Abstract
Determination of lipoprotein particle size and number using advanced lipoprotein tests (ALTs) is of particular importance to improve cardiovascular risk prediction. Here we present the Liposcale test, a novel ALT based on 2D diffusion-ordered 1H NMR spectroscopy. Our method uses diffusion coefficients to provide a direct measure of the mean particle sizes and numbers. Using 177 plasma samples from healthy individuals and the concentration of ApoB and ApoA from isolated lipoprotein fractions, our test showed a stronger correlation between the NMR-derived lipoprotein particle numbers and apolipoprotein concentrations than the LipoProfile® test commercialized by Liposcience. We also converted LDL particle numbers to ApoB equivalents (milligrams per deciliter) and our test yielded similar values of LDL-ApoB to the LipoProfile® test (absolute mean bias of 8.5 and 7.4 mg/dl, respectively). In addition, our HDL particle number values were more concordant with the calibrated values determined recently using ion mobility. Finally, principal component analysis distinguished type 2 diabetic patients with and without atherogenic dyslipidemia (AD) on a second cohort of 307 subjects characterized using the Liposcale test (area under the curve = 0.88) and showed concordant relationships between variables explaining AD. Altogether, our method provides reproducible and reliable characterization of lipoprotein particles and it is applicable to pathological states such as AD.
Keywords: low density lipoprotein particle number, cardiovascular risk, nuclear magnetic resonance, two-dimensional
Abnormal levels of blood lipids, such as high concentrations of LDL cholesterol (LDL-C) and low concentrations of HDL cholesterol (HDL-C), increase the risk of CVD, the first cause of death in developed countries (1). Accordingly, the European Atherosclerosis and Cardiology Societies and the National Cholesterol Education Program, through their third report of the Adult Treatment Panel, established LDL-C and nonHDL-C as the primary and secondary target, respectively, of cholesterol-lowering therapy to reduce cardiovascular risk (2). LDL-C-lowering therapy has been shown to reduce the rate of cardiovascular events in patients with or without cardiometabolic risk (CMR) (3, 4). However, a large proportion of patients under treatment, as well as undiagnosed individuals, may suffer cardiovascular events despite showing normal LDL-C levels. Cardiovascular events are more likely to occur in patients with diabetes and metabolic syndrome. These pathologies share a common phenotype characterized by a high content of triglycerides, a preponderance of small dense LDL particles, and low HDL levels. In individuals with this particular phenotype, LDL-C has been shown to be a poor predictor of cardiovascular risk, so standard lipid panels that measure the cholesterol or triglyceride content of lipoproteins seem to be insufficient to predict risk of CVD. To fill this gap, advanced lipoprotein tests (ALTs) (5) that allow for an extensive characterization of lipoprotein particles through a range of additional parameters, such as size and particle number, have been proposed for improving assessment of risk of CVD and for guiding lipid-lowering therapies (6).
NMR spectroscopy is a technique that enables analysis of lipoprotein particles (5). Briefly, depending on the size of the particle, the methyl moieties of the lipids in lipoprotein particles resonate at slightly different frequencies, the smaller particles resonating at lower frequencies. Therefore lipoproteins can be quantified either by decomposing the methyl signal of the core lipids into individual signals (7, 8) or using statistical methods on the entire methyl envelope to estimate lipid concentrations (9). Currently three methodologies use NMR to characterize lipoprotein particles. The method described by Jeyarajah, Cromwell, and Otvos (7) provides the size and particle number of the main lipoprotein classes (i.e., VLDL, LDL, and HDL) and the particle number of nine lipoprotein subclasses. This method is built on a library of 1D NMR spectra from previously isolated lipoprotein fractions and on an algorithm that fits their NMR methyl signal with those of lipoproteins from serum or plasma samples. The particle sizes of the isolated lipoprotein fractions were determined by transmission electron microscopy and gradient gel electrophoresis. A second method described by Kaess et al. (8) characterizes fifteen lipoprotein subclasses measuring the samples by magnetic field gradient intensities and temperatures. The latter NMR methodology was reported by Ala-Korpela and colleagues (9), and it estimates lipid content, size, and particle numbers of the main lipoprotein classes, as well as the particle numbers of fourteen lipoprotein subclasses based on regression models calibrated using the lipid content and size obtained by high performance liquid chromatography.
NMR-based ALTs have demonstrated that LDL and HDL particle numbers (LDL-P and HDL-P) are more powerful than classical cholesterol markers as indices of cardiovascular risk (10). For instance, LDL-P better indicated atherosclerotic risk than LDL-C in individuals with discordant LDL-P and LDL-C levels (11). This discordance is usually explained based on the large variability in the amount of cholesterol per LDL particle and to differences in LDL particle size. In another study, HDL-P, but not HDL-C, was inversely associated with carotid intima-media thickness after adjusting for covariates (12). Moreover, the use of NMR-derived lipoprotein subclasses improved risk stratification for subclinical atherosclerosis in comparison to conventional lipids (13). Altogether, these and other evidence have precipitated two significant events: 1) the 510(k) clearance from the US Food and Drug Administration (FDA) to market the Vantera® Clinical Analyzer commercialized by Liposcience, the first NMR-based diagnostic platform that determines LDL-P; and 2) the recommendation from the American Association for Clinical Chemistry Lipoprotein and Vascular Diseases Division Working Group on Best Practices to include LDL-P in the guidelines used to manage CMR based on the evidence collected from 25 clinical studies (14).
Despite these advances, there is still some controversy about the introduction of NMR-based ALTs into clinical practice, partly due to the fact that current methods do not provide a direct measure of lipoprotein sizes. As an alternative to current NMR methods, here we present the Liposcale test, a novel method for characterizing lipoprotein particles based on 2D diffusion-ordered 1H NMR spectroscopy (DOSY). DOSY allows measuring the diffusion coefficients and directly calculating lipoprotein sizes through the Stokes-Einstein equation (15). It is noteworthy that a direct measure of lipoprotein sizes is of particular importance because they are used to compute lipoprotein particle numbers by dividing the spatial volume of the total lipid molecules by the mean volume (i.e., size) of the lipoprotein particles. Our rationale is that using DOSY to directly calculate lipoprotein sizes should yield more accurate measurements of lipoprotein particle numbers than current NMR-based ALT methods. To develop our new DOSY-based ALT, we used a cohort of 177 healthy individuals and then we compared the lipoprotein particle numbers obtained using the Liposcale test with those obtained using the FDA-cleared ALT commercialized by Liposcience. Finally, we applied the Liposcale test to characterize a second cohort of 307 type 2 diabetic (T2DM) patients with and without atherogenic dyslipidemia (AD). Our results demonstrate that our methodology can be applied to study samples with aberrant lipid and lipoprotein concentrations and can add insight into the understanding of metabolic diseases.
MATERIALS AND METHODS
Study subjects
We used samples from the VITAGE project to develop the Liposcale test (16). Briefly, 177 healthy nonsmoking men (0 cigarettes/day for >6 months) were enrolled in Clermont-Ferrand (France) and Reus (Spain). Exclusion criteria were familial hypercholesterolemia, chronic diseases (including diabetes, cancer, cardiac insufficiency, inflammatory diseases, and unstable hypertension), or alcohol abuse. Mean age was 45.8 ± 15.5 years, including a minimum age of 19 years and a maximum age of 75 years. Fasting venous blood samples were collected in EDTA tubes and centrifuged immediately for 15 min at 4°C at 1,500 g. Plasma samples were then kept at −80°C until further analysis. The ethics committee of the two recruiting centers approved the study protocol, and written informed consent was obtained from all volunteers.
We used a second cohort to validate the results provided by the Liposcale test. Eligible participants were 307 T2DM men and women, ranging from 30 to 80 years of age, with (n = 91) or without (n = 216) AD. AD was defined as having triglyceride levels over 150 mg/dl and HDL-C levels under 40 mg/dl (men) or 50 mg/dl (women). Exclusion criteria were the presence of CVD and chronic hepatic or renal alterations. Patients under lipid-lowering medication entered a wash-out period of 4 weeks. Samples were kept at −80°C until NMR analysis. The study protocol was also approved by the ethics committee of the participating institution.
Lipid and lipoprotein measurements
Lipoproteins were separated by sequential preparative ultracentrifugation, using a Kontron 45.6 fixed-angle rotor in a Centrikon 75 (Kontron Instruments, Italy). The lipoprotein fractions isolated were VLDL (d < 1.006 g/ml), IDL (d = 1.006–1.019 g/ml), LDL (d = 1.019–1.063 g/ml), and HDL (d = 1.063–1.21 g/ml). Their cholesterol and triglyceride content was quantified using standard enzymatic assays adapted to the Cobas-Mira-Plus autoanalyzer (SPINREACT S.A.U., Spain) (17). The concentration of ApoB100 and ApoA1 in the ultra-centrifuged fractions was also quantified using an immunoturbidimetric assay adapted to the Cobas-Mira-Plus auto-analyzer (SPINREACT S.A.U.). The assay and the value of the calibrator concentration were standardized against the Certified Reference Material ApoA1 WHO/IFCC SP1-01 and ApoB100 WHO/IFCC SP3-07. Plasma samples were also analyzed by Liposcience (Raleigh, NC) to obtain reference values of VLDL, LDL, and HDL sizes and particle numbers (7).
Advanced lipoprotein testing using DOSY
2D DOSY.
Plasma samples were analyzed by NMR spectroscopy using a modified existing protocol (18). Briefly, 1H NMR spectra were recorded on a BrukerAvance III 600 spectrometer, operating at a proton frequency of 600.20 MHz (14.1 T), at 310 K. We used the double stimulated echo pulse program with bipolar gradient pulses and a longitudinal eddy current delay. The relaxation delay was 2 s, the finite impulse decays were collected into 64,000 complex data points and 32 scans were acquired on each sample. The gradient pulse strength was increased from 5 to 95% of the maximum strength of 53.5 Gauss cm−1 in 32 steps. The squared gradient pulse strength was linearly distributed.
Surface fitting.
The methyl signal was surface fitted using a previously reported procedure (18). The number of functions was increased to account for the nine lipoprotein subclasses. The uniqueness of the solutions was studied by fitting each sample 10 times with randomly chosen initial values of the signal intensities. As a result, we obtained unique solutions for all samples after 10 runs. Normalized root mean square errors of the fittings were calculated as previously described (18). The initial values of the signal intensities were taken to be the mean solution values of all samples.
Prediction of lipid concentration.
Partial least squares (PLS) regression models were calibrated to predict the cholesterol and triglyceride concentration of the main lipoprotein fractions (VLDL, LDL, and HDL). Validation performance of the PLS models were assessed by venetian blinds cross-validation splitting the data 10 times. Coefficients of determination between the predicted and reference concentrations ranged from 0.79 to 0.98 in the calibration step. The coefficients of determination of the validation step ranged from 0.81 to 0.98 and are in the range of other reported studies (19).
Lipoprotein sizing and particle number determination.
The NMR functions were associated with a given lipoprotein class (VLDL, LDL, or HDL) according to their associated NMR size. The main lipoprotein fractions were defined as VLDL (38.6–81.9 nm), LDL (18.9–26.5 nm), and HDL (7.8–11.5 nm). The mean particle size of every main fraction (VLDL, LDL, and HDL) was derived by averaging the NMR area of each fraction by its associated size. To obtain particle-weighted lipoprotein sizes, each NMR area was divided by its associated volume. Then a mean particle size was obtained for each lipoprotein class by multiplying the NMR lipoprotein particle sizes by their fractional particle concentration relative to the total particle concentration of a given class:
| (Eq. 1) |
where Ai and di are the area (au) and diameter (nm) of a given lipoprotein particle i. The particle numbers of each lipoprotein main fraction were calculated by dividing the lipid volume by the particle volume of a given class. The lipid volumes were determined by using common conversion factors to convert concentration units obtained from the PLS models into volume units (7). The relative areas of the lipoprotein components used to decompose the 2D spectra were used to derive the particle numbers of the nine lipoprotein subclasses.
Spiking experiments
We performed spiking experiments to validate the grouping of the NMR functions. Serum samples were obtained from three volunteers. Lipoprotein fractions (VLDL, LDL, and HDL) were obtained by sequential ultra-centrifugation as described above. Then, serum samples were spiked with the lipoprotein fractions from the same subjects one by one. A serum sample made up with buffer to equal volumes as the spiked samples was also analyzed.
Analytical performance
The analytical performance of the Liposcale test involved blood samples from two volunteers. Briefly, fasting venous blood samples were collected in EDTA tubes and centrifuged immediately for 15 min at 4°C at 1,500 g. Then, five aliquots were obtained for each subject and kept at −80°C until the NMR analysis. The within-assay precision of the method was studied based on the analyses of the five aliquots of the two subjects in the same day, while the inter-assay precision was based on the analyses of the five aliquots of the two subjects through three consecutive days.
Statistical analyses
The Bland-Altman plot was used to measure the agreement between both NMR techniques in assessing particle sizes and numbers (20). This plot uses the differences between observations made by the two methods on the same subjects and it analyzes the agreement between two methods in terms of bias and precision. When there is a relationship between difference and magnitude, the standard Bland-Altman analysis can be extended either with a logarithmic transformation approach or a more general regression approach. As a measure of agreement, we calculated the percentage error (PE) between both techniques, as described previously (21):
| (Eq. 2) |
where the coefficient of variation (CV) [CV (%) = (σ/μ) × 100] is defined as the ratio of the standard deviation σ to the mean μ of the population.
We also defined a maximum PE by using the CV of the old method twice in equation 2.
Spearman analysis was used to assess the relationship between continuous variables. All the analyses were performed with MATLAB version 7.10.0.499 R2010a (MathWorks). PLS and principal component analysis (PCA) models were built using PLS Toolbox (Eigenvector Research).
RESULTS
Assignment of DOSY NMR functions to the main lipoprotein classes
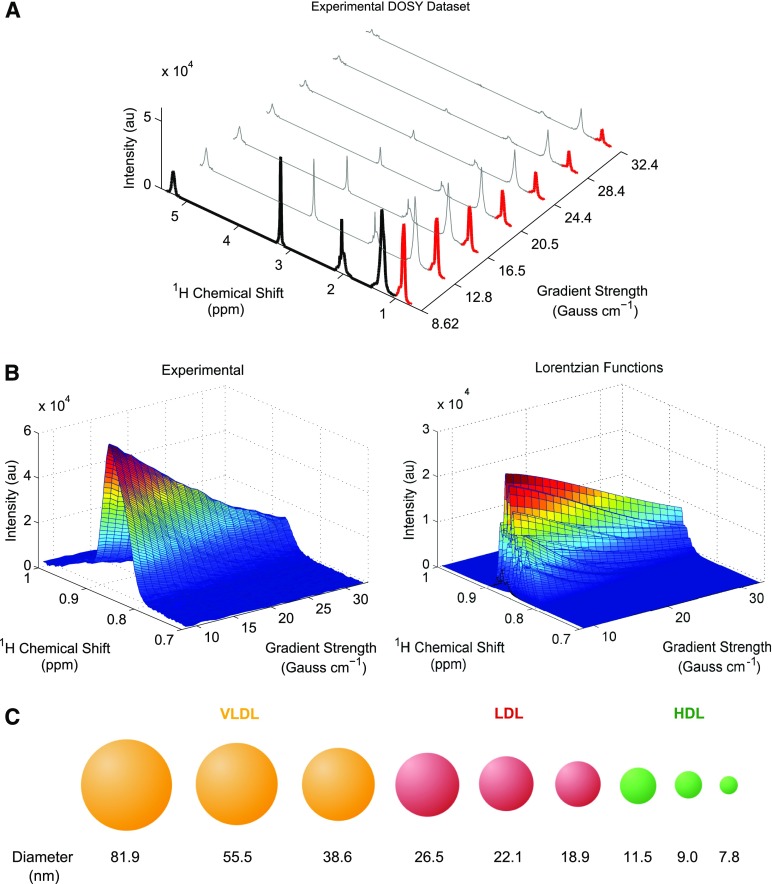
The Liposcale test provides lipid concentrations (i.e., triglycerides and cholesterol), sizes, and particle numbers for VLDL, LDL, and HDL classes, as well as the particle numbers of nine subclasses, namely large, medium, and small VLDL, LDL, and HDL, respectively. Figure 1 provides an overview of the Liposcale test showing the most important processes on which the characterization of lipoprotein classes is based. The use of the diffusion dimension represents the main difference between our approach and the NMR-based platform commercialized by Liposcience. The Liposcale test uses 2D spectra from DOSY experiments (Fig. 1A) to decompose the (CH3) proton resonances of the lipids in lipoprotein particles into nine Lorentzian functions (i.e., F1 to F9) (Fig. 1B). Of note, the cholesterol esters and triglycerides in the particle core contribute with three methyl groups each (5) and thus the total amount of all the methyl groups is mostly dependent on particle size (see Discussion).
Fig. 1.
Overview of the Liposcale test. A: Experimental DOSY spectra. Seven gradients out of 32 total are shown for visualization purposes. The first gradient has been filled in black to show the NMR regions of the spectra that are used to calibrate and predict cholesterol and triglyceride concentrations using PLS regression. In red, we show the attenuation of the methyl NMR signal along the gradient axis to show the region used to characterize the different lipoprotein subclasses. B: Surface fitting process for a given subject. The experimental attenuation of the methyl NMR signal (left) and the nine NMR functions used to fit the experimental surface (right). C: Using this methodology, we can elucidate nine lipoprotein subclasses, namely large, medium, and small VLDL, LDL, and HDL.
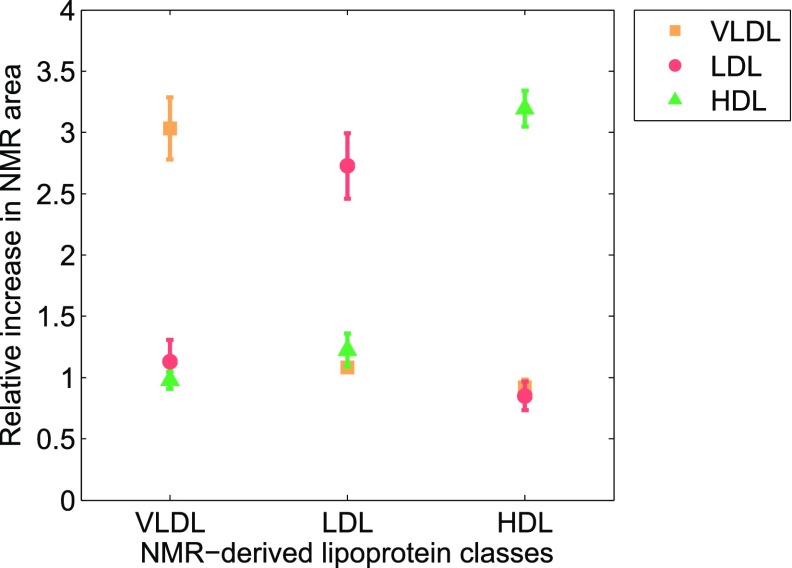
This approach largely prevents multiple solutions and, consequently, enhances robustness of the measurements. Indeed, we obtained an average normalized root mean square error for the surface fittings of less than 1.85% (see the Materials and Methods for details), which indicates that our deconvolution is highly reproducible. According to the Einstein-Stokes equation, the larger the diffusion coefficient, the smaller the size of a given lipoprotein particle will be. Thus, we associated the nine NMR functions with the three main lipoprotein classes based on their described size ranges (Fig. 1C) (22): F1 to F3 (38.6–81.9 nm) were associated with the VLDL particles, functions F4 to F6 (18.9–26.5 nm) with the LDL particles, and functions F7 to F9 (7.8–11.5 nm) with the HDL particles. These associations were validated using spike-in experiments. We isolated VLDL, LDL, and HDL particles from serum samples of three volunteers by ultracentrifugation (see the Materials and Methods for details), and each isolated lipoprotein fraction was mixed one by one with the serum sample of the same individual. Figure 2 shows the average relative change of every lipoprotein class in serum, represented as the total NMR area corresponding to VLDL (i.e., F1 to F3), LDL (F4 to F6), and HDL (F7 to F9) particles by applying our surface-fitting algorithm. Our results indicate that only the area of the lipoprotein fraction that has been spiked-in shows a substantial increment, demonstrating that our NMR functions are correctly assigned to VLDL, LDL, and HDL particles.
Fig. 2.
Spike-in experiments with isolated VLDL, LDL, and HDL particles in serum samples. Mean relative changes in the total area under the NMR functions for each lipoprotein class (VLDL = F1 + F2 + F3, LDL = F4 + F5 + F6, and HDL = F7 + F8 + F9). Each color represents spiked serum samples either with isolated VLDL, LDL, or HDL fractions.
Correlation between lipoprotein particle numbers and apolipoprotein concentrations
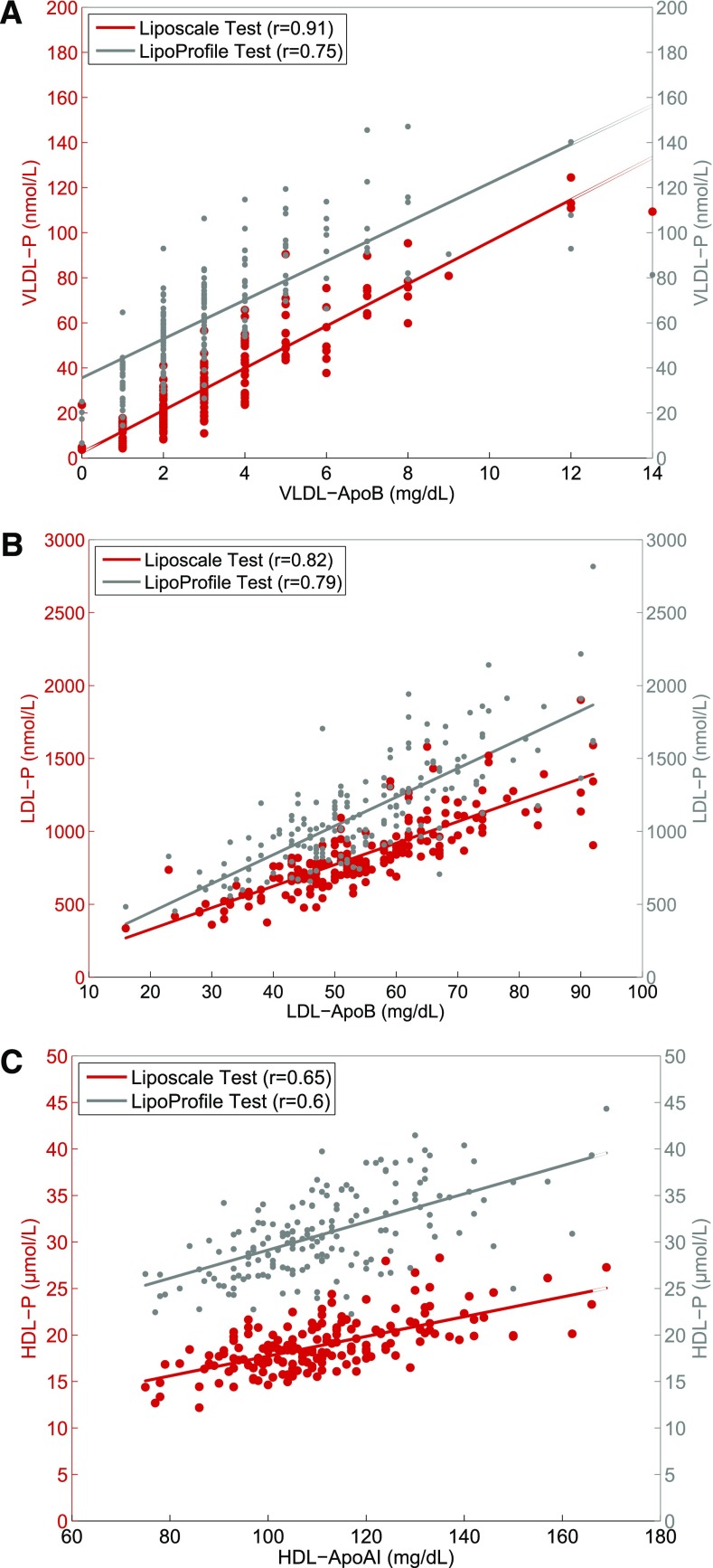
Next, we explored the degree of correlation between the VLDL-, LDL-, and HDL-Ps calculated using the Liposcale and the LipoProfile® tests, and the apoliproprotein content of each lipoprotein class, because the apolipoprotein concentrations in each class can serve as a surrogate of the number of particles. Figure 3A shows a strong positive linear relationship between the concentration of VLDL-ApoB determined using biochemical methods and the VLDL-P values determined using the Liposcale test (r = 0.91) and the LipoProfile® test (r = 0.75). Similarly, the correlation coefficients between LDL-P determined via the Liposcale and LipoProfile® tests and their respective apolipoprotein concentration were 0.82 and 0.78, respectively (Fig. 3B). Finally, the correlation coefficients between HDL-P and HDL-ApoA were 0.65 for the Liposcale test and 0.60 for the LipoProfile® test (Fig. 3C). Thus, the Liposcale test showed a stronger correlation between lipoprotein particle number and apolipoprotein concentration than the FDA-cleared NMR method that we took as the benchmark for the measurement of the number of particles in each lipoprotein class.
Fig. 3.
Regression analyses that examine the relationship among lipoprotein particle numbers using the Liposcale and LipoProfile® tests, and the apolipoprotein concentrations of VLDL (A), LDL (B), and HDL (C) particles.
Finally, we evaluated the effects of different proportions of triglycerides and cholesterol concentrations within the LDL lipoprotein particles on the correlations between LDL-ApoB measures and LDL-P concentrations. Subjects were divided into tertiles of the LDL-triglyceride to LDL-C ratio and the correlations between LDL-ApoB and LDL-P values determined by the two techniques were evaluated (see supplementary Fig. 1). The correlation coefficients between LDL-P determined via the Liposcale test and its respective apolipoprotein concentration increased across tertiles (0.84, 0.86, and 088, respectively), while the LipoProfile® test showed decreasing correlation coefficients across tertiles (0.83, 0.79, and 0.76, respectively).
LDL particle size and number agreement between the Liposcale and the LipoProfile® NMR tests
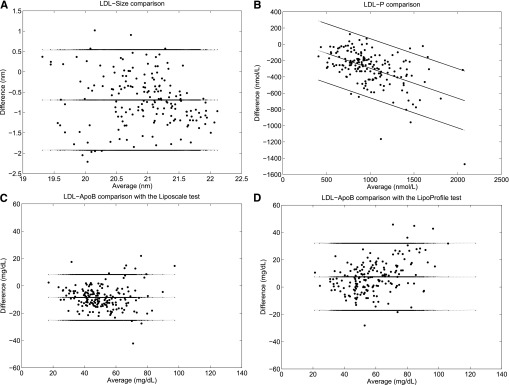
LDL-P is the most validated and clinically useful parameter that ALTs can determine. In this regard, three consensus reports (3, 23, 24) have all recommended that ApoB and/or measurements of LDL-P shall be incorporated into existing consensus guidelines for advanced CMR management. Thus, here we measured the agreement between our 2D-NMR test (i.e., Liposcale) and the 1D-NMR test (i.e., LipoProfile®) (see Fig. 4A, B). From the VITAGE cohort, the mean LDL-size and LDL-P were 20.6 nm and 843 nmol/l using Liposcale, and 21.3 nm and 1,133.1 nmol/l using LipoProfile®, respectively. This results in an average LDL-size and LDL-P difference (i.e., bias) between both methods of 0.7 nm and 288.1 nmol/l, respectively. We further analyzed the agreement between both methods using the Bland-Altman plot. The plot is characterized by a proportional bias in the form of a negative linear relationship between the two methods resulting from fewer small LDL particles using the Liposcale test in comparison with the LipoProfile® test. The logarithmic transformation of data did not correct this proportional error. Thus, to narrow down the limits of agreement, the difference between the methods was regressed on the average of the two methods (Fig. 4B).
Fig. 4.
Bland-Altman analyses to measure the agreement between both NMR tests in assessing particle sizes and numbers. Comparison of LDL-size (A) and LDL-P (B) as assessed by the Liposcale and the LipoProfile® tests. Comparison of LDL-ApoB concentrations measured from isolated LDL fractions using a classical biochemical method and the same concentration estimated using the Liposcale test (C) and the LipoProfile® test (D).
The CVs of LDL size and LDL-P were 3.14 and 31.0%, respectively, using the Liposcale test, and 3.4 and 32.3%, respectively, using the LipoProfile® test. The PE obtained between both methods was 4.6% for LDL size and 44.7% for LDL-P. In short, we consider that the agreement between both NMR tests is acceptable based on a PE below 5% for LDL-size and 50% for LDL-P.
The concentration of ApoB100 serves as a reference value of LDL-P due to the fact that each LDL particle contains only one molecule of ApoB100. In this regard, some authors have suggested that LDL-Ps (nanomoles per liter) should be converted to ApoB equivalents (milligrams per deciliter) to allow a direct comparison with an established biochemical parameter (14). Thus, to investigate which NMR test yielded more accurate LDL-P values, we converted LDL particle concentration (nanomoles per liter) into ApoB concentration (milligrams per deciliter) on the basis of a molecular mass for ApoB100 of 550 kDa (14). Once again, we assessed the agreement between the LDL-ApoB concentrations estimated by the Liposcale and LipoProfile® tests, and the same concentration assayed biochemically from LDL fractions isolated by ultracentrifugation (see Materials and Methods for details) using the Bland-Altman plot. The Bland-Altman analysis revealed a mean difference between the LDL-ApoB concentration estimated using the LipoProfile® test and biochemical methods of 7.4 mg/dl, while for Liposcale this mean difference was −8.5 mg/dl (Fig. 4C, D). The CV of LDL-ApoB concentration biochemically assayed was 26.5%, yielding a maximum PE of 40%. LDL-ApoB concentration obtained using the Liposcale and the LipoProfile® tests yielded CVs of 31.0 and 32.3%, respectively. Thus, the PE between the biochemical assay and Liposcale was 40.8%, and 41.8% when comparing with LipoProfile®.
Analytical performance of the Liposcale test
Next we assessed the precision of the Liposcale test understood as the ability of the assay to consistently reproduce the same result when different sample aliquots are taken from the same specimen. The within-assay precision of the method, calculated from the analysis of five different aliquots from two different subjects within the same day (i.e., n = 10), on the determination of cholesterol and triglyceride concentrations, and particle numbers for the LDL class and its small, medium, and large subclasses, was ≤5%. The inter-assay precision on the same parameters, calculated from the same five aliquots run on three consecutive days, was ≤8% (see supplementary Tables 1, 2). Similarly, the within-assay and inter-assay precision for cholesterol and triglyceride concentration, and VLDL- and HDL-P, were ≤6%. Finally, both within-assay and inter-assay precision for the mean particle size for every lipoprotein class were ≤1%. Overall, the precision values of our 2D-NMR method are within the range of those reported by Liposcience Inc. for the LipoProfile® test (7).
Characterization of AD in an independent cohort of diabetic patients
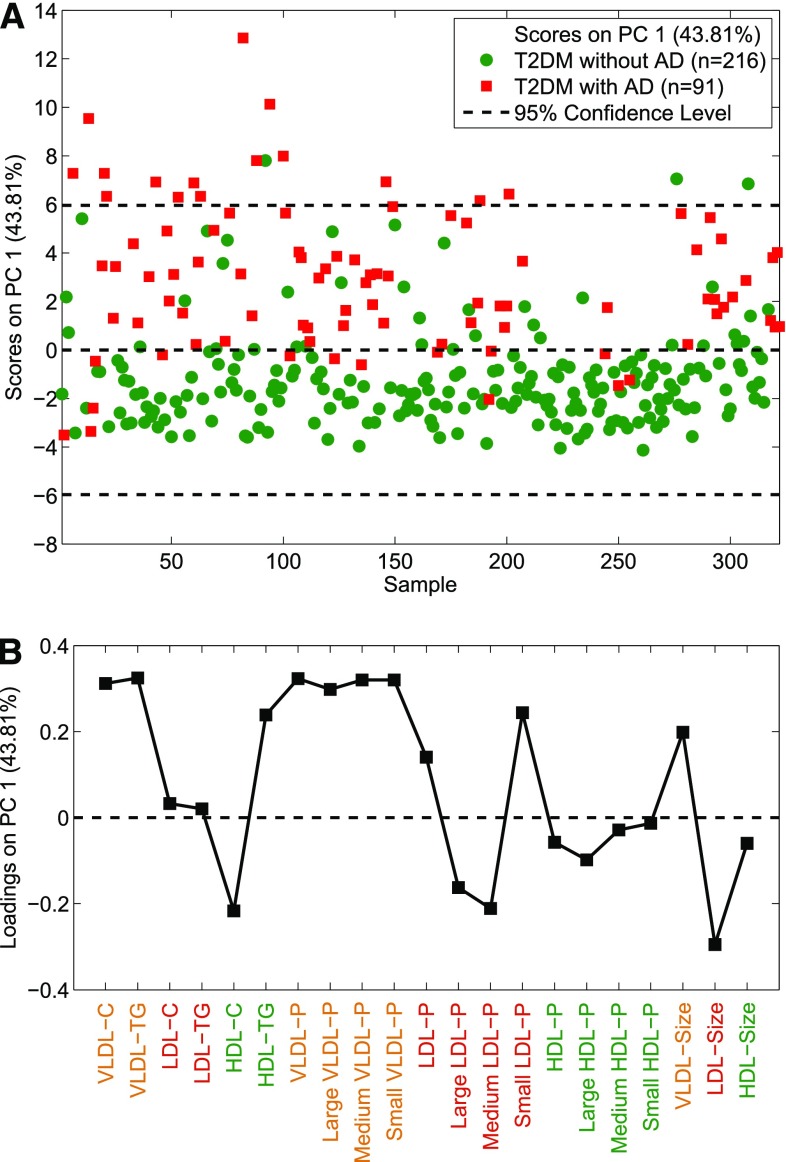
To demonstrate that the Liposcale test can be used to characterize and discriminate individuals with aberrant lipid and lipoprotein values, we implemented our 2D-NMR method on a cohort study of 307 subjects that included T2DM patients with (n = 91) and without (n = 216) AD. A PCA using the input variables cholesterol and triglyceride concentrations, mean sizes, and mean particle numbers of VLDL, LDL, and HDL classes, and particle numbers of nine subclasses of lipoproteins, shows two separate clusters along PC1 (43.81% of the variance) corresponding to the two patient groups, that is, T2DM and T2DM with AD (Fig. 5A). In order to evaluate which are the main variables responsible for the separation of the T2DM patients with and without AD, the resulting loadings were visualized in a variables-loading plot (Fig. 5B). The plot reveals that the triad of AD, namely increased blood concentrations of small dense LDL particles, decreased HDL particles, and increased triglyceride concentration, contributes to the separation of the two patient groups. Importantly, the loadings plot also shows that T2DM patients with AD have a smaller number of large and medium LDL and HDL particles relative to T2DM patients without AD. Moreover, patients with T2DM and AD have a greater number of total LDL and, in particular, of small LDL particles.
Fig. 5.
Scores plot (A) and loadings plot (B) for a PCA model built to discriminate between T2DM with and without AD.
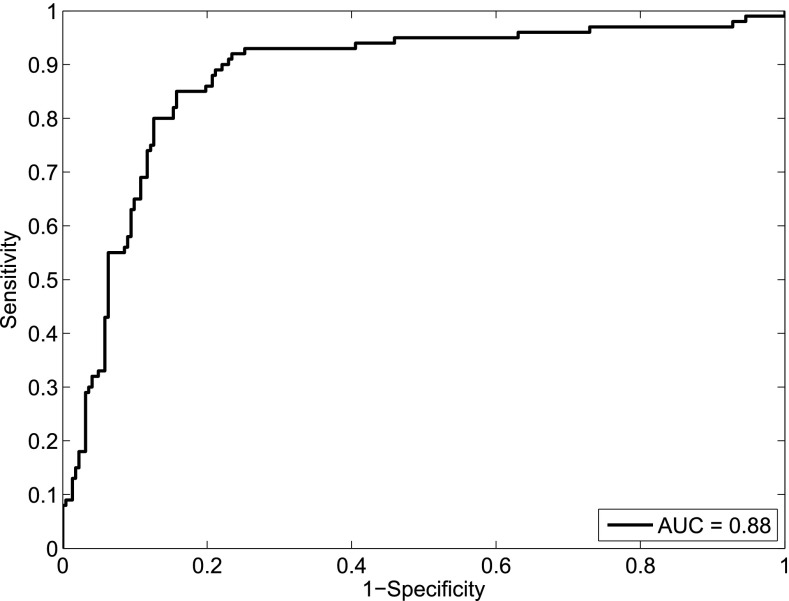
Considering the first principal component as the output of a putative classifier, we calculated the receiver-operating curve (ROC) in order to evaluate the classificatory power of our PCA model. As can be observed in the ROC curve depicted in Fig. 6, our approach showed an area under the curve (AUC) of 0.88, showing an excellent classification performance to discriminate between T2DM patients with and without AD. All together, these results demonstrate that the Liposcale test is clinically useful to classify individuals showing an abnormal lipid and lipoprotein pattern that is typical of AD, which has emerged as an important risk factor for myocardial infarction and CVD.
Fig. 6.
ROC curve evaluating the discriminatory power of the PCA model.
Finally, we validated the correlations between ApoB measures and lipoprotein particle concentrations at different triglyceride to cholesterol ratios on a group of 22 T2DM subjects with AD treated for 12 weeks with both fenofibrate and/or niacin (see supplementary Table 1) because these treatments may change the proportion of triglyceride and cholesterol concentrations within the lipoprotein particles. In this study, we used the total plasma concentration of ApoB as a reference value of total ApoB-containing lipoprotein particle concentration. Thus, we summed the particle numbers of the VLDL and LDL classes to obtain the total ApoB-containing lipoprotein particle numbers and then we converted these values into ApoB equivalents as described before. The results of the correlation analysis show a similar high degree of correlation between the sum of the concentrations of ApoB of the different lipoprotein fractions obtained by NMR and the total concentration of ApoB obtained biochemically at baseline (supplementary Fig. 3A, r = 0.83), after the fenofibrate intervention (supplementary Fig. 3B, r = 0.88), and after the niacin intervention (supplementary Fig. 3C, r = 0.89).
DISCUSSION
The methyl moieties of the lipids in lipoprotein particles resonate at slightly different frequencies depending on the size of the particle, the smaller particles resonating at lower frequencies (25). Though different lipoprotein subclasses may in theory be deconvoluted within the 1D-NMR spectrum according to their chemical shift positions, it has proven to be complicated in practice due to significant spectral overlap as different lipoprotein subclasses contribute to NMR resonance at the same frequency (18). This is of great importance for LDL subclasses, which are particularly difficult to characterize due to the effect of neighboring lipoprotein classes (i.e., VLDL and HDL) (26, 27). Moreover, because the cholesterol esters and triglycerides in the particle core contribute with three methyl groups each, the total amount of all the methyl groups is not affected by variations in core lipid compositions, i.e., variations in the cholesterol ester to triglyceride ratio driven by the cholesterol ester transfer protein (CETP), but is mostly dependent on particle size (28). Thus, the addition of a second dimension in the NMR spectrum by means of a diffusion experiment (i.e., DOSY) helps to better characterize the different lipoprotein subclasses. DOSY allows the separation of the lipoprotein subclasses according to their diffusion coefficient, and with the use of the Stokes-Einstein equation, DOSY NMR yields an objective separation of lipoprotein subclasses based on their size and favors the uniqueness of mathematical solutions compared with 1D-NMR.
In this study we present a novel ALT based on DOSY NMR spectroscopy called Liposcale, which has been compared with the established NMR method developed and commercialized by Otvos and colleagues and Liposcience (i.e., LipoProfile®) (7), respectively, which has become the only FDA-cleared blood test that directly quantifies LDL particles. Our study constitutes the first attempt to compare two 1H NMR methodologies, which is particularly important within the framework of standardized ALTs (14, 29) if LDL-P is intended to be used in cardiovascular risk management.
The agreement between the 2D-NMR (Liposcale) and 1D-NMR (LipoProfile®) methods for measuring LDL-P revealed a linear dependency between the difference and the magnitude of the measurements. However, the LDL-ApoB values obtained using our 2D-NMR method were more in conformity with biochemical values than those obtained using the FDA-cleared test. This disagreement between the two NMR methodologies may come partly from the small bias between the lipoprotein sizes determined by the two methods, through which our 2D-NMR method estimates smaller LDL particle sizes in comparison with the 1D-NMR method. This might be a consequence of the method used to derive the particle concentrations. While the LipoProfile® test computes mass-weighted particle numbers, our method computes particle-weighted particle numbers. From our point of view, the particle-weighted approach is more realistic because the concentration of small particles exceeds the concentration of larger ones. A previous study also reported a linear dependence between the difference and magnitude of LDL-size measured by Liposcience and gradient gel electrophoresis (30). On the other hand, no method-comparison study has evaluated different LDL-P assays. As well as the Liposcience test, ion mobility analysis has been shown to be an alternative method for directly measuring LDL-P (31). Although concerns were raised about its lack of accuracy (32), the authors further included correction factors which corrected the measurement of concentration of both nonHDL and HDL particles (33).
Concerning HDL particles, our 2D-NMR method showed a better correlation between HDL-P and HDL-ApoAI concentration relative to the LipoProfile® test. Even so, regardless of which NMR method is used, the correlation between HDL-P and HDL-ApoAI is not as good as when correlating LDL-P and the ApoB content. This could be partially explained by the lack of a direct relationship between HDL particles and ApoAI molecules. Unlike in LDL where each particle contains one molecule of ApoB, HDL particles contain between 2 and 4 molecules of ApoAI. A recent study has shown that the values of HDL-P estimated using an updated version of the Shen model, that relates the HDL-size and the ratio of HDL-C to ApoAI concentration, are approximately 50–60% lower than the HDL-P measurements obtained using the established NMR technique commercialized by Liposcience (34). Importantly, as can be seen in Fig. 3C, the Liposcale test estimated HDL-P values that are lower than the ones estimated by the LipoProfile® test, and consequently are more concordant with the number of HDL particles estimated by the modified Shen model and the calibrated ion mobility methodology reported recently by Hutchins et al. (34, 35).
Finally, the characterization of IDL lipoproteins by NMR spectroscopy is not straightforward due to i) its low concentration range compared with the other lipoproteins, and ii) its NMR response arises between the small VLDL and large LDL lipoproteins in terms of chemical shift. Taking this into account, we chose to be cautious and avoid the characterization of IDL, even in the event that this could appear as a limitation of our method. However, the isolation of the IDL fractions to obtain pure VLDL and LDL fractions allowed for a good characterization of the latter, which in turn are the most clinically useful together with the HDL fraction. Moreover, although in subjects with normal lipid levels the concentrations of VLDL-ApoB and IDL-ApoB might be low and therefore below the limit of detection of the immunoturbidimetric assay, in this circumstance the clinical utility of these parameters is minimal. However, in T2DM subjects or patients with AD, the concentrations of VLDL-ApoB and IDL-ApoB are higher and clinically relevant, and in this case the technique is very robust.
CONCLUSIONS
We evaluated a new methodology for quantifying lipoprotein subclasses based on 2D DOSY, which directly measures the sizes of lipoprotein particles. This methodology can provide the lipid concentration, lipoprotein size, and lipoprotein particle numbers of the main fractions and subclasses. We found very similar correlations between our test and a reference NMR technique, although our derived particle numbers measured yielded higher correlations with external validations, such as the concentration of VLDL-ApoB, LDL-ApoB, and HDL-ApoAI. Moreover, the characterization of AD on T2DM patients further demonstrated the applicability of our methodology in a population with pathological states.
Supplementary Material
Footnotes
Abbreviations:
- AD
- atherogenic dyslipidemia
- ALT
- advanced lipoprotein test
- AUC
- area under the curve
- CMR
- cardiometabolic risk
- CV
- coefficient of variation
- DOSY
- diffusion-ordered 1H NMR spectroscopy
- FDA
- US Food and Drug Administration
- HDL-C
- HDL cholesterol
- HDL-P
- HDL particle number
- LDL-C
- LDL cholesterol
- LDL-P
- LDL particle number
- PCA
- principal component analysis
- PE
- percentage error
- PLS
- partial least squares
- ROC
- receiver-operating curve
- T2DM
- type 2 diabetic
- VLDL-P
- VLDL particle number
This work was partially funded by CIBER de Diabetes y Enfermedades Metabólicas, an initiative of ISCIII (Ministerio de Ciencia e Innovación), as well as the FIS (Project PI081579). This work was also partly supported by the EU FP5 Program Quality of Life and Management of Living Resources, Key Action 1, Food, Nutrition, and Health, entitled Vitamin A, Vitamin E, and Carotenoid Status and Metabolism during Ageing: Functional and Nutritional Consequences, acronym VITAGE (Contract QLK1-CT- 1999-00830). R.M. and N.A. are employed by or have leadership roles in Biosfer Teslab. X.C. and L.M. have advisory roles at Biosfer Teslab. R.M., N.A., X.C., and L.M. are stock owners of Biosfer Teslab.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of text, two figures, and three tables.
REFERENCES
- 1.Gerszten R. E., Wang T. J. 2008. The search for new cardiovascular biomarkers. Nature. 451: 949–952. [DOI] [PubMed] [Google Scholar]
- 2.National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults (Adult Treatment Panel III). 2002. Third Report of the National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults (Adult Treatment Panel III) final report. Circulation. 106: 3143–3421. [PubMed] [Google Scholar]
- 3.Brunzell J. D., Davidson M., Furberg C. D., Goldberg R. B., Howard B. V., Stein J. H., Witztum J. L. 2008. Lipoprotein management in patients with cardiometabolic risk: consensus conference report from the American Diabetes Association and the American College of Cardiology Foundation. J. Am. Coll. Cardiol. 51: 1512–1524. [DOI] [PubMed] [Google Scholar]
- 4.Moin D. S., Rohatgi A. 2011. Clinical applications of advanced lipoprotein testing in diabetes mellitus. Clin. Lipidol. 6: 371–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mallol R., Rodriguez M. A., Brezmes J., Masana L., Correig X. 2013. Human serum/plasma lipoprotein analysis by NMR: application to the study of diabetic dyslipidemia. Prog. Nucl. Magn. Reson. Spectrosc. 70: 1–24. [DOI] [PubMed] [Google Scholar]
- 6.Stein J. H., McBride P. E. 2006. Should advanced lipoprotein testing be used in clinical practice? Nat. Clin. Pract. Cardiovasc. Med. 3: 640–641. [DOI] [PubMed] [Google Scholar]
- 7.Jeyarajah E. J., Cromwell W. C., Otvos J. D. 2006. Lipoprotein particle analysis by nuclear magnetic resonance spectroscopy. Clin. Lab. Med. 26: 847–870. [DOI] [PubMed] [Google Scholar]
- 8.Kaess B., Fischer M., Baessler A., Stark K., Huber F., Kremer W., Kalbitzer H. R., Schunkert H., Riegger G., Hengstenberg C. 2008. The lipoprotein subfraction profile: heritability and identification of quantitative trait loci. J. Lipid Res. 49: 715–723. [DOI] [PubMed] [Google Scholar]
- 9.Soininen P., Kangas A. J., Wurtz P., Tukiainen T., Tynkkynen T., Laatikainen R., Jarvelin M. R., Kahonen M., Lehtimaki T., Viikari J., et al. 2009. High-throughput serum NMR metabonomics for cost-effective holistic studies on systemic metabolism. Analyst. 134: 1781–1785. [DOI] [PubMed] [Google Scholar]
- 10.Sniderman A., Kwiterovich P. O. 2013. Update on the detection and treatment of atherogenic low-density lipoproteins. Curr. Opin. Endocrinol. Diabetes Obes. 20: 140–147. [DOI] [PubMed] [Google Scholar]
- 11.Otvos J. D., Mora S., Shalaurova I., Greenland P., Mackey R. H., Goff D. C., Jr 2011. Clinical implications of discordance between low-density lipoprotein cholesterol and particle number. J. Clin. Lipidol. 5: 105–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mackey R. H., Greenland P., Goff D. C., Jr, Lloyd-Jones D., Sibley C. T., Mora S. 2012. High-density lipoprotein cholesterol and particle concentrations, carotid atherosclerosis, and coronary events: MESA (multi-ethnic study of atherosclerosis). J. Am. Coll. Cardiol. 60: 508–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Würtz P., Raiko J. R., Magnussen C. G., Soininen P., Kangas A. J., Tynkkynen T., Thomson R., Laatikainen R., Savolainen M. J., Laurikka J., et al. 2012. High-throughput quantification of circulating metabolites improves prediction of subclinical atherosclerosis. Eur. Heart J. 33: 2307–2316. [DOI] [PubMed] [Google Scholar]
- 14.Cole T. G., Contois J. H., Csako G., McConnell J. P., Remaley A. T., Devaraj S., Hoefner D. M., Mallory T., Sethi A. A., Warnick G. R. 2013. Association of apolipoprotein B and nuclear magnetic resonance spectroscopy-derived LDL particle number with outcomes in 25 clinical studies: assessment by the AACC Lipoprotein and Vascular Diseases Division Working Group on Best Practices. Clin. Chem. 59: 752–770. [DOI] [PubMed] [Google Scholar]
- 15.Johnson C. S. 1999. Diffusion ordered nuclear magnetic resonance spectroscopy: principles and applications. Prog. Nucl. Magn. Reson. Spectrosc. 34: 203–256. [Google Scholar]
- 16.Rock E., Winklhofer-Roob B. M., Ribalta J., Scotter M., Vasson M. P., Brtko J., Brigelius-Flohe R., Bronner A., Azais-Braesco V. 2001. Vitamin A, vitamin E and carotenoid status and metabolism during ageing: functional and nutritional consequences (VITAGE PROJECT). Nutr. Metab. Cardiovasc. Dis. 11: 70–73. [PubMed] [Google Scholar]
- 17.Schumaker V. N., Puppione D. L. 1986. Sequential flotation ultracentrifugation. Methods Enzymol. 128: 155–170. [DOI] [PubMed] [Google Scholar]
- 18.Mallol R., Rodríguez M. A., Heras M., Vinaixa M., Cañellas N., Brezmes J., Plana N., Masana L., Correig X. 2011. Surface fitting of 2D diffusion-edited 1H NMR spectroscopy data for the characterisation of human plasma lipoproteins. Metabolomics. 7: 572–582. [Google Scholar]
- 19.Petersen M., Dyrby M., Toubro S., Engelsen S. B., Norgaard L., Pedersen H. T., Dyerberg J. 2005. Quantification of lipoprotein subclasses by proton nuclear magnetic resonance-based partial least-squares regression models. Clin. Chem. 51: 1457–1461. [DOI] [PubMed] [Google Scholar]
- 20.Bland J. M., Altman D. G. 1999. Measuring agreement in method comparison studies. Stat. Methods Med. Res. 8: 135–160. [DOI] [PubMed] [Google Scholar]
- 21.Cecconi M., Rhodes A., Poloniecki J., Della Rocca G., Grounds R. M. 2009. Bench-to-bedside review: the importance of the precision of the reference technique in method comparison studies–with specific reference to the measurement of cardiac output. Crit. Care. 13: 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okazaki M., Usui S., Fukui A., Kubota I., Tomoike H. 2006. Component analysis of HPLC profiles of unique lipoprotein subclass cholesterols for detection of coronary artery disease. Clin. Chem. 52: 2049–2053. [DOI] [PubMed] [Google Scholar]
- 23.Barter P. J., Ballantyne C. M., Carmena R., Cabezas M. C., Chapman M. J., Couture P., De Graaf J., Durrington P. N., Faergeman O., Frohlich J., et al. 2006. Apo B versus cholesterol in estimating cardiovascular risk and in guiding therapy: report of the thirty-person/ten-country panel. J. Intern. Med. 259: 247–258. [DOI] [PubMed] [Google Scholar]
- 24.Contois J. H., McConnell J. P., Sethi A. A., Csako G., Devaraj S., Hoefner D. M., Warnick G. R. 2009. Apolipoprotein B and cardiovascular disease risk: position statement from the AACC Lipoproteins and Vascular Diseases Division Working Group on Best Practices. Clin. Chem. 55: 407–419. [DOI] [PubMed] [Google Scholar]
- 25.Lounila J., Ala-Korpela M., Jokisaari J., Savolainen M. J., Kesaniemi Y. A. 1994. Effects of orientational order and particle size on the NMR line positions of lipoproteins. Phys. Rev. Lett. 72: 4049–4052. [DOI] [PubMed] [Google Scholar]
- 26.Dyrby M., Petersen M., Whittaker A. K., Lambert L., Norgaard L., Bro R., Engelsen S. B. 2005. Analysis of lipoproteins using 2D diffusion-edited NMR spectroscopy and multi-way chemometrics. Anal. Chim. Acta. 531: 209–216. [Google Scholar]
- 27.Ala-Korpela M., Lankinen N., Salminen A., Suna T., Soininen P., Laatikainen R., Ingman P., Jauhiainen M., Taskinen M. R., Heberger K., et al. 2007. The inherent accuracy of 1H NMR spectroscopy to quantify plasma lipoproteins is subclass dependent. Atherosclerosis. 190: 352–358. [DOI] [PubMed] [Google Scholar]
- 28.Otvos J. D., Jeyarajah E. J., Cromwell W. C. 2002. Measurement issues related to lipoprotein heterogeneity. Am. J. Cardiol. 90: 22i–29i. [DOI] [PubMed] [Google Scholar]
- 29.Ensign W., Hill N., Heward C. B. 2006. Disparate LDL phenotypic classification among 4 different methods assessing LDL particle characteristics. Clin. Chem. 52: 1722–1727. [DOI] [PubMed] [Google Scholar]
- 30.Witte D. R., Taskinen M. R., Perttunen-Nio H., van Tol A., Livingstone S., Colhoun H. M. 2004. Study of agreement between LDL size as measured by nuclear magnetic resonance and gradient gel electrophoresis. J. Lipid Res. 45: 1069–1076. [DOI] [PubMed] [Google Scholar]
- 31.Caulfield M. P., Li S., Lee G., Blanche P. J., Salameh W. A., Benner W. H., Reitz R. E., Krauss R. M. 2008. Direct determination of lipoprotein particle sizes and concentrations by ion mobility analysis. Clin. Chem. 54: 1307–1316. [DOI] [PubMed] [Google Scholar]
- 32.Otvos J. D., Rudel L. L., McConnell J. P. 2008. Concerns regarding lipoprotein particle measurement by ion mobility analysis. Clin. Chem. 54: 2086–2087. [DOI] [PubMed] [Google Scholar]
- 33.Caulfield M. P., Li S., Lee G., Blanche P. A., Salameh W. A., Benner W. H., Reitz R. E., Krauss R. M. 2008. In reply. Clin. Chem. 54: 2088–2089. [DOI] [PubMed] [Google Scholar]
- 34.Mazer N. A., Giulianini F., Paynter N. P., Jordan P., Mora S. 2013. A comparison of the theoretical relationship between HDL size and the ratio of HDL cholesterol to apolipoprotein A-I with experimental results from the Women’s Health Study. Clin. Chem. 59: 949–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hutchins P. M., Ronsein G. E., Monette J. S., Pamir N., Wimberger J., He Y., Anantharamaiah G. M., Kim D. S., Ranchalis J. E., Jarvik G. P., et al. 2014. Quantification of HDL particle concentration by calibrated ion mobility analysis. Clin. Chem. 60: 1393–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.