Summary
In the yeast Saccharomyces cerevisiae, the regulation of cell types by homeodomain transcription factors is a key paradigm; however, many questions remain regarding this class of developmental regulators in other fungi. In the human fungal pathogen Cryptococcus neoformans, the homeodomain transcription factors Sxi1α and Sxi2a are required for sexual development that produces infectious spores, but the molecular mechanisms by which they drive this process are unknown. To better understand homeodomain control of fungal development, we determined the targets of the Sxi2a-Sxi1α heterodimer using whole genome expression analyses paired with in silico and in vitro binding site identification methods. We identified Sxi-regulated genes that contained a site bound directly by the Sxi proteins that is required for full regulation in vivo. Among the targets of the Sxi2a-Sxi1α complex were many genes known to be involved in sexual reproduction, as well as several well-studied virulence genes. Our findings suggest that genes involved in sexual development are also important in mammalian disease. Our work advances the understanding of how homeodomain transcription factors control complex developmental events and suggests an intimate link between fungal development and virulence.
Keywords: Sexual development, fungal pathogenesis, transcriptional regulatory network, protein-DNA interactions, homeodomain transcription factors
Introduction
Transcriptional networks consisting of key regulators and their downstream targets control developmental processes in eukaryotes as diverse as embryogenesis in vertebrates, segmentation in fruit flies, and the specification of new cell types in yeast (Johnson, 1995; Schroeder et al., 2004; Sauka-Spengler and Bronner-Fraser, 2008). Fungi have proven to be very useful models for investigating the control of eukaryotic development, and one informative transcriptional circuit is that regulating cell type specification and sexual development. In the model yeast Saccharomyces cerevisiae, when cells of opposite mating types (a and α) fuse with one another, the mating type-specific homeodomain transcription factors a1 and α2 form a heterodimeric complex that binds to the promoters of haploid-specific genes through two individual half-sites and represses transcription (Galgoczy et al., 2004). This repression leads to the specification of the diploid state, rendering cells incapable of further mating and imbuing them with the capacity to undergo meiosis and sporulation. The paradigm of specifying cell type to establish developmental control is utilized across phylogenetically diverse fungi and frequently involves the heterodimerization of transcription factors similar to a1 and α2 (Schulz et al., 1990; Kües et al., 1994; Tsong et al., 2003).
Homeodomain proteins have been found in many eukaryotes, including fungi and humans, and are key nodes in transcriptional networks controlling developmental pathways (Gehring, 1993). The homeodomain itself consists of a 60 amino acid sequence that folds into a three helix bundle, with the third helix containing the DNA-contacting residues (Li et al., 1995). Many homeodomain proteins bind DNA in concert with other transcription factors that aid in the refinement of binding sites or facilitate nuclear localization (Spit et al., 1998).
In the human fungal pathogen Cryptococcus neoformans the homeodomain proteins Sxi2a and Sxi1α (Sex Inducer 2a and Sex Inducer 1α) control sexual development between a and α cells. Sxi2a (encoded by a cells) and Sxi1α (encoded by α cells) are both necessary and sufficient for inducing filamentation, a key step in sexual development, and previous studies have shown that they interact and are able to bind DNA in vitro (Hull et al., 2005; Stanton et al., 2009). However, relatively little is known about the in vivo molecular mechanisms by which they control gene expression during sexual development.
Sexual development of C. neoformans is of particular interest because the spores that result from this process cause disease in mice and are likely infectious particles in human disease (Giles et al., 2009; Velagapudi et al., 2009). C. neoformans is a global environmental pathogen that causes over a million cases of disease each year, primarily in those with compromised immune systems, and is a major cause of death for persons suffering from AIDS. In Sub-Saharan Africa it is responsible for the deaths of more than 600,000 HIV-infected individuals annually (Benjamin J Park et al., 2009). In this part of the world, genetic studies of the C. neoformans population indicate the presence of sexual recombination, and both mating types (a and α) have been isolated from clinical samples (Litvintseva et al., 2003). The Sxi proteins are key in governing spore formation during this form of sexual development. Thus, determining the precise genetic mechanisms employed by Sxi2a and Sxi1α to regulate development would greatly aid in our understanding not only of general developmental control mechanisms in eukaryotes but also of a specific process that results in the production of infectious particles (Giles et al., 2009; Velagapudi et al., 2009).
To determine how Sxi2a and Sxi1α control sexual development, we carried out a multi-pronged investigation involving in silico, in vitro, and in vivo approaches. We first identified DNA binding sites for Sxi2a and Sxi1α using an in vitro protein-binding array. We then identified likely direct targets of Sxi2a and Sxi1α in vivo using whole genome expression analyses in concert with an unbiased bioinformatic approach. We discovered that the Sxi proteins regulate the expression of over 375 genes, 50 of which contain a bipartite, conserved sequence in their predicted promoter regions. Strikingly, this sequence contained iterations of the individual Sxi2a and Sxi1α binding sites defined by the in vitro protein-binding array. This newfound binding site is responsible for the activation of genes in a Sxi-dependent manner in a 1-hybrid assay and in C. neoformans, indicating that these targets constitute at least a portion of the direct regulon for Sxi2a and Sxi1α. Furthermore, several of these direct targets of the Sxi proteins have been characterized previously based on their importance in processes required for virulence, such as capsule formation and melanin production (Zhu and Williamson, 2004; Panepinto et al., 2005). Our identification of the direct targets of Sxi2a-Sxi1α reveals a molecular intersection between development and virulence and supports the hypothesis that human pathogens can adapt the expression of common targets to accommodate disparate conditions in both the environment and the mammalian host.
Results
The homeodomain proteins Sxi2a and Sxi1α bind unique DNA sequences in vitro
To determine DNA binding sequences for both full-length Sxi proteins, we carried out a comprehensive in vitro protein-binding array. In this Cognate Site Identifier (CSI) analysis, the full-length Sxi proteins were produced using in vitro transcription/translation reactions in a wheat germ extract, labeled fluorescently, and bound either alone or together to arrays harboring all possible double-stranded 10-mer DNA sequences. The resulting binding profiles were normalized and ranked according to fluorescence intensity as a measure of relative binding affinity to determine all possible sequences to which the Sxi proteins could bind. We analyzed the top 1000 sequences (ranked from highest to lowest affinity) using the motif finding algorithm Multiple Em for Motif Elicitation (MEME).
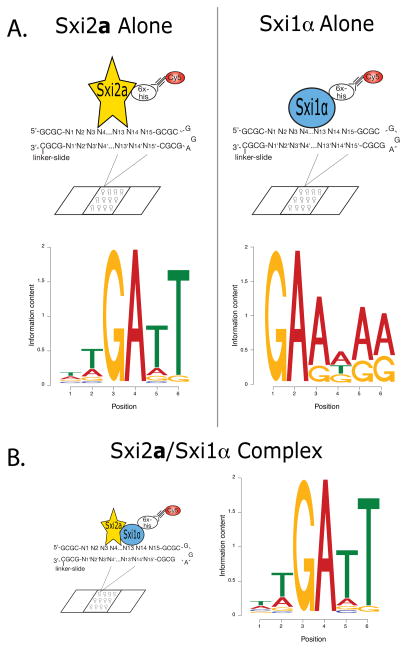
We discovered that the full length Sxi2a protein alone bound preferentially to a conserved 5′-TGATT-3′ sequence similar to a previously-described binding site for the homeodomain-only fragment of Sxi2a (5′-GATTG-3′) (Figure 1A) (Stanton et al., 2009). The Sxi1α protein alone bound a conserved 5′-GAA-3′ element (Figure 1B). When both proteins were incubated on the array at the same time, they bound sequences nearly identical to those bound by Sxi2a alone. This result was particularly informative because in this experiment we probed the array with versions of full-length Sxi proteins in which Sxi1α was fluorescently labeled and Sxi2a was not (Figure 1B). The simplest explanation for these data is that a complex of Sxi2a and Sxi1α bound DNA predominantly via the Sxi2a homeodomain.
Figure 1. In vitro binding site determination for Sxi2a and Sxi1α.
A. Individual in vitro binding site identification. Full-length Sxi2a (yellow star) and Sxi1α (blue oval) were labeled individually with Cy5 (red circle) and incubated separately with double-stranded 15-mer oligonucleotides spotted on glass slides (CSI arrays). Motifs shown represent the sequences with the highest binding affinities for each protein. B. Heterodimer in vitro binding site identification. Sxi1α was labeled with Cy5, and Sxi2a and Sxi1α were incubated together with the oligonucleotide array. The motif shown represents sequences to which the proteins bound with high relative affinity. In all motifs the height of each individual letter is representative of the conservation of that nucleotide.
While these data suggest strongly that Sxi2a and Sxi1α bind DNA as a heterodimer with each protein harboring the capacity to bind an individual consensus sequence, we could not determine the full size or orientation of a heterodimeric binding site in this experiment. Because the unique DNA sequences represented on the array were only 10 base pairs in length, we could not rule out that longer sequences could host a heterodimer consensus binding sequence containing each of the individual protein binding sites. Furthermore, efforts to use the in vitro binding sequences to identify high likelihood Sxi2a-Sxi1α heterodimer binding sites in the C. neoformans genome using the Regulatory Sequence Analysis Tools suite of programs (Turatsinze et al., 2008) were not fruitful (data not shown). While our CSI analyses resulted in the first description of in vitro binding sites for full-length Sxi2a and Sxi1α, in the absence of additional information, we were unable to determine the potential relevance of thousands of possible binding sites in the C. neoformans genome.
The Sxi proteins induce gene expression through a specific, bipartite DNA sequence
To identify sequences to which Sxi2a and Sxi1α likely bind to regulate genes in vivo, we first carried out whole-genome expression experiments to identify Sxi-regulated genes. We hypothesized that a subset of genes whose transcript levels changed in the presence or absence of the Sxi proteins would harbor binding sites for the Sxi2a-Sxi1α complex in their upstream regulatory regions. We carried out two independent whole-genome expression experiments comparing transcript levels in cells that either possessed or were lacking the Sxi proteins (Figure 2A). In the first experiment (Sxi +) we compared the expression profile of a haploid sxi1αΔ strain to that of a haploid strain expressing both SXI1α (under control of the GPD1 promoter) and an inducible copy of SXI2a (under control of the GAL7 promoter). When this strain was grown in galactose-containing liquid medium (YPGal), full sexual development was observed, including filamentation, basidia formation, and sporulation (Figure S1B). In the second experiment (Sxi −), we compared the expression profile of a wild type cross (JEC20 x JEC21) to the expression profile of a cross whose mating partners did not possess either of the transcription factors (sxi2aΔ x sxi1α Δ).
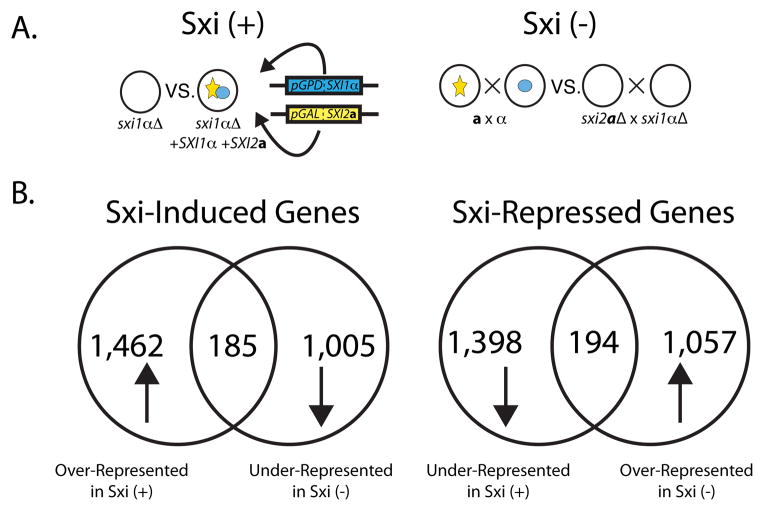
Figure 2. Sxi proteins regulate the transcript levels of over 350 genes.
A. Schematic representation of two independent Sxi regulation experiments. In the Sxi (+) experiment (left), SXI2a and SXI1α were expressed under the control of the GAL7 and GPD1 promoters, respectively, in a sxi1αΔ strain. Transcripts from this strain were compared with the sxi1αΔ strain expressing no Sxi proteins. In the Sxi (−) experiment (right), transcript abundance was compared between a wild type cross (a x α) and a cross between Sxi deletion strains (sxi2aΔ x sxi1αΔ). Sxi1α is represented in blue, and Sxi2a is in yellow. B. Classes of Sxi-regulated genes. Transcripts of Sxi-Induced genes were over-represented in the Sxi (+) experiment and under-represented in the Sxi (−) strain. Sxi-Repressed gene transcripts were under-represented in the Sxi (+) experiment and over-represented in the Sxi (−) experiment. Venn diagrams represent the total and the overlap in the number of genes from each experiment that displayed the indicated pattern of expression.
In both experiments RNA from each condition was harvested at early time points (Figure S1A) to facilitate the discovery of high-likelihood direct targets, labeled, and hybridized competitively to an oligonucleotide microarray representing the approximately 6,500 genes in the C. neoformans genome. Two biological replicates were carried out for both Sxi (+) and Sxi (−) experiments with each biological replicate consisting of 4 technical replicates for a total of 16 replicates between the two experiments (Tables SI and SII). From these data we defined two cohorts of regulated genes; Sxi-Induced and Sxi-Repressed. Sxi-Induced genes were defined as those whose transcript levels were overrepresented in the presence of Sxi2a and Sxi1α and underrepresented in their absence. Sxi-Repressed genes exhibited the opposite pattern (Figure 2B). We identified 185 Sxi-Induced genes and 194 Sxi-Repressed genes in total between both experiments (Tables SIII and SIV). The only previously described Sxi-regulated genes (CLP1 and CPR2) (Ekena et al., 2008; Hsueh et al., 2009) were both present in the Sxi-Induced gene cohort and were regulated at levels similar to those previously reported, indicating that changes in transcripts levels in the two experiments reflected biologically relevant changes in response to expression of the Sxi proteins. Using two independent expression datasets allowed us to define Sxi-regulated cohorts that constituted high likelihood target genes in a very stringent manner that would have not been possible with either dataset alone.
To determine whether any of these Sxi-regulated genes contained conserved DNA sequences in their predicted promoter regions that could act as binding sites for the Sxi proteins, we analyzed 1 kilobase of sequence upstream of the start codon of each of the Sxi-Induced and Sxi-Repressed genes using the motif finding algorithm MEME. The algorithm identified a single, conserved 21 bp motif upstream of a small group of Sxi-Induced genes (Figure 3). To probe the significance of this motif, we used the sequence identification program Motif Alignment & Search Tool (MAST) to search for the motif in upstream regions of genes both in the Sxi-Induced group and in several groups of 185 randomly selected genes from the genome. Highly conserved occurrences of the motif were identified upstream of Sxi-Induced genes; however, among groups of genes randomly selected from the genome, only poorly conserved occurrences were identified (data not shown). These findings indicate that the motif resides disproportionately in the predicted regulatory regions of Sxi-Induced genes.
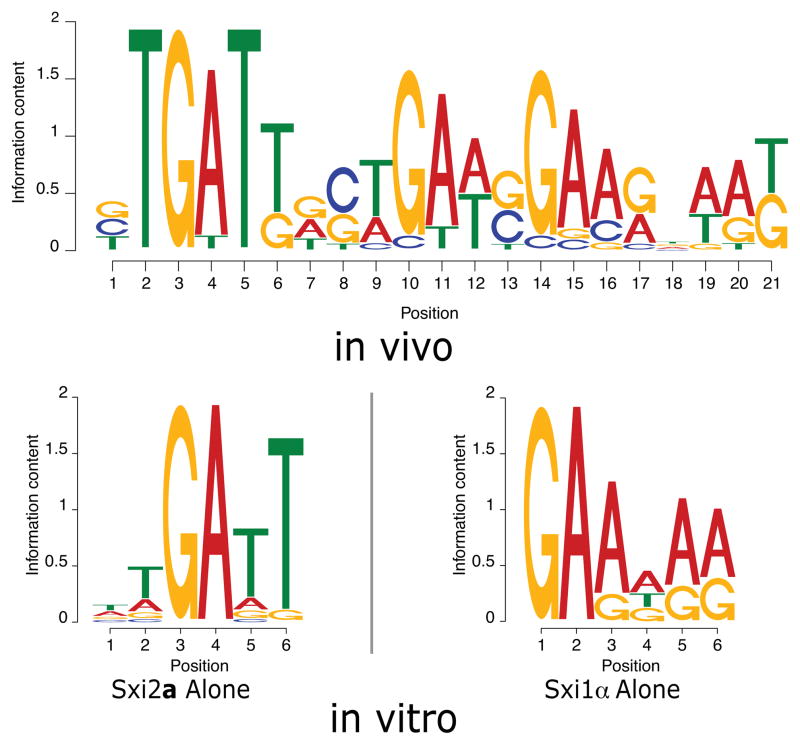
Figure 3. A conserved, bipartite sequence resides upstream of Sxi-Induced genes.
MEME analysis of upstream regions of regulated genes produced a conserved motif (top, in vivo). The individual in vitro binding sites for Sxi2a and Sxi1α are shown below (bottom, in vitro). In all motifs the height of each individual letter is representative of the conservation of that nucleotide.
Remarkably, the conserved Sxi-Induced gene motif contains the 5′-TGATT-3′ sequence bound by both Sxi2a and the Sxi2a-Sxi1α complex in vitro and the 5′-GAA-3′ sequence bound by Sxi1α in vitro (5′-GTGATTGCTGAAGGAAGGAAG-3′) (Figure 3). The discovery of sequences in this motif identical to those bound by the Sxi proteins in vitro was particularly striking because we identified the motif using an unbiased approach based solely on expression data and sequence analysis. Thus, two distinct approaches (in vitro and in vivo) converged on precisely the same sequences, providing confidence that the sequences identified are biologically relevant. Furthermore, the size of the motif (21bp) is consistent with known heterodimer binding sites in other eukaryotic systems (Brazas et al., 1995; Bradford et al., 1997; Galgoczy et al., 2004). Consistent with our hypothesis that at least some Sxi-regulated genes would harbor motifs to which the Sxi proteins could bind directly, we identified iterations of the motif upstream of both CLP1 (5′-CTGATTGCGCATTGACGGATG-3′) and CPR2 (5′-TTGATTGTTGATGGGCAAAAG-3′).
No Sxi-specific motifs were identified by MEME analysis of the Sxi-Repressed cohort of genes. While occurrences similar to the Sxi-Induced motif were located upstream of some Sxi-Repressed genes, almost all exhibited low sequence conservation. This contrasted with the occurrences from the Sxi-Induced genes that exhibited very high sequence conservation and were highly unlikely to have arisen by chance. The presence of the conserved motif in primarily induced genes suggests strongly that the Sxi2a-Sxi1α complex mediates transcriptional activation.
Sxi2a and Sxi1α work in concert to regulate transcription in a 1-hybrid assay
To evaluate the ability of Sxi2a and Sxi1α to bind specific occurrences of the Sxi-Induced motif, we assessed activation of a reporter gene using a yeast 1-hybrid assay. In this experiment, constructs expressing Sxi2a and/or Sxi1α fused with the S. cerevisiae Gal4 activation domain were transformed into S. cerevisiae along with a plasmid containing a pCYC1-lacZ reporter gene (Figure 4A). Three copies of the sites of interest from CLP1, CPR2, and two other Sxi-Induced genes (CNJ03000 and CNH01950) were each cloned into the CYC1 promoter. Transformed strains were evaluated for β-galactosidase (β-gal) activity. We found that strains containing the Sxi-Induced motif sequences showed a significant relative increase in β-gal activity only in the presence of Sxi2a and Sxi1α, implying a direct interaction between the Sxi proteins and the test sites. An increase in β-gal activity was not observed when a random 75 base pair sequence was cloned into the reporter, showing that binding is sequence-dependent (Figure 4A). Likewise, when only one of the transcription factors (either Sxi2a or Sxi1α) was transformed into the S. cerevisiae strain, no change in β-gal activity was observed, showing that both Sxi2a and Sxi1α are required for binding and activation (Figure 4B).
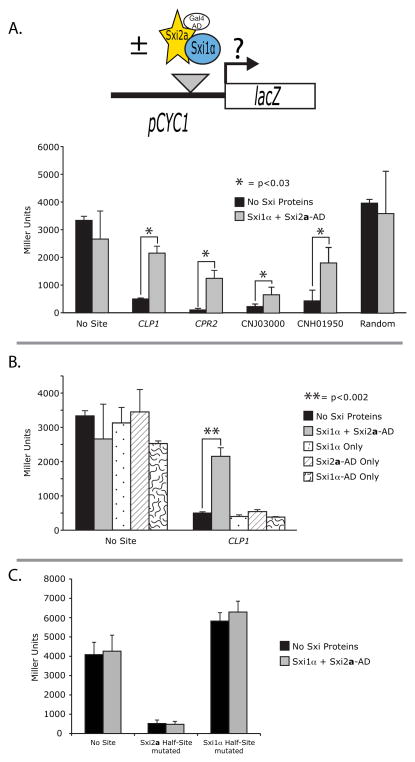
Figure 4. Individual occurrences of the Sxi-Induced motif were bound in a yeast 1-hybrid assay.
A. Reporter activation is dependent on Sxi-Induced sites. Top: Schematic of 1-hybrid binding experiment. Individual iterations of the Sxi-Induced motif were each cloned into a 1-hybrid reporter construct containing the S. cerevisiae CYC1 promoter driving lacZ expression. Reporter expression levels were assessed in S. cerevisiae, both in the absence and presence of the Sxi proteins with Sxi2a harboring the Gal4 Activation Domain (AD). Bottom: Reporter gene expression as a measure of β-galactosidase activity is shown for each construct tested: reporter with no site, three repeats of the CLP1, CPR2, CNJ03000, CNH01950 Sxi-Induced sites, or random polylinker sequence. Activity for each was assessed in the absence (black bars) or presence (gray bars) of Sxi1α and a Sxi2a-AD fusion protein. Stars represent statistical significance with p<0.03. B. Reporter activation is dependent on the presence of both Sxi2a and Sxi1α. Reporter constructs with no site or the CLP1 site were assessed for β-galactosidase activity in the presence of no Sxi proteins (black), both proteins as in A (gray), Sxi1α alone (dotted), Sxi2a-AD fusion alone (hatches), or Sxi1α-AD fusion alone (squiggles). Double stars represent statistical significance with p<0.002. C. Both half-sites are required for Sxi2a-Sxi1α Binding. Reporters were constructed containing mutant version of the CLP1 site in which the first half (Sxi2a half-site) or last half (Sxi1α half-site) of the sequence was mutated by converting purine bases to pyrimidines and vice versa. In the presence of either mutant half-site, there was no activation of the reporter construct by the Sxi proteins. In all graphs, activity is shown in Miller Units, and each assay was carried out in triplicate.
To test the contributions of each predicted half-site on binding and activation, we evaluated reporter constructs in which half of the binding site sequence containing either the Sxi2a or Sxi1α CSI binding sites were mutated. In the CLP1 sequence, we converted the first 10 or last 11 nucleotides from purines to pyrimidines and vice versa. We discovered that eliminating the Sxi-binding sequences from either portion of the sequence resulted in a complete loss of Sxi-dependent regulation (Figure 4C). When only the Sxi1α binding region of the CLP1 occurrence was mutated, or only the Sxi2a binding region of the CLP1 occurrence was mutated, β-gal levels remained unchanged in the presence and absence of the Sxi proteins, indicating that both half-sites are required for binding and activation in a 1-hybrid assay.
In our 1-hybrid assays we also observed changes in expression in the presence of the Sxi binding sequences independent of the Sxi proteins, suggesting that an endogenous protein of S. cerevisiae was mediating transcriptional repression through the Sxi1α half-site (Figure 4 A, B, C). We considered the possibility that the mating type-specific homeodomain proteins of S. cerevisiae could be interacting with the Sxi binding site; however, the S. cerevisiae reporter strain used was of the a mating type and does not contain α2, a1 does not bind DNA well on its own, and there are no sequences in the Sxi1α half-site resembling binding sites for homeodomain (or any other) transcription factors. It is also clear from the half-site analysis that the Sxi1α portion of the binding site mediates repression by the endogenous S. cerevisiae protein. Using the TOMTOM algorithm in the MEME suite, we attempted to identify candidate repressors, but no proteins in S. cerevisiae are obvious candidates for binding sequences in the occurrences tested. Regardless of the identity of the endogenous repressor, in the presence of Sxi2a and Sxi1α, we observed Sxi-dependent transcriptional activation, dependent on both half-sites in multiple 1-hybrid assays (Figure 4). These results indicate that both Sxi2a and Sxi1α make direct DNA contacts to fully bind and activate the reporter. We posit that Sxi2a and Sxi1α are physically interacting with one another and binding DNA in a sequence-specific manner to drive transcription through specific occurrences of the Sxi-Induced motif.
Sxi2a and Sxi1α mediate transcriptional activation in vivo through the Sxi Binding Site
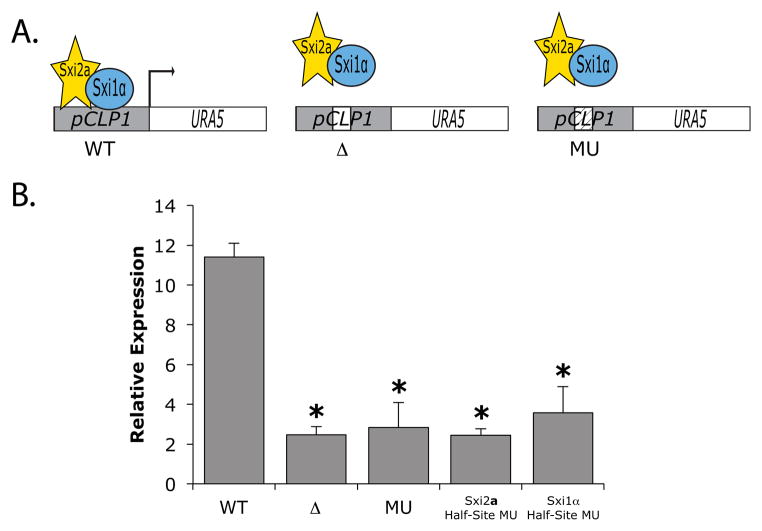
To determine whether a single occurrence of a Sxi-bound site was responsible for mediating regulation by Sxi2a and Sxi1α in vivo in C. neoformans, we evaluated the expression of a reporter plasmid under the control of the predicted promoter region of a Sxi-Induced gene. A plasmid harboring the URA5 open reading frame under the control of the predicted promoter of CLP1 was transformed into a C. neoformans strain harboring both a galactose-inducible SXI2a and a constitutively expressed SXI1α. URA5 gene expression was compared among three reporters that contained the native 21bp Sxi-Induced binding sequence (WT), a deletion of the 21bp sequence (Δ), or a 21bp sequence in which the native site was mutated by converting the pyrimidine nucleotides to purines and vice versa (MU) (Figure 5A). In the absence of the Sxi proteins, there was no statistically significant difference in URA5 transcript levels among the three reporter strains (data not shown). However, in galactose-inducing conditions, transcript levels were significantly higher in the wild-type reporter construct than in constructs harboring the binding site deletion (Δ) or the mutated site (MU) (Figure 5B). These data show that the Sxi binding site in the CLP1 promoter is in fact responsible for mediating Sxi-dependent activation in vivo and confirm a biological role for the newly identified Sxi Binding Site (SBS) in Sxi-dependent regulation of C. neoformans.
Figure 5. The Sxi-Induced site mediates expression of a target gene in vivo during development.
A. Schematic of three versions of an endogenous reporter plasmid. The CLP1 predicted promoter harboring its 21 bp Sxi-Induced binding site (left - WT), no binding site (middle - Δ), or a mutated site (right MU) were cloned upstream of the URA5 open reading frame and transformed into C. neoformans cells harboring inducible Sxi2a (yellow star) and Sxi1α (blue oval). B. Strains were evaluated for reporter gene transcript levels in the presence of Sxi2a and Sxi1α. Mutated sites consisted of purines converted to pyrimidines and vice versa for all nucleotides in the 21 bp site (MU), the first 10 bp of the site (Sxi2a Half-Site MU), and the last 11 bp of the site (Sxi1α Half-Site MU). URA5 expression was normalized to GPD1, and asterisks indicate p<0.003 for Δ and MU compared to the WT in the presence of the Sxi proteins.
To test the contribution of both half-sites to the overall levels of regulation conferred by the heterodimer of Sxi2a and Sxi1α in C. neoformans, we mutated the half-sites of the SBS in the in vivo reporter assay by again changing purines to pyrimidines and vice versa. We observed that both half-sites are required for full Sxi-mediated regulation in vivo, indicating that both proteins must interact with specific DNA sequences to activate transcription (Figure 5B).
Targets of Sxi2a and Sxi1α are associated with virulence
To identify the cohort of promoters to which the Sxi proteins bind directly in vivo, we attempted to carry out chromatin immunoprecipitations with antibodies against Sxi2a and Sxi1α. After numerous attempts using a diverse array of strains, antibodies, extracts, and approaches, satisfactory results were not obtained (data not shown), and this led us to take a bioinformatic approach to identify high likelihood direct targets of Sxi2a-Sxi1α. We used the sequence identification program MAST to probe the promoters of all 379 Sxi-Regulated genes for iterations of the SBS. We identified 32 and 18 genes in the Sxi-Induced and Sxi-Repressed groups, respectively, that contained at least one SBS in the 1 kb region upstream of the open reading frame (Table I and Table SV).
Table I.
Sxi Binding Sites Upstream of 32 Sxi-Induced Genes Ranked by Decreasing fold change
| Gene # | Sxi Binding Site (SBS) sequence | bp from ATG | strand +/− | SBS type | Gene Number | Gene Name/Description | Gene Group |
|---|---|---|---|---|---|---|---|
| 1 | TTGATTGTTGATGGGCAAAAG | −242 | + | I | CNB04250 | CPR2/non-MAT pheromone receptor | 1 |
| GTCTTGTGTGAAGCAGATTGG | −578 | − | III | ||||
| 2 | GTGATGGGAGATAGGGCAAGG | −530 | + | I | CNB01320 | hypothetical protein | 3 |
| 3 | CTGATTGCTGATCGACGGTAT | −641 | − | I | CNJ03000 | hypothetical protein | 3 |
| GTGATGACAGATGGCAGAAGT | −388 | + | I | ||||
| 4 | CTGATTGCGCATTGACGGATG | −331 | + | I | CNB01190 | CLP1/dikaryon regulator | 1 |
| 5 | CTGATTACTGAAGCAAACAAG | −171 | − | I | CNI00810 | hypothetical protein | 3 |
| 6 | GTGATTTCTGAACGAAGTTAT | −128 | − | I | CNH01950 | hypothetical protein | 3 |
| CTGATTGGTGAACGAAGATGT | −97 | − | I | ||||
| 7 | GTGATGTGACATGGCCGTAGG | −790 | + | I | CNF00230 | hypothetical protein | 3 |
| 8 | GTGATGACTGATGGAACGATG | −381 | − | I | CNL06750 | hypothetical protein | 3 |
| 9 | GTGATTTCTGAACGAAGTTAT | −331 | + | I | CNH01960 | hypothetical protein | 3 |
| CTGATTGGTGAACGAAGATGT | −362 | + | I | ||||
| 10 | CTGATTGCAGTACGACATAAG | −872 | − | I | CNG01240 | LAC1/diphenol oxidase | 4 |
| 11 | GTGTTGAGTGATGGAAGTGGT | −114 | − | II | CNJ02870 | hypothetical protein | 3 |
| 12 | TTGATTGCCCATTGAAGGAAG | −126 | − | I | CNI04000 | E3 ubiquitin-protein ligase | 2 |
| GTCATTTTACAACGAACAAGG | −323 | + | III | ||||
| 13 | GTGATTTCCGCTTGAACGAAT | −78 | − | I | CNA07770 | SPR1/exo-1,3-betaglucanase | 2 |
| CTGTTGGGAGAAAGGCGCGAT | −252 | − | II | ||||
| GTGTTGTCCGTAGGGAGCTAT | −766 | − | II | ||||
| 14 | TTGATTAGAGTTGGAGAGAAG | −518 | + | I | CNB04840 | COX11/cytochrome oxidase factor | 2 |
| 15 | TTGATGTGAGAAGGAAACAAG | −463 | − | I | CNB03490 | MED1/development regulator | 2 |
| 16 | GCGATCGTTGATGGGAGAAAT | −466 | + | III | CNF04150 | RPS2/ribosome subunit S2 | 2 |
| GTGATTTGCCAAGCGGGGTAT | −703 | + | I | ||||
| 17 | CTGATGAGAATATCAAGGAGG | −886 | − | III | CNE01390 | DED81/asparaginyl-tRNA synthetase | 2 |
| 18 | TGGATGGTAGATGGAGAGAAG | −34 | − | I | CNA00510 | CIT1/citrate synthase | 2 |
| 19 | TTGTTGGCTGTTTGGTGGAAG | −576 | − | II | CNA01120 | IMA5/alpha-glucosidase | 2 |
| 20 | CTGATTGGTATTCCAAGGGTG | −610 | − | I | CNI00410 | MPK1/MAP kinase | 2 |
| 21 | CTGATGGCAGAAGCAGCGTAG | −777 | − | I | CNJ02220 | ANT1/adenine nucleotide transporter | 2 |
| 22 | GAGATTTCTGTTGCAGAAAAG | −154 | − | III | CNJ00490 | AGO2/argonaute protein | 1 |
| 23 | TTGATGGCGGTACGTAGCAAT | −203 | − | I | CNC06460 | VAD1/DEAD box protein | 1,4 |
| GTGATTGGAGATCCAAAGGAA | −778 | + | I | ||||
| 24 | TAGATTTGTGATTGAGGGATT | −22 | − | III | CNA01190 | oxidoreductase | 2 |
| 25 | GTGATTGGCCCTCGGCGCAAG | −642 | − | I | CNJ00690 | DAL4/allantoin permease | 2 |
| 26 | GTGATTGGTGTACGAAAATGA | −344 | + | I | CNC01910 | DUR1/urea amidolyase | 2 |
| 27 | TTGAGGGCAGTTGGAGGAAAT | −693 | − | III | CNK02120 | CTR2/copper transporter | 2 |
| 28 | GTGCTGGCTGATGCGACTAAG | −144 | − | III | CNN01140 | CDC53/E3 ubiquitin ligase subunit | 2 |
| TTGTTGAGGCTTTGAAGGAGT | −886 | − | II | ||||
| 29 | TTCTTTTGAGAAGCCACAAGG | −168 | − | III | CNA05290 | SCC2/cohesin loading factor | 2 |
| TTGTTGATACATCGAAGGTGG | −904 | + | II | ||||
| 30 | CTGTTGTCTGATCCGCATCGG | −675 | − | II | CNA02070 | hypothetical protein | 3 |
| GGGATGGCTGATGGGAGAATG | −864 | + | III | ||||
| 31 | TGGATTATTGAACGCAAGTTG | −471 | − | III | CNF01940 | SSM4/E3 ubiquitin ligase | 2 |
| 32 | TTGTTGTGAGAACGATGGGAT | −743 | − | II | CNE03890 | CCP1/cytochrome c peroxidase | 2 |
Genes are listed in order of decreasing fold change ranging from 20-fold (#1) to less than 2-fold (#32) induction. Bolded sequences refer to conserved in vitro binding sequences for Sxi2a (TGAT) and Sxi1α (AAG or GAAG). SBS types are defined by the sequences of their Sxi2a half-sites as follows: I = TGAT, II = TGTT, III = all other half-sites. Gene groups: 1 = Sexual Development, 2 = Conserved Domains, 3 = Hypothetical Proteins, 4 = Virulence
Detailed analysis of the resulting SBSs revealed three classes of binding sites (I, II, or III) based on sequence composition of the Sxi2a half-site: Class I contains sites with 5′-TGAT-3′, Class II contains sites with 5′-TGTT-3′, and Class III contains the remaining sites. Significantly, binding sites harboring the Sxi2a binding sequence 5′-TGAT-3′ (Class I) correlated with the highest levels of activation in the gene expression analysis as the 10 highest Sxi-induced genes all contained at least one Class I binding site and over 50% of all Class I sites were in this top 1/3 of the regulated genes (Table I). In contrast, there were no correlations between the binding site sequences in repressed genes and levels of repression as the list 10 most Sxi-repressed genes only contained two Class I binding sites (Table SV). This further supports our hypothesis of a role for Sxi2a-Sxi1α in transcriptional activation. High activation sites were best represented by sequences containing a Sxi2a core binding sequence (5′-TGAT-3′), a variable spacer region of approximately 8 base pairs (ranging from 4 to 12 bp), and a Sxi1α core binding sequence (5′-GAAG-3′) (Table I).
The 32 Sxi-Induced genes harboring Sxi-binding sites fall into several gene groups based on protein sequence: 1) genes known to be involved in sexual development, 2) genes with predicted, conserved functions, 3) genes of unknown function, and 4) genes known to be involved in virulence. Genes previously implicated in sexual development include CPR2, CLP1, AGO2, and VAD1 (Ekena et al., 2008; Hsueh et al., 2009; Y-D Park et al., 2010; Xuying Wang et al., 2010). They all show phenotypes in the sexual development process, and CPR2 and CLP1 have been implicated as Sxi targets previously. Eighteen Sxi-Induced genes with SBSs have either clear homologs in S. cerevisiae or conserved protein domains. These fall roughly into categories of genes involved in protein degradation, carbohydrate catabolism, and general metabolism. Nine genes encode proteins with no predicted functions. Interestingly, 7 out of the 10 most regulated Sxi-Induced genes fall into this category, emphasizing the diverse nature of genes involved in developmental processes across fungi. Three genes, LAC1, VAD1, and MPK1 have been characterized previously as playing roles in virulence (Kraus et al., 2003; Zhu and Williamson, 2004; Panepinto et al., 2005). This last group of genes was somewhat surprising because there was no expectation a priori that Sxi2a-Sxi1α would directly regulate genes involved in virulence (Hull et al., 2004).
Sxi2a and Sxi1α indirectly regulate multiple biological processes
To infer the global biological processes controlled by the Sxi proteins, we evaluated all Sxi-Regulated genes using the Database for Annotation, Visualization, and Integrated Discovery (DAVID). We found that the Sxi-Induced cohort was highly enriched (p=1×10−6) for genes that possess Gene Ontology (GO) terms for catabolic processes, including β-oxidation. Other biological terms somewhat enriched (p ≥ 2.4×10−3) in the Sxi-Induced cohort include reproduction, nucleotide transport, and transition metal ion transport (data not shown). Interestingly, some of these enriched groups are known to be important biological processes used by the organism to survive in the host (Kronstad et al., 2012). The group of Sxi-Repressed genes was enriched for those involved in external encapsulating structure organization, NAD metabolic processes, and steroid metabolism.
While no genes encoding known DNA-binding proteins were determined to be direct targets of Sxi2a and Sxi1α, we did identify multiple putative transcription factors among the full cohort of Sxi-Regulated genes. These transcription factors could regulate subsequent stages of development; however, none of them is directly related to known developmental transcription factors in other systems. Overall, our data suggests that the Sxi proteins are responsible for promoting cellular events that consume metabolic stores, rearrange the cell-membrane and wall, and position the transcriptome for later events such as spore production.
Discussion
Sexual development in fungi is often controlled by compatible homeodomain transcription factors that heterodimerize and regulate the expression of genes required for changes in cell identity. Here we have shown that the Sxi2a-Sxi1α complex found in C. neoformans regulates the expression of over 375 genes during early development, and for approximately 50 of these genes, the Sxi proteins bind directly to a conserved, bipartite sequence in their promoters. While the binding sites recognized by the Sxi-heterodimer both in vitro and in vivo are similar to those used by other fungal homeodomain regulators, the downstream targets of Sxi2a and Sxi1α are strikingly different from those in other fungal systems. Interestingly, several of the Sxi2a-Sxi1α direct targets are genes with previously characterized roles in virulence, indicating that the gene network controlling fungal development intersects with fungal pathogenesis through the targets of the Sxi2a-Sxi1α heterodimer. These findings suggest that common factors involved in both development and pathogenesis are subject to similar selective pressures even though these processes take place in vastly different environments.
Sxi2a-Sxi1α binding sequences are nearly identical in vivo and in vitro
To determine the direct targets of Sxi2a and Sxi1α, we took a gene expression and bioinformatic approach in part to overcome challenges associated with ChIPs from cells undergoing sexual development. At least part of the difficulty surrounding ChIPs in C. neoformans during development appears to stem from unusually high protease activity in extracts, which might be explained by the fact that many of the direct targets of Sxi2a-Sxi1α are involved in ubiquitinylation and other proteasome-related activities (Table I). In the absence of direct in vivo binding data, we used particularly stringent criteria when analyzing our binding and expression data. For example, only the top 1,000 bound sites (out of 1.05×106) showing the highest affinity Sxi binding in the CSI experiment were carried forward in subsequent analyses. In addition, only those genes exhibiting opposing expression patterns in both Sxi-protein expression experiments (Figure 2) were considered Sxi-Regulated and analyzed further.
As a result of this stringency, we were comparing only the highest affinity binding sites from the CSI arrays to sites most likely to be associated with transcriptional regulation in vivo. We were somewhat surprised to see a correlation between the highest affinity binding sites in vitro and the regulatory sites used in vivo because we had considered that the highest affinity binding sites from the protein-binding arrays might not be the most effective regulatory sites in C. neoformans. In fact, among the E2F family of transcription factors, many in vivo binding sites are highly diverged from their consensus binding sequences in vitro. This difference is thought to result in lower affinity binding by E2Fs in vivo that facilitates a more flexible transcriptional response (Rabinovich et al., 2008). In contrast, for Sxi2a-Sxi1α the highest affinity sites in vitro correlate directly with the highest levels of regulation in vivo. This is similar to what has been observed for the S. cerevisiae transcription factor Gcn4, where high affinity binding in vivo is also linked to efficient regulation (Nutiu et al., 2011).
Another consequence of our approach is that some (or even many) direct targets of Sx2a-Sxi1α may have been excluded from our final list (Table I). Our analysis provides high confidence that we have captured bona fide direct Sxi2a-Sxi1α targets; however, we cannot rule out that there are direct targets in the Sxi-regulated gene pools that were not identified as being bound directly. The number of directly activated targets we identified (32) is consistent with the number of direct targets of the homologs of Sxi2a and Sxi1α in other systems (i.e. 19 in S. cerevisiae and 16 in the corn smut Ustilago maydis) (Galgoczy et al., 2004; Heimel et al., 2010), but advances in ChIP or other in vivo binding approaches will be necessary to fully identify all bound target genes.
Sxi2a-Sxi1α exhibit unique binding properties
The binding sites for the S. cerevisiae and C. neoformans cell type-specific homeodomain proteins are similar in sequence; however, the distance between the heterodimer half-sites is quite different. In S. cerevisiae, the spacing is absolutely conserved (GATGN9ACA), and reflects precise structural interactions between a1, α2, and DNA (Goutte and Johnson, 1994; Li et al., 1995; Galgoczy et al., 2004). However, in C. neoformans, we observed a variable distance between the Sxi2a and Sxi1α half-sites (TGATN4–12GAAG). The variability of this spacer region could be due to differences in the sizes of the homeodomain proteins. Sxi1α and Sxi2a are much larger proteins than a1 and α2 (432 and 699 amino acids vs. 126 and 210 amino acids, respectively), and we hypothesize that this increase in size could accommodate more flexible binding conformations of the heterodimer and lead to a greater variety of possible binding sequences.
Another difference between a1-α2 and Sxi2a-Sxi1α is the manner in which these heterodimers bind DNA. In S. cerevisiae, a1 does not bind DNA with high affinity in the absence of α2 (Goutte and Johnson, 1993); however, in C. neoformans, Sxi2a binds with nanomolar affinity to DNA in the absence of Sxi1α in vitro (Stanton et al., 2009). It is known that a1-α2 binding occurs through an interaction between the a1 homeodomain and the C-terminal tail of α2 (Stark and Johnson, 1994), and while the details of the Sxi2a-Sxi1α interaction are not known, our data indicate that both proteins must make sequence-specific DNA contacts in vivo to mediate regulation (Figure 4C and 5B). This suggests that Sxi1α facilitates Sxi2a binding to DNA only in vivo, whereas in S. cerevisiae, α2 is required for a1 binding both in vivo and in vitro. These findings are consistent with the a1-α2 model in which protein-protein interactions facilitate heterodimer binding via conformational and/or energetic changes in vivo.
Fungal homeodomain proteins regulate disparate targets among fungi
Of the genes we identified as Sxi2a-Sxi1α direct targets, only one is regulated by Sxi protein homologs in other organisms. In addition, none of the C. neoformans homologs of the 19 S. cerevisiae a1-α2 targets is found among the Sxi2a-Sxi1α list of direct targets, and only one a1-α2 target (STE4) shows a Sxi-dependent expression pattern (Sxi-Induced). This might not be surprising given the large phylogenetic distance between C. neoformans and S. cerevisiae and the stark differences in development between the two fungi; however, a similar lack of convergence occurs between direct targets of Sxi2a-Sxi1α and their homologs (bE-bW) in U. maydis, a more closely related fungus. Only one direct target of the bE-bW heterodimer, CLP1 (Scherer et al., 2006), is a Sxi target in C. neoformans, and ZNF2 (CNG02160), a homolog of RBF1 (the central target of bE-bW) is not regulated by the Sxi heterodimer either directly or indirectly. In this case, the lack of conservation between bE-bW and Sxi2a-Sxi1α targets is especially interesting because both homeodomain protein complexes are involved in the production of the same biological structure, a dikaryon. Taken together, it appears that sexual development under the control of homeodomain heterodimers in fungi has undergone transcriptional network rewiring: target genes are regulated via similar binding sites by the same class of transcription factors, but the genes themselves are largely unrelated.
While homeodomain targets are diverged between C. neoformans and other fungi, within C. neoformans the cohort of Sxi2a-Sxi1α targets contains many genes regulated in another form of sexual development, known as same-sex development (Lin et al., 2005). In this process, filaments, basidia, and spores are formed that appear nearly identical to those formed during opposite-sex development. Same-sex development is not dependent on either of the Sxi proteins and generally occurs in response to severe nutrient limitation and desiccation. A key transcription factor in same-sex development is Znf2 (Lin et al., 2010). Given the morphological similarities between opposite- and same-sex development, we hypothesized that Znf2 would regulate some of the same targets that Sxi2a-Sxi1α regulate during opposite-sex development. In fact, 14 of 17 genes induced by Znf2 during same-sex development were also induced by Sxi2a-Sxi1α. One of these genes, the non-mating type-specific pheromone receptor CPR2, contained an SBS. It is possible that the same-sex and opposite-sex developmental cascades converge at CPR2, linking these morphologically similar processes.
Sexual development and virulence intersect among the targets of Sxi2a-Sxi1α
An unexpected finding from our work was that the Sxi proteins regulate the expression of the known virulence genes LAC1, VAD1, and MPK1. Lac1 oxidizes diphenolic intermediates during melanin production and is required for dissemination in a mouse model of C. neoformans disease (Williamson, 1994; Salas et al., 1996). Vad1 is an RNA binding protein in the RCK/p54 family that regulates levels of multiple transcripts required for full virulence. Mpk1 is a Mitogen Activated Protein Kinase active during the response to cell wall stress that is also required for full virulence (Kraus et al., 2003). LAC1, VAD1, and MPK1 all contain SBSs in their predicted promoters (Table I).
Many previous studies of C. neoformans have shown an interaction between sexual development and virulence (Kwon-Chung et al., 1992; Alspaugh et al., 1997; Chang et al., 2001; Chang et al., 2003; Panepinto et al., 2005; Lin et al., 2006; Lin et al., 2010; Linqi Wang et al., 2012). A common theme between these processes is the requirement for stress response genes. The mammalian host is considered a harsh environment in which nutrient limitation is a barrier to pathogenic growth (Fleck et al., 2011). The ability of C. neoformans to survive and adapt to harsh environments via stress response pathways is a known requirement for virulence (Hu et al., 2008; Kronstad et al., 2012), and multiple studies have also shown a dependence on stress response factors for wild type levels of development (Alspaugh et al., 1997; Alspaugh et al., 2000; Jung and Bahn, 2009). Our data suggest that targets of the Sxi proteins (both direct and indirect) are involved in development and virulence, intersecting via nutritional stress responses. Many of these genes are involved in metabolism, including metal ion transport, β-oxidation, and flux through the TCA cycle. For example, VAD1 controls PCK1, a key component of gluconeogenesis (Panepinto et al., 2005) and LAC1 is upregulated after exposure to low levels of glucose similar to those present in the human brain (Zhu and Williamson, 2004).
Taken together, these results indicate that Sxi2a-Sxi1α regulates a diverse set of genes that include those at the junctions among sexual development, starvation, and virulence. Future studies will further elucidate the transcriptional network controlled by Sxi2a-Sxi1α and the roles that downstream Sxi-targets play in varied pathways. These insights will allow us to further understand morphological transitions during eukaryotic development and the connections between development and virulence in fungi.
Experimental Procedures
Strain manipulations and media
All strains used were serotype D in the JEC20 or JEC21 background and handled using standard techniques and media as described previously (Kwon-Chung et al., 1992; Kruzel et al., 2012). The inducible strain was constructed by transforming CHY2142 (α ura5 ade2 sxi1α::NAT) with two integrated constructs: pCH941 (pGPD1-SXI1α-URA5) and pCH948 (pGAL7-SXI2a-ADE2) to create CHY2228. The sxi1αΔ and sxi2aΔ strains were CHY2285 and CHY768, respectively, and have been described previously (Hull et al., 2002).
Sxi gene cloning and protein production
cDNAs for Sxi1α and Sxi2a were amplified from plasmids pCH286 and pCH287, respectively, via PCR and ligated into the pEU-E01-MCS vector at the SpeI site (CellFree Sciences). Preparations of the resulting vectors (pCH619 and pCH774) were purified and subjected to transcription and translation reactions in wheat germ extracts using the Premium Expression Kit, according to manufacturer’s instructions (CellFree Sciences). Protein production was confirmed using SDS-PAGE/Coomassie staining and DNA binding activity was confirmed using electrophoretic mobility shift assays (EMSAs).
Cognate Site Identifier analysis
Wheat germ extracts containing full-length, fluorescently labeled Sxi1α and/or Sxi2a were applied to 15-mer DNA Cognate Site Identifier (CSI) arrays, representing 10-mers of duplex DNA in every permutation (Carlson et al., 2010; Tietjen et al., 2011). Arrays were blocked in 2.5% non-fat dried milk for 1 hour at room temperature with gentle agitation prior to addition of recombinant proteins in wheat germ extracts. Binding assays [50 mM NaCl, 10 mM Tris-HCl pH 7.5, 1 mM MgCl2, 0.5 mM EDTA supplemented with bovine serum albumin (3 mg ml−1), non-fat dried milk (0.5%), DTT (0.5 mM), anti-6x-histidine Cyanine-5 conjugated antibody (Qiagen)] were incubated on ice for 40 minutes and were then incubated with the CSI array for 75 minutes at 4°C with gentle agitation. Fluorescence data were acquired using an Axon microarray scanner (Molecular Devices Corporation, Union City, CA). Data were analyzed using the GenePix Pro software, and statistical analysis was carried out as described previously (Carlson et al., 2010; Tietjen et al., 2011). Reported data are represented by at least three independent CSI arrays.
Whole genome transcript analysis
For Sxi protein induction, strains CHY610 and CHY2228 were grown in liquid culture to log phase in yeast peptone dextrose (YPD) medium at 30°C and then induced in galactose-containing medium (YPGal) for 6 hours at 22°C. Cells were pelleted and washed, and RNA was extracted from each sample using hot acid-phenol as described previously (Collart and Oliviero, 1993). For Sxi deletion crosses, JEC20 and JEC21 (or CHY768 and CHY2285) were mixed and plated onto V8 media (pH 7.0) and incubated at 22°C in the dark for approximately 16 hours. RNA was then extracted from the crosses using hot acid-phenol. All RNA samples were further purified using a Qiagen RNeasy midi column, and cDNA was synthesized using Cy3/5 labeled dCTP according to manufacturer’s instructions (GE Healthcare). Two biological replicates were carried out for both Sxi (+) and Sxi (−) experiments. Each biological replicate consisted of 4 technical replicates with 2 replicates per dye swap. Samples were competitively hybridized according to previously described methods to spotted oligonucleotide arrays from the Cryptococcus Community Microarray Consortium (Kruzel et al., 2012).
Arrays were scanned on a GenePix 400B scanner and the resulting spot intensities were extracted using GenePix Pro 4.0. Data were analyzed using Limma (Bioconductor) (Smyth and Speed, 2003; Ritchie et al., 2007; Smyth, 2004). Any genes exhibiting a nonzero expression change and possessing p ≤ 0.05 (T-test for differential expression) were considered either Sxi-Induced or Sxi-Repressed depending on the direction of their fold-change in each experiment. Three Sxi-regulated genes (two repressed and one induced) had their expression changes validated via quantitative Real-Time PCR. All array data have been deposited in the Gene Expression Omnibus and are accessible through GEO Series accession number GSE57287. Gene Ontology analysis of the Sxi-Induced and Sxi-Repressed groups was carried out using the web-based service DAVID (Huang et al., 2008; Huang et al., 2009).
MEME analysis
One thousand base pairs of sequence upstream of genes of interest were subjected to Multiple Em for Motif Elicitation (MEME) analysis in groups of 40 (Bailey and Elkan, 1994). The algorithm was programmed to identify motifs 6–50 nucleotides in length found any number of times in the sequences. Motifs of interest were subjected to Motif Alignment and Search Tool (MAST) analysis to identify occurrences of the motif in the entire Sxi-Induced or Sxi-Repressed cohorts (Bailey and Gribskov, 1998).
Yeast 1-hybrid assay
Oligonucleotides representing the 21-mer binding sites repeated three times were cloned into the Live Guarente vector (pCH563) at the SalI site (Guarente and Mason, 1983). For CLP1, CPR2, CNJ03000, and CNH01950, sequences were represented by oligos CHO3973/4, CHO4353/5, CHO4592/3, AND CHO4588/9, respectively. See Table SVII for oligonucleotide sequences. These constructs were transformed into S. cerevisiae strain EG123 along with plasmids expressing either a marker only or a Sxi transcription factor construct (Siliciano and Tatchell, 1986). Three independent transformants were evaluated for lacZ expression according to standard protocols (Stanton et al., 2009). Absorbance at 578nm was recorded and converted to Miller Units.
In vivo reporters
One kb of sequence upstream of the start codon for CLP1 was cloned upstream of the URA5 gene in pCH1184 to create pCH1294 (Kruzel et al., 2012). Overlap PCR was used to construct deletion (Δ) and mutated (MU) versions of the CLP1 upstream region (See Table SVII for oligos). Products of the overlap PCR were cloned into pCH1184 to create reporter constructs pCH1295, pCH1313, pCH1343, and pCh1344 (Δ, MU, Sxi1α Half-Site MU, Sxi2a Half-Site MU, respectively). Plasmids were linearized with I-SceI and transformed into CHY3389. URA5 transcripts from three independent transformants were evaluated by northern blot after the strain was grown on V8 plates supplemented with 0.026g L−1 uracil and 20g L−1 galactose for 6 hours.
Northern blot analysis
Northern blot analysis was carried out according to standard protocols using 10 μg of total RNA for each sample. PCR-generated probes were radiolabeled using the Rediprime II kit according to manufacturer’s instructions (GE Healthcare, see Table SV). Hybridizations and washes were carried out at 65°C as described previously (Brown and Mackey, 1997). Probes were constructed using genomic DNA in a PCR using oligos CHO805 and CHO806 for URA5 and oligos CHO651 and CHO652 for GPD1. Blots were exposed to a phosphor screen, imaged with a Typhoon FLA 9000 (GE Healthcare Life Sciences), and analyzed using the ImageQuant software package.
Supplementary Material
A. Northern blot analysis of SXI2a and SXI1α transcript levels in the Sxi (+) strain. RNA was harvested from cells before and during galactose induction in a liquid culture with gentle shaking. Black arrow indicates the 6 hr timepoint post-induction when RNA was harvested for the Sxi (+) array experiment and reflects the time immediately before maximum SXI2a and SXI1α expression. B. The Sxi (+) strain was grown in liquid YP+Galactose media for 24 hours with gentle shaking. Short filaments were observed that terminated in both naked basidia and basidia with 4 spores on their surfaces. Image reflects structures representing approximately 75% of the population at 24 hrs. White arrow basidium with spores. Black Arrow naked basidium.
Expression and P-values for the Sxi(+) Experiment
Expression and P-Values for the Sxi(−) Experiment
Expression and P-values for all Sxi-Induced Genes
Expression and P-values for all Sxi-Repressed Genes
Sxi Binding Sites Upstream of 18 Sxi-Repressed Genes
Strains and plasmids
Oligonucleotides
Acknowledgments
We thank Mingwei Huang, Naomi Walsh, Melissa M. Harrison, and Catherine A. Fox for discussion and critical comments on the manuscript. We thank Aseem Z. Ansari for assistance with the CSI arrays, Benjamin Mueller for bioinformatic support, and Alexander Dammann for technical support. This work was supported by NIH T32 AI55397 to MEM, NIH CA 133508 and HL099773 to AZA, and NIH R01 AI089370 to CMH.
References
- Alspaugh JA, Cavallo LM, Perfect JR, Heitman J. RAS1 regulates filamentation, mating and growth at high temperature of Cryptococcus neoformans. Molecular Microbiology. 2000;36:352–365. doi: 10.1046/j.1365-2958.2000.01852.x. [DOI] [PubMed] [Google Scholar]
- Alspaugh JA, Perfect JR, Heitman J. Cryptococcus neoformans mating and virulence are regulated by the G-protein alpha subunit GPA1 and cAMP. Genes & Development. 1997;11:3206–3217. doi: 10.1101/gad.11.23.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- Bailey TL, Gribskov M. Combining evidence using p-values: application to sequence homology searches. Bioinformatics. 1998;14:48–54. doi: 10.1093/bioinformatics/14.1.48. [DOI] [PubMed] [Google Scholar]
- Bradford AP, Wasylyk C, Wasylyk B, Gutierrez-Hartmann A. Interaction of Ets-1 and the POU-homeodomain protein GHF-1/Pit-1 reconstitutes pituitary-specific gene expression. Mol Cell Biol. 1997;17:1065–1074. doi: 10.1128/mcb.17.3.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazas RM, Bhoite LT, Murphy MD, Yu Y, Chen Y, Neklason DW, Stillman DJ. Determining the requirements for cooperative DNA binding by Swi5p and Pho2p (Grf10p/Bas2p) at the HO promoter. J Biol Chem. 1995;270:29151–29161. doi: 10.1074/jbc.270.49.29151. [DOI] [PubMed] [Google Scholar]
- Brown T, Mackey K. Analysis of RNA by Northern and Slot Blot Hybridization. Curr Protoc Mol Biol. 1997;37:4.9.1–4.9.16. doi: 10.1002/0471142727.mb0409s67. [DOI] [PubMed] [Google Scholar]
- Carlson CD, Warren CL, Hauschild KE, Ozers MS, Qadir N, Bhimsaria D, et al. Specificity landscapes of DNA binding molecules elucidate biological function. Proceedings of the National Academy of Sciences. 2010;107:4544–4549. doi: 10.1073/pnas.0914023107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YC, Miller GF, Kwon-Chung KJ. Importance of a Developmentally Regulated Pheromone Receptor of Cryptococcus neoformans for Virulence. Infection and Immunity. 2003;71:4953–4960. doi: 10.1128/IAI.71.9.4953-4960.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YC, Penoyer LA, Kwon-Chung KJ. The second STE12 homologue of Cryptococcus neoformans is MATa-specific and plays an important role in virulence. Proc Natl Acad Sci USA. 2001;98:3258–3263. doi: 10.1073/pnas.061031998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collart MA, Oliviero S. Preparation of Yeast RNA. Curr Protoc Mol Biol. 1993;23:13.12.1–13.12.5. doi: 10.1002/0471142727.mb1312s23. [DOI] [PubMed] [Google Scholar]
- Ekena JL, Stanton BC, Schiebe-Owens JA, Hull CM. Sexual Development in Cryptococcus neoformans Requires CLP1, a Target of the Homeodomain Transcription Factors Sxi1α and Sxi2a. Eukaryotic Cell. 2008;7:49–57. doi: 10.1128/EC.00377-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleck CB, Schöbel F, Brock M. Nutrient acquisition by pathogenic fungi: Nutrient availability, pathway regulation, and differences in substrate utilization. International Journal of Medical Microbiology. 2011;301:400–407. doi: 10.1016/j.ijmm.2011.04.007. [DOI] [PubMed] [Google Scholar]
- Galgoczy DJ, Cassidy-Stone A, Llinás M, O’Rourke SM, Herskowitz I, DeRisi JL, Johnson AD. Genomic dissection of the cell-type-specification circuit in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2004;101:18069–18074. doi: 10.1073/pnas.0407611102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring WJ. Exploring the homeobox. Gene. 1993;135:215–221. doi: 10.1016/0378-1119(93)90068-e. [DOI] [PubMed] [Google Scholar]
- Giles SS, Dagenais TRT, Botts MR, Keller NP, Hull CM. Elucidating the pathogenesis of spores from the human fungal pathogen Cryptococcus neoformans. Infection and Immunity. 2009;77:3491–3500. doi: 10.1128/IAI.00334-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goutte C, Johnson AD. Yeast a1 and alpha 2 homeodomain proteins form a DNA-binding activity with properties distinct from those of either protein. J Mol Biol. 1993;233:359–371. doi: 10.1006/jmbi.1993.1517. [DOI] [PubMed] [Google Scholar]
- Goutte C, Johnson AD. Recognition of a DNA operator by a dimer composed of two different homeodomain proteins. EMBO J. 1994;13:1434–1442. doi: 10.1002/j.1460-2075.1994.tb06397.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L, Mason T. Heme regulates transcription of the CYC1 gene of S. cerevisiae via an upstream activation site. Cell. 1983;32:1279–1286. doi: 10.1016/0092-8674(83)90309-4. [DOI] [PubMed] [Google Scholar]
- Heimel K, Scherer M, Vranes M, Wahl R, Pothiratana C, Schuler D, et al. The Transcription Factor Rbf1 Is the Master Regulator for b-Mating Type Controlled Pathogenic Development in Ustilago maydis. PLoS Pathog. 2010;6:e1001035. doi: 10.1371/journal.ppat.1001035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh YP, Xue C, Heitman J. A constitutively active GPCR governs morphogenic transitions in Cryptococcus neoformans. EMBO J. 2009;28:1220–1233. doi: 10.1038/emboj.2009.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Cheng PY, Sham A, Perfect JR, Kronstad JW. Metabolic adaptation in Cryptococcus neoformans during early murine pulmonary infection. Molecular Microbiology. 2008;69:1456–1475. doi: 10.1111/j.1365-2958.2008.06374.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2008;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Research. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull CM, Boily MJ, Heitman J. Sex-specific homeodomain proteins Sxi1alpha and Sxi2a coordinately regulate sexual development in Cryptococcus neoformans. Eukaryotic Cell. 2005;4:526–535. doi: 10.1128/EC.4.3.526-535.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull CM, Cox GM, Heitman J. The alpha-specific cell identity factor Sxi1alpha is not required for virulence of Cryptococcus neoformans. Infection and Immunity. 2004;72:3643– 3645. doi: 10.1128/IAI.72.6.3643-3645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull CM, Davidson RC, Heitman J. Cell identity and sexual development in Cryptococcus neoformans are controlled by the mating-type-specific homeodomain protein Sxi1alpha. Genes & Development. 2002;16:3046–3060. doi: 10.1101/gad.1041402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AD. Molecular Mechanisms of Cell-Type Determination in Budding Yeast. Curr Opin Genet Dev. 1995;5:552–558. doi: 10.1016/0959-437x(95)80022-0. [DOI] [PubMed] [Google Scholar]
- Jung KW, Bahn YS. The Stress-Activated Signaling (SAS) Pathways of a Human Fungal Pathogen, Cryptococcus neoformans. Mycobiology. 2009;37:161–170. doi: 10.4489/MYCO.2009.37.3.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus PR, Fox DS, Cox GM, Heitman J. The Cryptococcus neoformans MAP kinase Mpk1 regulates cell integrity in response to antifungal drugs and loss of calcineurin function. Molecular Microbiology. 2003;48:1377–1387. doi: 10.1046/j.1365-2958.2003.03508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronstad J, Saikia S, Nielson ED, Kretschmer M, Jung W, Hu G, et al. Adaptation of Cryptococcus neoformans to Mammalian Hosts: Integrated Regulation of Metabolism and Virulence. Eukaryotic Cell. 2012;11:109–118. doi: 10.1128/EC.05273-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruzel EK, Giles SS, Hull CM. Analysis of Cryptococcus neoformans Sexual Development Reveals Rewiring of the Pheromone-Response Network by a Change in Transcription Factor Identity. Genetics. 2012;191:435–449. doi: 10.1534/genetics.112.138958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kües U, Göttgens B, Stratmann R, Richardson WV, O’Shea SF, Casselton LA. A chimeric homeodomain protein causes self-compatibility and constitutive sexual development in the mushroom Coprinus cinereus. EMBO J. 1994;13:4054–4059. doi: 10.1002/j.1460-2075.1994.tb06722.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon-Chung KJ, Edman JC, Wickes BL. Genetic association of mating types and virulence in Cryptococcus neoformans. Infection and Immunity. 1992;60:602–605. doi: 10.1128/iai.60.2.602-605.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Stark MR, Johnson AD, Wolberger C. Crystal structure of the MATa1/MATalpha2 homeodomain heterodimer bound to DNA. Science. 1995;270:262–269. doi: 10.1126/science.270.5234.262. [DOI] [PubMed] [Google Scholar]
- Lin X, Huang JC, Mitchell TG, Heitman J. Virulence Attributes and Hyphal Growth of C. neoformans Are Quantitative Traits and the MATα Allele Enhances Filamentation. PLoS Genet. 2006;2:e187. doi: 10.1371/journal.pgen.0020187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Hull CM, Heitman J. Sexual reproduction between partners of the same mating type in Cryptococcus neoformans. Nature. 2005;434:1017–1021. doi: 10.1038/nature03448. [DOI] [PubMed] [Google Scholar]
- Lin X, Jackson JC, Feretzaki M, Xue C, Heitman J. Transcription factors Mat2 and Znf2 operate cellular circuits orchestrating opposite- and same-sex mating in Cryptococcus neoformans. PLoS Genet. 2010;6:e1000953. doi: 10.1371/journal.pgen.1000953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvintseva AP, Marra RE, Nielsen K, Heitman J, Vilgalys R, Mitchell TG. Evidence of Sexual Recombination among Cryptococcus neoformans Serotype A Isolates in Sub-Saharan Africa. Eukaryotic Cell. 2003;2:1162–1168. doi: 10.1128/EC.2.6.1162-1168.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutiu R, Friedman RC, Luo S, Khrebtukova I, Silva D, Li R, et al. Direct measurement of DNA affinity landscapes on a high-throughput sequencing instrument. Nature Biotechnology. 2011;29:659–664. doi: 10.1038/nbt.1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panepinto J, Liu L, Ramos J, Zhu X, Valyi-Nagy T, Eksi S, et al. The DEAD-box RNA helicase Vad1 regulates multiple virulence-associated genes in Cryptococcus neoformans. J Clin Invest. 2005;115:632–641. doi: 10.1172/JCI200523048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park BJ, Wannemuehler KA, Marston BJ, Govender N, Pappas PG, Chiller TM. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS. 2009;23:525–530. doi: 10.1097/QAD.0b013e328322ffac. [DOI] [PubMed] [Google Scholar]
- Park YD, Panepinto J, Shin S, Larsen P, Giles S, Williamson PR. Mating Pheromone in Cryptococcus neoformans Is Regulated by a Transcriptional/Degradative “Futile” Cycle. Journal of Biological Chemistry. 2010;285:34746–34756. doi: 10.1074/jbc.M110.136812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovich A, Jin VX, Rabinovich R, Xu X, Farnham PJ. E2F in vivo binding specificity: comparison of consensus versus nonconsensus binding sites. Genome Research. 2008;18:1763–1777. doi: 10.1101/gr.080622.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Silver J, Oshlack A, Holmes M, Diyagama D, Holloway A, Smyth GK. A comparison of background correction methods for two-colour microarrays. Bioinformatics. 2007;23:2700–2707. doi: 10.1093/bioinformatics/btm412. [DOI] [PubMed] [Google Scholar]
- Salas SD, Bennett JE, Kwon-Chung KJ, Perfect JR, Williamson PR. Effect of the laccase gene CNLAC1, on virulence of Cryptococcus neoformans. J Exp Med. 1996;184:377–386. doi: 10.1084/jem.184.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauka-Spengler T, Bronner-Fraser M. A gene regulatory network orchestrates neural crest formation. Nat Rev Mol Cell Biol. 2008;9:557–568. doi: 10.1038/nrm2428. [DOI] [PubMed] [Google Scholar]
- Scherer MM, Heimel KK, Starke VV, Kämper JJ. The Clp1 protein is required for clamp formation and pathogenic development of Ustilago maydis. Plant Cell. 2006;18:2388–2401. doi: 10.1105/tpc.106.043521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder MD, Pearce M, Fak J, Fan H, Unnerstall U, Emberly E, et al. Transcriptional Control in the Segmentation Gene Network of Drosophila. PLoS Biol. 2004;2:e271. doi: 10.1371/journal.pbio.0020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz B, Banuett F, Dahl M, Schlesinger R, Schäfer W, Martin T, et al. The b alleles of U. maydis, whose combinations program pathogenic development, code for polypeptides containing a homeodomain-related motif. Cell. 1990;60:295–306. doi: 10.1016/0092-8674(90)90744-y. [DOI] [PubMed] [Google Scholar]
- Siliciano PG, Tatchell K. Identification of the DNA sequences controlling the expression of the MATalpha locus of yeast. Proc Natl Acad Sci USA. 1986;83:2320–2324. doi: 10.1073/pnas.83.8.2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. Linear Models and Empirical Bayes Methods for Assessing Differential Expression in Microarray Experiments. Statistical Applications in Genetics and Molecular Biology. 2004;3(1):Article 3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Smyth GK, Speed T. Normalization of cDNA microarray data. Methods. 2003;31:265–273. doi: 10.1016/s1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- Spit A, Hyland RH, Mellor EJ, Casselton LA. A role for heterodimerization in nuclear localization of a homeodomain protein. Proc Natl Acad Sci USA. 1998;95:6228–6233. doi: 10.1073/pnas.95.11.6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton BC, Giles SS, Kruzel EK, Warren CL, Ansari AZ, Hull CM. Cognate Site Identifier analysis reveals novel binding properties of the Sex Inducer homeodomain proteins of Cryptococcus neoformans. Molecular Microbiology. 2009;72:1334–1347. doi: 10.1111/j.1365-2958.2009.06719.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark MR, Johnson AD. Interaction between two homeodomain proteins is specified by a short C-terminal tail. Nature. 1994;371:429–432. doi: 10.1038/371429a0. [DOI] [PubMed] [Google Scholar]
- Tietjen JR, Donato LJ, Bhimisaria D, Ansari AZ. Chapter 1 - Sequence-Specificity and Energy Landscapes of DNA-Binding Molecules. Methods in Enzymology. 2011;497:3–30. doi: 10.1016/B978-0-12-385075-1.00001-9. [DOI] [PubMed] [Google Scholar]
- Tsong AE, Miller MG, Raisner RM, Johnson AD. Evolution of a combinatorial transcriptional circuit: a case study in yeasts. Cell. 2003;115:389–399. doi: 10.1016/s0092-8674(03)00885-7. [DOI] [PubMed] [Google Scholar]
- Turatsinze JV, Thomas-Chollier M, Defrance M, Van Helden J. Using RSAT to scan genome sequences for transcription factor binding sites and cis-regulatory modules. Nat Protoc. 2008;3:1578–1588. doi: 10.1038/nprot.2008.97. [DOI] [PubMed] [Google Scholar]
- Velagapudi R, Hsueh YP, Geunes-Boyer S, Wright JR, Heitman J. Spores as Infectious Propagules of Cryptococcus neoformans. Infection and Immunity. 2009;77:4345–4355. doi: 10.1128/IAI.00542-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Zhai B, Lin X. The Link between Morphotype Transition and Virulence in Cryptococcus neoformans. PLoS Pathog. 2012;8:e1002765. doi: 10.1371/journal.ppat.1002765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Hsueh YP, Li W, Floyd A, Skalsky R, Heitman J. Sex-induced silencing defends the genome of Cryptococcus neoformans via RNAi. Genes & Development. 2010;24 :2566–2582. doi: 10.1101/gad.1970910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson PR. Biochemical and molecular characterization of the diphenol oxidase of Cryptococcus neoformans: identification as a laccase. J Bacteriol. 1994;176:656–664. doi: 10.1128/jb.176.3.656-664.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Williamson PR. Role of laccase in the biology and virulence of Cryptococcus neoformans. FEMS Yeast Research. 2004;5:1–10. doi: 10.1016/j.femsyr.2004.04.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A. Northern blot analysis of SXI2a and SXI1α transcript levels in the Sxi (+) strain. RNA was harvested from cells before and during galactose induction in a liquid culture with gentle shaking. Black arrow indicates the 6 hr timepoint post-induction when RNA was harvested for the Sxi (+) array experiment and reflects the time immediately before maximum SXI2a and SXI1α expression. B. The Sxi (+) strain was grown in liquid YP+Galactose media for 24 hours with gentle shaking. Short filaments were observed that terminated in both naked basidia and basidia with 4 spores on their surfaces. Image reflects structures representing approximately 75% of the population at 24 hrs. White arrow basidium with spores. Black Arrow naked basidium.
Expression and P-values for the Sxi(+) Experiment
Expression and P-Values for the Sxi(−) Experiment
Expression and P-values for all Sxi-Induced Genes
Expression and P-values for all Sxi-Repressed Genes
Sxi Binding Sites Upstream of 18 Sxi-Repressed Genes
Strains and plasmids
Oligonucleotides