG islands and transcription start sites (TSSs), and only a few recent studies investigated DNA methylation on other genomic features18–22. For example, gene body methylation can be tissue-specific and plays an important role in regulating intragenic promoter activity18. DNA methylation signatures of distal regulatory elements began to emerge from several recent studies18–23. These studies suggest that DNA methylation is important in regulating enhancers. Thus, accurate detection of differential DNA methylation status across different biological states could help identify and annotate functional enhancers in different cell types and developmental stages. Indeed, recent studies investigated the epigenetic dynamics including DNA methylation in a developmental context by differentiating human embryonic stem cells24,25. However, a thorough investigation on the relationship between DNA methylation and functional enhancers in live developing embryos is still lacking.
Here we use zebrafish to study DNA methylation changes during early development and investigate the potential biological function of developmental stage-specific differentially methylated regions (dsDMRs). Zebrafish have proven an outstanding model organism to study vertebrate development26. DNA methylation also plays an important role in normal zebrafish development. Previous studies have shown that perturbed DNA methylation in developing zebrafish embryos resulted in critical developmental defects27–29. Very recently, DNA methylation patterns of zebrafish gametes and early embryos were determined by using whole genome bisulfite sequencing (WGBS)30,31. These studies discovered global DNA methylation reprogramming differences between paternal and maternal genomes in very early zebrafish embryos. Interestingly, one study also reported that many differentially methylated regions were located in gene bodies, especially the first introns, and were overlapping with known enhancer regions, raising the possibility that those regions might include enhancers31. This result encouraged the thought that DNA methylation dynamics could potentially reveal important developmental regulatory elements during zebrafish development. Here, we generate comprehensive DNA methylation profiles of zebrafish developing embryos and sperm by using two complementary high-throughput sequencing technologies coupled with the state-of-art computational algorithms18,22,23,32,33. We identify thousands of differentially methylated DNA sequences in the zebrafish genome across different developmental stages. We find that a substantial portion of dsDMRs behave as developmental enhancers, a subset of which are successfully validated using in vivo transgenic zebrafish reporter assays. These dsDMRs strongly enrich for enhancer-associated histone modifications and for binding motifs of transcription factors involved in important developmental processes. Reconstruction of gene regulatory networks connecting these transcription factors and putative target genes with nearby dsDMRs demarcates epigenetic and regulatory events associated with organogenesis and signaling pathways in developing embryos. Our results suggest that the main targets of dynamic DNA methylation reprogramming in developing zebrafish embryos are developmental enhancers and that DNA methylation dynamics can be used to reliably discover functional enhancers.
RESULTS
Generation of DNA methylome maps of zebrafish embryogenesis
To understand how DNA methylation changes during zebrafish embryogenesis, we generated high-resolution DNA methylome maps of six developmental stages: sperm, 2.5 hour post fertilization (2.5 hpf, 256-cell), 3.5 hpf (MBT; high), 4.5 hpf (dome), 6 hpf (shield), and 24 hpf (Fig. 1a). For each developmental stage, we constructed two sequencing libraries using complementary technologies developed recently18: methylation dependent DNA immunoprecipitation followed by sequencing (MeDIP-seq) and methyl-sensitive restriction enzyme digestion followed by sequencing (MRE-seq). These libraries were sequenced to generate approximately 840 million reads in total (Supplementary Table 1), which were mapped to the zebrafish genome assembly (Zv9). As shown previously, MRE-seq scores were inversely correlated with MeDIP-seq scores and the two sequencing signals covered largely non-overlapping regions (Supplementary Figs 1–2).
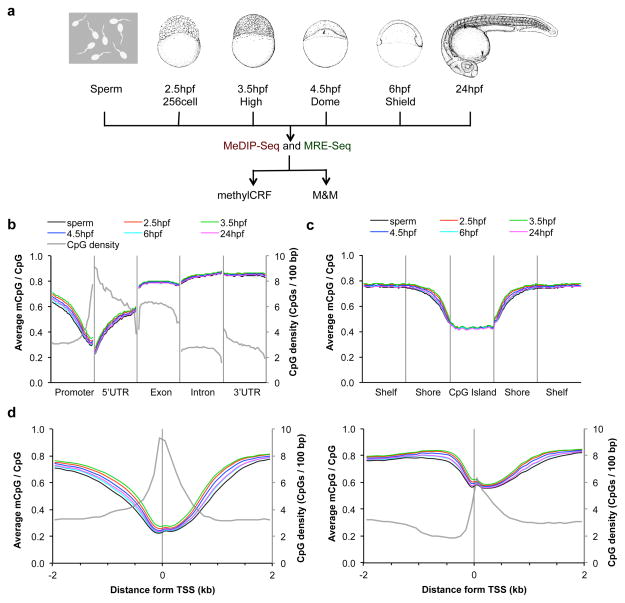
Figure 1. Global features of DNA methylation across zebrafish embryogenesis.
(a) A schematic representation of the study. MeDIP-seq and MRE-seq libraries were constructed from genomic DNA of six developmental stages: sperm, 2.5 hpf, 3.5 hpf, 4.5 hpf, 6 hpf and 24 hpf. Stage-specific drawings of representative embryos are adapted from ref 26 with permission from Wiley-Liss, Inc. © 1995. The two sequencing libraries for each developmental stage were further processed and analyzed using recently developed algorithms, methylCRF and M&M. Both algorithms integrate MeDIP-seq and MRE-seq data. (b–d) The average DNA methylation level across different genomic features: gene-associated regions (b), CpG islands and neighboring regions (c), and high and low CpG density promoters (d). Promoter was defined as 1 kb upstream from TSS in (b). CpG island shore was defined as 2 kb regions flanking a CpG island, and CpG shelf as a 2 kb region outside a CpG shore (away from the CGI). The average CpG densities (grey lines) over the regions was also plotted in (b) and (d).
Whole-genome methylation levels at single CpG resolution
We used methylCRF to estimate individual methylation levels of all CpGs in the zebrafish genome by combining MRE-seq and MeDIP-seq data for each developmental stage33. Briefly, methylCRF uses conditional random fields to integrate MeDIP- and MRE-seq data and predicts DNA methylation levels at single-CpG resolution. MethylCRF predictions were in high concordance with previously published predictions based on whole genome bisulfite sequencing (MethylC-seq)30,31 on the matched samples including sperm, 2.5 hpf and 3.5 hpf embryos (Supplementary Fig. 3a,b). Using previously developed benchmarking metrics33, we determined that methylCRF and MethylC-seq were about 91% concordant within a 0.25 difference (see Methods, Supplementary Fig. 3a,b). We found that the concordance increased with increasing MethylC-seq read depth (Supplementary Fig. 4), recapitulating what was reported previously33. Using bisulfite-sequencing based methods, we performed targeted validation for eight genomic regions where methylCRF and MethylC-seq predictions were discordant. The validation results agreed much better with methylCRF (Supplementary Fig. 5). Taken together, these data demonstrated the high quality of our DNA methylome maps.
To characterize the DNA methylation levels across different genic features, we plotted methylation levels across gene-associated regions including promoters, exons, introns, 5′ and 3′ untranslated regions (UTRs) (Fig. 1b). Overall, the average genic DNA methylation levels of each developmental stage were almost identical. We observed a negative correlation in the methylation level and the proximity to the transcription start site (TSS). The DNA methylation level increased in the 5′ UTR and stayed high in exons, introns and the 3′ UTR, which resembled the DNA methylation pattern over genic regions in the human embryonic stem cells34. We also investigated the DNA methylation levels across CpG islands (CGIs), and CGI shores and shelves. CpG islands were largely unmethylated and methylation levels increased with increasing distance from CpG islands (Fig. 1c).
Predicted DNA methylation levels were also plotted as an average profile throughout the protein-coding genes. The low DNA methylation around the TSS was observed again and the transcription termination sites (TES) had slightly higher DNA methylation levels (Supplementary Fig. 6a). The DNA methylation levels throughout long non-coding RNA genes looked very similar to those of protein-coding genes (Supplementary Fig. 6b). The DNA methylation levels across different genic and genomic features shown above were further confirmed using the recently published zebrafish MethylC-seq datasets (Supplementary Fig. 7).
The low DNA methylation level around the TSS was correlated with high CpG density around the TSS (Fig. 1b). We further investigated the relationship between the DNA methylation level and the CpG density of the promoters. We divided all protein-coding gene promoters into two categories, high CpG promoters (HCPs) and low CpG promoters (LCPs) (Supplementary Fig. 8a) and found that LCPs were more methylated than HCPs on average (Fig. 1d; Supplementary Fig. 8b). This confirms that the CpG density of the promoters is negatively correlated with the DNA methylation level in mammals35 and zebrafish30,31. Taken together, these results suggested that the global characteristic DNA methylation patterns over genic regions in zebrafish were established as early as 2.5 hpf and remained stable through early embryo development, and that the patterns were similar to those of other vertebrate animals including mouse and human.
Identification of differentially methylated regions
We next asked if there are regulated local DNA methylation changes across different developmental stages. We applied to our data a recently invented integrative statistical algorithm M&M23. Briefly, M&M integrates MeDIP-seq and MRE-seq by dynamically scaling, normalizing, and combining two datasets, and identifies differentially methylated regions (DMRs) between two samples. This allowed us to identify a total of 8225 DMRs between different developmental stages (see Methods, Fig. 2a; Supplementary Fig. 9a,b, Supplementary Data 1). It is noteworthy that the most dramatic differences were identified between 6 hpf and 24 hpf embryos where the majority of the over a thousand DMRs showed decreasing DNA methylation level from 6 hpf to 24 hpf embryos, suggesting that locus-specific demethylation process could be an important feature of early cell specification and differentiation.
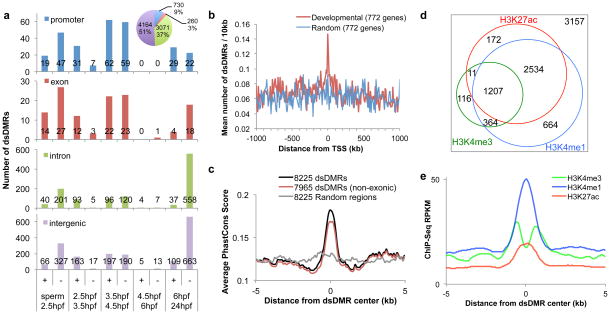
Figure 2. Identification of the developmental stage-specific differentially methylation regions (dsDMRs).
(a) The number of dsDMRs identified between neighboring developmental stages as indicated on x-axis. Plus sign (+) indicates dsDMRs with increasing DNA methylation levels with respect to step-wise developmental stages, and minus sign (−) indicates dsDMRs with decreasing DNA methylation. The genomic locations of dsDMRs were indicated by different colors. Pie chart: the genomic locations of all DMRs identified in pair-wise comparisons. (b) The distribution of dsDMRs around genes related to embryo development (red) or around random genes (blue). (c) Sequence conservation of dsDMRs. Vertebrate PhastCons scores of the dsDMRs and their flanking 10 kb regions were averaged and plotted. (d) A weighted Venn diagram of the number of dsDMRs overlapping with histone modification peaks from any developmental stages. (e) Histone modification signature of dsDMRs. Average histone modification ChIP-seq RPKM values from 24 hpf embryos were plotted over 10 kb regions centered on dsDMRs.
We then examined the enrichment of differentially methylated individual CpGs with respect to either hypermethylated or hypomethylated DMRs. Differences in methylation levels of individual CpGs showed clear separation between hypermethylated DMRs and hypomethylated DMRs (Supplementary Fig. 9c), confirming that our DMR set was highly specific. We thus termed these DMRs zebrafish developmental stage-specific DMRs (dsDMRs).
Genomic distribution of dsDMRs was highly non-random. About 10% of dsDMRs were located in the gene promoter regions, whereas the remaining dsDMRs were either in intergenic or intronic regions (Fig. 2a; Supplementary Fig. 9b). Most dsDMRs were located 10 to 50 kb away from its closest TSS, but the number of dsDMRs located less than 1kb from TSS was greater than expected (Supplementary Fig. 10a). This suggests that the promoters were enriched as a main target of DNA methylation changes during embryogenesis; however, the majority of DNA methylation changes occurred distal to gene promoters.
To investigate the relationship between dsDMRs and developmental processes, we asked if the dsDMRs were more enriched near a collection of 772 genes that were annotated as important for embryonic development (Methods). Strikingly, the distribution of dsDMRs was more significantly enriched near developmental genes than random expectation (Fig. 2b, Kolmogorov-Smirnov test, P-value < 1×10−14). Similarly, when we compared the number of dsDMRs located within 50kb from TSS of the developmental genes, we found that developmental genes had significantly higher number of dsDMRs than randomly chosen genes (Supplementary Fig. 10b). This suggests that the dsDMRs might have functions related to developmental processes.
To evaluate the functional potential of the dsDMRs, we asked whether their sequences were evolutionarily conserved by examining their PhastCons scores36 based on the 8-way vertebrate genome alignment with zebrafish from the UCSC Genome Browser37. A significant proportion of dsDMRs (1546 out of 8225) contained sequences that were evolutionarily conserved across vertebrates (hypergeometric test, P-value = 6.1×10−41). Average conservation scores of all dsDMRs were higher than their neighboring regions (Fig. 2c). This trend remained after we excluded DMRs that overlapped exonic sequences (Fig. 2c), suggesting many dsDMRs were contributed by conserved non-coding sequences.
We hypothesized that dsDMRs may encode important regulatory elements for developmental functions. We thus examined histone post-translational modifications on dsDMRs using publicly available ChIP-seq data on four zebrafish embryonic developing stages: dome (4.5 hpf), 80% epiboly (8.5 hpf), 24 hpf, and 48 hpf38. More than half of the dsDMRs (4952/8225) overlapped with at least one enhancer-associated histone ChIP-seq peak – histone H3 mono-methylated lysine 4 (H3K4me1) or histone H3 acetylated lysine 27 (H3K27ac) – from at least one of the four developmental stages (hypergeometric test, P-value = 4.7 × 10−1478; Fig. 2d; Supplementary Fig. 10c). Average ChIP-seq score over 10 kb regions around the DMRs identified strong peaks of H3K4me1 and H3K27ac centered on the DMRs (Fig. 2e; Supplementary Fig. 10d,e). This data revealed that dsDMRs were strongly associated with enhancer chromatin marks. Taken together, the genomic distribution, sequence conservation, and histone modification pattern of the dsDMRs we discovered suggest that they may be regulatory elements playing important roles in developmental processes.
Most dsDMRs are putative developmental enhancers
Across all developmental stage step-wise comparisons, the comparison between 6 hpf and 24 hpf embryos yielded a relatively large number of dsDMRs (a total of 1440), and interestingly, majority (1261, or 88 %) of them exhibited hypomethylation in 24 hpf embryos (Fig. 2a). Because many important developmental processes including cell specification occur during this time26, we hypothesized that these hypomethylated dsDMRs may represent distal enhancers that were dynamic during development20. Consistent with this hypothesis, we found that approximately 70% of these dsDMRs (880 out of 1261) directly overlapped with putative distal regulatory elements (PDREs) that were identified using enhancer histone marks38 (hypergeometric test, P-value = 1.9 × 10−790). Furthermore, this group of dsDMRs had the highest percentage of dsDMRs with conserved sequences (30%, 378 out of 1261; hypergeometric test, P-value = 6.1 × 10−52). The average conservation score over the 10kb regions surrounding dsDMRs revealed strong enrichment of conserved elements over the dsDMRs (Fig. 3a). When compared to the conservation profile of the PDREs identified in 24 hpf, the average PhastCons scores of the dsDMRs were even higher (Fig. 3a).
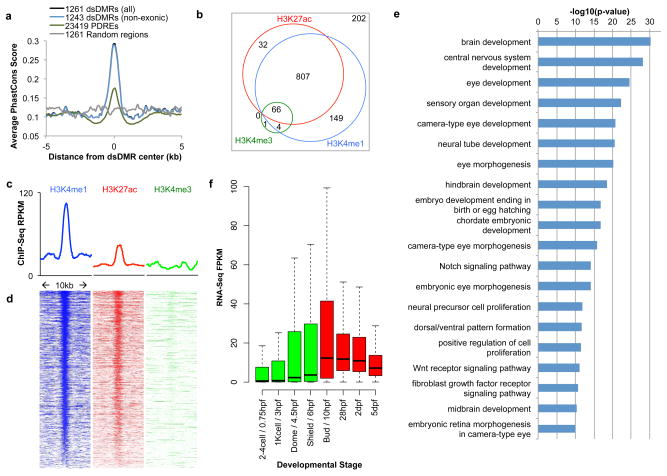
Figure 3. Most dsDMRs with decreasing DNA methylation level between 6 hpf and 24 hpf are developmental enhancers.
(a) Averaged vertebrate PhastCons scores of 10kb regions centered on dsDMRs were plotted. (b) A weighted Venn diagram of dsDMRs overlapping with different histone modification peaks from 24 hpf embryo. (c) Histone modification signature of dsDMRs. Average ChIP-seq RPKM values from 24 hpf embryos were plotted over 10 kb regions centered on the dsDMRs. (d) Heat maps of ChIP-seq signal over 10 kb regions centered on individual dsDMRs. (e) Enriched GO terms and their binomial p-values from analyzing dsDMRs using GREAT42. The top 20 GO terms were displayed here and the full list of GO terms enriched was in Supplementary Fig. 11a. (f) Expression profiles of genes associated with dsDMRs from GO enrichment analysis across different developmental stages.
The profiles of histone modification marks suggested that this set of dsDMRs strongly enriched for enhancer elements, much more than for promoters. 78% of these dsDMRs (988 out of 1261) overlapped with either or both of the two enhancer-associated marks H3K4me1 or H3K27ac peaks in the absence of the promoter-associated mark H3K4me3 peaks (hypergeometric test, P-value = 2.5 × 10−843), while a smaller but still significant portion of dsDMRs overlapped with H3K4me3 peaks (hypergeometric test, P-value = 9.6 × 10−9; Fig. 3b). Averaged ChIP-seq signals for the 10kb region around these dsDMRs showed strong enrichment of H3K4me1 and H3K27ac (Fig. 3c), and individual dsDMRs each displayed strong enhancer-associated signal (Fig. 3d).
To investigate whether the genes nearby these dsDMRs were enriched for any functional annotation, we performed Gene Ontology (GO) enrichment analysis on these dsDMRs using the tool GREAT39. This analysis revealed that genes associated with these dsDMRs strongly enriched for many biological functions involving brain and eye development, for instance, “brain development” and “central nervous system development” (Fig. 3e; Supplementary Fig. 11a,b; Supplementary Table 2 for the complete list). In addition, fibroblast growth factor receptor (FGFR), Notch and Wnt receptor signaling pathways were also enriched (Fig. 3e; Supplementary Fig. 11a,b).
To investigate whether the genes identified from GREAT analysis might be direct targets of dsDMRs, we asked if expression of these genes correlated with the methylation status of the dsDMRs by examining publically available expression profiles of developing zebrafish embryos40. The genes associated with these dsDMRs showed increased expression levels in 10 hpf (bud) or 28 hpf when compared to the levels in 6hpf (shield) (Fig. 3f; Supplementary Fig. 12). In particular, 72 genes had no or very little RNA expression (FPKM less than 1) in shield stage and were activated (FPKM greater than 1) in the later stages. Thus the expression dynamics of genes associated with this DMR set was consistent with the hypothesis that these dsDMRs were enhancers for regulating expression of their nearby genes.
Gene regulatory network derived from DNA methylome maps
Given the regulatory potential of dsDMRs, we hypothesized that these dsDMR enhancers coordinate many developmental processes by connecting upstream transcription factors (TFs) to their downstream target genes. To test this, we first identified the TF binding motifs enriched in these dsDMRs. We found that dsDMRs were highly enriched for binding motifs of many important developmental TFs, including Sox family and nuclear receptor family proteins (Fig. 4a; Supplementary Fig. 13). Zebrafish genes coding these TFs were also enriched for developmental processes such as “sensory organ development” and “embryonic morphogenesis” (Supplementary Fig. 14 and 15a; Methods). Thus, analysis of dsDMRs predicted many regulatory relationships among genes associated with development. We then asked whether these upstream TFs and downstream genes are known to be associated by querying databases of known TF-target genes (Methods). Indeed, we found a large number of connections among the TFs and genes (2964 edges with 233 nodes; Student’s t-test, P-value = 1.65 × 10−5; Supplementary Fig. 15b), suggesting a highly connected, non-random network among the TFs and genes identified from dsDMR analysis.
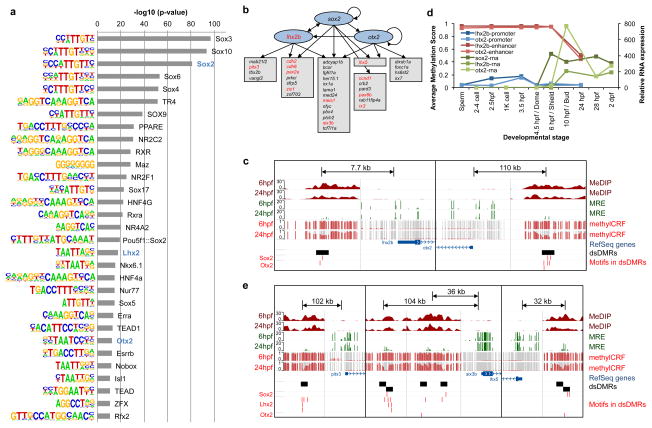
Figure 4. Gene regulatory network derived by dsDMRs.
(a) Enriched transcription factor binding motifs in dsDMRs and their hypergeometric p-values from HOMER62. On the left sequence logo of each motif was displayed. The three motifs used in eye development regulatory network construction were indicated by blue colored text. The motifs with p-values less than 10−10 were displayed here, and the full list of enriched motifs was in Supplementary Fig. 13. (b) The putative gene regulatory network of the eye development derived from dsDMR analysis. The blue ovals were transcription factors whose motifs were enriched in dsDMRs. The genes in the grey boxes were the target genes identified in GREAT analysis. Arrows indicates that the transcription factors had their binding motifs in neighboring dsDMRs of the target genes. The genes colored in red were known sox2 target genes identified from ChEA68. (c) The gene set view of 4 genomic regions (chr8:3242750–3245750, chr8:3250489–3253489, chr17:44294595–44297595, chr17:44405250–44408250) from the Epigenome browser69,70. The left panel displayed the regions around lhx2b promoter and its nearby dsDMR enhancer (indicated by the black box). The right panel displayed the regions around otx2 promoter and its nearby dsDMR enhancer. Both dsDMRs had Sox2 and Otx2 binding motifs (red ticks), suggesting that sox2 could be an upstream regulator of these two TFs. (d) The methylation profiles of the lhx2b and otx2 promoters and their neighboring dsDMR enhancers (blue and red lines, left y-axis) and the expression profile of the two genes (green lines, right y-axis). Each gene expression level was normalized to the expression level of 1K-cell stage. (e) The gene set view of 7 genomic regions (chr13:6556250–6559250, chr13:6658232–6661232, chr13:29912750–29915750, chr13:29935250–29940750, chr13:29993060–29996060, chr13:30062750–30065750, chr13:30077750–30080750) from the Epigenome browser69,70. The displays of pitx3 (left), six3b (middle) and lhx5 (right) promoters and their nearby dsDMR enhancers were as in (c).
Interestingly, we found that several GO terms (either the same or closely related terms) were enriched both in the TFs predicted to bind dsDMRs and in the genes predicted to be the targets of dsDMRs (Fig. 3e; Supplementary Fig. 11a, 14 and 15a). This result further supports that our method could link TFs to their targets in the context of a specific biological process. For example, we found that the GO terms “eye development” and “eye morphogenesis” were enriched in both TFs that were upstream of dsDMRs and in putative target genes that were downstream of dsDMRs. This result is consistent with the hypothesis that TFs involved in eye development bind to dsDMRs to regulate their downstream target genes.
To further test whether dsDMRs coordinate the expressions of genes associated with the eye development, we sought to construct a gene regulatory network by directly linking TFs and their target genes (Methods). We identified a total of 75 links between 3 TFs and their 35 target genes and constructed a putative gene regulatory network of eye development using these links (Fig. 4b). Interestingly, the majority of dsDMRs containing TF binding motifs were located far away from the TSS of target genes, highlighting the importance of these dynamically regulated distal enhancers in mediating eye development (Supplementary Fig. 16a). In this regulatory network, the TFs sox2, lhx2b and otx2 coordinated the expression of genes involved in eye development (Fig. 4b). All three TFs also had Sox2 motifs in their neighboring dsDMRs (Fig. 4c), suggesting that sox2 is the upstream regulator of this network. Further supporting this hypothesis, we found that transcription of sox2 started in 6 hpf (shield), which was followed by demethylation of dsDMRs that contained sox2 binding motif (Fig. 4d), The expression levels of lhx2b and otx2 were inversely correlated with the methylation levels of their neighboring dsDMRs, but were not correlated with the methylation levels of their own promoters (Fig. 4d), underscoring the regulatory potential of these dsDMRs. All downstream target genes had one or more binding motifs of the TFs sox2, lhx2b and otx2 in their neighboring dsDMRs (Fig. 4b,e) and the expression dynamics of these genes were correlated with methylation changes of neighboring dsDMRs (Fig 3f; Supplementary Fig. 16b). This data is consistent with the hypothesis that these genes are downstream target genes of the three TFs.
Our analysis not only provided regulatory connections among genes known to involve in a developmental process, but also brought novel genes into the network. For example, sema6a was not curated to share a GO term with eye developmental genes. However, we predicted that sema6a is an eye developmental gene because a dsDMR with binding motifs of the three TFs, Sox2, Lhx2, and Otx2, was associated with sema6a gene (Supplementary Fig. 17a). Indeed, sema6a is a transmembrane signaling protein which is important for mammalian retinal circuits assembly41 and is also required for zebrafish eye cohesion42. In addition, the expression levels of sema6a were inversely correlated with the methylation levels of their neighboring dsDMRs (Supplementary Fig. 17b). Thus, we were able to put sema6a in the eye development regulatory network. Similarly, we added cdon and ascl1b to the eye development network (Supplementary Fig. 17a,c,d). These two genes were not annotated by GO as eye developmental genes. However, cdon defines the correct proximo-distal patterning of the eye43; and Ascl1 is required for specification of the mammalian retina progenitors44. Thus, our dsDMR-based analysis was able to construct a high quality gene regulatory network for eye development.
In addition to eye development, we were also able to construct gene regulatory networks for other developmental processes. For example, we identified 33 links between 3 TFs and 12 target genes associated with central nervous system neuron differentiation and built a putative regulatory network (Supplementary Fig. 18). These findings highlight that the gene regulatory networks can be derived by analyzing epigenomic changes across developmental courses.
In vivo validation of dsDMRs as putative enhancer elements
We next tested whether the dsDMRs could function as developmental enhancers in live zebrafish. To this end, we chose 20 dsDMRs that were associated with 8 developmental genes. For this assay we chose dsDMRs far from the promoters (from at least 3kb away to more than 200kb away, Table 1; Supplementary Tables 3–5). We also included four dsDMRs that did not overlap with any conserved element in this test, and five dsDMRs that did not have overlapping enhancer histone peaks (Supplementary Table 4). We cloned these dsDMR sequences into a minimal promoter-driven green fluorescent protein (GFP) reporter cassette45,46 and injected zebrafish embryos with the reporter vector along with Tol2 transposase mRNA. In addition, as negative controls we also cloned 20 regions selected from within the vicinity of the tested dsDMRs but did not exhibit DNA methylation change (Supplementary Table 3). These non-dsDMR negative controls included regions that overlapped with conserved element or enhancer histone peaks (Supplementary Table 4). Each group of G0 embryos injected with a specific putative enhancer reporter construct expressed GFP in a specific pattern, suggesting that the transgene integration occurred early and often in embryonic development47. In contrast, G0 embryos injected with the negative controls showed, if any, inconsistent patterns of GFP expression (Supplementary Fig. 19). We were able to establish founder G1 transgenic fish expressing GFP for all 20 putative enhancer reporter constructs. Overall, the GFP expression pattern driven by each dsDMR enhancer recapitulated the expression pattern of its adjacent gene (Fig. 5; Supplementary Fig. 20; Supplementary Table 6). Interestingly, all of the four dsDMRs without conserved sequences (fgfr2-e1, fgfr2-e2, fgfr2-e4, and sox2-e2) showed in vivo enhancer activities (Fig. 5e; Supplementary Fig. 20i). We note that we paired each dsDMR enhancer with its closest putative target gene but the enhancer may have different or additional target genes. This explains the few, if any, differences between GFP expression patterns driven by dsDMR enhancers and those of the neighboring genes.
Table 1.
The dsDMR enhancers validated in vivo
| dsDMR enhancer | Adjacent gene | Location from TSS (kb) | Related enriched GO terms | Expression pattern of adjacent gene at 24hpf |
|---|---|---|---|---|
| six3a-e1 | six3a | −18.6 | brain development, central nervous system development | forebrain, midbrain hindbrain boundary, anterior neural tube |
| six3a-e2 | −50.4 | |||
|
| ||||
| six3b-e1 | six3b | −104.2 | brain development, central nervous system development, eye development, sensory organ development | immature eye, diencephalon, telencephalon, forebrain, optic vesicle |
| six3b-e2 | −35.6 | |||
|
| ||||
| fgfr2-e1 | fgfr2 | −3.7 | positive regulation of cell proliferation, fibroblast growth factor receptor signaling pathway | forebrain, hindbrain, diencephalon, central nervous system, solid lens vesicle |
| fgfr2-e2 | −8.6 | |||
| fgfr2-e3 | −38.8 | |||
| fgfr2-e4 | −107.5 | |||
|
| ||||
| fgf3-e1 | fgf3 | −54.2 | brain development, sensory organ development, dorsal/ventral pattern formation, cell fate specification, regulation of neurogenesis | forebrain, midbrain, midbrain hindbrain boundary, somite, diencephalon, optic stalk |
|
| ||||
| znf703-e1 | znf703 | −37.1 | brain development, central nervous system development, eye development, sensory organ development | hindbrain, midbrain, neural tube, somite, spinal cord, optic cup, vein |
| znf703-e2 | −74.7 | |||
| znf703-e3 | −78.8 | |||
|
| ||||
| sox2-e1 | sox2 | 219.2 | eye development, sensory organ development, dorsal/ventral pattern formation, regulation of transcription factor activity | immature eye, midbrain, neural tube, spinal cord, forebrain, hindbrain, telencephalon, cerebellum, octic vesicle |
| sox2-e2 | 68.0 | |||
| sox2-e3 | 64.5 | |||
| sox2-e4 | 20.3 | |||
| sox2-e5 | −47.1 | |||
|
| ||||
| wnt3-e1 | wnt3 | −3.1 | Wnt receptor signaling pathway | midbrain, hindbrain, cerebellum, diencephaoln |
|
| ||||
| wnt3a-e1 | wnt3a | 44.5 | brain development, neural tube development, dorsal/ventral pattern formation, Wnt receptor signaling pathway | neural crest cell, midbrain, cerebellum, diencephalon |
| wnt3a-e2 | 37.0 | |||
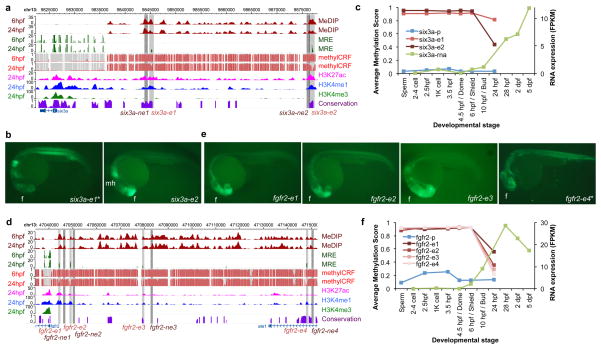
Figure 5. In vivo validation of dsDMR enhancers.
(a) Epigenome Browser69,70 view of the six3a gene, neighboring dsDMR enhancers (six3a-e1, and six3a-e2, grey boxes) and cloned region for a negative control (six3a-ne1, and six3a-ne2, dark grey boxes). (b) GFP expression driven by the dsDMR enhancers of the six3a gene. (c) The methylation profiles of the six3a promoter and neighboring dsDMR enhancers (blue and red lines, left y-axis) and the expression profile of the six3a gene (green line, right y-axis). (d) Epigenome Browser view of the fgfr2 gene, neighboring dsDMR enhancers (fgfr2-e1, fgfr2-e2, fgfr2-e3, and fgfr2-e4, grey boxes) and cloned regions for negative controls (fgfr2-ne1, fgfr2-ne2, fgfr2-ne3, and fgfr2-ne4, dark grey boxes). (e) GFP expression driven by the dsDMR enhancers of the fgfr2 gene. (f) The methylation profiles of the fgfr2 promoter and neighboring dsDMR enhancers (blue and red lines, left y-axis) and the expression profile of the fgfr2 gene (green line, right y-axis). The asterisk * indicates that only one G1 transgenic line was established. Forebrain (f); midbrain hindbrain boundary (mh).
We further illustrate our results with the following examples. The six3a and six3b genes play essential roles in the patterning of forebrain and eye development in many species including zebrafish48. The six3a gene is expressed in forebrain, midbrain hindbrain boundary, anterior neural tube, and medial longitudinal fasciculus, and the six3b gene is expressed in eyes, optic stalk, optic cup, forebrain, telencephalon, and diencephalon at 24 hpf (Supplementary Table 6). We tested two dsDMRs upstream of the six3a gene and two dsDMRs upstream of six3b. Each of these dsDMRs displayed enhancer activity in similar tissue-specific patterns at 24 hpf, driving expression patterns similar to that of the endogenous gene. However, each dsDMR enhancer drove distinct subset of the endogenous expression pattern, resulting in distinct GFP expression patterns. (Fig. 5a,b; Supplementary Fig. 20a,b). RNA expression analysis40 confirmed that these genes were mostly silenced from 2–4 cell stage until shield (~6 hpf, six3b) or bud (~10 hpf, six3a) and were activated between shield and 28 hpf (Fig. 5c; Supplementary Fig. 20c). Importantly, the promoter regions of both genes remained unmethylated across developmental stages; only the dsDMR enhancers exhibited dynamic DNA methylation changes which correlated well with gene expression. (Fig. 5c; Supplementary Fig. 20a). Thus, DNA methylation of these enhancers could potentially regulate the spatial expression of six3 genes.
The three dsDMRs upstream of the fgfr2 gene and one dsDMR upstream of the fgf3 gene also displayed enhancer activities in transgenic G1 fish (Fig. 5d,e,f; Supplementary Fig. 20d,e). FGFR signaling pathways play important roles in diverse developmental processes including cell growth, differentiation, patterning, or cell migration in vertebrates49. The fgfr2 gene is expressed in forebrain, hindbrain, diencephalon and solid lens vesicle, and the fgf3 gene is expressed in forebrain, midbrain and midbrain hindbrain boundary (Supplementary Table 6). The same overall expression patterns were observed in the G1 transgenic fish, but each enhancer drove only a subset of the full expression pattern (Fig. 5e; Supplementary Fig. 20e). Overall, expression patterns driven by the tested enhancers were similar to the expression patterns of their nearby putative target genes (Supplementary Fig. 20; Supplementary Table 6). Taken together, these data demonstrated that the dsDMR enhancers identified in this study were functional in a specific developmental stage; they were likely tissue-specific enhancers for spatial regulation of expression of important developmental genes.
DISCUSSION
Dynamic changes of DNA methylation play a key role in normal development and cell differentiation by defining cell type identities. DNA methylation has been extensively studied across developmental times, but until recently, most work has been focused on gene promoters and CpG islands and little is known about DNA methylation changes in intergenic regions. In this study, we comprehensively defined the genome-wide dynamics of DNA methylation in developing zebrafish embryos. We identified thousands of specific genomic regions (dsDMRs) whose methylation status underwent orchestrated changes across developmental stages. Surprisingly, the majority of dsDMRs were located outside gene promoters, CpG islands, and island shores. Although the primary signal we relied on was DNA methylation, dsDMRs we identified also strongly enriched for sequence conservation, enhancer histone marks, and association with developmental transcription factors and other developmental genes. Our transgenic zebrafish reporter assay provided strong evidence that these dsDMRs were functional developmental enhancers. Our study established that DNA methylation of enhancer elements is a major regulatory mechanism during vertebrate embryo development.
Many dsDMRs we identified were from the later developmental stages, such as 24 hpf embryos. Since the embryos at this stage are not homogeneous and have a diverse cell types and tissues, it is possible that dsDMRs we reported here reflect specificity of the dominant cell types or development of major structures such as neurons and the eye. As technologies to isolate individual cell types continue to advance, it would be interesting and important to revisit DNA methylation dynamics of developmental enhancers in a cell type-specific manner. The DNA methylome maps of whole developing embryos, together with previously published histone modification ChIP-seq41 and RNA-seq43 profiling of the whole embryos, will provide an invaluable resource and reference for current and future investigations.
Using MethylC-seq, Potok et al. identified 9013 DMRs in the earlier developmental stages, with many representing the difference between gametes (i.e., sperm and egg)31. Interestingly, these early developmental DMRs had only 431 overlaps with dsDMRs identified by our study. Majority of the early DMRs reported by Potok et al. did not undergo DNA methylation changes during later developmental stages profiled in our study (Supplementary Fig. 21a). The 431 overlapping regions (Supplementary Fig. 21b) showed DNA methylation changes mainly in 24 hpf embryos and the patterns were very similar to those between sperm and egg or between embryos and muscle (Supplementary Fig. 21c). Thus, the overlapping DMRs would mark regions associated with germline specification (hypermethylation in 24 hpf), or regions associated with muscle development (hypomethylation in 24 hpf). We conclude that our dsDMRs are overall distinct regions from the earlier-stage DMRs with a few expected overlaps.
It is also noteworthy that zebrafish might use different mechanisms of DNA methylation reprogramming in early versus late embryonic developing stages. It was reported that 5-hydroxymethylcytosine (5hmC), the intermediate catalyzed by the tet enzymes in DNA demethylation process, is not involved in the DNA reprogramming in earlier embryos30,31. The low expression levels of the 3 zebrafish tet genes across these developmental stages further supported this idea (Supplementary Fig. 22a). However, tet expression levels increased sharply between 6 hpf (shield) and 28 hpf and were inversely correlated with the methylation levels of dsDMRs analyzed in our study (Supplementary Fig. 22a,b). This raises the possibility that 5hmC mediates active DNA demethylation in later embryos, but not in earlier embryos. Further investigation will be required to elucidate the mechanisms of DNA methylation dynamics in zebrafish embryogenesis.
Previous studies have also identified several thousand putative developmental enhancers in zebrafish, named PDREs, by using three histone modifications: H3K4me1, H3K4me3 and H3K27ac38. The dsDMR enhancers we reported here had a significant overlap with the PDREs, but the two sets were not identical (Supplementary Fig. 23). Five of eighteen PDREs previously tested in stable transgenic assays were identified as dsDMRs in our study (regions 1, 2, 10, 11, and 12 in ref 38) and four of them were validated as functional enhancers in 24 hpf embryos. We tested the in vivo enhancer activity of twenty of our dsDMRs associated with eight important developmental genes. We were able to establish stable transgenic lines for 20 dsDMRs, and all 20 exhibited tissue-specific enhancer activities. Thus, our validation rate was higher than what was reported in the previous study (67%, ref 38). Although the number of validated enhancers remained small due to the low throughput nature of the transgenic experiment, our results suggest that DNA methylation dynamics can be a robust indicator of functional enhancers, possibly more specific than histone modification alone. Integrating DNA methylation, histone modification, and sequence conservation therefore could be a more effective strategy for defining cell type-specific or developmental stage-specific enhancers. Our study joins several recent works in supporting this idea. For example, by investigating intergenic hypomethylated regions in various human cell types, Schlesinger et al. suggested that de novo DNA demethylation defines distal regulatory elements50. Hon et al. pointed out that identifying tissue-specific DMRs (tsDMRs) can be an alternative strategy for finding putative regulatory elements51. The study showed that the enhancer-predicting resolution of tsDMRs were comparable to that of p300-binding sites and were much higher than that of using histone modifications. However, detecting putative enhancers based on differential DNA methylation requires CpGs in the primary sequences. The efficacy of such methods for CpG-poor enhancers remains to be examined.
Sequence conservation has long been used to predict functional elements including enhancers. For example, Pennacchio et al. tested in vivo enhancer activities of human conserved non-coding sequences in a transgenic mouse enhancer assay and found that 45% of tested sequences exhibit tissue-specific enhancer activities6. Similar results were reported in elsewhere52,53. However, it has been difficult to evaluate how many functional elements were missed by this type of approaches. Our results suggested that a large fraction of dsDMRs are not evolutionarily conserved at primary sequence level. However, they may still be functional developmental enhancers – we validated four such examples in this study (Supplementary Table 4).
DNA methylation at gene promoters has long been known to silence genes by blocking transcription54. In contrast, the relationship between DNA methylation at distal enhancers and gene expression remains less explored. In this study, we demonstrated that there is a positive correlation between DNA demethylation in enhancers and expression level of nearby genes (Fig. 3f). However, the impact of enhancer DNA methylation on gene expression is much more complex than a simple “on and off” switch. Instead, enhancer DNA methylation might act more similar to a “dial” that can fine-tune the expression of genes in a specific cell type and developmental time. For example, fgf3 gene expression levels across developmental courses did not correlate well with the DNA methylation status of the predicted enhancer (Supplementary Fig. 20d,f). This might be due to the fact that gene expression is regulated in a temporal- and spatial-specific manner but the existing RNA expression data were from the whole embryos. It is also possible that we did not identify the bona fide target genes for some enhancers, especially when some enhancers function independently of orientation and at great distances from their target genes. In addition, some enhancers validated in our study may have additional target genes.
Although DNA methylation has long been investigated in the context of gene promoters and CpG islands, only recently have studies emerged on DNA methylation signatures of enhancers. DNA hypomethylation has been observed in putative distal regulatory elements and tissue-specific enhancers are hypomethylated in a tissue-specific manner20,22–25,55,56. In line with these studies, our study provided the first evidence that functional enhancers undergo demethylation during embryogenesis. Importantly, to our knowledge, this is the first time the principle of enhancer-DNA methylation relationship is revealed in live developing embryos. These results led us further ask the relationship between DNA methylation and enhancer activity. A recent study showed that demethylation of distal enhancers induced by binding of glucocorticoid receptors leads to activation of the enhancer function21. Also, 5-hydoxymethylcytosine and the TET proteins are frequently found at distal enhancers, suggesting that demethylation of these elements is closely related to their function56,57. However, a recent study also suggested that some developmental enhancers remain hypomethylated but become inactive in adult tissues51. It is still unclear whether DNA hypomethylation of enhancers are necessary for their activities or hypomethylation is a result of enhancer-activation by another mechanism14. Further investigation will be required to elucidate the precise relationship between DNA methylation and enhancer activity.
METHODS
Collection of embryos and isolation of genomic DNA
All zebrafish were used in accordance with the protocols approved by the Washington University Animal Studies Committee (Protocol #20110236). Wild-type sjA strain zebrafish were maintained under standard conditions58. Sperm were collected from sexually mature zebrafish males by standard protocol58. Wild-type embryos were obtained through in vitro fertilization, grown in 28.5 °C egg water (60 μg ml−1 Ocean Sea Salts) to develop to the desired stages, and harvested at five different developmental stages: 256-cell (~2.5 hpf), 1Kcell-High (~3.5 hpf), dome (~4.5 hpf), shield (~6 hpf), and 24 hpf. Chorions were removed with pronase and yolk was removed with ice-cold deyolking buffer (55 mM NaCl, 1.8mM KCl, 1.25 mM NaHCO3). Embryos were snap-frozen in liquid nitrogen and stored at −80 °C until time of use. The genomic DNA was isolated by incubating embryos in genomic DNA extraction buffer (50 mM Tris, 1 mM EDTA, 0.5 % SDS, 1 mg/mL Proteinase K) followed by phenol-chloroform extraction. DNA concentrations and purity were measured using NanoVue (GE Healthcare Life Sciences) and DNA integrity was confirmed by gel electrophoresis.
MeDIP and MRE-seq library generation and sequencing
MeDIP and MRE sequencing libraries were generated as described previously18,59 with minor modifications. For MeDIP-seq, 500 ng of DNA isolated was sonicated to a fragment size of 100–500 bp, end-processed, and ligated to paired-end adapters. After agarose gel size-selection of 166–566 bp, DNA was heat denatured and then immunoprecipitated using a mouse monoclonal anti-methylcytidine antibody (1 μg of antibody per 1 μg of DNA, Eurogentec) in 500 μl of immunoprecipitation buffer (10 μM sodium phosphate, pH 7.0, 140 mM sodium chloride and 0.05% Triton X-100) overnight at 4 °C. Antibody/DNA complexes were isolated by addition of 1 μl of rabbit anti-mouse IgG secondary antibody (2.4 mg ml−1, Jackson Immunoresearch) and 100 μl protein A/G agarose beads (Pierce Biotechnology) for 2 h at 4 °C. Beads were washed six times with immunoprecipitation buffer and then DNA was eluted in TE buffer with 0.25% SDS and 0.25 mg ml−1 of proteinase K for 2 h at 50 °C. DNA was then purified with QIAquick purification kit (Qiagen) and eluted in 30 μl EB buffer. DNA was amplified by 12 cycles of PCR with the standard Illumina index primers, and size-selected (220–620 bp) by agarose gel electrophoresis.
For MRE-seq, 5 parallel digests (HpaII, Hin6I, SsiI, BstUI, and HpyCH4IV; New England Biolabs) were performed, each with 200 ng of DNA. Five units of enzyme were initially incubated with DNA for 3 h and then additional five units of enzyme were added to the digestion for a total of 6 h of digestion time. DNA was purified by phenol/chloroform/isoamyl alcohol extraction, followed by chloroform extraction using phase lock gels. Digested DNA from the different reactions was combined and precipitated with 1/10 volume of 3 M sodium acetate (pH 5.2) and 2.5 volumes of ethanol. The purified DNA was size-selected (100–500 bp), end-processed, and ligated to single-end adapters. After the second size-selection (166–566 bp), DNA was amplified by 15 cycles of PCR and size-selected (220–620 bp) by agarose gel electrophoresis. MeDIP and MRE libraries were sequenced on Illumina HiSeq machine with a total number of approximately 540 million MeDIP-seq reads and 300 million MRE-seq reads. These reads were mapped to the latest zebrafish genome assembly (Zv9) by using Novoalign (Novocraft). The complete list of dsDMRs are accessible at http://epigenome.wustl.edu/Zebrafish_DNAme/.
Estimation of methylation levels at single CpG resolution
Methylation levels at single CpG resolution were estimated by using methylCRF with default parameteres33,59. The methylCRF scores of 24,222,562 CpGs in 26 chromosomes were calculated and 1,028,605 CpGs in scaffolds were excluded. Concordance between MethylC-seq and methylCRF predictions was measured by calculating the differences of predictions on the same CpG sites from the matched samples. The CpGs whose methylation levels do not exist in MethylC-seq were excluded in concordance calculation.
The average DNA methylation levels over different genic or genomic features were calculated per CpG in 30 bins per genic or genomic feature. Gene coordinates were obtained from zebrafish gene set of Ensembl release 71 and promoter was defined as 1 kb upstream region from the TSS. CpG island annotation was obtained from the UCSC Genome Browser CpG islands track. CGI shores and CGI shelves were defined as regions 2 kb up and downstream of the CGIs and CGI shores, respectively. Long non-coding RNA gene annotations were obtained from the recent zebrafish lncRNA study40.
To classify promoters in two categories, HCPs and LCPs, we first determined the GC content and the ratio of observed versus expected (o/e) CpG in sliding 500-bp windows with 5-bp offset for each 1 kb upstream TSS region. The o/e CpG ratio was calculated by the following formula: (number of CpGs times number of bp)/(number of Cs times number of Gs) 35. The promoters were classified as HCPs if the promoters contain a subsequence with an o/e CpG ratio of ≥ 0.65 and a GC content of > 0.30, and were classified as LCPs if they did not meet these criteria. More than half of the promoters were classified as HCPs (Supplementary Fig. 8a). The average DNA methylation levels over 4 kb regions around the TSS were calculated per CpG in 100-bp bins (Supplementary Fig. 8b).
Targeted validation of the methylation levels
Genomic DNA underwent bisulfite conversion using EpiTect Bisulfite Kit (Qiagen) following the manufacturer’s instructions. Regions of interest were amplified with specific PCR primers (Supplementary Table 7) using HiFi HotStart Uracil+ ReadyMix (Kapa Biosystems). For clonal Sanger sequencing, amplified regions were cloned into pCR4-TOPO vector (Invitrogen) and more than 15 bacterial clones were used for Sanger sequencing. For pyrosequencing, amplified regions were subjected to pyrosequencing reaction in PyroMark Q24 (Qiagen) with specific sequencing primers (Supplementary Table 7) following the manufacturer’s instructions. The data were analyzed with the PyroMark Q24 software (Qiagen).
Analysis of differentially methylated regions
Differentially methylated regions (DMRs) between developmental stages were identified by using methylMnM package23,59 with the default parameters in R environment. Briefly, the coverage of MeDIP and MRE sequencing data and genomic CpG information were calculated in each 500-bp genomic bin. Scaffolds and the mitochondrial genome were excluded from the analysis. DMRs with a Q-value less than 1×10−5 were selected for analysis. The average methylation level of CpGs in each DMR was calculated using methylCRF scores, and the differences of methylation scores between two developmental stages were calculated.
To annotate dsDMRs, we first divided the zebrafish genome into the four non-overlapping genomic features based on the following criteria. Gene annotation was obtained from zebrafish gene set of Ensembl release 71. The longest transcript was chosen from multiple transcripts for each protein-coding gene. Promoters were defined as 1.5 kb upstream and 0.5 kb downstream region from TSS of each protein-coding gene. Exons and introns were defined as annotated in Ensembl release 71, excluding regions that were defined as promoters. All the remaining genomic regions were defined as intergenic. Each dsDMR was annotated as one of the four genomic features if the center of the dsDMR resides within the corresponding genomic feature.
To investigate the distribution of dsDMRs around developmental genes, we first chose the 772 genes under the Gene Ontology term “GO0009790: embryo development”. We also randomly chose the same number of protein-coding genes. We calculated the number of dsDMRs in a 10 kb window upstream of the TSS of each gene selected. We then calculated the mean number of dsDMRs for each 10 kb windows over the 772 genes and generated the average dsDMR distribution plot.
For evolutionary conservation analysis, the vertebrate PhastCons score based on the 8-way vertebrate genome alignment was obtained from the UCSC Genome Browser PhastCons conservation track. Histone modification ChIP-seq data were obtained from the recent study38.
Analysis of dsDMRs as developmental enhancers
Gene Ontology enrichment was performed using the GREAT analysis tool39. Gene regulatory domains were defined by default as the following: basal regulatory domain was defined as a region spanning 5 kb upstream of and 1 kb downstream from the TSS; the distal regulatory domain was then extended in both directions to the nearest gene’s basal domain but no more than 1000 kb in one direction. The GO terms that have a minimum of 2.5-fold region-based enrichment and that have a false discovery rate below 0.05 were reported.
For gene expression analysis, the RNA-seq data were obtained from a recent study40. The RNA-seq reads were mapped to the zebrafish genome assembly Zv9 by using tophat60 and FPKM values were calculated for each gene by using cufflinks61.
Transcription factor binding motif enrichment analysis
Motif enrichment analysis in dsDMRs was performed using the HOMER tool62. HOMER scanned the sequences of dsDMRs for known motifs including HOMER-provided motifs library and JASPAR core vertebrate motifs63, and calculated enrichment score P-values using hypergeometric test. DAVID tool64 was used to functionally annotate the transcription factors whose motifs are enriched in dsDMRs. The zebrafish Ensemble gene IDs corresponding to the enriched motifs were used as input for DAVID tool. The entire zebrafish genome was first used as background for DAVID. Since many GO terms related to transcription activities were enriched against this background (Supplementary Fig. 14), all TFs from motif enrichment analysis were also used as background for DAVID (Supplementary Fig. 15a).
Construction of gene regulatory network
To obtain the regulatory interactions among the putative target genes and TFs identified from dsDMRs, these genes and TFs were used as nodes in the UCSC Interaction Browser65 as described previously66 with minor modifications as detailed below. First, 215 genes were identified as putative targets of dsDMR enhancers by their association with dsDMRs from GREAT analysis. Then 49 TFs whose motifs were enriched in dsDMRs were included as additional nodes. Two network collection databases were used to query for interactions among the genes: GEA_CLR network connecting TF to targets67 and ChEA transcription factor network68. To compute statistical significance, 10 sets of 215 random genes and 49 random TFs were used as nodes and an expected distribution of connections among the given number of genes was obtained (mean of 1886 and standard deviation of 260; Supplementary Fig. 15b).
To build putative regulatory network of the eye development, TFs were first linked to the target genes. TFs related with the eye development were selected from the TFs with enriched motifs. Target genes related with the eye development were selected from the gene list identified from GREAT analysis. A TF whose motif is in a dsDMR was linked to a gene to which GREAT associate the dsDMR. The known target genes of sox2 were from the ChEA system68.
Zebrafish in vivo enhancer assay
Putative enhancer sequences were PCR-amplified with specific primers (Supplementary Table 3) and TOPO-cloned into an entry vector (pCR8/GW/TOPO, Invitrogen) of the Gateway cloning system. The cloned plasmid was then recombined with the destination vector (pGW_cfosEGFP, generous gift from Shannon Fisher) to generate the desired reporter plasmid. Prepared plasmid DNAs for microinjection were further purified with QIAquick spin columns (Qiagen). Tol2 transposase mRNA was transcribed in vitro from the pCS-Tp vector46 (a generous gift from Shannon Fisher), using the mMessage mMachine Sp6 kit (Ambion). Embryos at one-cell or two-cell stages were injected with transposase mRNA, transposon plasmid DNA, and phenol red solution as described previously45. At least 100 embryos were injected for each construct. The reporter expression patterns were analyzed at 24 hpf stage. If more than 10 % of embryos exhibit the consistent GFP expression pattern, the construct was considered as a positive enhancer. The embryos with a specific expression patterns were selected and raised to sexual maturity. Sexually mature G0 adults were crossed with sjA wild-type strain to obtain germline transmission. Two or more independent G1 transgenic lines were established for each construct, unless otherwise indicated. G1 transgenic fish embryos were photographed at 24 hpf stage.
Supplementary Material
Acknowledgments
We thank Shannon Fisher for generously providing enhancer reporter plasmid and transposase expression plasmid. We thank Brian Stephens and Rachel Kerstetter for fish husbandry and technical assistance. We are grateful to everyone in Wang laboratory for helpful advice and discussions. H.J.L. is a McDonnell International Scholar (Washington University in St. Louis) and a Kwanjeong Educational Foundation Abroad Scholar (South Korea). R.F.L. is supported by the NSF Graduate Research Fellowship Program DGE-1143954. B.Z. is supported by NIDA’s R25 program DA027995. S.L.J. is supported by RO1GM05698. T.W. is supported by Basil O’Connor Starter Scholar Research Award Grant no. 5-FY10-491 from the March of Dimes Foundation, American Cancer Society grant RSG-14-049-01-DMC, and NIH grants RO1HG007354, RO1HG007175, and RO1ES024992.
Footnotes
AUTHOR CONTRIBUTIONS
H.J.L., S.L.J. and T.W. conceived and designed the study. H.J.L. collected samples and generated sequencing libraries. H.J.L., R.F.L., B.M., S.L.J. and T.W. analyzed the data. B.Z., M.S. and D.L. contributed to computational algorithms. H.J.L. and T.W. wrote the manuscript. All authors discussed the results and contributed to writing the manuscript.
Accession codes: The MeDIP-seq and MRE-seq data from this study have been deposited to the NCBI Gene Expression Omnibus (GEO) under accession number GSE52706, containing the Subseries GSE52703 (MeDIP-seq) and GSE52704 (MRE-seq).
Competing financial interests: The authors do not have conflicts of interests to declare.
References
- 1.Banerji J, Rusconi S, Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981;27:299–308. doi: 10.1016/0092-8674(81)90413-x. [DOI] [PubMed] [Google Scholar]
- 2.Ong CT, Corces VG. Enhancers: emerging roles in cell fate specification. EMBO Rep. 2012;13:423–430. doi: 10.1038/embor.2012.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Visel A, Rubin EM, Pennacchio LA. Genomic views of distant-acting enhancers. Nature. 2009;461:199–205. doi: 10.1038/nature08451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bejerano G, et al. Ultraconserved elements in the human genome. Science. 2004;304:1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- 5.Woolfe A, et al. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 2005;3:e7. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pennacchio LA, et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- 7.Blow MJ, et al. ChIP-Seq identification of weakly conserved heart enhancers. Nat Genet. 2010;42:806–810. doi: 10.1038/ng.650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 9.Visel A, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shen Y, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Visel A, Minovitsky S, Dubchak I, Pennacchio LA. VISTA Enhancer Browser--a database of tissue-specific human enhancers. Nucleic Acids Res. 2007;35:D88–92. doi: 10.1093/nar/gkl822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pennacchio LA, Bickmore W, Dean A, Nobrega MA, Bejerano G. Enhancers: five essential questions. Nat Rev Genet. 2013;14:288–295. doi: 10.1038/nrg3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 15.Kitamura E, et al. Analysis of tissue-specific differentially methylated regions (TDMs) in humans. Genomics. 2007;89:326–337. doi: 10.1016/j.ygeno.2006.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Illingworth R, et al. A novel CpG island set identifies tissue-specific methylation at developmental gene loci. PLoS Biol. 2008;6:e22. doi: 10.1371/journal.pbio.0060022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 18.Maunakea AK, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shukla S, et al. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479:74–79. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stadler MB, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- 21.Wiench M, et al. DNA methylation status predicts cell type-specific enhancer activity. EMBO J. 2011;30:3028–3039. doi: 10.1038/emboj.2011.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xie M, et al. DNA hypomethylation within specific transposable element families associates with tissue-specific enhancer landscape. Nat Genet. 2013;45:836–841. doi: 10.1038/ng.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang B, et al. Functional DNA methylation differences between tissues, cell types, and across individuals discovered using the M&M algorithm. Genome Res. 2013;23:1522–1540. doi: 10.1101/gr.156539.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gifford CA, et al. Transcriptional and epigenetic dynamics during specification of human embryonic stem cells. Cell. 2013;153:1149–1163. doi: 10.1016/j.cell.2013.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie W, et al. Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell. 2013;153:1134–1148. doi: 10.1016/j.cell.2013.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 27.Martin CC, Laforest L, Akimenko MA, Ekker M. A role for DNA methylation in gastrulation and somite patterning. Dev Biol. 1999;206:189–205. doi: 10.1006/dbio.1998.9105. [DOI] [PubMed] [Google Scholar]
- 28.Rai K, et al. Zebra fish Dnmt1 and Suv39h1 regulate organ-specific terminal differentiation during development. Mol Cell Biol. 2006;26:7077–7085. doi: 10.1128/MCB.00312-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rai K, et al. Dnmt3 and G9a cooperate for tissue-specific development in zebrafish. J Biol Chem. 2010;285:4110–4121. doi: 10.1074/jbc.M109.073676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang L, et al. Sperm, but not oocyte, DNA methylome is inherited by zebrafish early embryos. Cell. 2013;153:773–784. doi: 10.1016/j.cell.2013.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Potok ME, Nix DA, Parnell TJ, Cairns BR. Reprogramming the maternal zebrafish genome after fertilization to match the paternal methylation pattern. Cell. 2013;153:759–772. doi: 10.1016/j.cell.2013.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harris RA, et al. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat Biotechnol. 2010;28:1097–1105. doi: 10.1038/nbt.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stevens M, et al. Estimating absolute methylation levels at single-CpG resolution from methylation enrichment and restriction enzyme sequencing methods. Genome Res. 2013;23:1541–1553. doi: 10.1101/gr.152231.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lister R, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weber M, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 36.Siepel A, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meyer LR, et al. The UCSC Genome Browser database: extensions and updates 2013. Nucleic Acids Res. 2013;41:D64–69. doi: 10.1093/nar/gks1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bogdanovic O, et al. Dynamics of enhancer chromatin signatures mark the transition from pluripotency to cell specification during embryogenesis. Genome Res. 2012;22:2043–2053. doi: 10.1101/gr.134833.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McLean CY, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pauli A, et al. Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res. 2012;22:577–591. doi: 10.1101/gr.133009.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun LO, et al. On and off retinal circuit assembly by divergent molecular mechanisms. Science. 2013;342:1241974. doi: 10.1126/science.1241974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ebert AM, Childs SJ, Hehr CL, Cechmanek PB, McFarlane S. Sema6a and Plxna2 mediate spatially regulated repulsion within the developing eye to promote eye vesicle cohesion. Development. 2014;141:2473–2482. doi: 10.1242/dev.103499. [DOI] [PubMed] [Google Scholar]
- 43.Cardozo MJ, et al. Cdon acts as a Hedgehog decoy receptor during proximal-distal patterning of the optic vesicle. Nat Commun. 2014;5:4272. doi: 10.1038/ncomms5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brzezinski JA, 4th, Kim EJ, Johnson JE, Reh TA. Ascl1 expression defines a subpopulation of lineage-restricted progenitors in the mammalian retina. Development. 2011;138:3519–3531. doi: 10.1242/dev.064006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fisher S, et al. Evaluating the biological relevance of putative enhancers using Tol2 transposon-mediated transgenesis in zebrafish. Nat Protoc. 2006;1:1297–1305. doi: 10.1038/nprot.2006.230. [DOI] [PubMed] [Google Scholar]
- 46.Hu G, Goll MG, Fisher S. PhiC31 integrase mediates efficient cassette exchange in the zebrafish germline. Dev Dyn. 2011;240:2101–2107. doi: 10.1002/dvdy.22699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tryon RC, Higdon CW, Johnson SL. Lineage relationship of direct-developing melanocytes and melanocyte stem cells in the zebrafish. PLoS One. 2011;6:e21010. doi: 10.1371/journal.pone.0021010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Serikaku MA, O’Tousa JE. sine oculis is a homeobox gene required for Drosophila visual system development. Genetics. 1994;138:1137–1150. doi: 10.1093/genetics/138.4.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thisse B, Thisse C. Functions and regulations of fibroblast growth factor signaling during embryonic development. Dev Biol. 2005;287:390–402. doi: 10.1016/j.ydbio.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 50.Schlesinger F, Smith AD, Gingeras TR, Hannon GJ, Hodges E. De novo DNA demethylation and non-coding transcription define active intergenic regulatory elements. Genome Res. 2013;23:1601–1614. doi: 10.1101/gr.157271.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hon GC, et al. Epigenetic memory at embryonic enhancers identified in DNA methylation maps from adult mouse tissues. Nat Genet. 2013;45:1198–1206. doi: 10.1038/ng.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Prabhakar S, et al. Close sequence comparisons are sufficient to identify human cis-regulatory elements. Genome Res. 2006;16:855–863. doi: 10.1101/gr.4717506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Visel A, et al. Ultraconservation identifies a small subset of extremely constrained developmental enhancers. Nat Genet. 2008;40:158–160. doi: 10.1038/ng.2007.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kass SU, Landsberger N, Wolffe AP. DNA methylation directs a time-dependent repression of transcription initiation. Curr Biol. 1997;7:157–165. doi: 10.1016/s0960-9822(97)70086-1. [DOI] [PubMed] [Google Scholar]
- 55.Ziller MJ, et al. Charting a dynamic DNA methylation landscape of the human genome. Nature. 2013;500:477–481. doi: 10.1038/nature12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stroud H, Feng S, Morey Kinney S, Pradhan S, Jacobsen SE. 5-Hydroxymethylcytosine is associated with enhancers and gene bodies in human embryonic stem cells. Genome Biol. 2011;12:R54. doi: 10.1186/gb-2011-12-6-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Williams K, et al. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bradford Y, et al. ZFIN: enhancements and updates to the Zebrafish Model Organism Database. Nucleic Acids Res. 2011;39:D822–829. doi: 10.1093/nar/gkq1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li D, Zhang B, Xing X, Wang T. Combining MeDIP-seq and MRE-seq to investigate genome-wide CpG methylation. Methods. 2014 doi: 10.1016/j.ymeth.2014.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mathelier A, et al. JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res. 2014;42:D14–147. doi: 10.1093/nar/gkt997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wong CK, et al. The UCSC Interaction Browser: multidimensional data views in pathway context. Nucleic Acids Res. 2013;41:W218–224. doi: 10.1093/nar/gkt473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lowdon RF, et al. Regulatory network decoded from epigenomes of surface ectoderm-derived cell types. Nat commun. 2014;5:5442. doi: 10.1038/ncomms6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Faith JJ, et al. Large-scale mapping and validation of Escherichia coli transcriptional regulation from a compendium of expression profiles. PLoS Biol. 2007;5:e8. doi: 10.1371/journal.pbio.0050008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lachmann A, et al. ChEA: transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics. 2010;26:2438–2444. doi: 10.1093/bioinformatics/btq466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhou X, et al. The Human Epigenome Browser at Washington University. Nat Methods. 2011;8:989–990. doi: 10.1038/nmeth.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhou X, et al. Exploring long-range genome interactions using the WashU Epigenome Browser. Nat Methods. 2013;10:375–376. doi: 10.1038/nmeth.2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.