Abstract
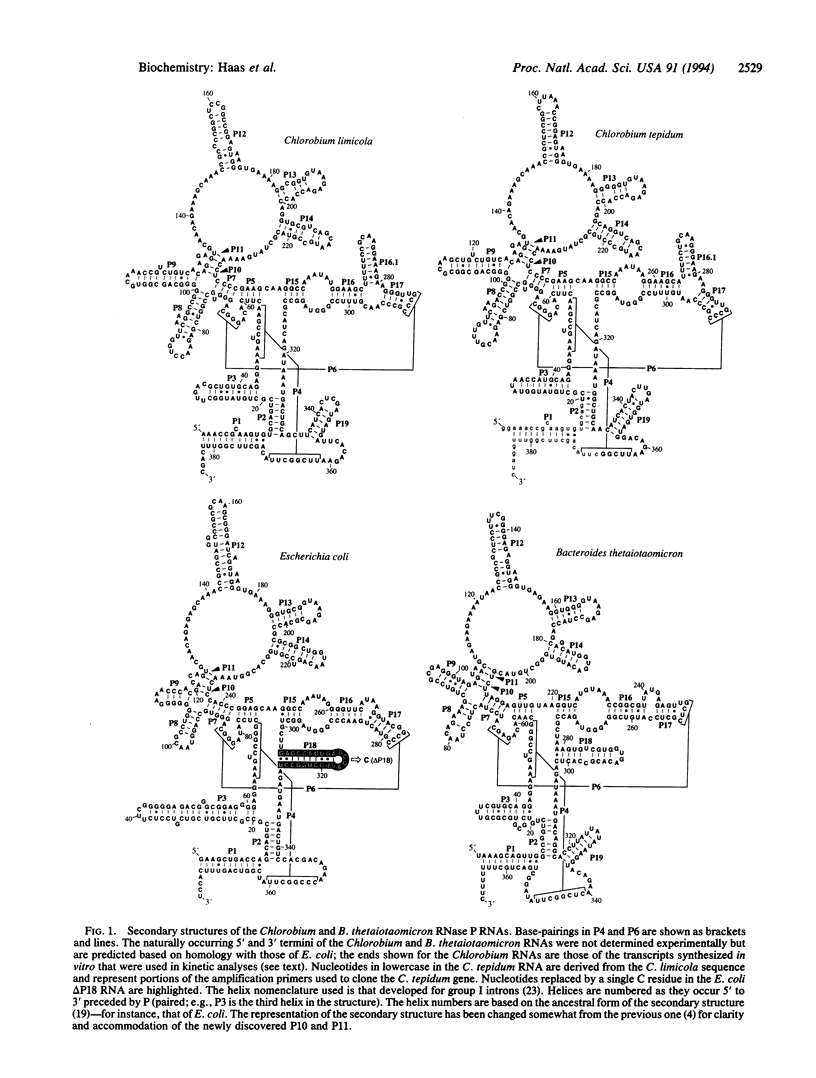
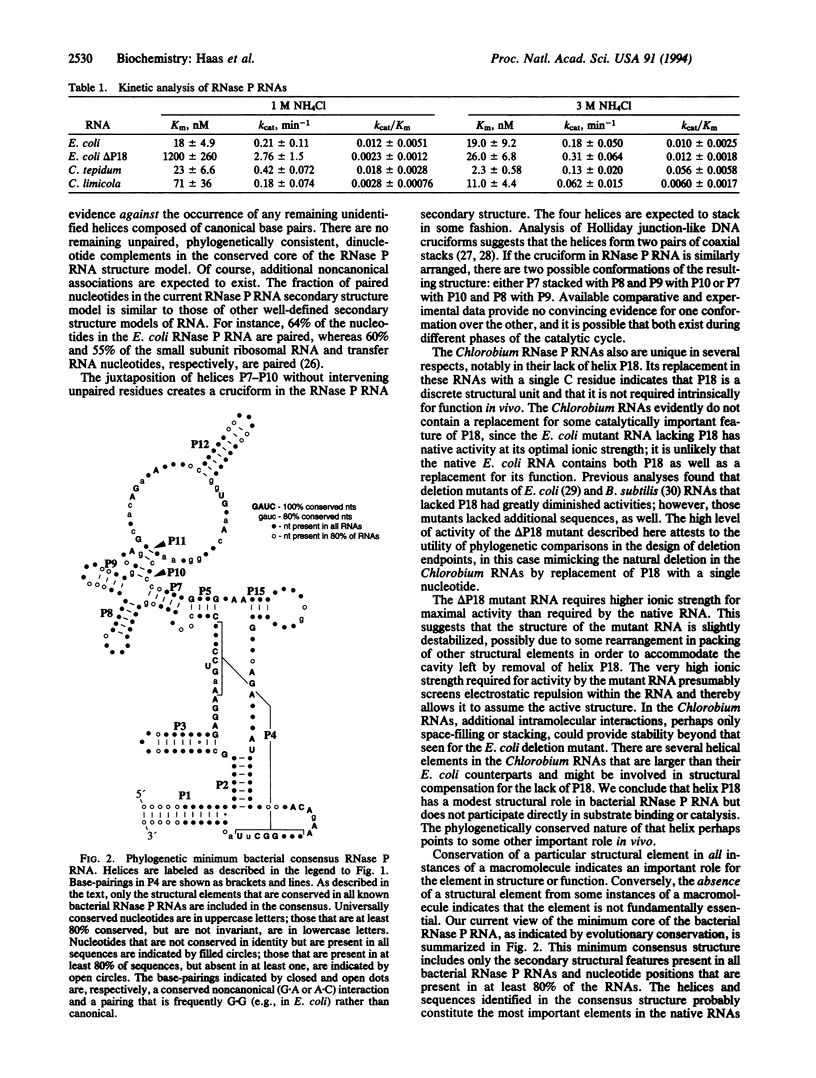
Phylogenetic comparative analyses of RNase P RNA-encoding gene sequences from Chlorobium limicola, Chlorobium tepidum, Bacteroides thetaiotaomicron, and Flavobacterium yabuuchiae refine the secondary structure model of the general (eu)bacterial RNase P RNA and show that a highly conserved feature of that RNA is not essential. Two helices, comprised of 2 base pairs each, are added to the secondary structure model and form part of a cruciform in the RNA. Novel sequence variations in the B. thetaiotaomicron and F. yabuuchiae RNA indicate the likelihood that all secondary structure resulting from canonical base-pairing has been detected: there are no remaining unpaired, contiguous, canonical complementarities in the structure model common to all bacterial RNase P RNAs. A nomenclature for the elements of the completed secondary structure model is proposed. The Chlorobium RNase P RNAs lack a stem-loop structure that is otherwise universally present and highly conserved in structure in other (eu)bacterial RNase P RNAs. The Chlorobium RNAs are nevertheless catalytic, with kinetic properties similar to those of RNase P RNAs of Escherichia coli and other Bacteria. Removal of this stem-loop structure from the E. coli RNA affects neither its affinity for nor its catalytic rate for cleavage of a precursor transfer RNA substrate. These results show that this structural element does not play a direct role in substrate binding or catalysis.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altman S. Ribonuclease P. Postscript. J Biol Chem. 1990 Nov 25;265(33):20053–20056. [PubMed] [Google Scholar]
- Angert E. R., Clements K. D., Pace N. R. The largest bacterium. Nature. 1993 Mar 18;362(6417):239–241. doi: 10.1038/362239a0. [DOI] [PubMed] [Google Scholar]
- Banta A. B., Haas E. S., Brown J. W., Pace N. R. Sequence of the ribonuclease P RNA gene from the cyanobacterium Anacystis nidulans. Nucleic Acids Res. 1992 Feb 25;20(4):911–911. doi: 10.1093/nar/20.4.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. W., Haas E. S., James B. D., Hunt D. A., Liu J. S., Pace N. R. Phylogenetic analysis and evolution of RNase P RNA in proteobacteria. J Bacteriol. 1991 Jun;173(12):3855–3863. doi: 10.1128/jb.173.12.3855-3863.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. W., Haas E. S., Pace N. R. Characterization of ribonuclease P RNAs from thermophilic bacteria. Nucleic Acids Res. 1993 Feb 11;21(3):671–679. doi: 10.1093/nar/21.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. W., Pace N. R. Ribonuclease P RNA and protein subunits from bacteria. Nucleic Acids Res. 1992 Apr 11;20(7):1451–1456. doi: 10.1093/nar/20.7.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgin A. B., Pace N. R. Mapping the active site of ribonuclease P RNA using a substrate containing a photoaffinity agent. EMBO J. 1990 Dec;9(12):4111–4118. doi: 10.1002/j.1460-2075.1990.tb07633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke J. M., Belfort M., Cech T. R., Davies R. W., Schweyen R. J., Shub D. A., Szostak J. W., Tabak H. F. Structural conventions for group I introns. Nucleic Acids Res. 1987 Sep 25;15(18):7217–7221. doi: 10.1093/nar/15.18.7217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper J. P., Hagerman P. J. Geometry of a branched DNA structure in solution. Proc Natl Acad Sci U S A. 1989 Oct;86(19):7336–7340. doi: 10.1073/pnas.86.19.7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darr S. C., Brown J. W., Pace N. R. The varieties of ribonuclease P. Trends Biochem Sci. 1992 May;17(5):178–182. doi: 10.1016/0968-0004(92)90262-8. [DOI] [PubMed] [Google Scholar]
- Darr S. C., Zito K., Smith D., Pace N. R. Contributions of phylogenetically variable structural elements to the function of the ribozyme ribonuclease P. Biochemistry. 1992 Jan 21;31(2):328–333. doi: 10.1021/bi00117a003. [DOI] [PubMed] [Google Scholar]
- Duckett D. R., Murchie A. I., Diekmann S., von Kitzing E., Kemper B., Lilley D. M. The structure of the Holliday junction, and its resolution. Cell. 1988 Oct 7;55(1):79–89. doi: 10.1016/0092-8674(88)90011-6. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C., Altman S. M1 RNA with large terminal deletions retains its catalytic activity. Cell. 1986 Apr 25;45(2):177–183. doi: 10.1016/0092-8674(86)90381-8. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C., Gardiner K., Marsh T., Pace N., Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983 Dec;35(3 Pt 2):849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- Haas E. S., Morse D. P., Brown J. W., Schmidt F. J., Pace N. R. Long-range structure in ribonuclease P RNA. Science. 1991 Nov 8;254(5033):853–856. doi: 10.1126/science.1719634. [DOI] [PubMed] [Google Scholar]
- James B. D., Olsen G. J., Liu J. S., Pace N. R. The secondary structure of ribonuclease P RNA, the catalytic element of a ribonucleoprotein enzyme. Cell. 1988 Jan 15;52(1):19–26. doi: 10.1016/0092-8674(88)90527-2. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Pace N. R., Smith D. Ribonuclease P: function and variation. J Biol Chem. 1990 Mar 5;265(7):3587–3590. [PubMed] [Google Scholar]
- Reich C., Gardiner K. J., Olsen G. J., Pace B., Marsh T. L., Pace N. R. The RNA component of the Bacillus subtilis RNase P. Sequence, activity, and partial secondary structure. J Biol Chem. 1986 Jun 15;261(17):7888–7893. [PubMed] [Google Scholar]
- Reich C., Olsen G. J., Pace B., Pace N. R. Role of the protein moiety of ribonuclease P, a ribonucleoprotein enzyme. Science. 1988 Jan 8;239(4836):178–181. doi: 10.1126/science.3122322. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallsjö A., Svärd S. G., Kufel J., Kirsebom L. A. A novel tertiary interaction in M1 RNA, the catalytic subunit of Escherichia coli RNase P. Nucleic Acids Res. 1993 Aug 25;21(17):3927–3933. doi: 10.1093/nar/21.17.3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vioque A. Analysis of the gene encoding the RNA subunit of ribonuclease P from cyanobacteria. Nucleic Acids Res. 1992 Dec 11;20(23):6331–6337. doi: 10.1093/nar/20.23.6331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waugh D. S., Pace N. R. Gap-scan deletion analysis of Bacillus subtilis RNase P RNA. FASEB J. 1993 Jan;7(1):188–195. doi: 10.1096/fasebj.7.1.7678561. [DOI] [PubMed] [Google Scholar]
- Woese C. R., Kandler O., Wheelis M. L. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]


