Abstract
Flowering time is an important ecological trait that determines the transition from vegetative to reproductive growth. Flowering time in cotton is controlled by short-day photoperiods, with strict photoperiod sensitivity. As the CO-FT (CONSTANS-FLOWER LOCUS T) module regulates photoperiodic flowering in several plants, we selected eight CONSTANS genes (COL) in group I to detect their expression patterns in long-day and short-day conditions. Further, we individually cloned and sequenced their homologs from 25 different cotton accessions and one outgroup. Finally, we studied their structures, phylogenetic relationship, and molecular evolution in both coding region and three characteristic domains. All the eight COLs in group I show diurnal expression. In the orthologous and homeologous loci, each gene structure in different cotton species is highly conserved, while length variation has occurred due to insertions/deletions in intron and/or exon regions. Six genes, COL2 to COL5, COL7 and COL8, exhibit higher nucleotide diversity in the D-subgenome than in the A-subgenome. The Ks values of 98.37% in all allotetraploid cotton species examined were higher in the A-D and At-Dt comparison than in the A-At and D-Dt comparisons, and the Pearson’s correlation coefficient (r) of Ks between A vs. D and At vs. Dt also showed positive, high correlations, with a correlation coefficient of at least 0.797. The nucleotide polymorphism in wild species is significantly higher compared to G. hirsutum and G. barbadense, indicating a genetic bottleneck associated with the domesticated cotton species. Three characteristic domains in eight COLs exhibit different evolutionary rates, with the CCT domain highly conserved, while the B-box and Var domain much more variable in allotetraploid species. Taken together, COL1, COL2 and COL8 endured greater selective pressures during the domestication process. The study improves our understanding of the domestication-related genes/traits during cotton evolutionary process.
Introduction
The taxonomic and evolutionary history of the cotton genus (Gossypium) extends back approximately 10 million years [1–3]. The cotton genus currently includes 50 species distributed in arid and semi-arid regions of the tropics and subtropics. Most of these species are diploid (n = 13), while five are allopolyploid (AD-genome; n = 26) [2,4]. Gossypium tomentosum is endemic to the Hawaiian Islands [5], while G. mustelinum is restricted to a relatively small region of northeast Brazil [6] and G. darwinii is native to the Galapagos Islands [7]. In addition to these three true wild species, G. barbadense and G. hirsutum are two cultivated allopolyploid species that have been independently domesticated over a vast geographical area, with a wealth of morphological forms spanning the wild to domesticated continuum [2,8–10]. Due to human-mediated influences and agronomic improvement, domesticated G. barbadense and G. hirsutum have been modified by parallel changes and exhibit extraordinary morphological variation, e.g., the loss of photoperiod sensitivity, transformation from perennial shrubs and small trees to more compact, highly productive annual plants, evolving seeds bearing vastly elongated, abundant, single-celled hairs, and a reduction in seed dormancy [11,12]. As a consequence of human selection and crop improvement, G. hirsutum has been domesticated for its dramatically high fiber yields and expanded planting area. Seven races of G. hirsutum have been identified to date, including ‘yucatanense’, ‘punctatum’, ‘palmeri’, ‘latifolium’, ‘mariegalante’, ‘morrilli’, and ‘richmondii’ [10]. Among these, ‘latifolium’ is considered to be the progenitor of the modern cultivated Upland cotton [13]. Semi-domesticated species of G. barbadense include several races such as ‘peruvianum’, ‘vitifolium’, and ‘brasiliense’; however, the origins of modern cultivated G. barbadense are complex and somewhat obscure [14]. In Gossypium, wild forms genetically close to the actual ancestors and domesticated species both exist, providing opportunities to study target genes selected during domestication by comparing both types of cotton and their parallel evolution [12].
Many crop species are subjected to similar evolutionary constraints and human involvement, which cause phenotypic changes that are common among crop species, namely, “domestication syndrome” (e.g., changes in flowering time) [12,15]. Flowering time is a common, important ecological trait that determines the transition from vegetative to reproductive growth and is regulated by four pathways including the autonomous, gibberellin, photoperiod, and vernalization pathways [16–19]. In particular, photoperiodic sensitivity is considered to be the most important factor in determining flowering time and thus ensures crop adaptation to specific growing seasons, cultivation areas, and natural environmental variation [20–22].
The CONSTANS (CO) transcription factor is a central regulator of the photoperiod pathway, which functions by mediating between the circadian clock and floral integrators [23,24], and CONSTANS LIKE (COL) genes are members of a recently identified family of plant zinc finger proteins. CO was first identified from an Arabidopsis thaliana mutant exhibiting late flowering, specifically under long-day photoperiodic conditions (LD) [25]. Subsequently, the Hd1 (Heading date 1) gene of rice (Oryza sativa), which is homologous to CO, was also shown to be responsible for flowering time in rice [26]. Previous reports have shown that COL genes in several dicots, such as Brassica napus BnCOa1 [27,28] and Pharbitis nil PnCO, could complement the function of Arabidopsis CO when introduced into a co mutant of Arabidopsis. Moreover, three CO/Hd1 homologs of hexaploid wheat in monocots identified through sequence similarity analysis could complement the function of Hd1 when transformed into a rice line deficient in Hd1 [29]. The common function of these genes in different crops demonstrates that CO is involved in a conserved pathway regulating flowering in plants.
Since the release of a large number of publicly available sequences and the complete whole-genome sequences of some plants, genome-wide analyses of the COL gene family have been performed. There are 17, 17, and nine COL family members in Arabidopsis, rice, and barley, respectively [30–31]. Two conserved domains, including a zinc finger domain at the N-terminus that resembles B-boxes and a CCT (CO, COL, TOC1) domain at the C-terminus, are strictly conserved in these genes, while there is a more variable domain in the middle region. Phylogenetic analysis revealed that the COLs can be divided into four major groups. In detail, type I COL genes include two B-boxes and CCT domains, with a single intron located between the B-box and the CCT domains, which includes AtCO and AtCOL1 to AtCOL5 in Arabidopsis and OsA to OsG in rice. Type II COL genes, with only one B-box and a CCT domain with one intron, include AtCOL6 to AtCOL8 and AtCOL16 in Arabidopsis and OsJ to OsL in rice. Type III COL genes, with one full B-box, a second diverged zinc finger, and a CCT domain, include OsM to OsP, Loc_Os06g01340, Loc_Os09g33550, and Loc_Os07g047140, which are similar to AtCOL9 to AtCOL15 genes in Arabidopsis, containing three introns. OsH and OsI, which contain one intron, belong to type IV and are novel, as they lack B-box domains but have a COL CCT domain, this group of genes was recently designated the CCT MOTIF FAMILY (CMF) [22,30,32]. Mutants in B-boxes and CCT conserved domains display a severe late flowering phenotype [33]. Lagercrantz and Axelsson reported that the COLs evolve rapidly, particularly the variable domain in the middle region, which is the most diverged and most rapidly evolving, but there also are fixed residues that show significant conservation [34].
Flowering in an optimal condition could avoid stress damages and balance resource distribution, and further improve crop yield and quality [35]. The wild species of cotton is controlled by short-day photoperiods, with strict photoperiod sensitivity, while domesticated G. barbadense and G. hirsutum lose the photoperiod sensitivity [11,12]. So photoperiod sensitivity is considered as an important factor in determining flowering time in short-day photoperiods in cotton. Due to the conserved function of CO related to photoperiodic flowering in several plants and the limited information in cotton, we selected CO family members to study their structural and evolutionary characterization in Gossypium. Totally, we identified 23 putative COL genes in G. raimondii genome based on sequence data from G. raimondii (http://www.phytozome.net) [36] and divided these genes into three subfamilies, as reported by Griffiths et al. (2003) [30]. We focused on the eight genes in group I, which are also clustered in the same group with the Arabidopsis CO and rice Hd1 flowering time loci. Diurnal expression were performed to analyze their function in respose to light and dark treatment, and we futher studied their sequence, structure, and molecular evolutionary rate variation in 25 cotton accessions, including the Old World diploids G. herbaceum L. and G. raimondii (which are considered to be extant relatives of the A- and D-genome diploid ancestors, respectively, of the allotetraploid lineage), three wild allotetraploid species, and 20 New World allotetraploid accessions (including 11 in G. hirsutum, with seven semi-domesticated species [races] and four cultivated accessions, and nine in G. barbadense, with three semi-domesticated species [races] and six cultivated accessions), as well as Thespesia populneoides (Roxb.) Kostel as a phylogenetic outgroup. We tested the footprints of selection sigatures for the eight COL genes by investigating the nucleotide diversity and neutrality test in allotetraploid cotton species. The study improves our understanding that the domestication-related genes have enhanced cotton adaptation and diversification during the evolutionary process, and the domestication and selection of COL genes might also contribute to the improvement of yield and quality in cotton.
Materials and Methods
Plant materials
Eight cotton genes among 23 GrCOLs were isolated from 25 different cotton accessions and one outgroup, which are listed in Table 1. One diploid A-genome species and one diploid D-genome species were chosen, which represent the best living models of the A- and D-genome donor, respectively. A total of 23 allopolyploid accessions involved in five cotton species were examined, including three wild allopolyploid species, 11 accessions from G. hirsutum species (seven semi-domesticated races and four cultivated accessions), and nine accessions from G. barbadense species (three semi-domesticated races and six cultivated accessions). Thespesia populneoides (Roxb.) Kostel was chosen as the outgroup. Cultivated G. hirsutum and G. barbadense accessions were sampled from the Jiangpu Experimental Station at Nanjing Agricultural University, Nanjing, Jiangsu, China. Other wild, semi-domesticated cotton species and the outgroup was collected from the National Wild Cotton Plantation at Hainan Island, China. All necessary permits for collecting the wild, semi-domesticated cotton species and the outgroup were obtained from the National Wild Cotton Plantation at Hainan Island, Cotton Research Institute, Chinese Academy of Agricultural Sciences, China. Genomic DNA was isolated from young leaves using methods reported previously [37].
Table 1. Cotton accessions used in the study with their geographic distribution in Gossypium.
| No. | Species | Genome | Geographic distribution |
|---|---|---|---|
| 1 | G. herbaceum var. Africanum | A1 | Africa |
| 2 | G. raimondii Ulbr | D5 | Peru |
| 3 | G. hirsutum acc. TM-1 | AD1 | America |
| 4 | G. hirsutum cv. ZMS12 | AD1 | China |
| 5 | G. hirsutum cv. SM3 | AD1 | China |
| 6 | G. hirsutum cv. JM | AD1 | China |
| 7 | G. hirsutum race punctatum | AD1 | Mexico |
| 8 | G. hirsutum race morrilli | AD1 | Mexico |
| 9 | G. hirsutum race yucatanense | AD1 | Mexico |
| 10 | G. hirsutum race richmondii | AD1 | Mexico |
| 11 | G. hirsutum race marie-galante | AD1 | Mexico |
| 12 | G. hirsutum race latifolium | AD1 | Mexico |
| 13 | G. hirsutum race palmeri | AD1 | Mexico |
| 14 | G. barbadense cv. Hai7124 | AD2 | China |
| 15 | G. barbadense cv. Pima-1 | AD2 | America-Egypt |
| 16 | G. barbadense cv. Junhai | AD2 | China |
| 17 | G. barbadense cv. Giza36 | AD2 | Egypt |
| 18 | G. barbadense cv. 3–79 | AD2 | America-Egypt |
| 19 | G. barbadense acc. Mit-Afifi | AD2 | Egypt |
| 20 | G. barbadense race peruvianum 1 | AD2 | Peru |
| 21 | G. barbadense race peruvianum 2 | AD2 | Peru |
| 22 | G. barbadense race brasiliense | AD2 | Brazil |
| 23 | G. tomentosum Nutt. Ex Seem. | AD3 | Hawaii |
| 24 | G. mustelinum Miers ex Watt | AD4 | Brazil |
| 25 | G. darwinii G. Watt | AD5 | Galapagos Islands |
Identification of new CO family genes in cotton
COL protein sequences in Arabidopsis and rice referred to Griffiths et al. 2003 [22] and the Plant Transcription Factor Database (http://plntfdb.bio.uni-potsdam.de/v3.0/), reported by Wu et al. 2013 [30]. To identify COL transcription factor genes in the cotton genome, CO genes in Arabidopsis were used as queries to screen the Pfam database. Then, the Pfam database providing the B-box (PF00643) or CCT (PF06203) domain seed file was compared with the diploid cotton (Gossypium raimondii)_221_protein transcript database. The protein-coding genes with both B-box (PF00643) and CCT (PF06203) domains were manually examined as putative new members of CO in cotton (designated GrCOLs). Multiple alignments were then further performed with Cluster 1.83 and examined manually to confirm the correction. Finally, BLASTP was performed to search the diploid cotton (G. raimondii) genome database with an e-value cutoff of 1×10–15, and the genomic sequences of the GrCOLs were obtained.
PCR amplification, cloning, and sequencing
Gene-specific PCR primers for eight COLs in group I were designed according to the predicted sequences in the diploid cotton (G. raimondii) genome database by Primer 5.0 (S2 Table). All PCR amplification products were extracted using an Axyprep DNA Gel Extraction Kit, cloned into the pMD19-T Vector (TaKaRa) according to the manufacturer’s instructions, and sequenced. In cases of apparent PCR-mediated recombination detection in allopolyploid cotton [38], at least 10 clones per gene were randomly sequenced, with at least three clones per subgenome, and these recombinant clones were omitted to confirm the sequence correction for each duplicated copy and to obtain the homeologs of both the A- and D-subgenomes by comparing sequences from their diploids using the Neighbor-Joining method in Clustal 1.83. The genomic sequences were compared with the cDNA sequences to determine the sizes of exons and introns.
RNA isolation and qRT-PCR analysis
For diurnal expression pattern analysis, G. hirsutum acc. TM-1 were grown in LD (16h light/8h dark) or SD (8h light /16h dark) conditions respectively, and harvested the young leaves fully open every 4 h for 48h during the seeding period when the third leaf fully open, and put them in the liquid nitrogen immediately for use. Total RNA was extracted from leaves according to the method of Jiang and Zhang [39], and 10μl cDNA was synthesized using 500ng RNA with the HiScript Q RT SuperMix for qPCR (+gDNA wiper) (Vazyme), and the gene-specific primers used for qRT-PCR were designed by Primer 5.0 and are listed in (S2 Table). qRT-PCR (20μl reaction volume with 1μl cDNA, 0.5μM each gene-specific primer and FastStart Universal SYBR Green Master(ROX) (Roche) 10ul) were performed by ABI 7500 real-time PCR system. The Histone3 (AF024716) gene (forward primer 5′-GAAGCCTCATCGATACCGTC-3′ and reverse primer squences 5′-CTACCACTACCATCATGG-3′ respectively) was used as the control. The expression level of COL genes was analyzed according to the relative quantification method [40].
Data analysis
The corresponding subgenome sequences of each gene with the same order were combined, and the combined sequence data were used to conduct phylogenetic analyses using the Maximum Likelihood (ML) method provided by MEGA5.1, with 1,000 bootstrap replicates. A ML tree of CONSTANS-like proteins from Arabidopsis, rice, and cotton was also constructed to determine the evolutionary relationships between GrCOL gene family members and those of Arabidopsis and rice.
DnaSP 5.0 was used to estimate the total nucleotide diversity for the genomic sequence of each data set (π), and the nucleotide diversity of all site (πtotal), synonymous (πs) and nonsynonymous site(πa) for the entire coding region and separately for the B-box, Var, and CCT domains of each gene were also analyzed. The synonymous substitution rates (Ks) and nonsynonymous substitution rates (Ka) among 25 cotton accessions and one outgroup for the entire coding region of each gene, and neutral tests of Tajima’s D, Fu and Li’s D and F were also estimated by DnaSP 5.0 [41].
Results
Isolation and characterization of COL family genes in cotton
To identify genes encoding COL in the cotton genome, the primary HMM profiles using the B-box (PF00643) and CCT (PF06203) domains as the seed file, which were retrieved from the Pfam database [42], were used to search the diploid cotton (Gossypium raimondii)_221_protein transcript database (http://www.phytozome.net), and those with both domains (i.e., the candidate COL genes in G. raimondii) were manually selected.
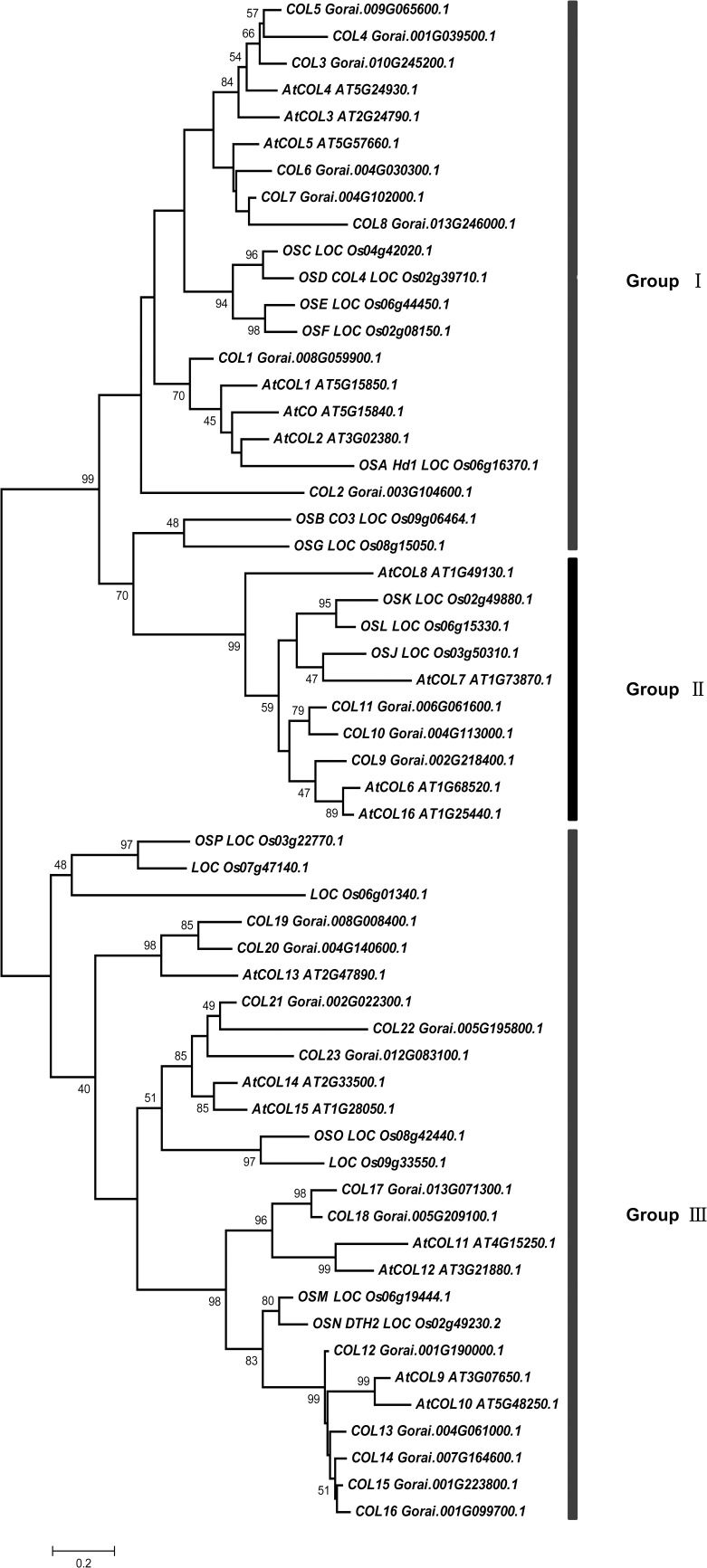
We identified 23 genes encoding both B-box and CCT domains, which were designated COL1-23 (S1 Table). To clarify the phylogenetic relationship between COL family genes, we constructed phylogenetic trees using amino acid sequences of the COLs in Arabidopsis, rice, and cotton using the maximum likelihood (ML) method based on multiple alignment analyses. As shown in Fig. 1, three major clades are indicated in the tree, and COLs in cotton were classified into groups I, II, and III, which are similar to the groups identified for the dicot plant Arabidopsis and the monocot plant rice. These results indicate that divergence of COLs from different species occurred earlier than the divergence of monocots and dicots. There are eight genes in group I, which were predicted to encode two B-box and one CCT domain, except for COL8, encoding a protein with one intact B-box, one incomplete B-box, and one CCT domain. Three genes in group II were predicted to encode proteins containing one B-box and one CCT domain. The remaining 12 genes, which are in group III, encode one B-box, a second diverged zinc finger, and one CCT domain. Furthermore, we cloned the eight genes in group I, designated COL1 to COL8, and studied these genes via phylogenetic and evolutionary analysis.
Fig 1. Maximum likelihood tree of CONSTANS-like proteins in Arabidopsis, rice and cotton.
CO-like proteins in Arabidopsis and rice are based on Griffiths et al. (2003) and the Plant Transcription Factor Database (http://plntfdb.bio.uni-potsdam.de/v3.0/), and their amino acid sequences were obtained from the Plant Transcription Factor Database.
Using gene-specific primers (S2 Table), we performed full-length PCR cloning and sequencing of the eight genes in 25 cotton accessions and one outgroup (Table 1), which led to the identification of eight COL genes present in one copy in the diploid cotton species and two copies in the allotetraploids. The results of structural characterization of the eight genes in the 25 cotton accessions are summarized in Table 2. The eight genes are highly conserved, and their full-length genomic DNA sequences are ranging from 1,030 bp (COL6) to 1,611 bp (COL1), with exception of frame-shift mutation for 1bp deletion in few species. Multiple alignments of the genomic and cDNA sequences showed that all genes share the same one-intron structure. This intron ranges from 77 to 680 bp in length, with the longest intron present in COL1, COL2, and COL8 compared with that of the other family members. For the same subgenome in different cotton accessions, insertion/deletion events occurred in introns or exon II of COL2, COL6, and COL8, leading to their length variation, while the remaining five genes had the same length in the same subgenomes of different cotton accessions. The structures of A- and D- homeologs from the same gene were further analyzed. Length differences were present in homeologs of COL4 and COL7, which were caused by insertions/deletions in exon I or II. There are two distinct homeologs for all genes in each allotetraploid cotton accession, while there is a single type of COL3 in the outgroup Thespesia populneoides (Roxb.) Kostel. Sequence information for these eight genes in the 25 cotton accessions and one outgroup has been submitted to GenBank (accession numbers: KM201660-KM202059).
Table 2. Structural analysis of eight COL genes in 25 cotton accessions.
| Gene | Full length/length of ORF(bp)/number of AA/length of extron1/length of intron/length of extron2 | Accession numbers |
|---|---|---|
| COL1 | 1611/1125/374/774/486/351 | KM201660-KM201709 |
| COL2 | 1 a ,14–22 a , and 24–25 a At b :1547/1125/374/790/422/335;3–13 a At b :1550/1128/375/790/422/338;23 a At b :1502/1080/359/790/422/290;2 Dt c :1543/1122/373/790/421/332;3–25 a Dt c :1546/1125/374/790/421/335 | KM201710-KM201759 |
| COL3 | 1094/1017/338/699/77/318 | KM201760-KM201809 |
| COL4 | 23 a At b : 342/342/113/342;1 a ,3–22 a ,and 24–25 a At b :1071/981/326/697/90/284; Dt c :1090/999/332/697/91/302 | KM201810-KM201859 |
| COL5 | 1103/1008/335/693/95/315 | KM201860-KM201909 |
| COL6 | At b :1033/945/314/626/88/319;2 Dt c :1030/942/313/626/88/316;3–23 a and 25 a Dt c :1031/942/313/626/89/316; 24 a Dt c :1032/942/313/626/90/316 | KM201910-KM201959 |
| COL7 | At b :1206/1110/369/713/96/397; Dt c :1203/1107/368/710/96/397 | KM202010-KM202059 |
| COL8 | 14–19 a and 24 a At b : 1559/882/293/516/677/366;1 a ,3 a ,11 a ,13 a and 20–23 a At b :1565/888/295/516/677/372;12 a At b : 1568/888/295/516/680/372;14–22 a and 24 a Dt c :1519/882/293/516/637/366;9–10 a ,12–13 a Dt c :1548/888/295/516/660/372; 23 a Dt c :1549/891/296/516/658/375; 3–6 a Dt c :1552/894/297/516/658/378;8 a Dt c :1553/894/297/516/659/378;7 a ,11 a and 25 a Dt c :1554/894/297/516/660/378;2 a Dt c :1360/684/227/516/676/168 | KM201960-KM202009 |
a Code designations are the same as in Table 1.
b At = A-subgenome from tetraploid cotton species.
c Dt = D-subgenome from tetraploid cotton species.
Diurnal expression pattern in light/dark cycles of the eight COL genes
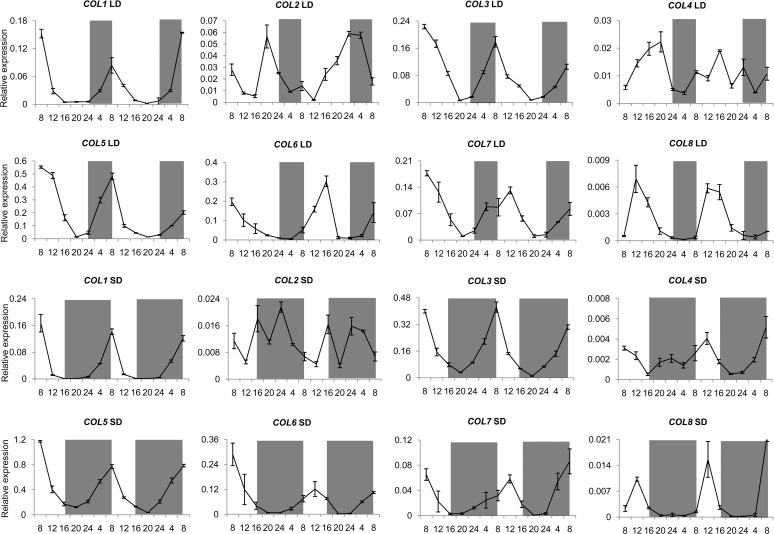
To examine the circadian rhythm of the candidate COL genes in cotton, we designed the gene-specific primers for qRT-PCR according to D-genome sequences (S2 Table), and investigated the expression level in the seedling leaves when the third leaf fully open under long-day (LD) (16h light/8h dark) or short-day (SD) (8h light/ 16h dark) condition respectively. The eight COL genes all showed diurnal expression patterns (Fig. 2). COL1, COL3 and COL5 exhibited the similar diurnal expression patterns under LD and SD conditions, the expression peaked in the dawn and started to decrease rapidly to the lowest at the end of light, then started to accumulate until the next dawn. COL6 and COL7 also had cycled with the light/dark induction treatment, but with the highest level 4 h later after the dawn and with lowest 8 h later. COL8 expression started to accumulate after dawn with the peak 4 h later, and then declined quickly in the both photoperiodic conditions. COL2 and COL4 showed different expression patterns in the two photoperiodic conditions. The expression pattern of COL4 was similar to COL6 and COL7 in SD condition, while there was no obvious diurnal expression pattern in LD condition. The expression of COL2 started to accumulate at 4 h after dawn with expression peaking at dusk, and then declined during the dark in LD condition. However, COL2 peaked twice in SD condition, its first peak occurred at the dusk, and reached the second peak 8h later. The diurnal expression patterns of the eight COLs suggest their conserved function in regulating the light signaling pathway in cotton.
Fig 2. Diurnal expression patterns of eight COL genes in TM-1 leaves under LD or SD conditions.
The X-axin represent time point (hours) and Y-axin indicates relative expression levels with cotton histone3 (AF024716) as the control. Mean values ± SD were obtained from three biological repeats. The gray bars over each chart represent dark periods.
Eight COL homeologs from allotetraploid species showed independent evolution after polyploid formation
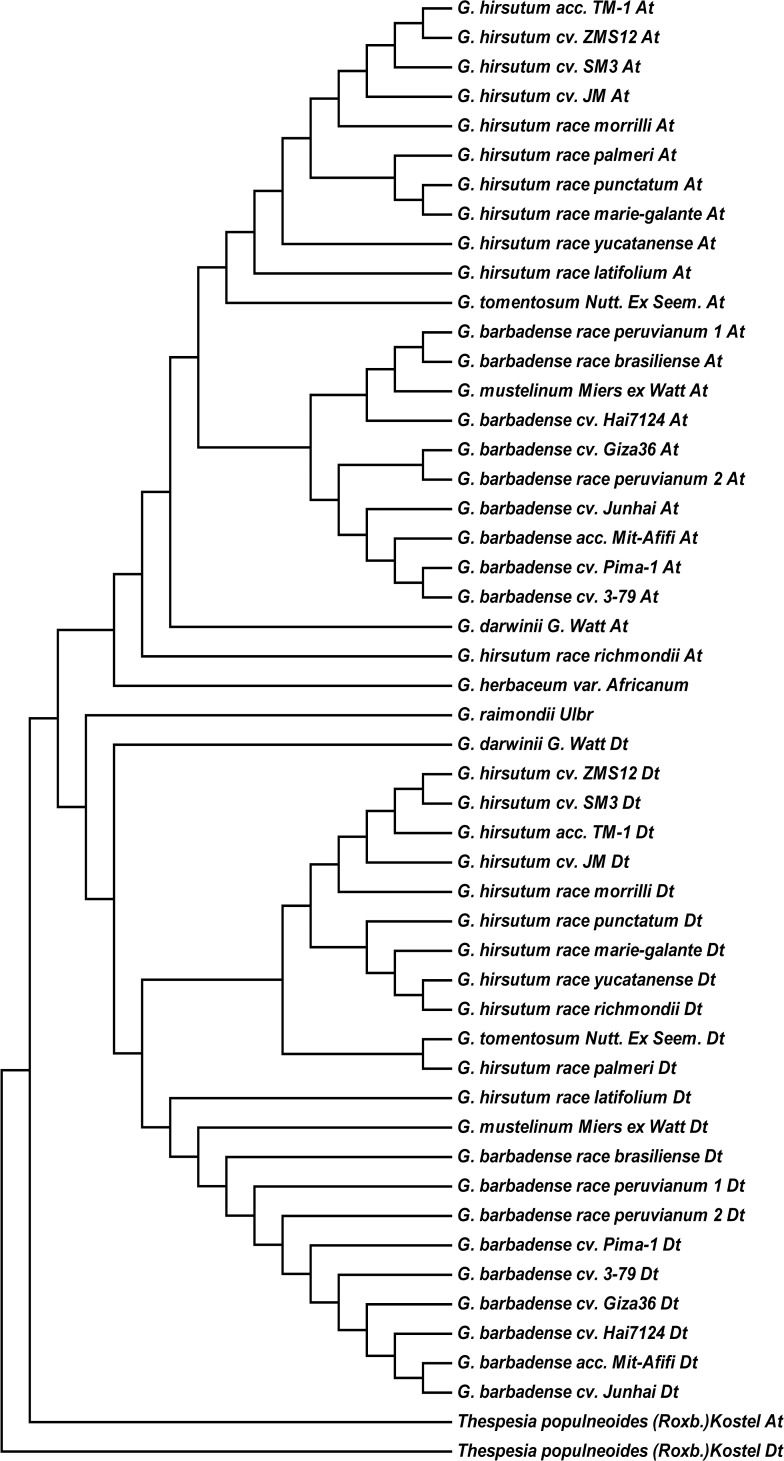
The sequences of the eight genes from the same subgenome were combined in order for 25 cotton accessions and one outgroup, and a phylogenetic tree was constructed using the ML method (Fig. 3). The outgroup Thespesia populneoides (Roxb.) Kostel was the most divergent member of this group and clustered into an individual clade, while the other members were divided into two principal clades; the A-genome and A-subgenomes in the tetraploid cotton accessions comprised one monophyletic clade, while the D-genome and D-subgenomes represented another monophyletic clade. Furthermore, the A-genome group was divided into two main subgroups; one included the A-subgenomes of G. hirsutum semi-domesticated, cultivated accessions and G. tomentosum and the other included the A-subgenomes of G. barbadense semi-domesticated, cultivated accessions and G. mustelinum species. G. darwinii and G. hirsutum race richmondii were clustered below in the A-genome group. Similarly, two main subgroups for G. barbadense and G. hirsutum were divided in the D-genome group, with the closest relationship between G. hirsutum and G. tomentosum and between G. barbadense and G. mustelinum. The exception was G. darwinii, the lone member of the D-genome group.
Fig 3. Phylogenetic tree based on the combined sequence of eight CO-like genes by the maximum likelihood method.
Bootstrap values (%) based on 1000 replicates are indicated beside the nodes.
Using G. herbaceum and G. raimondii as controls for comparisons of their orthologs, we calculated the synonymous substitution rates (Ks) of each tested gene between orthologs (A vs. D, A vs. At, and D vs. Dt) and between homeologs (At vs. Dt) in the 25 accessions based on their coding regions (S3 Table). Of the 184 pairs compared (eight pairwise comparisons × 23 allotetraploid accessions) in allotetraploid species, the Ks values of 98.37% of the genes were higher in the A-D and At-Dt comparison than in the A-At and D-Dt comparisons. Furthermore, the Pearson’s correlation coefficient (r) of Ks between A vs. D and At vs. Dt also showed positive, high correlations, with correlation coefficients of at least 0.797 (Table 3).
Table 3. Correlation analysis between A-D and At-Dt comparisons for each allotetraploid accession.
| Pairwise comparison | r of Ks |
|---|---|
| A-D vs 3 At-Dt | 0.919** |
| A-D vs 4 At-Dt | 0.919** |
| A-D vs 5 At-Dt | 0.919** |
| A-D vs 6 At-Dt | 0.894** |
| A-D vs 7 At-Dt | 0.868** |
| A-D vs 8 At-Dt | 0.922** |
| A-D vs 9 At-Dt | 0.917** |
| A-D vs 10 At-Dt | 0.842** |
| A-D vs 11 At-Dt | 0.902** |
| A-D vs 12 At-Dt | 0.939** |
| A-D vs 13 At-Dt | 0.869** |
| A-D vs 14 At-Dt | 0.890** |
| A-D vs 15 At-Dt | 0.900** |
| A-D vs 16 At-Dt | 0.888** |
| A-D vs 17 At-Dt | 0.888** |
| A-D vs 18 At-Dt | 0.889** |
| A-D vs 19 At-Dt | 0.889** |
| A-D vs 20 At-Dt | 0.905** |
| A-D vs 21 At-Dt | 0.896** |
| A-D vs 22 At-Dt | 0.903** |
| A-D vs 23 At-Dt | 0.797* |
| A-D vs 24 At-Dt | 0.873** |
| A-D vs 25 At-Dt | 0.871** |
Code designations are the same as in Table 1.
r: Correlation coefficient.
** Correlation is significant at P<0.01 (2-tailed).
* Correlation is significant at P<0.05 (2-tailed).
Taken together, these results suggest that A-D divergence for the eight COLs occurred well before the formation of the polyploids, and duplicated genes of A- and D- subgenomes from allotetraploid species evolve independently after the formation of the polyploids.
Nucleotide diversity of the eight COLs showed different homoelogous evolutionary rate in allotetraploid species
Pairwise comparisons of nucleotide diversity (π) for the combined sequence of the eight COL genes and each gene between subgenomes within each allotetraploid accession was performed, respectively (Table 4). The average π value of the combined sequence in the D vs Dt (0.01051) were significantly greater than the value in A vs At (0.00586) (P = 4.9E-21). Among the 184 pairwise comparisons, 76.63% (141) harbored greater nucleotide diversity in the D-subgenome than that in the A-subgenome in the allotetraploid accessions. In detail, six genes, including COL2 to COL5, COL7 and COL8, showed significantly higher nucleotide diversity in the D-subgenome than in the A-subgenome of the allotetraploid accessions examined. However, COL6 showed significantly higher nucleotide diversity in the A-subgenome than in the D-subgenome. There was no significant difference in the A vs At and D vs Dt in COL1. These results indicate that the eight COLs in group I harbor different evolutionary rates between homeologs of the allotetraploid accessions, and most genes of the D-subgenomes have been evolving more rapidly than those of the A-subgenomes.
Table 4. Estimates of nucleotide diversity of A vs At and D vs Dt for eight COL genes in cotton according to their genomic sequences.
| Gene | A1-At | D5-Dt |
|---|---|---|
| combined sequence | 0.00586 | 0.01051** |
| COL1 | 0.00605 | 0.00532 |
| COL2 | 0.00624 | 0.00848** |
| COL3 | 0.0039 | 0.00827** |
| COL4 | 0.00678 | 0.01201** |
| COL5 | 0.00161 | 0.00584** |
| COL6 | 0.01124** | 0.00536 |
| COL7 | 0.0048 | 0.00575** |
| COL8 | 0.00631 | 0.02956** |
| average | 0.00587 | 0.01007 |
Taxa include 23 allotetraploids accessions and the two genome donors to the allotetraploid accessions. At = A genome from the allotetraploid cotton species; Dt = D genome from the allotetraploid cotton species; D5 = G. raimondii; A1 = G. herbaceum.
** P<0.01
Nucleotide diversity of the eight COLs showed different evolutionary rate in different cotton species and different domains
To further explore the domestication forces acting on allotetraploid species, we divided the tested allotetraploid accessions into three types, including tetraploid wild species, semi-domesticated and domesticated species of G. hirsutum, semi-domesticated and domesticated species of G. barbadense. Their nucleotide diversity (π) was estimated respectively for synonymous, nonsynonymous and the total sites of each data set with the ORF of each gene (Table 5). Generally speaking, the nucleotide diversity at synonymous substitution sites (πs) was significantly higher than that at non-synonymous substitution sites (πa) (0.00451 vs 0.00179)(P = 0.0001), and the three wild allotetraploid species possessed significally higher nucleotide diversity of πtotal than G. hirsutum (0.00369 vs 0.00139) (P = 0.001), and G. barbadense (0.00369 vs 0.00035) (P = 7.3E-7), suggesting a genetic bottleneck associated with the domesticated cotton species.
Table 5. Nucleotide polymorphism and neutrality tests of eight COL genes in cotton according to their ORF sequences.
| Gene | Accession | πtotal | πs | πa | TD | FD | FF |
|---|---|---|---|---|---|---|---|
| COL1At | total | 0.00226 | 0.00479 | 0.00147 | −2.21** | −3.62** | −3.73** |
| G. hirsutum | 0.00304 | 0.00734 | 0.0017 | −2.00* | −2.33** | −2.54** | |
| G. barbadense | 0.00059 | 0.00083 | 0.00052 | −1.51 | −1.68 | −1.82 | |
| wild | 0.00178 | 0.00748 | 0 | nd | Nd | nd | |
| COL1Dt | total | 0.00385 | 0.00752 | 0.00272 | −1.01 | 0.06 | −0.31 |
| G. hirsutum | 0.00252 | 0.00477 | 0.00183 | −1.34 | −1.47 | −1.63 | |
| G. barbadense | 0.00198 | 0.00396 | 0.00136 | 0.03 | −0.29 | −0.23 | |
| wild | 0.0077 | 0.01998 | 0.0039 | nd | Nd | nd | |
| COL2At | total | 0.00335 | 0.00698 | 0.00232 | −0.47 | −1.52 | −1.41 |
| G. hirsutum | 0.00045 | 0 | 0.00059 | −0.78 | −0.33 | −0.49 | |
| G. barbadense | 0.00089 | 0 | 0.00115 | 1.23 | 1.06 | 1.22 | |
| wild | 0.00681 | 0.01451 | 0.00462 | nd | Nd | nd | |
| COL2Dt | total | 0.00232 | 0.00481 | 0.00161 | −0.13 | −1.57 | −1.33 |
| G. hirsutum | 0.00081 | 0.00144 | 0.00063 | −1.79* | −2.13* | −2.30* | |
| G. barbadense | 0 | 0 | 0 | nd | Nd | nd | |
| wild | 0.00533 | 0.01055 | 0.00385 | nd | Nd | nd | |
| COL3At | total | 0.00274 | 0.00214 | 0.00294 | 0.10 | −0.58 | −0.44 |
| G. hirsutum | 0.00097 | 0 | 0.00127 | 1.34 | 1.00 | 1.21 | |
| G. barbadense | 0.00055 | 0 | 0.00072 | 1.40 | 0.84 | 1.07 | |
| wild | 0.00393 | 0.00277 | 0.00431 | nd | Nd | nd | |
| COL3Dt | total | 0.00282 | 0.00559 | 0.00198 | 0.20 | −0.58 | −0.41 |
| G. hirsutum | 0.00136 | 0.00366 | 0.00066 | −0.73 | −0.76 | −0.85 | |
| G. barbadense | 0 | 0 | 0 | nd | Nd | nd | |
| wild | 0.00459 | 0.00278 | 0.00517 | nd | Nd | nd | |
| COL4At | total | 0.00198 | 0.00114 | 0.00224 | −1.40 | −2.41 | −2.46 |
| G. hirsutum | 0.00089 | 0.00158 | 0.00068 | −1.32 | −1.21 | −1.39 | |
| G. barbadense | 0.00113 | 0 | 0.00148 | 0.03 | 0.23 | 0.20 | |
| wild | 0.00544 | 0.00291 | 0.00623 | nd | Nd | nd | |
| COL4Dt | total | 0.00017 | 0 | 0.00023 | −1.51 | −2.13 | −2.26 |
| G. hirsutum | 0.00018 | 0 | 0.00024 | −1.13 | −1.29 | −1.40 | |
| G. barbadense | 0 | 0 | 0 | nd | Nd | nd | |
| wild | 0.00067 | 0 | 0.00087 | nd | Nd | nd | |
| COL5At | total | 0.00172 | 0.00287 | 0.0013 | 0.20 | −0.95 | −0.71 |
| G. hirsutum | 0.00069 | 0 | 0.0009 | 0.04 | −0.33 | −0.27 | |
| G. barbadense | 0 | 0 | 0 | nd | Nd | nd | |
| wild | 0.00265 | 0.00833 | 0.00087 | nd | Nd | nd | |
| COL5Dt | total | 0.00225 | 0.00434 | 0.00161 | −1.05 | −1.55 | −1.63 |
| G. hirsutum | 0.00152 | 0.00076 | 0.00176 | −0.40 | −0.08 | −0.18 | |
| G. barbadense | 0 | 0 | 0 | nd | Nd | nd | |
| wild | 0.00463 | 0.01674 | 0.00087 | nd | Nd | nd | |
| COL6At | total | 0.00236 | 0.00669 | 0.00106 | −0.87 | −2.25 | −2.15 |
| G. hirsutum | 0.00166 | 0.00399 | 0.00096 | −1.40 | −1.23 | −1.44 | |
| G. barbadense | 0 | 0 | 0 | nd | Nd | nd | |
| wild | 0.00353 | 0.01209 | 0.00092 | nd | Nd | nd | |
| COL6Dt | total | 0.00081 | 0.00308 | 0.00012 | −0.83 | −1.80 | −1.76 |
| G. hirsutum | 0.00019 | 0 | 0.00025 | −1.13 | −1.29 | −1.40 | |
| G. barbadense | 0 | 0 | 0 | nd | Nd | nd | |
| wild | 0.00142 | 0.00612 | 0 | nd | Nd | nd | |
| COL7At | total | 0.00191 | 0.00618 | 0.00066 | 0.37 | −0.66 | −0.42 |
| G. hirsutum | 0.00033 | 0 | 0.00043 | −1.43 | −1.66 | −1.80 | |
| G. barbadense | 0.0005 | 0 | 0.00065 | 1.40 | 0.84 | 1.07 | |
| wild | 0.0018 | 0.00529 | 0.00078 | nd | nd | nd | |
| COL7Dt | total | 0.00274 | 0.00564 | 0.00188 | −1.08 | −2.20 | −2.18 |
| G. hirsutum | 0.00312 | 0.0046 | 0.00267 | −1.44 | 1.71 | −1.86 | |
| G. barbadense | 0 | 0 | 0 | nd | nd | nd | |
| wild | 0.0012 | 0.00264 | 0.00078 | nd | nd | nd | |
| COL8At | total | 0.00341 | 0.00671 | 0.00241 | 0.37 | −1.08 | −0.76 |
| G. hirsutum | 0.00119 | 0.00203 | 0.00093 | −1.46 | −1.44 | −1.63 | |
| G. barbadense | 0 | 0 | 0 | nd | nd | nd | |
| wild | 0.00378 | 0.00647 | 0.00299 | nd | nd | nd | |
| COL8Dt | total | 0.00394 | 0.00366 | 0.00405 | 0.57 | 0.51 | 0.62 |
| G. hirsutum | 0.00324 | 0.00637 | 0.00227 | 1.59 | 1.33 # | 1.57 # | |
| G. barbadense | 0 | 0 | 0 | nd | nd | nd | |
| wild | 0.00378 | 0 | 0.00498 | nd | nd | nd | |
| average | total | 0.00241 | 0.00451 | 0.00179 | −0.54725 | −1.39592 | −1.33363 |
| G. hirsutum | 0.00139 | 0.00228 | 0.00111 | −0.83652 | −0.72036 | −1.02529 | |
| G. barbadense | 0.00035 | 0.00030 | 0.00037 | 0.16132 | 0.06295 | 0.09409 | |
| wild | 0.00369 | 0.00742 | 0.00257 | nd | nd | nd |
TD, the Tajima’s D; FD, Fu and Li’s D; FF, Fu and Li’s F.
nd, not determined (not implemented yet).
# P<0.1
* P<0.05
** P<0.01.
As reported by Robson et al. (2001) [33] and Griffiths et al. (2003) [30], the B-box and CCT domains are two conserved domains of CO proteins that are required for the promotion of flowering. To further explore the evolutionary rate of the three characteristic domains, we further analyzed the nucleotide diversity of the three domains of each data set for the eight COL genes respectively (Table 6). The nucleotide diversity of most genes was significantly lower in the CCT domain, while the B-box domain and the Var domain possessed relatively high rates of replacement substitutions. There were no diffrences in π between B-box and the Var domain (P = 0.285), and the two domains evolved signifacally faster than the CCT domain (0.00284 vs 0.00119, P = 0.028 for B-box and 0.00243 vs 0.00119, P = 0.014 for Var domain, respectively). These results demonstrate that the B-box and Var domains have been quite variable, while the CCT domain is highly conserved in sequence and function among 25 cotton accessions.
Table 6. Nucleotide polymorphism of B-box, Var and CCT domains for eight COL genes in allotetraploid cotton.
| Gene | B-box | Var | CCT |
|---|---|---|---|
| COL1At | 0.00104 | 0.00213 | 0.00251 |
| COL1Dt | 0.0016 | 0.00493 | 0.00576 |
| COL2At | 0.00696 | 0.00314 | 0 |
| COL2Dt | 0.00311 | 0.00269 | 0 |
| COL3At | 0.00406 | 0.00145 | 0.00067 |
| COL3Dt | 0.00357 | 0.002 | 0.00135 |
| COL4At | 0.00435 | 0.0014 | 0.00067 |
| COL4Dt | 0.00035 | 0.00016 | 0 |
| COL5At | 0.00069 | 0.00285 | 0 |
| COL5Dt | 0.0027 | 0.00149 | 0.00202 |
| COL6At | 0.00073 | 0.00306 | 0.00533 |
| COL6Dt | 0.00073 | 0.00151 | 0 |
| COL7At | 0.00315 | 0.00236 | 0 |
| COL7Dt | 0.00248 | 0.00376 | 0 |
| COL8At | 0.0072 | 0.00022 | 0.00067 |
| COL8Dt | 0.00267 | 0.00579 | 0 |
| average | 0.00284 | 0.00243 | 0.00119 |
Neutrality tests of the eight COL genes reveal three selected genes during domestication
To test the departure from neutrality, Tajima’s D (1989) and Fu and Li’s D and F (1993) [43–44] were estimated to test whether the nucleotide polymorphism data of the eight COL genes fit the neutral model (Table 5). We showed that both COL1 A-subgenome and COL2 D-subgenome in G. hirsutum significantly deviated from the neutral expectation with a negative value, indicating an excess of low frequency alleles. And the negative values are consistent with the possibility of recent positive selection in G. hirsutum. Fu and Li’ D and F were significantly positive in COL8 D-subgenome of G. hirsutum at P<0.1, this result suggest that the allele of COL8 maintained a high frequency variants and might experience balance selection [45–46]. Taken together, COL1, COL2 and COL8 endured greater selective pressures during the domestication process.
Discussion
The homeologs of eight COL genes are evolving independently at the allopolyploid level
Allotetraploids originate from an interspecific hybridization event between diploid A- and D-genome species. Here, we performed ML analysis of the eight genes among 26 accessions, including 25 cotton accessions and one outgroup, to help elucidate the relationship between the homeologs at the allopolyploid level. The phylogenetic analysis showed that the outgroup Thespesia populneoides (Roxb.) Kostel was quite distant from the other allotetraploid cotton species and clustered into an individual clade, while the others were divided into two major clades, each containing the At or Dt subgroup with their corresponding diploid ancestral species. The results show that homeologs of the eight genes are evolving independently in the tetraploid accessions examined, including wild, semi-domesticated, and cultivated species. Furthermore, 98.37% of the Ks values were higher in the A-D and At-Dt comparisons than in the A-At and D-Dt comparisons, and the Pearson’s correlation coefficient for the A-At and D-Dt comparisons of the eight genes of the diploid and all of the allotetraploid accessions exhibited a significant positive correlation (r 2 = 0.797). This observation indicates that the A-D divergence occurr well before the formation of the polyploids, and duplicated genes of At and Dt of eight COL genes from allotetraploid species evolve independently after the formation of the polyploids. These results are in agreement with the results of previous reports [47–49]. From the study, G. tomentosum (from the Hawaiian Islands) had a closer relationship with G. hirsutum, while G. mustelinum was closer to G. barbadense than to G. darwinii. Similarly, the D and Dt clade yielded similar results to those of the A and At clade. These results are also largely in agreement with those of previous studies [2,5].
COL transcription factors have conserved functions among different plant species
COL transcription factors play important roles in regulate flowering time in the photoperiod signaling pathway, which coordinates light and circadian clock inputs (primarily in leaves) to induce the expression of the florigen gene FLOWERING LOCUS T (FT) [50–51]. These proteins are widely present among species, from lower plants such as mosses [52–53] to algae (which exhibit strong photoperiod responses [54–55]) to higher flowering plants including monocots and dicots. These transcription factor genes include CO in Arabidopsis, Hd1 in rice, and its homologs in barley, ryegrass, sugar beet, and soybean [25,26,56–59]. The CO-FT module is conserved in all known plant species, although it has different modes of action in different species. CO promotes the expression of FT under LD conditions in Arabidopsis thaliana [25,33], while Hd1, the ortholog of CO in rice, functions in the promotion of Hd3a (the FT ortholog) expression under SD conditions and as a repressor under non-inductive long day conditions [50,60]. CO is a central regulator of the photoperiod pathway, triggering the production of the mobile florigen hormone FT, which induces flower differentiation. The homologs of COL3 and COL5 have previously been cloned in cotton, and qRT-PCR analysis shows that the expression of the COL5 homolog is controlled by daily oscillations and exhibits a diurnal rhythm, with higher expression levels observed in the dark than in the light [61]; this expression pattern is similar to that of CO in Arabidopsis and COL in other plants [22,31,56,62,63]. COL genes in group I of cotton harbored two B-box and a CCT conserved domains with the same to that in other plants, and expression analysis indicated that all the eight genes showed a diurnal rhythm expression pattern in TM-1. COL1, COL3, and COL5-COL7 showed similar diurnal expression patterns under both LD and SD conditions, and the expression peak were present in the dawn or 4h later, and declined rapidly to the lowest until dusk, with similar to AtCOL1 and AtCOL2 in Arabidopsis, GmCOL1 and GmCOL2 in soybean, OsB, OsE and OsD in rice [31,33,56]. COL8 showed similar diurnal expression pattern with OsG, which was also a special gene with internal deletion of the second B-box domain [31]. COL4 was one unique gene that the diurnal expression pattern were more evident under SD than in LD condition just like ZCN8 in maize[64], and COL4 might perceive SD signal in TM-1, but not responsive to LD regulation. The COL2 expression in LD condition peaked once per 24h-period and twice in SD condition. The diurnal expression analysis indicated that the COL gene family in group I was potentially involved in regulating the light signaling pathway or photoperiodic flowering in cotton as other plants, but more detailed functional analyses are needed for further study.
Selection signatures of eight COL genes in coding region and domains in the allotetraploid species
Nucleotide polymorphisms of the eight COL genes show that most COL genes in wild allopolyploid species possess significantly higher nucleotide diversity than that of G. hirsutum and G. barbadense, this reduction of diversity could result from genetic bottlenecks during various stages of domestication. The limited genetic diversity of cultivated G. hirsutum had been observed in previous studies [65–66]. The neutrality test showed that COL1 A-subgenome and COL2 D-subgenome of G. hirsutum significantly deviated from zero with a negative value, implying an excess of low frequency alleles. This was consistent with the possibility of recent positive selection in G. hirsutum [3,11]. COL1 displays diurnal expression patterns with similar to AtCOL1 and AtCOL2 in Arabidopsis, the result indicated that COL1 in cotton may play conserved function in light input pathway but not affect flowering time [62]. While COL2, orthologous to Hd1 in rice, exhibits distinct diurnal expression in LD and SD conditions, indicating that COL2 was potentially regulating photoperiodic flowering in cotton, with similar function as Hd1 [26]. So, COL1 and COL2 genes were the potential target of positive selection in light signal or photoperiodic flowering pathway of G. hirsutum. Especially, nucleotide diversity of COL2 D-subgenome of G. hirsutum is approximately sixfold lower than the wild allopolyploid species, so COL2 is expected to be the better selected CO gene in cotton. Interestingly, COL8, with one intact B-box domain similar to that of OsB/OsCO3 and OsG [30], evolved faster among the tested eight genes. OsB/OsCO3 was reported to regulate negatively the photoperiodic flowering in rice [67], and COL8 showed similar diurnal expression pattern with OsG [31]. So COL8 might also involve in the photoperiodic flowering in cotton. The neutrality tests of COL8 D-subgenome were significantly positive with P<0.1 in G. hirsutum, indicating an excess of higher frequency alleles, and balancing selection is expected to act on COL8 [45]. Higher frequency variants in COL8 D-subgenome may contribute to satisfy the need of multiple environments and better adaptation of cotton, and promote the evolution of photoperiod sensitivity in G. hirsutum. Taken together, selection acted on the three potential target COL genes in G. hirsutum might be responsibe for the wide adaptation of G. hirsutum [2]. In other plants such as rice and maize, selection of COL homologs appears to be common during parallel adaptation [12,22,68–70].
CO is a typical transcription factor with three characteristic domains, including the B-box, Var domain, and CCT domain, which indicates that it is a unique type of transcriptional regulator present only in the plant kingdom [25]. COL genes within the Brassicaceae family are evolving rapidly, and different domains in the COL genes are heterogeneous [34]. We analyzed the nucleotide diversity of B-box domain, the Var domain and the CCT domain in cotton respectively. The results suggested that the nucleotide diversity of most genes were significantly lower in the CCT domain, indicated that the CCT domain is highly conserved, possibly due to high functional evolutionary constraints acting on this domain. Natural variation within genes with CCT domains has previously been reported, including COL, PRR (PSEUDO RESPONSE REGULATORs), and CMF genes, which are critical to the control of plant flowering [24–26,32,71–73]. The CCT domain shows homology to the NF-YA1/2 domains of HAP2, which help form the trimeric CO/At HAP3/At HAP5 complex and bind to CCAAT boxes in eukaryotic promoters to regulate flowering of Arabidopsis through the expression of FT [74], as well as interacting with the ubiquitin ligase COP1 [75] and nuclear localization signal [30,33]. Therefore, the strong conservation of the CCT domain is thought to be necessary for its role in the control of photoperiodic flowering. B-box domain is involved in DNA binding and protein-protein interactions, as plants with mutations in this region display severe late flowering phenotypes [33,76]. Most genes with B-box domains display a divergent diurnal expression pattern, indicating that this domain functions in the light signaling pathway [31,61,62,77]. The Var domain, with a lower degree of conservation in amino acid sequence among the COLs, activates transcription, as demonstrated by yeast-two hybrid assays [78], although its fixed residues are significantly conserved. It is recently shown that DTH2, which encodes a COL protein in rice, and two functional nucleotide polymorphisms (FNPs) in the B-box and Var domain, respectively, are associated with the changes in flowering time and increased reproductive fitness that have occurred during the northward expansion of rice cultivation [22]. In this study, the B-box and the Var domain evolve signifacally faster than the CCT domain among the eight COL genes, indicating the two domains endure relax evolutionary constraint, and may be associated with the changes in flowering time of cotton. Higher nucleotide diversity in the two domains may enable cotton to form a diversity of habitats to adapt the variable environments and expansion of the cultivation area.
Conclusions
CO-FT is conserved and plays important roles in the photoperiodic regulation of flowering time in plants. We revealed that eight COL homeologs from allotetraploid accessions have evolved independently after polyploid formation. COL1, COL2 and COL8 are potential selected genes during domestication, with strong conservation on the CCT domain and great diversity on the B-box and Var domains. This study provides valuable information that increases our understanding of the dynamic evolutionary of the COL gene family in cotton and the potential target COL genes during the domestication and adaptation of cotton.
Supporting Information
(XLS)
(XLS)
(XLS)
Acknowledgments
This program was financially supported in part by The State Key Basic Research and Development Plan of China (2011CB109300), and a project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions and JCIC-MCP. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This program was financially supported in part by The State Key Basic Research and Development Plan of China (2011CB109300), and a project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions and JCIC-MCP. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Cronn RC, Small RL, Haselkorn T, Wendel JF. Rapid diversification of the cotton genus (Gossypium: Malvaceae) revealed by analysis of sixteen nuclear and chloroplast genes. Am J Bot. 2002; 89: 707–725. 10.3732/ajb.89.4.707 [DOI] [PubMed] [Google Scholar]
- 2. Wendel JF, Cronn RC. Polyploidy and the evolutionary history of cotton. Adv Agron. 2003; 78: 139–186. [Google Scholar]
- 3. Wendel JF, Brubaker CL, Seelanan T. The origin and evolution of Gossypium In: Physiology of cotton. Edited by Stewart JM, Oosterhuis D, Heitholt JJ, Mauney JR. (Eds.).Netherlands: Springer; 2010; 1–18. [Google Scholar]
- 4. Buriev ZT, Saha S, Shermatov SE, Jenkins JN, Abdukarimov A, Stelly DM, et al. Molecular evolution of the clustered MIC-3 multigene family of Gossypium species. Theor Appl Genet. 2011; 123: 1359–1373. 10.1007/s00122-011-1672-y [DOI] [PubMed] [Google Scholar]
- 5. Dejoode DR, Wendel JF. Genetic diversity and origin of the Hawaiian Islands Cotton, Gossypium tomentosum . Am J Bot. 1992; 79: 1311–1319. [Google Scholar]
- 6. Wendel JF, Rowley R, Stewart JM. Genetic diversity in and phylogenetic relationships of the Brazilian endemic cotton, Gossypium mustelinum (Malvaceae). Plant Syst Evol. 1994; 192: 49–59. [Google Scholar]
- 7. Wendel JF, Percy RG. Allozyme diversity and introgression in the Galapagos-Islands endemic Gossypium Darwinii and its relationship to continental Gossypium barbadense . Biochem Syst Ecol. 1990; 18: 517–528. [Google Scholar]
- 8. Brubaker CL, Wendel JF. Reevaluating the origin of domesticated cotton (Gossypium hirsutum Malvaceae) using nuclear restriction fragment length polymorphisms (RFLPs). Am J Bot. 1994; 81: 1309–1326. [Google Scholar]
- 9. Brubaker CL, Wendel JF. RFLP diversity in cotton In: Genetic improvement of cotton: Emerging technologies. Edited by Jenkins JN, Saha S. (Eds.) USA: Science Publishers Inc Enfield NH; 2001; 81–102. [Google Scholar]
- 10. Hutchinson JB. Intra-specific differentiation in Gossypium hirsutum . Heredity. 1951; 5: 161–193. [Google Scholar]
- 11. Bao Y, Hu G, Flagel LE, Salmon A, Bezanilla M, Paterson AH, et al. Parallel up-regulation of the profilin gene family following independent domestication of diploid and allopolyploid cotton (Gossypium). Proc Natl Acad Sci USA. 2011; 108: 21152–21157. 10.1073/pnas.1115926109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Olsen KM, Wendel JF. Crop plants as models for understanding plant adaptation and diversification. Front Plant Sci. 2013; 4: 290 10.3389/fpls.2013.00290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lubbers EL., Chee PW. The worldwide gene pool of G. hirsutum and its improvement In: Genetics and Genomics of Cotton. Edited by Paterson AH. (Ed.). New York: Springer; 2009; 23–52. [Google Scholar]
- 14. Percy RG. The worldwide gene pool of Gossypium barbadense L. and its improvement . In: Genetics and Genomics of Cotton. Edited by Paterson AH. (Ed.) New York: Springer; 2009; 53–68. [Google Scholar]
- 15. Harlan JR. Crops & man 2nd edn. Madison, Wis., USA: American Society of Agronomy: Crop Science Society of America; 1992. [Google Scholar]
- 16. Mouradov A, Cremer F, Coupland G. Control of flowering time: Interacting pathways as a basis for diversity. Plant Cell. 2002; 14: 111–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Boss PK, Bastow RM, Mylne JS, Dean C. Multiple pathways in the decision to flower: Enabling, promoting, and resetting. Plant Cell. 2004; 16: 18–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jack T. Molecular and genetic mechanisms of floral control. Plant Cell. 2004; 16 Suppl: S1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Baurle I, Dean C. The timing of developmental transitions in plants. Cell. 2006; 125: 655–664. [DOI] [PubMed] [Google Scholar]
- 20. Izawa T. Adaptation of flowering-time by natural and artificial selection in Arabidopsis and rice. J Exp Bot. 2007; 58: 3091–3097. [DOI] [PubMed] [Google Scholar]
- 21. Tsuji H, Taoka K, Shimamoto K. Regulation of flowering in rice: two florigen genes, a complex gene network, and natural variation. Curr Opin Plant Biol. 2011; 14: 45–52. 10.1016/j.pbi.2010.08.016 [DOI] [PubMed] [Google Scholar]
- 22. Wu W, Zheng XM, Lu G, Zhong Z, Gao H, Chen L, et al. Association of functional nucleotide polymorphisms at DTH2 with the northward expansion of rice cultivation in Asia. Proc Natl Acad Sci USA. 2013; 110: 2775–2780. 10.1073/pnas.1213962110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Suarez-Lopez P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis . Nature. 2001; 410: 1116–1120. [DOI] [PubMed] [Google Scholar]
- 24. Cheng XF, Wang ZY. Overexpression of COL9, a CONSTANS-LIKE gene, delays flowering by reducing expression of CO and FT in Arabidopsis thaliana . Plant J. 2005; 43: 758–768. [DOI] [PubMed] [Google Scholar]
- 25. Putterill J, Robson F, Lee K, Simon R, Coupland G. The CONSTANS gene Of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc-finger transcription factors. Cell. 1995; 80: 847–857. [DOI] [PubMed] [Google Scholar]
- 26. Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, Fuse T, et al. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS . Plant Cell. 2000; 12: 2473–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Robert LS, Robson F, Sharpe A, Lydiate D, Coupland G. Conserved structure and function of the Arabidopsis flowering time gene CONSTANS in Brassica napus . Plant Mol Biol. 1998; 37: 763–772. [DOI] [PubMed] [Google Scholar]
- 28. Liu JY, Yu JP, McIntosh L, Kende H, Zeevaart JAD. Isolation of a CONSTANS ortholog from Pharbitis nil and its role in flowering. Plant Physiol. 2001; 125: 1821–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nemoto Y, Kisaka M, Fuse T, Yano M, Ogihara Y. Characterization and functional analysis of three wheat genes with homology to the CONSTANS flowering time gene in transgenic rice. Plant J. 2003; 36: 82–93. [DOI] [PubMed] [Google Scholar]
- 30. Griffiths S, Dunford RP, Coupland G, Laurie DA. The evolution of CONSTANS-like gene families in barley, rice, and Arabidopsis. Plant Physiol. 2003; 131: 1855–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Huang J, Zhao X, Weng X, Wang L, Xie W. The rice B-box zinc finger gene family: genomic identification, characterization, expression profiling and diurnal analysis. PloS One. 2012; 7: e48242 10.1371/journal.pone.0048242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cockram J, Thiel T, Steuernagel B, Stein N, Taudien S, Bailey PC, et al. Genome dynamics explain the evolution of flowering time CCT domain gene families in the Poaceae. PloS One. 2012; 7: e45307 10.1371/journal.pone.0045307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Robson F, Costa MMR, Hepworth SR, Vizir I, Pineiro M, Reeves PH, et al. Functional importance of conserved domains in the flowering-time gene CONSTANS demonstrated by analysis of mutant alleles and transgenic plants. Plant J. 2001; 28: 619–631. [DOI] [PubMed] [Google Scholar]
- 34. Lagercrantz U, Axelsson T. Rapid evolution of the family of CONSTANS LIKE genes in plants. Mol Biol Evol. 2000; 17: 1499–1507. [DOI] [PubMed] [Google Scholar]
- 35. Roux F, Touzet P, Cuguen J, Le Corre V. How to be early flowering: An evolutionary perspective. Trends Plant Sci. 2006; 11: 375–381. [DOI] [PubMed] [Google Scholar]
- 36. Paterson AH, Wendel JF, Gundlach H, Guo H, Jenkins J, Jin D, et al. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature. 2012; 492: 423–427. 10.1038/nature11798 [DOI] [PubMed] [Google Scholar]
- 37. Paterson AH, Brubaker CL, Wendel JF. A rapid method for extraction of cotton (Gossypium spp.) genomic DNA suitable for RFLP or PCR analysis. Plant Mol Biol Rep. 1993; 11: 122–127. [Google Scholar]
- 38. Cronn R, Cedroni M, Haselkorn T, Grover C, Wendel JF. PCR-mediated recombination in amplification products derived from polyploid cotton. Theor Appl Genet. 2002; 104: 482–489. [DOI] [PubMed] [Google Scholar]
- 39. Jiang J, Zhang T. Extraction of totol RNA in cotton tissues with CTAB/acidic phenolic method. Cotton Sci. 2003; 15: 166–167. [Google Scholar]
- 40. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods. 2001; 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 41. Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009; 25: 1451–1452. 10.1093/bioinformatics/btp187 [DOI] [PubMed] [Google Scholar]
- 42. Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, et al. The Pfam protein families database. Nucleic Acids Res. 2012; 40: 290–301. 10.1093/nar/gkr717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tajima F. Statistical method for testing the neutral mutationhypothesis by DNA polymorphism. Genetics. 1989; 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fu YX, Li WH. Statistical tests of neutrality of mutations. Genetics. 1993; 133: 693–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hudson RR, Kaplan NL. The coalescent process in models with selection and recombination. Genetics. 1988; 120: 831–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wright SI, Gaut BS. Molecular population genetics and the search for adaptive evolution in plants. Mol Biol Evol. 2005; 22: 506–519. [DOI] [PubMed] [Google Scholar]
- 47. Cronn RC, Small RL, Wendel JF. Duplicated genes evolve independently after polyploid formation in cotton. Proc Natl Acad Sci USA. 1999; 96: 14406–14411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Guo WZ, Fang D, Yu WD, Zhang TZ. Sequence divergence of microsatellites and phylogeny analysis in tetraploid cotton species and their putative diploid ancestors. J Integr Plant Biol. 2005; 47: 1418–1430. [Google Scholar]
- 49. Zhu H, Lv J, Zhao L, Tong X, Zhou B, Zhang T, et al. Molecular evolution and phylogenetic analysis of genes related to cotton fibers development from wild and domesticated cotton species in Gossypium . Mol Phylogenet Evol. 2012; 63: 589–597. 10.1016/j.ympev.2012.01.025 [DOI] [PubMed] [Google Scholar]
- 50. Valverde F. CONSTANS and the evolutionary origin of photoperiodic timing of flowering. J Exp Bot. 2011; 62: 2453–2463. 10.1093/jxb/erq449 [DOI] [PubMed] [Google Scholar]
- 51. Turck F, Fornara F, Coupland G. Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu Rev Plant Biol. 2008; 59: 573–594. 10.1146/annurev.arplant.59.032607.092755 [DOI] [PubMed] [Google Scholar]
- 52. Shimizu M, Ichikawa K, Aoki S. Photoperiod-regulated expression of the PpCOL1 gene encoding a homolog of CO/COL proteins in the moss Physcomitrella patens . Biochem Biophys Res Commun. 2004; 324: 1296–1301. [DOI] [PubMed] [Google Scholar]
- 53. Zobell O, Coupland G, Reiss B. The family of CONSTANS-like genes in Physcomitrella patens. Plant Biol. 2005; 7: 266–275. [DOI] [PubMed] [Google Scholar]
- 54. Romero JM, Valverde F. Evolutionarily conserved photoperiod mechanisms in plants: when did plant photoperiodic signaling appear? Plant Signal Behav. 2009; 4: 642–644. 10.1016/j.cub.2009.01.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Serrano G, Herrera-Palau R, Romero JM, Serrano A, Coupland G, Valverde F. Chlamydomonas CONSTANS and the evolution of plant photoperiodic signaling. Curr Biol. 2009; 19: 359–368. 10.1016/j.cub.2009.01.044 [DOI] [PubMed] [Google Scholar]
- 56. Fan C, Hu R, Zhang X, Wang X, Zhang W, Zhang Q, et al. Conserved CO-FT regulons contribute to the photoperiod flowering control in soybean. BMC Plant Biol. 2014; 14: 9 10.1186/1471-2229-14-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kikuchi R, Kawahigashi H, Oshima M, Ando T, Handa H. The differential expression of HvCO9, a member of the CONSTANS-like gene family, contributes to the control of flowering under short-day conditions in barley. J Exp Bot. 2012; 63: 773–784. 10.1093/jxb/err299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Martin J, Storgaard M, Andersen CH, Nielsen KK. Photoperiodic regulation of flowering in perennial ryegrass involving a CONSTANS-like homolog. Plant Mol Biol. 2004; 56: 159–169. [DOI] [PubMed] [Google Scholar]
- 59. Chia TY, Muller A, Jung C, Mutasa-Gottgens ES. Sugar beet contains a large CONSTANS-LIKE gene family including a CO homologue that is independent of the early-bolting (B) gene locus. J Exp Bot. 2008; 59: 2735–2748. 10.1093/jxb/ern129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hayama R, Yokoi S, Tamaki S, Yano M, Shimamoto K. Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature. 2003; 422: 719–722. [DOI] [PubMed] [Google Scholar]
- 61. Gu C, Lv XC, Zhang K, Cui BM, Huang XZ. Cloning and expression of GbCO gene in Gossypium barbadense L. XinJiang Agricultural Sciences. 2013; 50: 214–222. [Google Scholar]
- 62. Ledger S, Strayer C, Ashton F, Kay SA, Putterill J. Analysis of the function of two circadian-regulated CONSTANS-LIKE genes. Plant J. 2001; 26: 15–22. [DOI] [PubMed] [Google Scholar]
- 63. Shin BS, Lee JH, Lee JH, Jeong HJ, Yun CH, Kim JK. Circadian regulation of rice (Oryza sativa L.) CONSTANS-like gene transcripts. Mol Cells. 2004; 17: 10–16. [PubMed] [Google Scholar]
- 64. Meng X, Muszynski MG, Danilevskaya ON. The FT-like ZCN8 gene functions as a floral activator and is involved in photoperiod sensitivity in maize. Plant Cell. 2011; 23: 942–960. 10.1105/tpc.110.081406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wendel JF, Brubaker CL, Percival AE. Genetic diversity in Gossypium hirsutum and the origin of upland cotton. Am J Bot. 1992; 79: 1291–1310. [Google Scholar]
- 66. Iqbal MJ, Reddy OUK, El-Zik KM, Pepper AE. A genetic bottleneck in the ‘evolution under domestication’ of upland cotton Gossypium hirsutum L. examined using DNA fingerprinting. Theor Appl Genet. 2001; 103: 547–554. [Google Scholar]
- 67. Kim SK, Yun CH, Lee JH, Jang HY, Park HY, Kim JK. OsCO3, a CONSTANS-LIKE gene, controls flowering by negatively regulating the expression of FT-like genes under SD conditions in rice. Planta. 2008; 228: 355–365. 10.1007/s00425-008-0742-0 [DOI] [PubMed] [Google Scholar]
- 68. Huang CL, Hung CY, Chiang YC, Hwang CC, Hsu TW, Huang CC, et al. Footprints of natural and artificial selection for photoperiod pathway genes in Oryza . Plant J. 2012; 70: 769–782. 10.1111/j.1365-313X.2012.04915.x [DOI] [PubMed] [Google Scholar]
- 69. Takahashi Y, Shimamoto K. Heading date 1 (Hd1), an ortholog of Arabidopsis CONSTANS, is a possible target of human selection during domestication to diversify flowering times of cultivated rice. Genes & Genet Syst. 2011; 86: 175–182. [DOI] [PubMed] [Google Scholar]
- 70. Hung HY, Shannon LM, Tian F, Bradbury PJ, Chen C, Flint-Garcia SA, et al. ZmCCT and the genetic basis of day-length adaptation underlying the postdomestication spread of maize. Proc Natl Acad Sci USA. 2012; 109: 1913–1921. 10.1073/pnas.1117158109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Murakami M, Matsushika A, Ashikari M, Yamashino T, Mizuno T. Circadian-associated rice pseudo response regulators (OsPRRs): insight into the control of flowering time. Biosci Biotechnol Biochem. 2005; 69: 410–414. [DOI] [PubMed] [Google Scholar]
- 72. Turner A, Beales J, Faure S, Dunford RP, Laurie DA. The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science. 2005; 310: 1031–1034. [DOI] [PubMed] [Google Scholar]
- 73. Murphy RL, Klein RR, Morishige DT, Brady JA, Rooney WL, Miller FR, et al. Coincident light and clock regulation of pseudoresponse regulator protein 37 (PRR37) controls photoperiodic flowering in sorghum. Proc Natl Acad Sci USA. 2011; 108: 16469–16474. 10.1073/pnas.1106212108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wenkel S, Turck F, Singer K, Gissot L, Le Gourrierec J, Samach A, et al. CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis . Plant Cell. 2006; 18: 2971–2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Jang S, Marchal V, Panigrahi KC, Wenkel S, Soppe W, Deng XW, et al. Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J. 2008; 27: 1277–1288. 10.1038/emboj.2008.68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Khanna R, Kronmiller B, Maszle DR, Coupland G, Holm M, Mizuno T, et al. The Arabidopsis B-box zinc finger family. Plant Cell. 2009; 21: 3416–3420. 10.1105/tpc.109.069088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kumagai T, Ito S, Nakamichi N, Niwa Y, Murakami M, Yamashino T, et al. The common function of a novel subfamily of B-box zinc finger proteins with reference to circadian-associated events in Arabidopsis thaliana . Biosci Biotechnol Biochem. 2008; 72: 1539–1549. [DOI] [PubMed] [Google Scholar]
- 78. Ben-Naim O, Eshed R, Parnis A, Teper-Bamnolker P, Shalit A, Coupland G, et al. The CCAAT binding factor can mediate interactions between CONSTANS-like proteins and DNA. Plant J. 2006; 46: 462–476. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLS)
(XLS)
(XLS)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.