Abstract
Objective
It is important to identify separate publications that report outcomes from the same underlying clinical trial, in order to avoid over-counting these as independent pieces of evidence.
Methods
We created positive and negative training sets (comprised of pairs of articles reporting on the same condition and intervention) that were, or were not, linked to the same clinicaltrials.gov trial registry number. Features were extracted from MEDLINE and PubMed metadata; pairwise similarity scores were modeled using logistic regression.
Results
Article pairs from the same trial were identified with high accuracy (F1 score = 0.843). We also created a clustering tool, Aggregator, that takes as input a PubMed user query for RCTs on a given topic, and returns article clusters predicted to arise from the same clinical trial.
Discussion
Although painstaking examination of full-text may be needed to be conclusive, metadata are surprisingly accurate in predicting when two articles derive from the same underlying clinical trial.
Keywords: evidence-based medicine, clinical trials, systematic reviews, bias, information retrieval, informatics
INTRODUCTION
The road from clinical trial to clinical practice is long and slippery. New treatments are tested in clinical trials (of varying size and design), which may be registered in formal clinical trial registries (or not), and which may be published in the peer-reviewed literature (or not) [1]. Systematic reviews and meta-analyses are written to assess the published evidence in a standardized and comprehensive manner, and to reach conclusions concerning efficacy and safety that are as free from bias as possible [2].
An important type of bias occurs when one clinical trial gives rise to multiple publications that are erroneously counted as arising from independent sources of evidence, which can lead to inaccurate estimates of efficacy [3, 4]. This is a challenging problem requiring close reading of the full text (plus other information such as writing to authors), made worse by the fact that multiple publications often do not cite each other, may have completely non-intersecting sets of authors, and may not contain clinical trial registry numbers [5, 6]. In some ways, the problem of deciding whether two articles belong to the same clinical trial is similar to the problem of deciding whether two articles bearing the same author name were written by the same individual. A machine learning model based on MEDLINE metadata features is highly accurate in collecting together all articles written by the same author-individual [7, 8] and we hypothesized that a machine learning approach might aggregate together the articles that arise from the same underlying clinical trial.
In this paper, we have analyzed multiple publications in a well-defined corpus, namely, the set of MEDLINE articles that contain one or more NCT registry numbers and that are linked automatically to registered trials within clinicaltrials.gov. We created a positive training set consisting of pairs of clinical trial articles that were linked to the same registry entry, and a balanced negative training set consisting of pairs of articles that shared the same condition and intervention, but that were linked to different registry entries. We examined various MEDLINE and PubMed metadata to assess which had discriminative value in positive vs. negative sets, and built a machine learning model based on pairwise similarity scores that estimated the probability that any given pair of articles come from the same registry entry. The model was then extended to identify clusters of related randomized controlled trial articles retrieved from PubMed queries. The results show that metadata features can identify multiple publications deriving from the same underlying trial with high accuracy.
METHODS
ClinicalTrials.gov (http://clinicaltrials.gov/), founded in 2000, contains 145,300 registered trials (May 2013) [9–11]. Studies are conducted in all 50 states and in 185 countries. About 8,900 trials include results submitted by the trial organizers, and 4,824 trials have a total of 7,827 linked publications, of which 7,501 have links to PubMed. Most publications are manually submitted by trial organizers; these may not necessarily represent trial outcomes (e.g., some are review articles that explain the motivation for conducting clinical trials on the topic). 2,665 publications contain registry (NCT) numbers indexed in a special MEDLINE field, which are automatically linked to ClinicalTrials.gov [12].
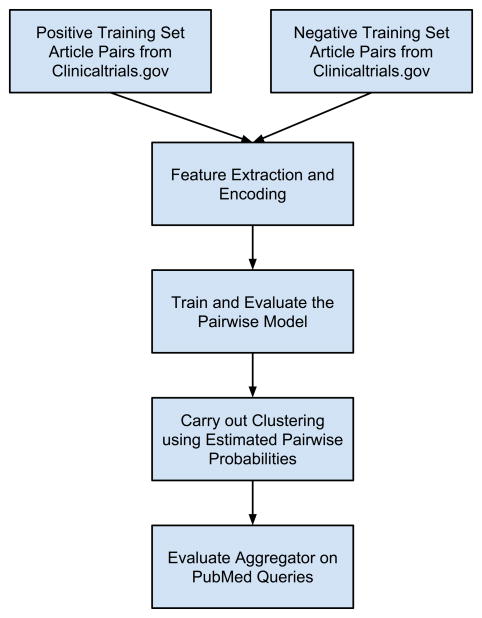
As outlined in Figure 1, we first constructed and trained a pairwise model to predict whether two articles arise from the same clinical trial. Then we built a clustering tool, Aggregator, using the pairwise prediction model and a hierarchical clustering strategy. Each of these will be described and discussed in turn.
Figure 1.
Diagram outlining the workflow of model building and evaluation.
See Methods for details.
1. A pairwise model to predict whether two articles arise from the same clinical trial
1.1 Choosing articles for the positive and negative gold standard training sets
The following rules were used to choose articles:
Articles chosen were linked to clinicaltrials.gov registry entries (April 2013), written in English, published after 1987, and contained abstracts.
The NCT number in the MEDLINE record matched the registry number in clinicaltrials.gov.
Articles having multiple registry numbers (113 of 3083) were excluded. We also cleansed some cases of typographical errors and author errors in assigning the registry number.
Articles were indexed as clinical trials in the Publication Type field. (The publication type (PT) includes six types of clinical trial articles. Articles whose publication type contains the word trial or multicenter study are included here.)
From all the clinical trials that had multiple articles that satisfy the above constraints, we randomly chose pairs of articles and put them in our positive set, choosing at most 3 pairs of articles from any single trial (this prevents trials having dozens of articles from dominating the dataset). This gave 438 positive pairs. To build the negative set, for each trial in clinicaltrials.gov that had automatically linked publications, we searched clinicaltrials.gov for additional trials that shared the same condition and intervention. Finally, we chose a pair of articles linked to different trials within the set of identified additional trials (but sharing the same condition and intervention). This gave 488 negative pairs.
1.2 Feature selection and scoring
Pair modeling for supervised learning is performed by treating each pair as a positive or negative sample, and the model features are defined as functions on the pair of publication data. A variety of pairwise similarity features were computed from MEDLINE and PubMed metadata and were examined for discriminative power in the positive vs. negative training sets. We began by considering all Medline metadata fields attached to articles, plus a few others that we thought of heuristically (e.g. sponsor names or all-Capitalized words). We then examined the distribution of pairwise similarity feature scores in our positive vs. negative sets [often, multiple ways of encoding the similarity scores were tested for each feature] and kept those features and encoding schemes that appeared to have the best discriminative value. We threw out publication date difference, total number of authors listed on the paper, similarity of words in the abstract, number of shared names in the investigator field, number of shared MeSH terms, shared sponsor names and match on journal name. A total of 12 features were finally employed in the model (Table 1) [8,13–16].
Table 1.
Features employed in the pairwise similarity model.
|
1.3 Machine learning algorithms
MATLAB (2013a release) decision tree library, SVM library, and multinomial logistic regression library were used with default parameters.
1.4 Creating and evaluating the pairwise machine learning model
Positive and negative training sets were based on pairs of clinical trial articles linked to clinicaltrials.gov. After carrying out feature selection and pairwise similarity scoring, three different machine learning algorithms (decision tree, SVM and logistic regression) were tested for their performance in predicting whether pairs belonged to the same trial or not, using 10-fold cross-validation on the training sets. That is, we chose one-tenth of the pairs at random as the test set and trained the model on the remaining 90% of pairs, and did this ten times, to arrive at the average performance. This is a standard way of evaluating machine learning performance, especially when the size of the training sets is not large enough to reserve a large number as a separate test set. All three approaches gave comparable predictive performance, but logistic regression was favored as simple and providing a confidence score between 0 and 1 that served as a directly interpretable estimate of probability.
2. Aggregator: a clustering tool that uses the pairwise prediction model and a hierarchical clustering strategy
2.1. Clustering algorithm
The pairwise model was used to get the pairwise similarity scores and confidence scores between each pair of publications. Articles were then clustered using hierarchical agglomerative clustering with average link similarity. We chose 0.5 as a natural stopping criterion (i.e. when the maximum estimated probability that an article belonged to any existing cluster ≤ 0.5 we stopped merging). Pairwise scores were iteratively corrected for triplet violations as described [7, 8]. However, this had no appreciable effect on performance and was not employed in the final model.
2.2. Calculation of performance indices for evaluating the clustering solution
To examine performance of the clustering tool, 20 PubMed queries (based on well-studied medical conditions) and 22 PubMed queries (based on a specified condition and a specified intervention for which clinical trials have been performed) were formulated. The queries were restricted to articles written in English, after 1987, indexed as randomized controlled trials (Table 2). Each of these queries returned hundreds of PubMed articles, a small subset of which contained NCT numbers. The clustering algorithm was applied on all the returned PubMed articles for each query, but we only used the small set of NCT-containing publications as a tagged gold standard to evaluate the clustering result. We calculated the average split rate of the clustering solution (i.e. how often two tagged articles that belong to the same trial were placed in separate clusters) and the average purity of the solution (i.e. a measure of the proportion of articles in one cluster that belonged together). These evaluations were made for each of the query results, and then averaged.
Table 2.
PubMed queries used for evaluating Aggregator.
| condition queries: | condition/intervention queries: |
|---|---|
| Abdominal Neoplasms | Abdominal Neoplasms and Carboplatin |
| Brain Infarction | Brain Infarction and aspirin |
| Colonic Neoplasms | Colonic Neoplasms and oxaliplatin |
| Parkinson disease | Parkinson disease and IPX066 |
| Arbovirus Infections | Arbovirus Infections and vaccine |
| dental caries | dental caries and fluoride |
| Femoral Fractures | Femoral Fractures and (plate or plates) |
| Corneal Diseases | Corneal Diseases and riboflavin |
| Corneal Diseases and hyaluronate | |
| carcinoma, small-cell lung | carcinoma, non-small-cell lung and paclitaxel |
| carcinoma, non-small-cell lung | |
| Acne Vulgaris | Acne Vulgaris and Retin-A |
| Substance Withdrawal Syndrome | Substance Withdrawal Syndrome and Lofexidine |
| Substance Withdrawal Syndrome and patch | |
| aids-related opportunistic infections | aids-related opportunistic infections and sulfamethoxazole- trimethoprim |
| Biliary Tract Diseases | Biliary Tract Diseases and surgery |
| Acquired Immunodeficiency Syndrome | Acquired Immunodeficiency Syndrome and behavior |
| AIDS-Related Complex | Diabetic Angiopathies[mh] and foot |
| Malaria, Falciparum | obesity[mh] and adolescent |
| collagen diseases | obesity[mh] and bariatric surgery |
| Anemia, Aplastic | bone diseases, endocrine and somatropin |
| Lupus Erythematosus, Systemic | Rheumatoid Arthritis and adalimumab |
| Rheumatoid Arthritis and etanercept |
Twenty common conditions were queried in PubMed, restricted to randomized controlled trials [publication type], for which hundreds (but not thousands) of articles were retrieved. We also queried 22 condition/specific intervention pairs for which numerous randomized controlled trials were retrieved. As far as possible, the condition/intervention pairs were matched to the simple condition queries. Capitalized terms are MeSH headings, but all terms were queried as shown (restricted to MeSH where [mh] is indicated).
For the average split rate, we calculate the split rate of each true cluster (based on tagged NCT-containing articles within the retrieved set), and take the average.
We also calculated the harmonic mean of split rate and purity to give a F1 score: F1 = 2*((1−split)*purity)/((1−split)+purity).
RESULTS
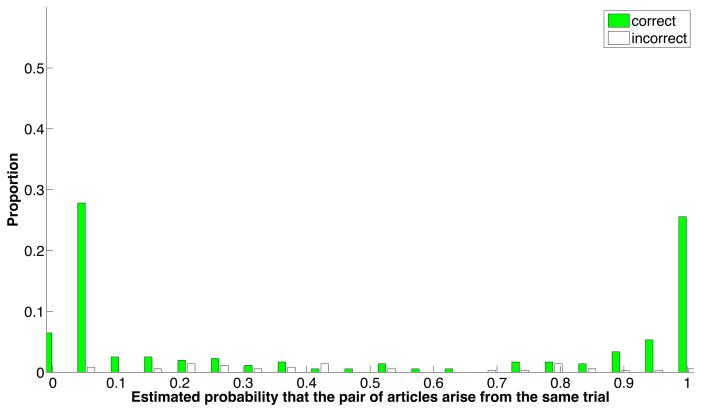
Performance of the pairwise model is shown in Figure 2. On the training dataset, the precision is 0.881, recall = 0.813, accuracy =0.859, and F1 = 0.843. Limiting performance is the fact that not all articles expressed all features used in similarity scoring, and to a lesser extent, that these features were not sufficient to make accurate discriminations in all cases.
Figure 2. Performance of the pairwise similarity model.
The positive and negative training sets were mixed and evaluated pairwise using 10-fold cross-validation. Shown are the proportions of correct and incorrect predictions, as a function of the confidence score for each pair of articles. Most correct predicted probability estimates were very definitive (i.e., less than 0.1 or greater than 0.9). In contrast, the incorrect estimates were scattered between 0 and 1, but particularly below 0.5. This suggests that the biggest limitation to performance is due to features missing from articles, causing some positive pairs to receive low predicted probability estimates.
Several kinds of errors were observed: A) Splitting errors: The model sometimes failed to predict that two articles studying entirely different topics belonged together. For example, PMID 21473976 and 19502645 are linked to the same trial (NCT00006305) but article 21473976 describes primary clinical outcomes whereas 19502645 studies the relationship between race/ethnicity and baseline clinical parameters. B) Lumping errors: The model sometimes put a pair of articles together inappropriately that shared many authors and studied almost the same topic. For example, PMID 20140214 reports a phase I trial of a malarial vaccine whereas article 21916638 is from the phase II trial of the same vaccine (registered under a different number). Such errors are minor, since they would not lead systematic reviewers to view these articles as separate pieces of evidence.
To test the robustness of these findings using a different type of gold standard, and to assess whether additional training data would be likely to improve the model further, we added training data from a new dataset. Specifically, we took five Drug Effectiveness Review Project (DERP) systematic reviews covering diverse topics, in which the reviewers had manually identified cases in which two or more included PubMed citations derived from the same underlying trial. 59 positive pairs of citations from the five queries were added to the original positive training set, and 9 negative pairs chosen randomly were added to the negative set (so that the positive and negative training sets both contained 497 instances). On this larger training set, using 10-fold cross-validation, the precision is 0.877, recall = 0.833, accuracy =0.858, and F1 = 0.854. These parameters are similar to that observed in the original clinicaltrials.gov training set discussed above.
2.2. Evaluation of Aggregator
The pairwise article similarity scores, together with the confidence scores used as a proxy for estimated probability, were used to create a tool that takes as input a set of articles retrieved from PubMed, and aggregates together articles that are predicted to arise from the same underlying trial. The tool was evaluated on articles that are not necessarily linked to clinicaltrials.gov by formulating a total of 42 PubMed queries (20 on a range of specified medical conditions and 22 on a condition plus a specified intervention), restricting queries to articles that are indexed as randomized clinical trials (Table 2). The articles retrieved from each query were then clustered by Aggregator. A subset of these articles which had NCT numbers were used as tagged gold standards.
The most important error is “splitting”, i.e. the proportion of articles that belong to the same trial but are predicted to reside in distinct clusters. This needs to be minimized so that users will not falsely regard different studies as independent. The purity parameter measures “lumping” of articles that arise from distinct studies, but are clustered together; this is less important since it should be easier for systematic reviewers to manually separate articles that are placed together in one cluster than to identify and link related articles that are placed in separate clusters.
As shown in Table 3, the best average performance was seen for the condition+intervention queries (split rate = 0.0%, F1 = 0.91). This was somewhat better than observed for the queries based on condition alone (split rate = 10.5%, F1 = 0.79). This probably reflects the fact that the condition+intervention articles are a more topically homogeneous set, and thus resembled more closely the positive vs. training sets that were used in the model.
Table 3.
Performance of Aggregator.
| Set of Articles to be Clustered | split | purity | F1 |
|---|---|---|---|
| Conditions queries, all retrieved articles | 0.105 | 0.74 | 0.79 |
| Conditions queries, only NCT-containing articles | 0.107 | 0.72 | 0.77 |
| Condition+Intervention query, all retrieved articles | 0.00 | 0.86 | 0.91 |
| Condition+Intervention queries, only NCT-containing articles | 0.0078 | 0.79 | 0.86 |
A series of 20 PubMed queries were carried out on various conditions (see Methods) and 22 queries were carried out on similar conditions plus specific interventions (Table 2). We used Aggregator either to cluster all articles retrieved by these searches, or only clustered the subset of articles that contained NCT numbers. Note that performance was somewhat better when all retrieved articles were clustered than when only the subset of NCT-containing articles was clustered. This indicates that the clustering process, by taking into account multiple interactions among a larger number of articles, improved upon the predictions based on the pairwise similarity model alone.
DISCUSSION
Multiple publications that report clinical outcomes from the same clinical trial can bias the apparent effect size calculated in systematic reviews and meta-analyses [3, 4]. This is a well-recognized problem that is generally handled by time-consuming manual effort [5, 6]. Document clustering is a mature field, and several other studies have clustered PubMed articles according to, e.g., textual and semantic similarity [17–20] or clustered clinicaltrials.gov registry entries by shared clinical trial eligibility criteria [21]. However, to our knowledge, ours is the first study that has investigated the ability of MEDLINE and PubMed metadata to identify articles that derive from the same underlying trial. There is some ambiguity in defining the “same clinical trial” because two different publications might follow the same underlying study design, yet might describe results that were carried out on different or overlapping sets of subjects. One paper might only describe females, while another arising from the same trial might describe elderly patients (regardless of gender) in the same cohort. In our analysis, we defined a trial as one given a unique registry number in Clinicaltrials.gov.
A pairwise similarity model incorporating 12 features had surprisingly good accuracy, of which shared author names, and high similarity in the PubMed Related Articles function [13] were the most important. Some features were highly predictive (e.g. match on all-capitalized words in the abstract) but were present in only a small proportion of articles. A clustering tool, Aggregator, gave superb performance in predicting which topically related RCTs retrieved from PubMed queries belong to the same underlying trial (F1 = 0.91).
Our study had several limitations: The model was only designed to evaluate clinical trial articles that are topically similar and PubMed indexed. It was not specifically designed to detect cases of plagiarism (by different authors), identical publication of the same study in different journals (by the same authors), or situations where trial organizers have deliberately attempted to obscure relationships among their publications, e.g., by using ghostwriters, failing to cite each other, or omitting registry information. Further research is indicated to evaluate these situations as well as more heterogeneous gold standard training sets, e.g., lists of manually disambiguated trial articles in published systematic reviews. Aggregator can achieve its highest performance under field conditions only when the articles to be evaluated are not missing features used in the model (e.g. are not missing abstracts) and when the type of articles is similar to that used for training data (e.g., consist of a set of RCTs carried out on the same condition and intervention). Full text features (e.g., shared sponsor names, blocks of text, acknowledgements, citations, etc.) may be helpful as well. We plan to implement Aggregator as one piece of an overall pipeline of informatics tools that can accelerate the process of writing systematic reviews [22–25].
Highlights.
A single clinical trial can generate numerous published articles
Identifying articles that derive from the same underlying trial is important
A pairwise model predicts when two articles derive from the same underlying trial
Aggregator identifies articles that derive from the same trial in PubMed searches
Acknowledgments
FUNDING
This work was supported by NIH grants R01 LM010817 and P01 AG039347. The study sponsor had no role in study design; in the collection, analysis and interpretation data; in the writing of the report; or in the decision to submit the paper for publication.
Footnotes
COMPETING INTERESTS
The authors declare that they have no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ross JS, Tse T, Zarin DA, Xu H, Zhou L, Krumholz HM. Publication of NIH funded trials registered in ClinicalTrials.gov: cross sectional analysis. BMJ. 2012;344:d7292. doi: 10.1136/bmj.d7292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cook DJ, Mulrow CD, Haynes RB. Systematic Reviews: Synthesis of Best Evidence for Clinical Decisions. Ann Intern Med. 1997;126:376–380. doi: 10.7326/0003-4819-126-5-199703010-00006. [DOI] [PubMed] [Google Scholar]
- 3.Tramèr MR, Reynolds DJ, Moore RA, McQuay HJ. Impact of covert duplicate publication on meta-analysis: a case study. BMJ. 1997;315:635–640. doi: 10.1136/bmj.315.7109.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thornton A, Lee P. Publication bias in meta-analysis: its causes and consequences. J Clin Epidemiol. 2000;53:207–216. doi: 10.1016/s0895-4356(99)00161-4. [DOI] [PubMed] [Google Scholar]
- 5.von Elm E, Poglia G, Walder B, Tramèr MR. Different patterns of duplicate publication: an analysis of articles used in systematic reviews. JAMA. 2004;291:974–980. doi: 10.1001/jama.291.8.974. [DOI] [PubMed] [Google Scholar]
- 6.Wilhelmus KR. Redundant publication of clinical trials on herpetic keratitis. Am J Ophthalmol. 2007;144:222–226. doi: 10.1016/j.ajo.2007.04.047. [DOI] [PubMed] [Google Scholar]
- 7.Torvik VI, Weeber M, Swanson DR, Smalheiser NR. A probabilistic similarity metric for Medline records: A model for author name disambiguation. Journal of the American Society for Information Science and Technology. 2005;56:140–158. [Google Scholar]
- 8.Torvik VI, Smalheiser NR. Author Name Disambiguation in MEDLINE. ACM Trans Knowl Discov Data. 2009;3:3. doi: 10.1145/1552303.1552304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bourgeois FT, Murthy S, Mandl KD. Comparative effectiveness research: an empirical study of trials registered in ClinicalTrials.gov. PLoS One. 2012;7:e28820. doi: 10.1371/journal.pone.0028820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Califf RM, Zarin DA, Kramer JM, Sherman RE, Aberle LH, Tasneem A. Characteristics of clinical trials registered in ClinicalTrials.gov, 2007–2010. JAMA. 2012;307:1838–1847. doi: 10.1001/jama.2012.3424. [DOI] [PubMed] [Google Scholar]
- 11.Roumiantseva D, Carini S, Sim I, Wagner TH. Sponsorship and design characteristics of trials registered in ClinicalTrials.gov. Contemp Clin Trials. 2013;34:348–355. doi: 10.1016/j.cct.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 12.Huser V, Cimino JJ. Precision and negative predictive value of links between ClinicalTrials.gov and PubMed. AMIA Annu Symp Proc. 2012:400–408. [PMC free article] [PubMed] [Google Scholar]
- 13.Lin J, Wilbur WJ. PubMed related articles: a probabilistic topic-based model for content similarity. BMC Bioinformatics. 2007;8:423. doi: 10.1186/1471-2105-8-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. [accessed July 2013];Authority: tools for identifying Medline articles written by a particular author. http://arrowsmith.psych.uic.edu/arrowsmith_uic/author2.html.
- 15.Zhou W, Torvik VI, Smalheiser NR. ADAM: another database of abbreviations in MEDLINE. Bioinformatics. 2006;22:2813–2818. doi: 10.1093/bioinformatics/btl480. [DOI] [PubMed] [Google Scholar]
- 16. [accessed July 2013];ADAM: Another Database of Abbreviations in MEDLINE. doi: 10.1093/bioinformatics/btl480. http://arrowsmith.psych.uic.edu/arrowsmith_uic/adam.html. [DOI] [PubMed]
- 17.Boyack KW, Newman D, Duhon RJ, Klavans R, Patek M, Biberstine JR, Schijvenaars B, Skupin A, Ma N, Börner K. Clustering more than two million biomedical publications: comparing the accuracies of nine text-based similarity approaches. PLoS One. 2011;6:e18029. doi: 10.1371/journal.pone.0018029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gu J, Feng W, Zeng J, Mamitsuka H, Zhu S. Efficient Semisupervised MEDLINE Document Clustering With MeSH-Semantic and Global-Content Constraints. IEEE Transactions On Cybernetics. 2013;43:1265–1276. doi: 10.1109/TSMCB.2012.2227998. [DOI] [PubMed] [Google Scholar]
- 19.Zhang X, Jing L, Hu X, Ng M, Xia J, Zhou X. Medical Document Clustering Using Ontology-Based Term Similarity Measures. International Journal of Data Warehousing and Mining (IJDWM) 2010;4:62–73. [Google Scholar]
- 20.Lin Y, Li W, Chen K, Liu Y. A document clustering and ranking system for exploring MEDLINE citations. J Am Med Inform Assoc. 2007;14:651–661. doi: 10.1197/jamia.M2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hao T, Rusanov A, Boland MR, Weng C. Clustering clinical trials with similar eligibility criteria features. J Biomed Informat. doi: 10.1016/j.jbi.2014.01.009. Available online 1 February 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sampson M, Shojania KG, Garritty C, Horsley T, Ocampo M, Moher D. Systematic reviews can be produced and published faster. J Clin Epidemiol. 2008;61:531–536. doi: 10.1016/j.jclinepi.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 23.Cohen AM, Adam CE, Davis JM, et al. Evidence-based medicine, the essential role of systematic reviews, and the need for automated text mining tools. Proceedings of the 1st ACM International Health Informatics Symposium. 2010:376–380. doi: 10.1145/1882992.1883046. [DOI] [Google Scholar]
- 24.Smalheiser NR, Lin C, Jia L, Jiang Y, Cohen AM, Yu C, Davis JM, Adams CE, McDonagh MS, Meng W. Design and implementation of Metta, a metasearch engine for biomedical literature intended for systematic reviewers. Health Information Science and Systems. 2014;2:1. doi: 10.1186/2047-2501-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cohen AM, Smalheiser NR, McDonagh MS, Yu C, Adams CE, Davis JM, Yu PS. Automated confidence ranked classification of randomized controlled trial articles: An aid to evidence-based medicine. JAMIA. 2014 doi: 10.1093/jamia/ocu025. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]