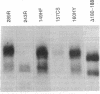
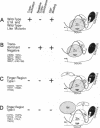
Abstract
The 289R E1A protein of adenovirus transactivates a variety of viral and cellular promoters through protein-protein interactions. In earlier studies, mutational analyses of the E1A transactivating domain identified residues that are critical for transactivation and implied that the zinc finger region of the transactivating domain binds a transcription factor. Also, the E1A activation domain was found to bind to the TATA box binding protein (TBP) in vitro. Here, we tested the significance of the E1A-TBP interaction for E1A transactivation by analyzing the effects of conservative substitutions at each of the 49 residues of the E1A activation domain. Seven of the substitutions significantly diminished TBP binding in vitro. All of these were in the zinc finger region and were defective for transactivation in vivo. The perfect correlation between reduced TBP binding and transactivation argues strongly that a direct interaction between the E1A activation domain and TBP is critical to the mechanism of E1A activation. This genetic analysis leads us to further suggest that another factor, which is limiting, is also necessary for E1A-mediated transactivation.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berk A. J. Adenovirus promoters and E1A transactivation. Annu Rev Genet. 1986;20:45–79. doi: 10.1146/annurev.ge.20.120186.000401. [DOI] [PubMed] [Google Scholar]
- Boyer T. G., Berk A. J. Functional interaction of adenovirus E1A with holo-TFIID. Genes Dev. 1993 Sep;7(9):1810–1823. doi: 10.1101/gad.7.9.1810. [DOI] [PubMed] [Google Scholar]
- Brou C., Chaudhary S., Davidson I., Lutz Y., Wu J., Egly J. M., Tora L., Chambon P. Distinct TFIID complexes mediate the effect of different transcriptional activators. EMBO J. 1993 Feb;12(2):489–499. doi: 10.1002/j.1460-2075.1993.tb05681.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S., Zhou H. Transcription factor IID mutants defective for interaction with transcription factor IIA. Science. 1992 Feb 28;255(5048):1130–1132. doi: 10.1126/science.1546314. [DOI] [PubMed] [Google Scholar]
- Chatterjee P. K., Bruner M., Flint S. J., Harter M. L. DNA-binding properties of an adenovirus 289R E1A protein. EMBO J. 1988 Mar;7(3):835–841. doi: 10.1002/j.1460-2075.1988.tb02882.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatton B., Bocco J. L., Gaire M., Hauss C., Reimund B., Goetz J., Kedinger C. Transcriptional activation by the adenovirus larger E1a product is mediated by members of the cellular transcription factor ATF family which can directly associate with E1a. Mol Cell Biol. 1993 Jan;13(1):561–570. doi: 10.1128/mcb.13.1.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culp J. S., Webster L. C., Friedman D. J., Smith C. L., Huang W. J., Wu F. Y., Rosenberg M., Ricciardi R. P. The 289-amino acid E1A protein of adenovirus binds zinc in a region that is important for trans-activation. Proc Natl Acad Sci U S A. 1988 Sep;85(17):6450–6454. doi: 10.1073/pnas.85.17.6450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field J., Nikawa J., Broek D., MacDonald B., Rodgers L., Wilson I. A., Lerner R. A., Wigler M. Purification of a RAS-responsive adenylyl cyclase complex from Saccharomyces cerevisiae by use of an epitope addition method. Mol Cell Biol. 1988 May;8(5):2159–2165. doi: 10.1128/mcb.8.5.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint J., Shenk T. Adenovirus E1A protein paradigm viral transactivator. Annu Rev Genet. 1989;23:141–161. doi: 10.1146/annurev.ge.23.120189.001041. [DOI] [PubMed] [Google Scholar]
- Glenn G. M., Ricciardi R. P. An adenovirus type 5 E1A protein with a single amino acid substitution blocks wild-type E1A transactivation. Mol Cell Biol. 1987 Mar;7(3):1004–1011. doi: 10.1128/mcb.7.3.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruda M. C., Zabolotny J. M., Xiao J. H., Davidson I., Alwine J. C. Transcriptional activation by simian virus 40 large T antigen: interactions with multiple components of the transcription complex. Mol Cell Biol. 1993 Feb;13(2):961–969. doi: 10.1128/mcb.13.2.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemeier C., Walker S., Caswell R., Kouzarides T., Sinclair J. The human cytomegalovirus 80-kilodalton but not the 72-kilodalton immediate-early protein transactivates heterologous promoters in a TATA box-dependent mechanism and interacts directly with TFIID. J Virol. 1992 Jul;66(7):4452–4456. doi: 10.1128/jvi.66.7.4452-4456.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisatake K., Hasegawa S., Takada R., Nakatani Y., Horikoshi M., Roeder R. G. The p250 subunit of native TATA box-binding factor TFIID is the cell-cycle regulatory protein CCG1. Nature. 1993 Mar 11;362(6416):179–181. doi: 10.1038/362179a0. [DOI] [PubMed] [Google Scholar]
- Hisatake K., Roeder R. G., Horikoshi M. Functional dissection of TFIIB domains required for TFIIB-TFIID-promoter complex formation and basal transcription activity. Nature. 1993 Jun 24;363(6431):744–747. doi: 10.1038/363744a0. [DOI] [PubMed] [Google Scholar]
- Horikoshi N., Maguire K., Kralli A., Maldonado E., Reinberg D., Weinmann R. Direct interaction between adenovirus E1A protein and the TATA box binding transcription factor IID. Proc Natl Acad Sci U S A. 1991 Jun 15;88(12):5124–5128. doi: 10.1073/pnas.88.12.5124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W. S., Kao C. C., Bryant G. O., Liu X., Berk A. J. Adenovirus E1A activation domain binds the basic repeat in the TATA box transcription factor. Cell. 1991 Oct 18;67(2):365–376. doi: 10.1016/0092-8674(91)90188-5. [DOI] [PubMed] [Google Scholar]
- Lieberman P. M., Berk A. J. The Zta trans-activator protein stabilizes TFIID association with promoter DNA by direct protein-protein interaction. Genes Dev. 1991 Dec;5(12B):2441–2454. doi: 10.1101/gad.5.12b.2441. [DOI] [PubMed] [Google Scholar]
- Lillie J. W., Green M. R. Transcription activation by the adenovirus E1a protein. Nature. 1989 Mar 2;338(6210):39–44. doi: 10.1038/338039a0. [DOI] [PubMed] [Google Scholar]
- Liu F., Green M. R. A specific member of the ATF transcription factor family can mediate transcription activation by the adenovirus E1a protein. Cell. 1990 Jun 29;61(7):1217–1224. doi: 10.1016/0092-8674(90)90686-9. [DOI] [PubMed] [Google Scholar]
- Martin K. J., Lillie J. W., Green M. R. Evidence for interaction of different eukaryotic transcriptional activators with distinct cellular targets. Nature. 1990 Jul 12;346(6280):147–152. doi: 10.1038/346147a0. [DOI] [PubMed] [Google Scholar]
- Meisterernst M., Roy A. L., Lieu H. M., Roeder R. G. Activation of class II gene transcription by regulatory factors is potentiated by a novel activity. Cell. 1991 Sep 6;66(5):981–993. doi: 10.1016/0092-8674(91)90443-3. [DOI] [PubMed] [Google Scholar]
- Montell C., Fisher E. F., Caruthers M. H., Berk A. J. Resolving the functions of overlapping viral genes by site-specific mutagenesis at a mRNA splice site. Nature. 1982 Feb 4;295(5848):380–384. doi: 10.1038/295380a0. [DOI] [PubMed] [Google Scholar]
- Nikolov D. B., Hu S. H., Lin J., Gasch A., Hoffmann A., Horikoshi M., Chua N. H., Roeder R. G., Burley S. K. Crystal structure of TFIID TATA-box binding protein. Nature. 1992 Nov 5;360(6399):40–46. doi: 10.1038/360040a0. [DOI] [PubMed] [Google Scholar]
- Ptashne M. How eukaryotic transcriptional activators work. Nature. 1988 Oct 20;335(6192):683–689. doi: 10.1038/335683a0. [DOI] [PubMed] [Google Scholar]
- Pugh B. F., Tjian R. Diverse transcriptional functions of the multisubunit eukaryotic TFIID complex. J Biol Chem. 1992 Jan 15;267(2):679–682. [PubMed] [Google Scholar]
- Ruppert S., Wang E. H., Tjian R. Cloning and expression of human TAFII250: a TBP-associated factor implicated in cell-cycle regulation. Nature. 1993 Mar 11;362(6416):175–179. doi: 10.1038/362175a0. [DOI] [PubMed] [Google Scholar]
- Stringer K. F., Ingles C. J., Greenblatt J. Direct and selective binding of an acidic transcriptional activation domain to the TATA-box factor TFIID. Nature. 1990 Jun 28;345(6278):783–786. doi: 10.1038/345783a0. [DOI] [PubMed] [Google Scholar]
- Tanese N., Pugh B. F., Tjian R. Coactivators for a proline-rich activator purified from the multisubunit human TFIID complex. Genes Dev. 1991 Dec;5(12A):2212–2224. doi: 10.1101/gad.5.12a.2212. [DOI] [PubMed] [Google Scholar]
- Webster L. C., Ricciardi R. P. trans-dominant mutants of E1A provide genetic evidence that the zinc finger of the trans-activating domain binds a transcription factor. Mol Cell Biol. 1991 Sep;11(9):4287–4296. doi: 10.1128/mcb.11.9.4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster L. C., Zhang K., Chance B., Ayene I., Culp J. S., Huang W. J., Wu F. Y., Ricciardi R. P. Conversion of the E1A Cys4 zinc finger to a nonfunctional His2,Cys2 zinc finger by a single point mutation. Proc Natl Acad Sci U S A. 1991 Nov 15;88(22):9989–9993. doi: 10.1073/pnas.88.22.9989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zawel L., Reinberg D. Advances in RNA polymerase II transcription. Curr Opin Cell Biol. 1992 Jun;4(3):488–495. doi: 10.1016/0955-0674(92)90016-6. [DOI] [PubMed] [Google Scholar]
- Zhou Q., Lieberman P. M., Boyer T. G., Berk A. J. Holo-TFIID supports transcriptional stimulation by diverse activators and from a TATA-less promoter. Genes Dev. 1992 Oct;6(10):1964–1974. doi: 10.1101/gad.6.10.1964. [DOI] [PubMed] [Google Scholar]