Background: Cdc7 is a kinase that, together with its activator Dbf4, regulates the initiation of DNA replication.
Results: Drosophila Cdc7 is a functional ortholog of Cdc7 that is required for endoreplication and gene amplification, although Chiffon/Dbf4 is not essential for endoreplication.
Conclusion: Cdc7 can function during endoreplication independently of Chiffon.
Significance: Either Cdc7 functions alone during endocycling or undiscovered Cdc7 activator(s) exist in Drosophila.
Keywords: Cell Cycle, DNA Replication, Drosophila, Gene Amplification, Phosphorylation, Cdc7, Dbf4, Endoreplication, Serine-Threonine Protein Kinase
Abstract
Cdc7 is a serine-threonine kinase that phosphorylates components of the pre-replication complex during DNA replication initiation. Cdc7 is highly conserved, and Cdc7 orthologs have been characterized in organisms ranging from yeast to humans. Cdc7 is activated specifically during late G1/S phase by binding to its regulatory subunit, Dbf4. Drosophila melanogaster contains a Dbf4 ortholog, Chiffon, which is essential for chorion amplification in Drosophila egg chambers. However, no Drosophila ortholog of Cdc7 has yet been characterized. Here, we report the functional and biochemical characterization of a Drosophila ortholog of Cdc7. Co-expression of Drosophila Cdc7 and Chiffon is able to complement a growth defect in yeast containing a temperature-sensitive Cdc7 mutant. Cdc7 and Chiffon physically interact and can be co-purified from insect cells. Cdc7 phosphorylates the known Cdc7 substrates Mcm2 and histone H3 in vitro, and Cdc7 kinase activity is stimulated by Chiffon and inhibited by the Cdc7-specific inhibitor XL413. Drosophila egg chamber follicle cells deficient for Cdc7 have a defect in two types of DNA replication, endoreplication and chorion gene amplification. However, follicle cells deficient for Chiffon have a defect in chorion gene amplification but still undergo endocycling. Our results show that Cdc7 interacts with Chiffon to form a functional Dbf4-dependent kinase complex and that Cdc7 is necessary for DNA replication in Drosophila egg chamber follicle cells. Additionally, we show that Chiffon is a member of an expanding subset of DNA replication initiation factors that are not strictly required for endoreplication in Drosophila.
Introduction
DNA replication initiates from many genomic loci, termed origins of replication, during S phase of the cell cycle. To ensure that replication initiates only once from each origin per cell cycle during normal mitosis, replication initiation is divided into two distinct temporal phases as follows: origin licensing in late M/G1 phase and origin firing during S phase (1). During origin licensing, the pre-replicative complex (pre-RC),2 consisting of the origin recognition complex (ORC), Cdt1, and Cdc6, assembles on origins where it functions to load the Mcm2-7 DNA helicase onto chromatin in an inactive state. The subsequent activation of the Mcm2-7 DNA helicase is required for origin firing and the initiation of DNA replication, and this activation is controlled by a series of phosphorylation events catalyzed by the cell cycle kinases cyclin-dependent kinase and Cdc7 (2, 3). Together, cyclin-dependent kinase and Cdc7 phosphorylate components of the pre-RC and other replication initiation factors, leading to the recruitment of additional replisome components to the pre-RC, the formation of an active Cdc45·Mcm2-7·GINS (CMG) helicase complex, and the bidirectional progression of active replication forks away from the origin. The timing of both origin licensing and origin firing is tightly regulated by cell cycle-dependent oscillations of cyclin-dependent kinase and Cdc7 kinase activity (4, 5).
Whereas origins fire only once per cell cycle during normal mitosis, specialized tissues in both plants and animals can undergo a form of genome replication termed endoreplication, which consists of alternating G/S phases without any intervening mitotic division. Endoreplication in terminally differentiated cells such as ovarian nurse and follicle cells, and larval tissues in Drosophila, has been proposed to support growth and development by increasing metabolic capacity (6). Additionally, endoreplication may play a role in continued cell growth and repair in cells that are aging or undergoing other forms of genotoxic stress (7, 8). In humans, polyploidization is commonly observed in many types of cancer (8, 9), and endoreplication may be a contributing factor to both cancer development and resistance to chemotherapeutic agents (7, 10). During endoreplication in Drosophila, alternating G and S phases are controlled by oscillations in Cdk2-cyclin E activity (11–14). Because late replication patterns of DNA synthesis, including replication of heterochromatic sequences, are not observed in endoreplicating egg chamber cells, the Drosophila endocycle is assumed to have no S phase checkpoint ensuring that genomic replication is complete (12). In addition, an increasing body of evidence suggests that the initiation of DNA replication may be fundamentally different between mitotically proliferating and endocycling cells, as several of the Drosophila licensing factors necessary for mitotic proliferation, including several ORC proteins (15, 16), Mcm2 (17), and Mcm4/dpa (18), are not strictly required for endocycling. For example, endoreplication continues in the absence of Drosophila ORC proteins, although the resulting genomic ploidy is reduced, and the pattern of genomic regions fully replicated during endocycling is altered (15, 16). This suggests that disruption of these pre-RC components leads to alteration, but not complete loss, of endocycling. In contrast, other pre-RC components, such as Cdt1/dup and Mcm6, are required for both endocycling and mitotic DNA replication (15, 19). Although Cdc7 plays a critical role in regulating the timing of origin firing during normal mitotic proliferation, its role in regulating endoreplication has not been characterized.
During mitosis, Cdc7 kinase activity peaks during the late G1/S phase of the cell cycle due to its association with a cyclin-like regulatory subunit, Dbf4 (20). The Cdc7-Dbf4 (Dbf4-dependent kinase (DDK)) complex is capable of phosphorylating every component of the Mcm2-7 helicase in vitro, with the exception of Mcm5 (21, 22). Additionally, DDK-mediated phosphorylation of Mcm4 stimulates association of Cdc45 with Mcm2-7 (23, 24) and relieves an inhibitory activity within the Mcm4 N-terminal serine/threonine-rich domain (25). Thus, DDK-mediated phosphorylation of the Mcm2-7 helicase induces both direct conformational changes and creates new binding sites for regulatory factors, which together result in activation of the helicase and the initiation of DNA replication. In addition to Mcm2-7, DDK phosphorylates other substrates that might also regulate efficient origin firing. In yeast, DDK has been shown to phosphorylate Thr-45 of histone H3, which is a histone modification associated with proper DNA replication (26). Because two of the major targets of DDK, Mcm2 and Mcm4, are not required for endocycling in Drosophila (17, 18), it has been unclear as to whether DDK would be required for origin firing during this specialized form of DNA replication. Prior to this study, it was not possible to examine the role for DDK in endoreplication because although Cdc7 orthologs had been identified in other eukaryotes ranging from yeast to humans (27–36), and a Dbf4 subunit had been identified in Drosophila (37), the Drosophila ortholog of Cdc7 had not been experimentally identified.
Here, we identify the previously uncharacterized kinase lethal(1)G0148 as the Drosophila ortholog of Cdc7. We show that Drosophila DDK, composed of l(1)G0148 (Cdc7) and Chiffon, is capable of functionally complementing a Cdc7 deficiency in Saccharomyces cerevisiae. We further show that Cdc7 and Chiffon physically associate and that recombinant DDK phosphorylates the known Cdc7 substrates Mcm2 and histone H3 in vitro. Finally, we demonstrate that Cdc7 is necessary for both chorion gene amplification and endoreplication. Surprisingly, however, we find that although Chiffon is necessary for gene amplification, endoreplication in Drosophila continues in the absence of Chiffon, albeit with altered timing. We conclude that Drosophila Cdc7 is an ortholog of Cdc7 that is necessary for multiple forms of DNA replication in Drosophila and that although Chiffon/Dbf4 may play some role in the Drosophila endocycle, it is not essential for endoreplication to occur.
EXPERIMENTAL PROCEDURES
Construction of Alignment and Phylogenetic Tree
Cdc7 amino acid sequences were aligned using ClustalOmega, and a phylogenetic tree was constructed from the aligned sequences using the neighbor-joining method (MAFFT version 7). Bootstrap analysis of the predicted tree was performed, and confidence values were calculated.
Yeast Strains and Manipulation
The Drosophila Gene Collection (DGC) clone AT30978 was used as a template for PCR cloning of Drosophila cdc7 (l(1)G0148, CG32742; FBgn0028360). CG5790 (FBgn0032677) was amplified from OregonR genomic DNA. The cDNA for chiffon (CG5813; FBgn0000307) was amplified from OregonR 0–12-h embryo cDNA. Coding sequences for S. cerevisiae CDC7, Drosophila cdc7, and CG5790 were cloned into the galactose-inducible yeast expression vector pRS313gal (38). Full-length chiffon and chiffonN (N-terminal 1–400 amino acids) coding sequences were cloned into pRS316gal (38). Constructs were co-transformed into cdc7-90 S. cerevisiae (AKY1664 MATa HMRae** cdc7-90 ade2-1 his3 leu2-3,112 ura3 can1-100) (39). Assays were performed to assess the growth of yeast strains at permissive and restrictive temperatures. Briefly, yeast strains were grown overnight in selective media with 2% glucose, subcultured to A600 = 0.5, and grown to A600 ∼1.0. Cells were then diluted to A600 = 1.0, spotted on pre-warmed selective media containing 2% galactose and 1% raffinose as a 10-fold serial dilution, and incubated at permissive (23 °C) or restrictive (30 °C) temperatures. For Western analysis of ectopic protein expression, ∼3.6 × 108 cells were harvested from liquid cultures induced with 2% galactose. For co-immunoprecipitation, ∼1.5 × 1010 cells were harvested from liquid cultures induced with 2% galactose, washed in PBS, and resuspended in Yeast Lysis Buffer (50 mm HEPES-NaOH, pH 7.5, 150 mm NaCl, 10% glycerol, 0.1% Triton X-100, 5 mm EDTA, supplemented with protease and phosphatase inhibitors). Cells were mechanically lysed in the presence of 0.5-mm diameter zirconia/silica beads, and lysates were clarified by centrifugation at 17,500 × g at 4 °C. 3 mg of each lysate was incubated with 25 μl of anti-HA affinity gel (Sigma) for 2 h at 4 °C with rotation, washed four times with Yeast Lysis Buffer, and analyzed by SDS-PAGE and Western blotting.
Western Blots
The following antibodies were used: HA-HRP (Roche Applied Science); FLAG-HRP (Sigma); rabbit histone H3 (Active Motif). Western blots were developed using Luminata Crescendo HRP substrate (Millipore), imaged on a Bio-Rad ChemiDocXRS, and analyzed using ImageLab (version 5.0) software.
Mutagenesis of cdc7
A D269N substitution mutant of cdc7 was generated using the QuikChange system (Stratagene). pRS313gal-Cdc7 was used as a template for mutagenesis PCR with the following primers: 5′ CGG CGA GAG TTT CTC CTC GTT AAC TTC GGT CTG GCC CAG CAT GTG 3′ and 5′ CAC ATG CTG GGC CAG ACC GAA GTT AAC GAG GAG AAA CTC TCG CCG 3′. Mutagenesis products were verified by sequencing.
Cloning and Purification of Mcm2-GST and Mcm4-GST
The following DGC clones were used as templates for PCR cloning: LD47441 for Mcm2 (CG7538, FBgn0014861) and RE04051 for Mcm4 (disc proliferation abnormal, CG1616, FBgn0015929). The cDNA regions encoding the N-terminal 1–279 amino acids of Mcm2 and the N-terminal 1–233 amino acids of Mcm4 were cloned into pGex6P1, expressed, and purified from Escherichia coli, and dialyzed into Elution Buffer II (50 mm HEPES, pH 7.5, 150 mm NaCl, 10% glycerol).
Cloning and Purification of Recombinant Cdc7 and DDK from Insect Cells
Coding sequences for Cdc7 and ChiffonN (N-terminal 1–400 amino acids) were cloned into pBACPAK8 vectors with N-terminal His-FLAG or 2×HA epitope tags, respectively. pBACPAK8-HisFLAG-Cdc7 and pBACPAK8-HA-ChiffonN were transfected into Sf21 cells, and viral supernatants were generated as described previously (40). Sf21 cells were infected with HisFLAG-Cdc7 alone to purify Cdc7 or with HisFLAG-Cdc7 and HA-ChiffonN to purify DDK, and recombinant protein or complex was purified using tandem affinity chromatography. Briefly, cells were lysed in Lysis Buffer (50 mm HEPES, pH 7.5, 2 mm MgCl2, 10% glycerol, 0.2% Triton X-100, 20 mm imidazole, supplemented with complete protease inhibitor mixture (Roche Applied Science)) containing either 500 mm (Cdc7) or 300 mm (DDK) NaCl, and the soluble supernatant was incubated with Ni-NTA-agarose (Cdc7 and DDK), washed in lysis buffer, and eluted in Ni-NTA Elution Buffer I (50 mm HEPES, pH 7.5, 300 mm NaCl, 10% glycerol, 250 mm imidazole). Ni-NTA elutions were pooled and bound to either anti-FLAG affinity gel (Sigma; Cdc7) or anti-HA affinity gel (Sigma; DDK). Beads were washed twice with High Salt Wash Buffer (50 mm HEPES, pH 7.5, 10% glycerol, 300 mm NaCl) and once with Low Salt Wash Buffer (50 mm HEPES, pH 7.5, 150 mm NaCl, 10% glycerol), and Cdc7 or DDK was eluted in Elution Buffer II (50 mm HEPES, pH 7.5, 150 mm NaCl, 10% glycerol, supplemented with complete protease inhibitor mixture (Roche Applied Science)) containing either 0.5 mg/ml 3×FLAG peptide or 0.2 mg/ml 3×HA peptide.
Kinase Assays
In vitro kinase assays were performed as described previously (41) with the following modifications. Briefly, 25-μl reaction volumes containing a final concentration of 40 mm HEPES-NaOH, pH 7.5, 0.5 mm EDTA, 0.5 mm EGTA, 1 mm β-glycerophosphate, 1 mm NaF, 2 mm DTT, 10 mm magnesium acetate, 0.1 mm unlabeled ATP, and 2 μCi of [32P]ATP (PerkinElmer Life Sciences, catalog no. BLU502A250UC), substrate (0.5 μg of Mcm4-GST or Mcm2-GST or 2 μg of purified HeLa core histones), and Cdc7 (6, 12, or 24 ng) or DDK (6, 12, 24 ng Cdc7 with co-purified ChiffonN) were incubated at 30 °C for 30 min (Mcm2/4 as substrate) or 60 min (core histones as substrate). The XL413 inhibitor was synthesized by us as described previously (42) and solubilized in DMSO. Reaction mixtures were separated by SDS-PAGE (10% gel, Mcm2/4; 15% gel, HeLa core histones), stained with Coomassie, imaged, dried, and exposed to a phosphor screen (GE Healthcare). Screens were scanned on a Typhoon phosphorimager, and images were analyzed using ImageQuant TL software.
Drosophila Stocks
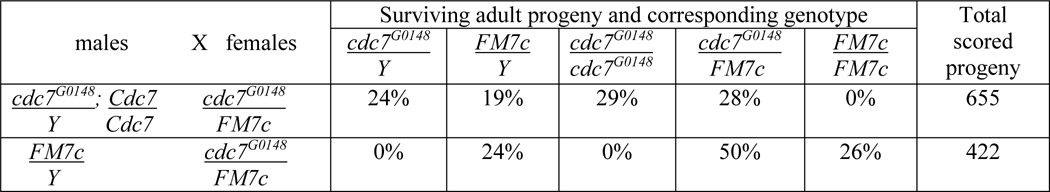
cdc7G0148 flies (w67c23 P{w+mC = lacW}cdc7G0148/FM7c) were obtained from the Bloomington Drosophila Stock Center (stock number 11937). chifETBE3 flies (chifETBE3/CyO) were kindly provided by John Towe, and contain an ∼6-kb genomic deletion in chiffon together with a mutation in a second gene, cactus (37). For the generation of Cdc7 rescue flies, the genomic region containing the coding sequence and regulatory elements for cdc7 (chromosome X, 6548176-6554728) consisting of −1007 bp from the start codon (ATG, +1) of cdc7 (within the adjacent gene CG3226) to +5546 bp (within the adjacent gene CG3224) was cloned into the pCa4B vector and integrated into the attP40 site using site-directed transgenesis (43, 44) to generate w1118; P{w+mC = Cdc7}attP40 flies. The Cdc7 rescue stock was crossed with w67c23 P{w+mC = lacW}cdc7G0148/FM7c, P{w+mC = GAL4-twi.G}108.4, P{UAS-2×EGFP}AX flies using standard genetic techniques to generate w67c23 P{w+mC = lacW}cdc7G0148; P{w+mC = Cdc7}attP40 flies, which are homozygous viable (Table 1).
TABLE 1.
Lethality of cdc7 flies is rescued by expression of Cdc7
Male cdc7G0148 flies carrying the Cdc7 genomic rescue construct on the second chromosome or male FM7c flies without the genomic rescue construct were crossed with female cdc7G0148/FM7c flies, and the surviving adult progeny was scored for presence of the FM7c balancer chromosome using the visible Bar eye marker. The percentage of flies corresponding to each genotype and the total number of scored progeny for each cross are indicated.
Immunohistochemistry
Somatic clones were induced in egg chambers from w67c23 P{w+mC = lacW}cdc7G0148, P{w+mW.hs = FRT(whs)}101/y1 w67c23, P{w+mC = Ubi-GFP.D}ID-1, P{w+mW.hs = FRT(whs)}101; P{w+mW.hs = en2.4-GAL4}e22c, P{w+mC = UAS-FLP1.D}JD1 or y1 w1118, P{ry+t7.2 = 70FLP}3F; chifETBE3, P{ry+t7.2 = neoFRT}40A/P{w+mC = Ubi-GFP(S65T)nls}2L, P{ry+t7.2 = neoFRT}40A adult females. As a control, wild-type clones were induced in egg chambers from y1 w* v24 P{w+mW.hs = FRTw(hs)}101/y1 w67c23, P{w+mC = Ubi-GFP.D}ID-1, P{w+mW.hs = FRT(whs)}101; P{w+mW.hs = en2.4-GAL4}e22c, P{w+mC = UAS-FLP1.D}JD1 adult females. Clones were induced without heat shock for cdc7G0148 and wild-type FRT101 controls and with 40 min of heat shock at 37 °C on the day prior to ovary dissection for chifETBE3 females. Ovaries were dissected from adult females at 3 days post-eclosion, labeled with 5-bromo-2-deoxyuridine (BrdU), fixed, and immunostained as described previously (45). BrdU-labeled egg chambers were immunostained with mouse anti-BrdU antibody (Pharmingen) followed by anti-mouse Alexa568 antibody (Invitrogen) and stained with DAPI. A Nikon A1R-MP microscope was used to image immunostained egg chambers, and images were analyzed using NIS-Elements software. Confocal images are presented as reconstructed maximum intensity projections of multiple z-stack planes for each egg chamber.
RESULTS
Drosophila Contains Two Cdc7 Homologs
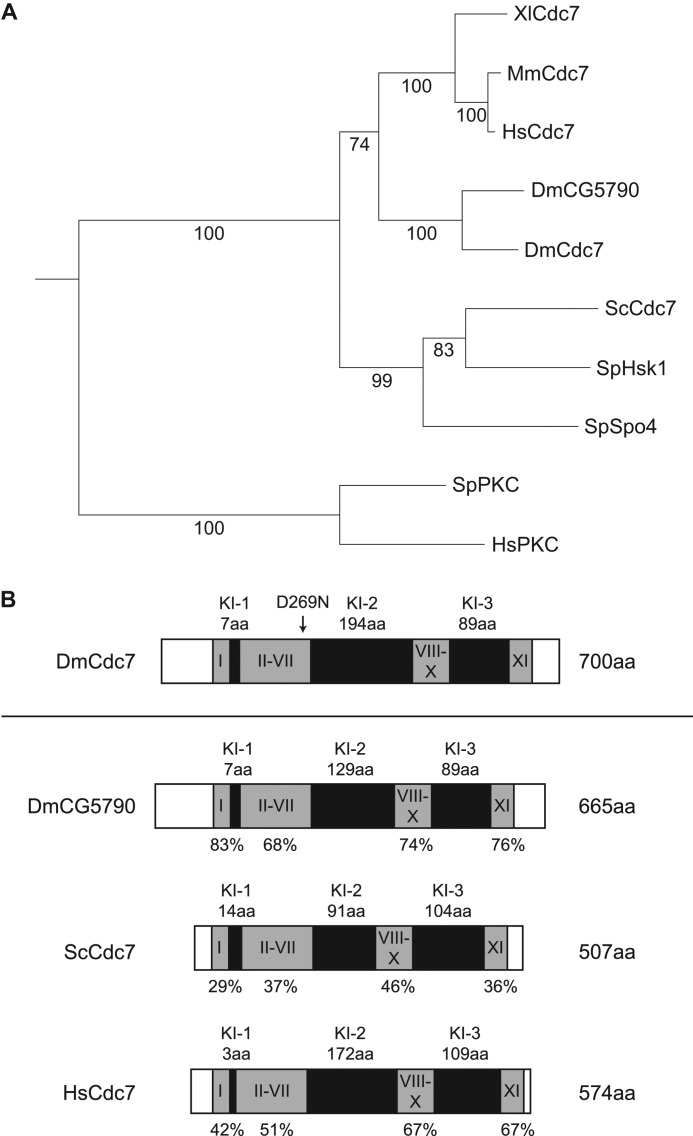
Cdc7 orthologs have been identified and characterized from numerous model organisms (27–36), but no characterization of a Drosophila Cdc7 ortholog had been reported prior to this study. However, because the Cdc7-Dbf4 (DDK) kinase plays a highly conserved role in the initiation of DNA replication, and because a Dbf4 ortholog, Chiffon, has previously been identified in Drosophila (37), we predicted that the Drosophila genome would also encode a homolog of Cdc7. We identified two potential candidates for Drosophila Cdc7 by BLAST search, lethal(1)G0148 and CG5790, and compared these proteins to known Cdc7 orthologs using phylogenetic analysis (Fig. 1A). l(1)G0148 and CG5790 share 26–29% overall amino acid identity with S. cerevisiae Cdc7, S. pombe Hsk1 and Spo4, and 35–36% amino acid identity with Xenopus, mouse, and human Cdc7 orthologs. In addition, l(1)G0148 and CG5790 are 44% identical overall and are more closely related to each other than to any other Cdc7 ortholog.
FIGURE 1.
Phylogenetic and schematic comparison of Drosophila Cdc7 orthologs. A, phylogenetic tree comparing Cdc7 orthologs and two putative Drosophila melanogaster Cdc7 orthologs, l(1)G0148 (Cdc7) and CG5790. The phylogenetic tree was constructed from aligned protein sequences using the neighbor-joining method, and confidence values from bootstrap analysis of the predicted tree are indicated on tree branches. S. pombe protein kinase C and Homo sapiens protein kinase Cϵ type are included as an out-group. Accession numbers are as follows; Xenopus laevis Cdc7, AAD21532.1; Mus musculus Cdc7, AAH80702.1; H. sapiens Cdc7, AAC52080.1; D. melanogaster CG5790, NP_609876.2; D. melanogaster lethal(1)G0148 (Cdc7), AAF46180.2; S. cerevisiae Cdc7, CAA98574.1; S. pombe Hsk1, NP_596328.1; S. pombe Spo4, NP_596598; S. pombe PKC, AAA35323.1; H. sapiens PKCϵ type, NP_005391.1. B, schematic comparison of D. melanogaster Cdc7 with D. melanogaster CG5790, S. cerevisiae Cdc7, and H. sapiens Cdc7. The locations of conserved kinase domains I to XI are shown in gray, and the relative positions and lengths of Cdc7-specific kinase insert (KI) domains 1–3 are indicated in black. The percent identity of conserved kinase domains in each Cdc7 ortholog relative to the corresponding domains in DmCdc7 is indicated below each domain. The location of a mutation in a conserved kinase motif, D269N, is indicated with an arrow above Drosophila Cdc7. aa, amino acids.
Cdc7 is known to contain characteristic kinase insert domains, which are large sequence inserts between the conserved kinase catalytic subdomains VII and VIII (kinase insert 2) and X and XI (kinase insert 3) (28–30, 32–34, 36, 46). To further characterize l(1)G0148 and CG5790, we compared the amino acid sequences of l(1)G0148 and CG5790 with S. cerevisiae Cdc7 and human Cdc7, and we found that l(1)G0148 and CG5790 both contain kinase insert domains at the same positions relative to the catalytic subdomains (Figs. 1B and 2). These results strongly suggest that l(1)G0148 and CG5790 are homologous to Cdc7, and thus, we conclude that l(1)G0148 and CG5790 are paralogs of Cdc7.
FIGURE 2.
Protein alignment of Cdc7 orthologs. Alignment of Drosophila Cdc7 with Cdc7 orthologs as follows: S. cerevisiae Cdc7, CAA98574.1; S. pombe Hsk1, NP_596328.1; S. pombe Spo4, NP_596598; X. laevis Cdc7, AAD21532.1; M. musculus Cdc7, AAH80702.1; H. sapiens Cdc7, AAC52080.1; D. melanogaster CG5790, NP_609876.2; and D. melanogaster lethal(1)G0148 (Cdc7), AAF46180.2. The locations of conserved kinase subdomains I to XI are noted, along with the locations of Cdc7-specific kinase insert domains 1–3. Identical residues are indicated by an asterisk, and highly similar residues are indicated by a colon. An arrow indicates the location of a substitution mutation, D269N, in a conserved kinase motif within Cdc7.
Because there are two paralogs of Cdc7 in Drosophila, we reasoned that one of these proteins might have developed an evolutionarily diverged function relative to other Cdc7 homologs. To determine which Drosophila Cdc7 paralog was most likely to function in mitotic proliferation, we examined the expression pattern of l(1)G0148 and CG5790. The lethal(1)G0148 (FBgn0028360) gene encodes a 700-amino acid protein with a predicted mass of 79 kDa, whereas CG5790 (FBgn0032677) encodes a 665-amino acid protein with a predicted mass of 75 kDa. High throughput tissue-specific transcript expression data shows that l(1)G0148 is highly expressed in adult Drosophila ovaries and testes and has low to moderate expression in most other tissues throughout development (47). In contrast, CG5790 is expressed almost exclusively in adult male testes, with moderate expression in imaginal discs (47). Two DDK complexes have previously been reported in another species. In S. pombe, Hsk1-Him1/Dfp1 controls the initiation of DNA replication during the S phase of mitosis (33, 48, 49), whereas Spo4-Spo6 executes a function unrelated to DNA replication during meiosis (34). Because l(1)G0148 is expressed ubiquitously, and CG5790 is testis-specific, CG5790 is unlikely to be required for DNA replication in all tissues. Furthermore, a large scale RNA-mediated interference screen in cultured Drosophila cells showed that knockdown of l(1)G0148 resulted in delays in S phase progression (50), consistent with a potential role for l(1)G0148 in origin firing during mitotic proliferation. We have refrained from naming CG5790, despite its homology to Cdc7, because the function of this protein in DNA replication or meiosis is unclear. However, for the sake of clarity, we will hereafter refer to the second more widely expressed Cdc7 paralog, l(1)G0148, as Cdc7 based on the results presented below.
Expression of Drosophila DDK Restores Growth in Yeast Containing the Temperature-sensitive cdc7-90 Allele
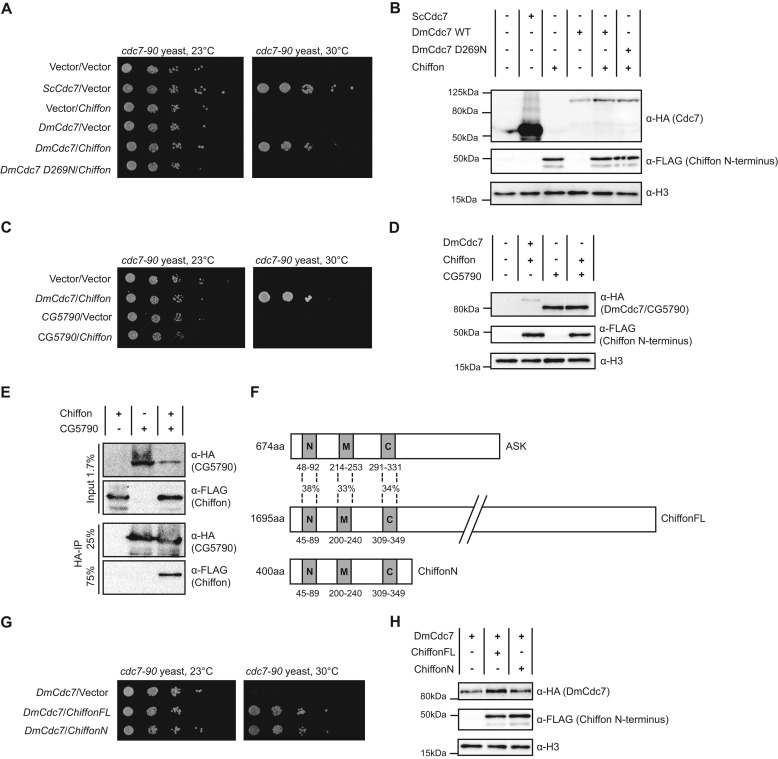
It has been previously been shown that the human DDK complex is capable of functionally complementing a yeast strain deficient for DDK (51). We therefore asked whether the expression of Drosophila Cdc7 could similarly complement a yeast strain lacking functional DDK. We expressed Drosophila Cdc7 alone, or in combination with Chiffon, in a strain of S. cerevisiae that contains the temperature-sensitive cdc7-90 allele (39). At the restrictive temperature, cdc7-90 yeast fail to grow (Fig. 3A, vector/vector control), but growth is restored by the expression of S. cerevisiae Cdc7 (Fig. 3A, ScCdc7/vector). Notably, we observed that whereas expression of Drosophila Cdc7 alone could not restore growth at the restrictive temperature, co-expression of DmCdc7 and Chiffon complemented the temperature-sensitive growth defect (Fig. 3A, DmCdc7/Chiffon). These results are similar to those previously observed by Davey et al. (51) in which complementation of a nonfunctional yeast DDK required the expression of both the Cdc7 and Dbf4 (ASK) subunits of the human DDK complex. Importantly, a Cdc7 substitution mutant, DmCdc7 D269N, which contains a mutation in a conserved DFG motif critical for phosphorylation (52), failed to complement the cdc7-90 temperature-sensitive growth defect when co-expressed with Chiffon (Fig. 3A, DmD269Cdc7/Chiffon), even though its expression was detectable at similar levels as wild-type DmCdc7 (Fig. 3B). This finding indicates that the complementation of the cdc7-90 growth defect is dependent on the kinase activity of Drosophila DDK. Notably, we failed to detect expression of full-length Chiffon in rescue strains at its expected size of 188 kDa; however, we consistently detected an N-terminal ∼50-kDa fragment of Chiffon in these strains (Fig. 3B). This finding indicates that full-length Chiffon may be subject to proteolytic cleavage but that the N-terminal region of Chiffon is sufficient for both the interaction with and activation of Cdc7. We have also observed a similar truncation product when full-length Chiffon is expressed in Sf21 cells and in Drosophila S2 cells, indicating that Chiffon might also be subject to cleavage in vivo (data not shown).
FIGURE 3.
Drosophila DDK complex can rescue a temperature-sensitive growth defect in cdc7-90 yeast. A, growth assay showing that co-expression of D. melanogaster Cdc7 and Chiffon restores growth at the restrictive temperature (30 °C) in yeast carrying the temperature-sensitive cdc7-90 allele, whereas the expression of DmCdc7 with a mutation in a conserved kinase motif (D269N) does not. cdc7-90 yeast were co-transformed with the indicated plasmid combinations, and yeast strains were plated on selective media and incubated at permissive (23 °C) or restrictive (30 °C) temperatures. B, Western blots verifying the expression of Cdc7 and Chiffon in the cdc7-90 rescue strains shown in A. Cdc7 and Chiffon were expressed as fusion proteins with N-terminal epitope tags as follows: HA, Cdc7; FLAG, Chiffon. Histone H3 was analyzed as a loading control. C, growth assay showing that co-expression of D. melanogaster CG5790 and Chiffon does not restore growth of yeast carrying the temperature-sensitive cdc7-90 allele at the restrictive temperature (30 °C), as described in A. Plasmid combinations contained within each yeast strain are as indicated. D, Western blots verifying the expression of Cdc7, CG5790, and Chiffon in the cdc7-90 rescue strains shown in C, as described in B. CG5790 was expressed as a fusion protein with an N-terminal HA epitope tag. E, HA-CG5790 co-immunoprecipitates Chiffon in lysates from cdc7-90 yeast strains ectopically expressing epitope-tagged CG5790 and Chiffon as described in C. Lysates were incubated with anti-HA affinity resin, and co-immunoprecipitating proteins were analyzed by SDS-PAGE and Western blotting. F, schematic comparing the locations of conserved N, M, and C domains in the Dbf4 orthologs ASK, Chiffon, and an N-terminal Chiffon truncation, ChiffonN (amino acids 1–400). The percent identity of the N, M, and C domains in human ASK and Chiffon is indicated. G, growth assay showing that co-expression of DmCdc7 with either full-length Chiffon (ChiffonFL) or ChiffonN restores growth at the restrictive temperature (30 °C) in yeast carrying the temperature-sensitive cdc7-90 allele as described in A. H, Western blots verifying the expression of Cdc7 and full-length and truncated Chiffon in the cdc7-90 rescue strains shown in G, as described in B.
We next asked whether CG5790 could also complement the mitotic growth defect in cdc7-90 yeast. We expressed CG5790 alone, or in combination with Chiffon, in cdc7-90 yeast as described above. However, in contrast to Cdc7, co-expression of CG5790 and Chiffon failed to rescue the cdc7-90 temperature-sensitive growth defect (Fig. 3C, CG5790/chiffon). To address the possibility that Chiffon may not interact with and activate CG5790, we examined whether CG5790 and Chiffon interact in cdc7-90 yeast strains expressing both Drosophila proteins. To do this, we immunoprecipitated HA-tagged CG5790 from yeast lysates using antibodies against HA, and we examined the co-immunoprecipitating proteins for the presence of FLAG-tagged Chiffon by Western blotting. We observe that the N-terminal portion of Chiffon co-immunoprecipitates with CG5790 (Fig. 3E), indicating that, although CG5790 cannot complement the mitotic growth defect of cdc7-90 yeast, Chiffon does interact with CG5790 in cdc7-90 yeast strains. Thus, we conclude that CG5790 does not functionally complement the mitotic growth requirement for S. cerevisiae Cdc7, suggesting that this Cdc7 paralog may have an alternative function in Drosophila. Together, these findings support our conclusion that l(1)G0148 is the Drosophila ortholog of Cdc7 that is involved in DNA replication initiation during mitotic proliferation and that CG5790 might have a non-DNA replication function specific to highly meiotic tissues, such as the testis. Additionally, our results suggest that CG5790 function in Drosophila testis could be regulated by Chiffon.
N-terminal Region of Chiffon Is Sufficient for the Interaction with and Activation of Cdc7
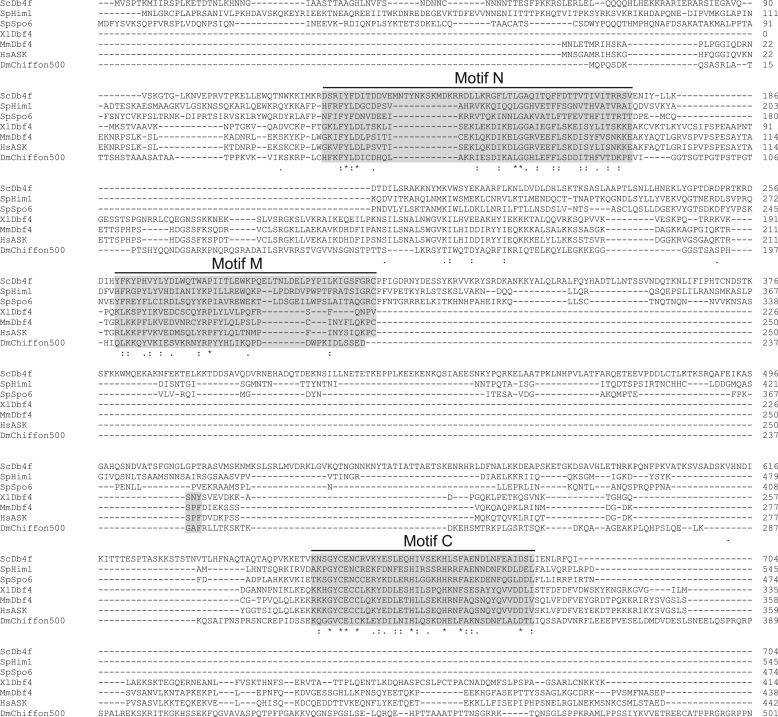
Because the N-terminal ∼50-kDa degradation fragment of Chiffon appeared to be sufficient to activate Drosophila Cdc7 activity in the yeast rescue experiment (Fig. 3, A and B), we sought to determine whether the N-terminal region of Chiffon was indeed sufficient to interact with and activate Cdc7. The primary sequence of Dbf4 orthologs are generally poorly conserved, with the exception of three short regions as follows: the N, M, and C motifs (53, 54). Motifs M and C have been previously identified as being necessary for the binding and activation of Cdc7 by Dbf4 (55–57). We aligned Chiffon with previously characterized Dbf4 orthologs (20, 37, 48, 49, 58–66) to identify the N, M, and C motifs (Figs. 3F and 4). Motifs N and C in Chiffon were previously described as the CDDN2 and CDDN1 domains, respectively (37). Because the M and C motifs are contained within the N-terminal first 400 amino acids of Chiffon, we co-expressed a truncated version of Chiffon, ChiffonN, that consists of the first 400 amino acids, together with Cdc7 in cdc7-90 yeast, and examined growth at the restrictive temperature. Co-expression of Cdc7 with the N-terminal truncation of Chiffon, ChiffonN, restored growth at the restrictive temperature, similar to expression of full-length Chiffon (Fig. 3G). Furthermore, the same ∼50-kDa degradation fragment of Chiffon was detected in yeast expressing full-length Chiffon or ChiffonN (Fig. 3H), indicating that ChiffonN contains all the sequence present in the full-length Chiffon-presumed proteolytic cleavage product. We therefore conclude that the N-terminal 400 amino acids of Chiffon containing the N, M, and C motifs are sufficient for the ability of Cdc7 to rescue the mitotic growth defect of cdc7-90 yeast in vivo.
FIGURE 4.
Protein alignment of Dbf4 orthologs. Alignment of the first 500 amino acids of D. melanogaster Chiffon with Dbf4 orthologs as follows: S. cerevisiae Dbf4, CAA98869.1; S. pombe Him1, O59836.1; X. laevis Dbf4, BAC76421.1; M. musculus Dbf4, Q9QZ41.1; H. sapiens ASK, Q9UBU7.1; D. melanogaster Chiffon, AAD48779.1. The locations of the conserved Dbf4 N, M, and C motifs are indicated. Identical residues are indicated by an asterisk, and highly similar residues are indicated by a colon.
We next sought to determine whether the N-terminal region of Chiffon was sufficient to interact with Cdc7 in vitro. To accomplish this, we co-expressed epitope-tagged Cdc7 and ChiffonN in Sf21 insect cells, and we purified the DDK complex using tandem affinity chromatography against both epitope tags. Using this approach, we purified Cdc7 alone (Fig. 5A, lane 1) or bound to ChiffonN (Fig. 5A, DDK, lane 2). These results indicate that Cdc7 and ChiffonN interact directly to form the DDK complex and that the N-terminal region of Chiffon is sufficient for this interaction. We note that ChiffonN co-purified with Cdc7 appears as a diffuse smear when separated by SDS-PAGE (Fig. 5, A and B). This result is consistent with observations of the human DDK complex, in which ASK appears as a smear on SDS-polyacrylamide gels due to autophosphorylation of DDK (41). Taken together, our results indicate that the N-terminal region of Chiffon is sufficient for both its interaction with Cdc7 and for rescue of the mitotic growth defect of cdc7-90 yeast in vivo.
FIGURE 5.
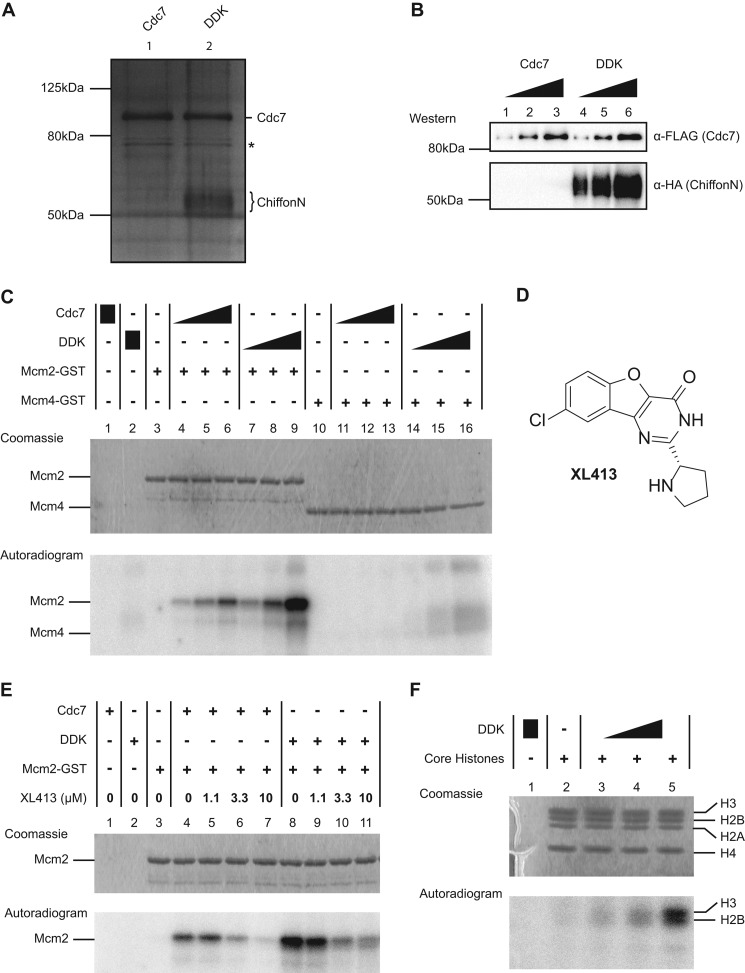
Drosophila Cdc7 phosphorylates Mcm2 and histone H3, Chiffon stimulates Cdc7 kinase activity, and XL413 inhibits Cdc7 kinase activity. A, silver-stained SDS-polyacrylamide gel showing purified recombinant Drosophila Cdc7 or DDK complex. Cdc7 and ChiffonN were expressed in Sf21 cells as fusion proteins with N-terminal epitope tags as follows: His-FLAG, Cdc7; HA, ChiffonN. Cdc7 was expressed alone or co-expressed with ChiffonN. Cdc7 was purified by sequential Ni-NTA-agarose chromatography followed by FLAG-agarose chromatography, and DDK was purified using sequential nickel-agarose chromatography, followed by HA-agarose chromatography. Lane 1, Cdc7 purification; lane 2, Cdc7 + ChiffonN (DDK) co-purification. ChiffonN appears as a diffuse band (brace) due to phosphorylation by Cdc7. A nonspecific contaminant present at low levels in Cdc7 and DDK purifications is marked by an asterisk. B, Western analysis of Cdc7 and DDK used for kinase assay in C. Increasing amounts of Cdc7 (6, 12, or 24 ng) or DDK (6, 12, 24 ng of Cdc7, with co-purified ChiffonN) were resolved by SDS-PAGE and detected by Western blotting with antibodies against FLAG (Cdc7) and HA (ChiffonN). C, Chiffon stimulates Cdc7 kinase activity. In vitro kinase assays were performed in the presence of [γ-32P]ATP at 30 °C for 30 min. Increasing amounts of Cdc7 (6, 12, and 24 ng) or DDK complex (6, 12, and 24 ng of Cdc7, with co-purified ChiffonN) were incubated with 0.5 μg of Mcm2(N1–279)-GST or Mcm4(N1–233)-GST. Reactions were separated by SDS-PAGE, stained with Coomassie (upper panel), and exposed to a phosphor screen (lower panel). D, chemical structure of the Cdc7-specific inhibitor XL413. E, Cdc7 kinase activity is inhibited by the Cdc7-specific inhibitor XL413. Kinase assays were performed as in C. Cdc7 (12 ng) or DDK (12 ng Cdc7) was incubated with [γ-32P]ATP and 0.5 μg of Mcm2-GST in the presence of either vehicle (DMSO) or increasing concentrations of XL413 (1.1, 3.3, and 10 μm, respectively). F, DDK phosphorylates histones H3 and H2B. Increasing amounts of DDK (6, 12, and 24 ng of Cdc7) were incubated with 2 μg of core histones in the presence of [γ-32P]ATP at 30 °C for 60 min. Reactions were separated and analyzed as in C.
Chiffon Stimulates Cdc7 Kinase Activity, Which Is Inhibited by XL413
The DDK complex is proposed to regulate the initiation of DNA replication through phosphorylation of the Mcm2-7 helicase (2, 3, 67, 68). Because Dbf4 is required to activate Cdc7 kinase activity, the activity of DDK varies throughout the cell cycle because of oscillating Dbf4 protein levels, resulting in the activation of Cdc7 kinase activity specifically at late G1/S phase (20, 21, 58, 69–71). Because Chiffon interacts with Drosophila Cdc7 in vitro, and is able to rescue the growth defect of cdc7-90 yeast when co-expressed with Cdc7, we next sought to determine whether Chiffon activates the kinase activity of Cdc7. To accomplish this, we performed in vitro kinase assays with recombinant DDK (Cdc7 with co-purified ChiffonN) or Cdc7 alone on N-terminal fragments of Drosophila Mcm2 and Mcm4, which are known substrates of Cdc7 (24, 36, 41, 72, 73). In contrast to previous in vitro studies with human Cdc7 (41, 64), we found that Drosophila Cdc7 was capable of phosphorylating Mcm2 in a dose-dependent manner in the absence of Chiffon (Fig. 5C). However, incubation of Mcm2 with DDK yielded substantially increased phosphorylation of Mcm2 (Fig. 5C), even though Cdc7 was present in DDK at similar amounts as compared with those lanes containing Cdc7 alone (Fig. 5B). These results indicate that Cdc7 alone is capable of phosphorylating Mcm2 but show that Chiffon stimulates Cdc7 kinase activity in vitro. We did not detect phosphorylation of Mcm4 in the presence of Cdc7 alone, and we detected only very weak phosphorylation of Mcm4 in the presence of the highest amounts of DDK (Fig. 5C). These results are consistent with previous in vitro kinase assays performed with the human DDK complex, which showed that Mcm2 is a better in vitro substrate for DDK phosphorylation than Mcm4 (41).
Recently, a small molecule inhibitor of Cdc7, XL413 (Fig. 5D), was synthesized, tested for inhibition of human Cdc7 in vitro and in vivo, and advanced into clinical trials as a potential chemotherapeutic agent (42). To determine whether Drosophila Cdc7 is relevant as a model for human Cdc7 function, we tested whether XL413 could inhibit the kinase activity of Drosophila Cdc7 in vitro. To accomplish this, we performed in vitro kinase assays using either Cdc7 or DDK with Mcm2 as substrate in the presence of increasing concentrations of XL413, relative to vehicle alone (DMSO). We observed a dose-dependent inhibition of Mcm2 phosphorylation by both Cdc7 and DDK in the presence of increasing concentrations of XL413 (Fig. 5E). We therefore conclude that XL413, a selective inhibitor of human Cdc7, inhibits the kinase activity of Drosophila Cdc7.
Drosophila DDK Phosphorylates Histones H3 and H2B
In addition to the Mcm2-7 complex, Thr-45 of histone H3 has been identified as a substrate for Cdc7 phosphorylation in S. cerevisiae (26). We therefore asked whether Drosophila DDK could also phosphorylate histone H3. We performed kinase assays in which DDK was incubated with purified HeLa core histones as substrate. We observed substantial phosphorylation of histone H3 in the presence of increasing amounts of DDK in vitro, and unexpectedly, we also observed lower levels of phosphorylation of histone H2B (Fig. 5F). Phosphorylation of histone H2B was not observed by Baker et al. (26). Thus, this activity could reflect either the expanded activity of DDK in vitro on core histones relative to nucleosomes or a novel biological target in Drosophila relative to yeast.
Cdc7 Is Required for DNA Replication in Drosophila Egg Chamber Follicle Cells
Depletion or disruption of DDK has been shown to inhibit DNA replication in a variety of metazoan systems (61, 64, 74–78). Thus, we sought to examine the effect of disrupting Cdc7 activity on DNA replication in Drosophila. We identified a fly line containing a P-element transposon insertion in the fourth intron of the X-linked cdc7 gene, cdc7G0148 (79), which is lethal in the homozygous state such that no cdc7G0148 male flies are observed (Table 1). This lethality is rescued by expression of cdc7 from a genomic rescue construct inserted on another chromosome, such that viable cdc7G0148; P{w+mC = Cdc7}attP40 adult progeny can be obtained in the expected ratios in both male and female adult progeny (Table 1). This finding indicates that proper Cdc7 function is required for viability in Drosophila. However, we note that cdc7G0148 is unlikely to be a null allele because of the intronic position of the transposon insertion and because we can obtain, albeit rarely, somatic clones of this allele, suggesting that some low level of Cdc7 protein expression remains in this mutant genotype.
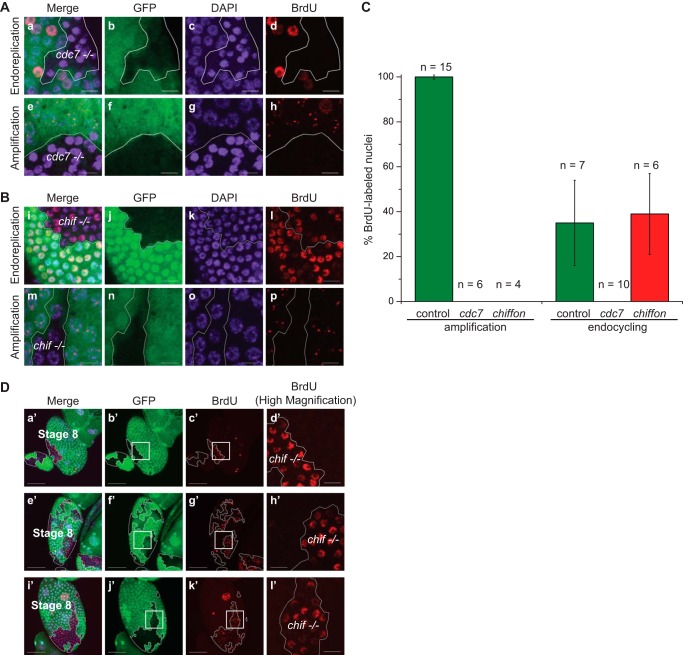
To determine whether Cdc7 is required for DNA replication, we examined the egg chamber follicle cells of the ovary, which are a well characterized system for studying DNA replication in Drosophila (45, 80). Follicle cells in Drosophila egg chambers undergo mitotic divisions early in egg chamber development, followed by three endocycles in which the entire genome is replicated without any intervening cell division, leading to polyploidy. These endocycles are then followed by an amplification phase, during which the genomic loci encoding the chorion eggshell protein are over-replicated relative to the rest of the genome. Importantly, chorion loci amplification requires all known components of the DNA replication machinery and is recognized as a sensitive model system for identifying factors that are required for proper DNA replication (45). We used the FLP/FRT genetic mosaic system to generate somatic clones that were homozygous mutant for Cdc7 in Drosophila egg chambers (81, 82). Homozygous cdc7G0148/cdc7G0148 egg chamber clones were generated in females that were otherwise heterozygous for the strong hypomorphic cdc7G0148 allele. DNA replication in egg chambers was then visualized through incorporation of BrdU followed by immunostaining with antibodies against BrdU. Whereas BrdU incorporation in wild-type (GFP-positive) follicle cells is observed in both the endocycle and amplification stage, we do not observe BrdU incorporation in homozygous cdc7G0148/cdc7G0148 egg chamber clones (GFP-negative, outlined in white) in either the endocycle (Fig. 6A, panels a–d) or amplification stage follicle cells (Fig. 6A, panels e–h). We also observe smaller, condensed DAPI-positive nuclei in homozygous cdc7G0148/cdc7G0148 amplification stage clones, which together with the lack of BrdU incorporation is consistent with a failure of these cells to undergo proper DNA endoreplication. The more intense DAPI staining in these small cdc7G0148 nuclei is indicative of increased chromatin density, possibly due to chromatin condensation, but does not represent canonical apoptosis as determined by immunostaining with antibodies against activated caspase-3 (data not shown). We quantified these data by determining the percentage of BrdU-labeled, DAPI-positive nuclei in cdc7G0148 clones relative to control wild-type FRT101 clones (Fig. 6C). Multiple clones were analyzed from individual animals (n) for each genotype where possible. However, we note that very few clones (∼1 clone per two animals examined) were able to be identified for cdc7G0148 relative to both chiffon and the wild-type control. This observation is consistent with the hypomorphic nature of the cdc7G0148 allele and indicates that Cdc7 function is usually required for the mitotic proliferation necessary for clone formation. Thus, we conclude that Cdc7 is required for DNA replication both in endocycling and in amplification stage cells. These observations suggest that Cdc7 is generally required for DNA replication in Drosophila and is likely also to be required for mitotic proliferation.
FIGURE 6.
Mutations in DDK differentially disrupt DNA replication in Drosophila egg chamber follicle cells. A, endocycle and amplification stage DNA replication is eliminated in egg chamber follicle cells lacking Cdc7. Mosaic egg chambers were generated using the FLP/FRT system. BrdU incorporation is shown in egg chamber follicle cells, and DAPI staining is shown for comparison. Somatic cdc7G0148 clones are marked by the absence of GFP (outlined in white). A representative endocycle stage egg chamber (panels a–d) and representative amplification stage egg chamber (panels e–h) are shown. Scale, 10 μm. B, amplification stage DNA replication is eliminated in egg chamber follicle cells lacking Chiffon, but the endocycle stage replication persists. Egg chambers carrying somatic chifETBE3 clones were generated using the FLP/FRT system and BrdU and DAPI analyzed as in A. A representative endocycle stage egg chamber (panels i–l) and a representative amplification stage egg chamber (panels m–p) are shown. Scale, 10 μm. C, quantification of DNA replication defects observed in cdc7G0148 and chifETBE3 somatic clones shown in A and B. The percentage of BrdU-labeled nuclei in clones deficient for cdc7 or chiffon was compared with control somatic clones (FRT101), in which the FLP/FRT system was used to generate egg chambers mosaic for GFP expression in an otherwise wild-type genetic background. Mean percentages ± S.D. are shown for the number of animals (n) examined for each genotype and developmental stage. D, chifETBE3 somatic clones continue endocycling after endoreplication in nonclonal regions of the egg chamber has ceased. Egg chambers carrying somatic chifETBE3 clones were generated using the FLP/FRT system, and BrdU and DAPI were analyzed as in A. Representative images from three different animals (panels a′–d′, panels e′–h′, and panels i′–l′) are shown. Scale, 50 μm (Merge, GFP, BrdU) or 10 μm (BrdU high magnification).
Chiffon Is Essential for Amplification Stage, but Not Endocycle Stage, of DNA Replication
Because Dbf4 is required for the activity of Cdc7 during late G1 and S phase, we next sought to determine whether Chiffon is required for DNA replication by Cdc7 in both the endocycle and amplification stage egg chambers. Mutations in chiffon result in a thin and fragile chorion and reduce amplification of chorion loci DNA, consistent with a requirement for Chiffon in DNA replication in developing egg chambers (37, 83). However, the direct effect of chiffon mutations on DNA replication in endocycling follicle cells had not previously been examined. Thus, we used the FLP/FRT system to generate somatic clones homozygous for the chifETBE3 null allele (37), and we examined BrdU incorporation in Drosophila egg chambers. Whereas BrdU incorporation in wild-type (GFP-positive) follicle cells is observed in both the endocycle and amplification stage, we found that chorion amplification was eliminated in clones homozygous null for chiffon (Fig. 6B, panels m–p), consistent with previous findings (37, 83). However, nuclei within the homozygous chifETBE3/chifETBE3 clones appear to be the same size as nuclei in follicle cells outside of the clone (Fig. 6B, panel o). We therefore examined BrdU incorporation in endocycle stage clones homozygous for the null chiffon allele. Unexpectedly, examination of chifETBE3/chifETBE3 clones in endocycle stage egg chambers revealed that endocycle stage DNA replication continued in clones lacking Chiffon (Fig. 6B, panels i–l). We quantified these data by determining the percentage of BrdU-labeled nuclei in chiffon clones relative to control wild-type clones (Fig. 6C). These data show that whereas BrdU incorporation is never observed in chiffon amplification stage follicle cells, endocycling chiffon clones exhibit a similar fraction of nuclei incorporating BrdU to that observed in wild-type control clones. Although DNA replication is not eliminated in chifETBE3/chifETBE3 endocycling cells, we did observe differences in the timing of endocycling in the chiffon clones relative to the surrounding wild-type cells. Notably, BrdU incorporation is still observed in several of the chiffon clones after endoreplication in nonclonal regions of the egg chamber has largely ceased (Fig. 6D, representative images from three individual animals shown). Our data do not support the conclusion that chiffon clones undergo an extra endocycle, because post-endocycle nuclei within chiffon clones appear to be the same size as nuclei in the rest of the egg chamber in amplification stage egg chambers (Fig. 6B, panel o). In addition, we do not observe endocycling patterns of BrdU incorporation in amplification stage egg chambers in the chiffon clones, suggesting that the developmental transition from endocycling to amplification is not affected by loss of Chiffon. Rather, the continued BrdU incorporation in chiffon mutant clones in stage 8 egg chambers is most consistent with a delayed exit of the chiffon cells from the endocycle S phase relative to their wild-type counterparts. This observation indicates that although Chiffon is not necessary for endoreplication within egg chamber follicle cells, Chiffon might still function in the proper timing or progression of endoreplication. Thus, together our findings suggest that the DDK complex containing Cdc7 and Chiffon is required for the initiation of DNA replication during chorion gene amplification, although Cdc7 can function independently of Chiffon during the Drosophila endocycle.
DISCUSSION
Drosophila Contains Two Cdc7 Homologs
We have identified two Drosophila genes that encode proteins with homology to Cdc7 as follows: the ubiquitously expressed gene l(1)G0148 (cdc7), and the testis-specific gene CG5790. The only other organism in which two Cdc7 homologs have been identified is S. pombe, which encodes two DDK complexes with distinct functions and regulatory subunits, Hsk1-Him1/Dfp1, which regulate S phase DNA replication initiation during mitotic proliferation (33, 48, 49), and Spo4-Spo6, which is dispensable for mitosis but essential for the progression of meiosis II (34, 66). S. cerevisiae DDK has also been found to regulate several aspects of meiosis (84), suggesting that DDK-type complexes might play a general role in regulating meiosis in multiple organisms. The expression patterns of Drosophila cdc7 and CG5790, and the inability of CG5790 to complement the cdc7-90 temperature-sensitive growth defect in S. cerevisiae, suggest that, of the two paralogs, Cdc7 is more likely to play a general role in S phase DNA replication during mitotic proliferation. Furthermore, the testis-specific expression pattern of CG5790 would be most consistent with a role for this putative kinase in meiosis. We predict that Chiffon could act as the regulatory subunit for CG5790 because Chiffon is the only protein encoded by the Drosophila genome that has any significant homology to known Dbf4 orthologs and because Chiffon has been identified in an enhancer-trap screen for genes that are strongly expressed in post-mitotic spermatogonia and spermatocytes in Drosophila testis (85). Additionally, our observation that the N terminus of Chiffon, which contains the canonical Cdc7-interacting motifs, co-immunoprecipitates CG5790 from yeast lysates, indicates that Chiffon and CG5790 are capable of interacting. This supports a potential role for CG5790 and Chiffon in Drosophila meiosis. However, because cdc7 and CG5790 are both expressed in Drosophila testis, the meiotic role of each Cdc7 paralog and the identity of the in vivo regulatory Dbf4-like subunit for CG5790 remain to be determined.
Cdc7 and Chiffon Are Differentially Required for Endocycling
The results from our analysis of somatic mosaic egg chambers show that although Cdc7 is required for both endocycling and follicle cell amplification, Chiffon is required for amplification but is not essential for endocycling. Consistent with our observations, Chiffon was previously found to be required for amplification of the chorion loci in egg chamber follicle cells (37, 83), and examination of DAPI-stained chiffon null egg chambers suggested that follicle cells could complete endocycling normally in the absence of Chiffon (86). Together, these results indicate that Cdc7 can initiate DNA replication in endocycling cells independently of Chiffon.
How might Cdc7 function independently of Chiffon during the endocycle? One potential explanation is that an alternative Dbf4-like activating subunit exists within Drosophila that serves to activate Cdc7 specifically during the endocycle. Multiple Dbf4 paralogs that regulate the same Cdc7 ortholog at distinct developmental stages have been identified in both human and Xenopus (87–90). In Xenopus egg extracts, depletion of the Cdc7-Drf1 DDK complex severely impairs DNA synthesis, whereas depletion of the alternative Cdc7-Dbf4 DDK complex has no effect on replication (90, 91). Cdc7-Drf1 and Cdc7-Dbf4 are developmentally regulated, as Cdc7-Drf1 is the primary DDK complex in Xenopus oocytes and early embryos, and Cdc7-Dbf4 predominates at later stages of embryonic development. It is possible that Drosophila Cdc7 is similarly regulated by multiple activating subunits during different developmental stages, with Chiffon being required for the activation and targeting of Cdc7 to replication origins during gene amplification, and an as-of-yet undiscovered Dbf4-like regulator(s) functioning during endoreplication and mitotic proliferation. Although Chiffon is the only protein within Drosophila with significant homology to known Dbf4 orthologs, several findings support the possibility that additional Cdc7 activating or targeting subunits could exist in Drosophila. First, examination of Xenopus egg extracts found that a substantial portion of Cdc7 is not complexed with either Drf1 or Dbf1, and extracts immunodepleted for Drf1 and Dbf1 retained some capacity for supporting DNA replication (90). Second, the observation that chiffon appears to be dispensable for mitotic proliferation, because chiffon null mutants are viable (37) and we were able to recover large chiffon clones in mosaic Drosophila egg chambers, suggests that Drosophila could contain an undiscovered Cdc7 activator for mitotic proliferation. Together, these observations suggest that an additional Cdc7 regulating subunit, which may potentially bear little similarity to Dbf4, remains to be discovered. Because this putative Cdc7 regulatory subunit may be present in both Xenopus and Drosophila, it is intriguing to speculate that it could also function in humans to target or activate Cdc7 in a Dbf4-independent manner.
Another possibility is that Cdc7 can phosphorylate and activate the pre-RC during endocycling in the absence of any regulatory subunit. Although there are few reported cases of Cdc7 functioning independently of a Dbf4 subunit, our in vitro kinase results, which are consistent with observations of yeast Cdc7 activity (72), show that Drosophila Cdc7 does possess some ability to phosphorylate Mcm2 in the absence of Chiffon. However, it is possible that Cdc7 can phosphorylate its substrates independently of Chiffon in vitro but that Chiffon or another protein is required to target its activity in vivo. This explanation would be consistent with the inability of Cdc7 to rescue the mitotic growth defect of cdc7-90 yeast in the absence of Chiffon. A recent study has shown that Dbf4 knockdown has no effect on Cdc7-dependent induction of smooth muscle cell differentiation in murine cell lines, which is instead dependent on interactions between Cdc7 and the SMAD3 transcriptional modulator (92). Thus, there is a precedent for the ability of Cdc7 to function in specific developmental processes independently of Dbf4, potentially through interactions with other unrelated factors.
An additional possibility is that the unique nature of the replication initiation machinery during the endocycle might lead to a partial bypass of the requirement for DDK for the initiation of DNA replication. Previously, endoreplication in Drosophila has been shown to continue in the absence of several pre-RC components that are otherwise essential for mitotic proliferation, including dpa/Mcm4 (18), ORC proteins 1, 2, and 5 (15), Mcm2 (17), and Mcm5, although this last factor is disputed (15, 93). We note that although endoreplication continues in these mutants, there is evidence that the pattern and level of replication is altered from that of the wild type (15, 16). Two distinct Mcm complexes are present in Drosophila that contain either Mcm2 and Mcm4 or Mcm5 (94). The key substrate of DDK for mitotic replication initiation in S. cerevisiae is Mcm4, and deletion of an inhibitory domain in the Mcm4 N-terminal tail bypasses the cell cycle requirement for DDK (25). Because dpa/Mcm4 and Mcm2 may not strictly be required for endocycling, and because Mcm complexes exist that lack Mcm4, this key mitotic target of DDK might not be required for the initiation of DNA replication in endocycling cells.
What then might be the primary target of Cdc7 in endocycling cells? Because Cdc7 interacts directly with Mcm5 (95), and because both yeast Cdc7 (72) and Drosophila Cdc7 exhibit kinase activity toward Mcm2 in the absence of Dbf4, it is plausible that other subunits of the Mcm2-7 helicase might be the primary targets of Cdc7 in endocycling cells that are critical for DNA replication. Mcm6 could be a potential target for Cdc7 phosphorylation during endocycling because Mcm6 is phosphorylated by DDK in vitro and in vivo, and there is evidence that the phosphorylation of Mcm2, Mcm4, and Mcm6 serves a redundant purpose during proliferation (23, 96, 97). Consistent with this idea, loss of Mcm6 (19) or knockdown of the Mcm2-7 loading factor Cdt1 (15) severely impairs endocycling, suggesting that the Mcm2-7 helicase, in one form or another, does play a critical role in endoreplication.
Prolonged Endocycling Occurs in the Absence of Chiffon
Although our analysis shows that Chiffon is not necessary for endocycling, we did observe prolonged BrdU labeling in chiffon mutant endocycle stage clones, which appears to be most consistent with increased length of the endocycle S phase in these cells. Prolonged endocycling due to increased time in S phase has been previously observed in egg chamber follicle cells deficient for Dacapo (98), which is a cyclin-dependent kinase inhibitor of cyclin E/cdk2 that promotes Mcm2 association with chromatin during endocycling (99). Similarly, it has been hypothesized that the direct disruption of cyclin E levels also leads to an elongated endocycle S phase, because the replication of late-replicating heterochromatic DNA sequences that usually become under-represented during endoreplication is restored in a cycE hypomorph (12). It is possible that although Chiffon is not necessary for Cdc7 activity in endocycling cells, it still plays a role in efficient origin licensing or firing, and therefore S phase timing, during endoreplication.
Examining the replication of heterochromatic sequences in chiffon mutants may be helpful in future studies to determine whether Chiffon, similar to cyclin E (12) and ORC (16), although not necessary for endoreplication, does influence S phase length and the overall pattern of genomic replication during endoreplication.
C-terminal Extension of Chiffon Is Not Required for DDK Function
Although full-length Chiffon consists of 1695 amino acids, we found that the N-terminal 400 amino acids of Chiffon were sufficient to activate Cdc7 in the cdc7-90 yeast strain, interact with Cdc7 in vitro, and stimulate Cdc7 kinase activity. This raises the question as to the function of the large C-terminal extension present in Drosophila Chiffon, which does not appear to be present in the mammalian and yeast Dbf4 orthologs. Mammalian ASK does contain a modest C-terminal extension that is not present in yeast Dbf4 (Figs. 3 and 4) and that is implicated in an autoinhibition of Cdc7 that is relieved through interactions with LEDGF (100, 101). However, there is no detectable sequence similarity between the C-terminal portions of Chiffon and ASK. Instead, the large C-terminal extension of Chiffon appears to be an insect-specific variant of the Dbf4 protein (86). Our results indicate that full-length Chiffon might be regulated by proteolytic cleavage because both full-length recombinant Chiffon and Chiffon expressed in yeast appear as an N-terminal 50-kDa truncation fragment that is sufficient to interact with Cdc7. Additionally, an ∼50-kDa N-terminal truncation product of Chiffon was also observed when epitope-tagged Chiffon was transiently expressed in cultured S2 cells (data not shown). Dbf4 is known to be a substrate of the anaphase-promoting complex (21), and it is possible that Chiffon is regulated by the anaphase-promoting complex during the cell cycle. However, the conservation of the C-terminal extension of Chiffon in insect species indicates that the C-terminal region of Chiffon, although not required for DDK activity, might be required for some unknown function that could potentially be distinct from DDK activity.
Conclusion
In summary, we report the characterization of Drosophila Cdc7, a functional homolog of Cdc7 that is necessary for multiple forms of DNA replication in Drosophila. Surprisingly, although Cdc7 is essential for endoreplication in ovarian follicle cells, its activating subunit Chiffon is not. Although not necessary for endoreplication, Chiffon might still play a role in the timing and progression of S phase during the endocycle. The involvement of Cdc7 in polyploidization, which may be a route to aneuploidy and drug resistance in cancer (7), may increase its importance as a target of novel chemotherapeutic agents. Additionally, because Cdc7 appears to function during endocycling in the absence of Chiffon, future studies on the role of Cdc7 in endocycling cells might yield the identification of novel activators of Cdc7 and provide further insight into the specialized mode of replication initiation that occurs during endoreplication.
Acknowledgments
The chifETBE3 flies were kindly provided by John Tower, and the cdc7-90 yeast strain was kindly provided by Ann Kirchmaier. We thank Jie Li and Xiaoqi Liu for assistance with the kinase assays. Fly stocks were from the Bloomington Drosophila Stock Center (supported by National Institutes of Health Grant P40OD018537); plasmids were from the Drosophila Genomics Resource Center (supported by National Institutes of Health Grant OD010949-10), and information was from FlyBase and FlyMine. NMR, mass spectrometry, and DNA sequencing data were acquired by core facilities supported by National Institutes of Health Grant P30 CA023168.
This work was supported by American Cancer Society Institutional Research Grant 58-006-53 (to the Purdue University Center for Cancer Research) and by Purdue University Center for Cancer Research Small Grants Program.
- pre-RC
- pre-replicative complex
- DDK
- Dbf4-dependent kinase
- Ni-NTA
- nickel-nitrilotriacetic acid
- ORC
- origin recognition complex.
REFERENCES
- 1. Sacco E., Hasan M. M., Alberghina L., Vanoni M. (2012) Comparative analysis of the molecular mechanisms controlling the initiation of chromosomal DNA replication in yeast and in mammalian cells. Biotechnol. Adv. 30, 73–98 [DOI] [PubMed] [Google Scholar]
- 2. Labib K. (2010) How do Cdc7 and cyclin-dependent kinases trigger the initiation of chromosome replication in eukaryotic cells? Genes Dev. 24, 1208–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tanaka S., Araki H. (2010) Regulation of the initiation step of DNA replication by cyclin-dependent kinases. Chromosoma 119, 565–574 [DOI] [PubMed] [Google Scholar]
- 4. Depamphilis M. L., de Renty C. M., Ullah Z., Lee C. Y. (2012) “The Octet”: eight protein kinases that control mammalian DNA replication. Front. Physiol. 3, 368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Diffley J. F. (2004) Regulation of early events in chromosome replication. Curr. Biol. 14, R778–R786 [DOI] [PubMed] [Google Scholar]
- 6. Edgar B. A., Orr-Weaver T. L. (2001) Endoreplication cell cycles: more for less. Cell 105, 297–306 [DOI] [PubMed] [Google Scholar]
- 7. Fox D. T., Duronio R. J. (2013) Endoreplication and polyploidy: insights into development and disease. Development 140, 3–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Storchova Z., Pellman D. (2004) From polyploidy to aneuploidy, genome instability and cancer. Nat. Rev. Mol. Cell Biol. 5, 45–54 [DOI] [PubMed] [Google Scholar]
- 9. Davoli T., de Lange T. (2011) The causes and consequences of polyploidy in normal development and cancer. Annu. Rev. Cell Dev. Biol. 27, 585–610 [DOI] [PubMed] [Google Scholar]
- 10. Lee H. O., Davidson J. M., Duronio R. J. (2009) Endoreplication: polyploidy with purpose. Genes Dev. 23, 2461–2477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Knoblich J. A., Sauer K., Jones L., Richardson H., Saint R., Lehner C. F. (1994) Cyclin E controls S phase progression and its down-regulation during Drosophila embryogenesis is required for the arrest of cell proliferation. Cell 77, 107–120 [DOI] [PubMed] [Google Scholar]
- 12. Lilly M. A., Spradling A. C. (1996) The Drosophila endocycle is controlled by Cyclin E and lacks a checkpoint ensuring S-phase completion. Genes Dev. 10, 2514–2526 [DOI] [PubMed] [Google Scholar]
- 13. Weiss A., Herzig A., Jacobs H., Lehner C. F. (1998) Continuous cyclin E expression inhibits progression through endoreduplication cycles in Drosophila. Curr. Biol. 8, 239–242 [DOI] [PubMed] [Google Scholar]
- 14. Follette P. J., Duronio R. J., O'Farrell P. H. (1998) Fluctuations in cyclin E levels are required for multiple rounds of endocycle S phase in Drosophila. Curr. Biol. 8, 235–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Park S. Y., Asano M. (2008) The origin recognition complex is dispensable for endoreplication in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 105, 12343–12348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sher N., Bell G. W., Li S., Nordman J., Eng T., Eaton M. L., Macalpine D. M., Orr-Weaver T. L. (2012) Developmental control of gene copy number by repression of replication initiation and fork progression. Genome Res. 22, 64–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Treisman J. E., Follette P. J., O'Farrell P. H., Rubin G. M. (1995) Cell proliferation and DNA replication defects in a Drosophila MCM2 mutant. Genes Dev. 9, 1709–1715 [DOI] [PubMed] [Google Scholar]
- 18. Feger G., Vaessin H., Su T. T., Wolff E., Jan L. Y., Jan Y. N. (1995) dpa, a member of the MCM family, is required for mitotic DNA replication but not endoreplication in Drosophila. EMBO J. 14, 5387–5398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schwed G., May N., Pechersky Y., Calvi B. R. (2002) Drosophila minichromosome maintenance 6 is required for chorion gene amplification and genomic replication. Mol. Biol. Cell 13, 607–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jackson A. L., Pahl P. M., Harrison K., Rosamond J., Sclafani R. A. (1993) Cell cycle regulation of the yeast Cdc7 protein kinase by association with the Dbf4 protein. Mol. Cell. Biol. 13, 2899–2908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Weinreich M., Stillman B. (1999) Cdc7p-Dbf4p kinase binds to chromatin during S phase and is regulated by both the APC and the RAD53 checkpoint pathway. EMBO J. 18, 5334–5346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lei M., Kawasaki Y., Young M. R., Kihara M., Sugino A., Tye B. K. (1997) Mcm2 is a target of regulation by Cdc7-Dbf4 during the initiation of DNA synthesis. Genes Dev. 11, 3365–3374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Masai H., Taniyama C., Ogino K., Matsui E., Kakusho N., Matsumoto S., Kim J. M., Ishii A., Tanaka T., Kobayashi T., Tamai K., Ohtani K., Arai K. (2006) Phosphorylation of MCM4 by Cdc7 kinase facilitates its interaction with Cdc45 on the chromatin. J. Biol. Chem. 281, 39249–39261 [DOI] [PubMed] [Google Scholar]
- 24. Sheu Y. J., Stillman B. (2006) Cdc7-Dbf4 phosphorylates MCM proteins via a docking site-mediated mechanism to promote S phase progression. Mol. Cell 24, 101–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sheu Y. J., Stillman B. (2010) The Dbf4-Cdc7 kinase promotes S phase by alleviating an inhibitory activity in Mcm4. Nature 463, 113–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Baker S. P., Phillips J., Anderson S., Qiu Q., Shabanowitz J., Smith M. M., Yates J. R., 3rd, Hunt D. F., Grant P. A. (2010) Histone H3 Thr 45 phosphorylation is a replication-associated post-translational modification in S. cerevisiae. Nat. Cell Biol. 12, 294–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bahman M., Buck V., White A., Rosamond J. (1988) Characterisation of the CDC7 gene product of Saccharomyces cerevisiae as a protein kinase needed for the initiation of mitotic DNA synthesis. Biochim. Biophys. Acta 951, 335–343 [DOI] [PubMed] [Google Scholar]
- 28. Faul T., Staib C., Nanda I., Schmid M., Grummt F. (1999) Identification and characterization of mouse homologue to yeast Cdc7 protein and chromosomal localization of the cognate mouse gene Cdc7l. Chromosoma 108, 26–31 [DOI] [PubMed] [Google Scholar]
- 29. Guo B., Lee H. (1999) Cloning and characterization of Chinese hamster CDC7 (ChCDC7). Somatic Cell Mol. Genet. 25, 159–171 [DOI] [PubMed] [Google Scholar]
- 30. Hess G. F., Drong R. F., Weiland K. L., Slightom J. L., Sclafani R. A., Hollingsworth R. E. (1998) A human homolog of the yeast CDC7 gene is overexpressed in some tumors and transformed cell lines. Gene 211, 133–140 [DOI] [PubMed] [Google Scholar]
- 31. Hollingsworth R. E., Jr., Sclafani R. A. (1990) DNA metabolism gene CDC7 from yeast encodes a serine (threonine) protein kinase. Proc. Natl. Acad. Sci. U.S.A. 87, 6272–6276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kim J. M., Sato N., Yamada M., Arai K., Masai H. (1998) Growth regulation of the expression of mouse cDNA and gene encoding a serine/threonine kinase related to Saccharomyces cerevisiae CDC7 essential for G1/S transition. Structure, chromosomal localization, and expression of mouse gene for S. cerevisiae Cdc7-related kinase. J. Biol. Chem. 273, 23248–23257 [DOI] [PubMed] [Google Scholar]
- 33. Masai H., Miyake T., Arai K. (1995) hsk1+, a Schizosaccharomyces pombe gene related to Saccharomyces cerevisiae CDC7, is required for chromosomal replication. EMBO J. 14, 3094–3104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nakamura T., Nakamura-Kubo M., Nakamura T., Shimoda C. (2002) Novel fission yeast Cdc7-Dbf4-like kinase complex required for the initiation and progression of meiotic second division. Mol. Cell. Biol. 22, 309–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Patterson M., Sclafani R. A., Fangman W. L., Rosamond J. (1986) Molecular characterization of cell cycle gene CDC7 from Saccharomyces cerevisiae. Mol. Cell. Biol. 6, 1590–1598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sato N., Arai K., Masai H. (1997) Human and Xenopus cDNAs encoding budding yeast Cdc7-related kinases: in vitro phosphorylation of MCM subunits by a putative human homologue of Cdc7. EMBO J. 16, 4340–4351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Landis G., Tower J. (1999) The Drosophila chiffon gene is required for chorion gene amplification, and is related to the yeast Dbf4 regulator of DNA replication and cell cycle. Development 126, 4281–4293 [DOI] [PubMed] [Google Scholar]
- 38. Sikorski R. S., Hieter P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Axelrod A., Rine J. (1991) A role for CDC7 in repression of transcription at the silent mating-type locus HMR in Saccharomyces cerevisiae. Mol. Cell. Biol. 11, 1080–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Clontech Laboratories, Inc. (2007) BacPAK Baculovirus Expression System User Manual, Clontech Laboratories, Inc., Mountain View, CA [Google Scholar]
- 41. Masai H., Matsui E., You Z., Ishimi Y., Tamai K., Arai K. (2000) Human Cdc7-related kinase complex. In vitro phosphorylation of MCM by concerted actions of Cdks and Cdc7 and that of a critical threonine residue of Cdc7 bY Cdks. J. Biol. Chem. 275, 29042–29052 [DOI] [PubMed] [Google Scholar]
- 42. Koltun E. S., Tsuhako A. L., Brown D. S., Aay N., Arcalas A., Chan V., Du H., Engst S., Ferguson K., Franzini M., Galan A., Holst C. R., Huang P., Kane B., Kim M. H., Li J., Markby D., Mohan M., Noson K., Plonowski A., Richards S. J., Robertson S., Shaw K., Stott G., Stout T. J., Young J., Yu P., Zaharia C. A., Zhang W., Zhou P., Nuss J. M., Xu W., Kearney P. C. (2012) Discovery of XL413, a potent and selective CDC7 inhibitor. Bioorg. Med. Chem. Lett. 22, 3727–3731 [DOI] [PubMed] [Google Scholar]
- 43. Bischof J., Maeda R. K., Hediger M., Karch F., Basler K. (2007) An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. U.S.A. 104, 3312–3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Markstein M., Pitsouli C., Villalta C., Celniker S. E., Perrimon N. (2008) Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat. Genet. 40, 476–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Calvi B. R., Spradling A. C. (1999) Chorion gene amplification in Drosophila: a model for metazoan origins of DNA replication and S-phase control. Methods 18, 407–417 [DOI] [PubMed] [Google Scholar]
- 46. Hanks S. K., Quinn A. M., Hunter T. (1988) The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science 241, 42–52 [DOI] [PubMed] [Google Scholar]
- 47. Graveley B. R., Brooks A. N., Carlson J. W., Duff M. O., Landolin J. M., Yang L., Artieri C. G., van Baren M. J., Boley N., Booth B. W., Brown J. B., Cherbas L., Davis C. A., Dobin A., Li R., Lin W., Malone J. H., Mattiuzzo N. R., Miller D., Sturgill D., Tuch B. B., Zaleski C., Zhang D., Blanchette M., Dudoit S., Eads B., Green R. E., Hammonds A., Jiang L., Kapranov P., Langton L., Perrimon N., Sandler J. E., Wan K. H., Willingham A., Zhang Y., Zou Y., Andrews J., Bickel P. J., Brenner S. E., Brent M. R., Cherbas P., Gingeras T. R., Hoskins R. A., Kaufman T. C., Oliver B., Celniker S. E. (2011) The developmental transcriptome of Drosophila melanogaster. Nature 471, 473–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Brown G. W., Kelly T. J. (1998) Purification of Hsk1, a minichromosome maintenance protein kinase from fission yeast. J. Biol. Chem. 273, 22083–22090 [DOI] [PubMed] [Google Scholar]
- 49. Takeda T., Ogino K., Matsui E., Cho M. K., Kumagai H., Miyake T., Arai K., Masai H. (1999) A fission yeast gene, him1(+)/dfp1(+), encoding a regulatory subunit for Hsk1 kinase, plays essential roles in S-phase initiation as well as in S-phase checkpoint control and recovery from DNA damage. Mol. Cell. Biol. 19, 5535–5547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bettencourt-Dias M., Giet R., Sinka R., Mazumdar A., Lock W. G., Balloux F., Zafiropoulos P. J., Yamaguchi S., Winter S., Carthew R. W., Cooper M., Jones D., Frenz L., Glover D. M. (2004) Genome-wide survey of protein kinases required for cell cycle progression. Nature 432, 980–987 [DOI] [PubMed] [Google Scholar]
- 51. Davey M. J., Andrighetti H. J., Ma X., Brandl C. J. (2011) A synthetic human kinase can control cell cycle progression in budding yeast. G3 1, 317–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Endicott J. A., Noble M. E., Johnson L. N. (2012) The structural basis for control of eukaryotic protein kinases. Annu. Rev. Biochem. 81, 587–613 [DOI] [PubMed] [Google Scholar]
- 53. Masai H., Arai K. (2000) Dbf4 motifs: conserved motifs in activation subunits for Cdc7 kinases essential for S-phase. Biochem. Biophys. Res. Commun. 275, 228–232 [DOI] [PubMed] [Google Scholar]
- 54. Matthews L. A., Guarné A. (2013) Dbf4: the whole is greater than the sum of its parts. Cell Cycle 12, 1180–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hughes S., Elustondo F., Di Fonzo A., Leroux F. G., Wong A. C., Snijders A. P., Matthews S. J., Cherepanov P. (2012) Crystal structure of human CDC7 kinase in complex with its activator DBF4. Nat. Struct. Mol. Biol. 19, 1101–1107 [DOI] [PubMed] [Google Scholar]
- 56. Kitamura R., Fukatsu R., Kakusho N., Cho Y. S., Taniyama C., Yamazaki S., Toh G. T., Yanagi K., Arai N., Chang H. J., Masai H. (2011) Molecular mechanism of activation of human Cdc7 kinase: bipartite interaction with Dbf4/activator of S phase kinase (ASK) activation subunit stimulates ATP binding and substrate recognition. J. Biol. Chem. 286, 23031–23043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ogino K., Takeda T., Matsui E., Iiyama H., Taniyama C., Arai K., Masai H. (2001) Bipartite binding of a kinase activator activates Cdc7-related kinase essential for S phase. J. Biol. Chem. 276, 31376–31387 [DOI] [PubMed] [Google Scholar]
- 58. Chapman J. W., Johnston L. H. (1989) The yeast gene, DBF4, essential for entry into S phase is cell cycle regulated. Exp. Cell Res. 180, 419–428 [DOI] [PubMed] [Google Scholar]
- 59. Furukohri A., Sato N., Masai H., Arai K., Sugino A., Waga S. (2003) Identification and characterization of a Xenopus homolog of Dbf4, a regulatory subunit of the Cdc7 protein kinase required for the initiation of DNA replication. J. Biochem. 134, 447–457 [DOI] [PubMed] [Google Scholar]
- 60. Jares P., Luciani M. G., Blow J. J. (2004) A Xenopus Dbf4 homolog is required for Cdc7 chromatin binding and DNA replication. BMC Mol. Biol. 5, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Jiang W., McDonald D., Hope T. J., Hunter T. (1999) Mammalian Cdc7-Dbf4 protein kinase complex is essential for initiation of DNA replication. EMBO J. 18, 5703–5713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Johnston L. H., Thomas A. P. (1982) A further two mutants defective in initiation of the S phase in the yeast Saccharomyces cerevisiae. Mol. Gen. Genet. 186, 445–448 [DOI] [PubMed] [Google Scholar]
- 63. Kitada K., Johnston L. H., Sugino T., Sugino A. (1992) Temperature-sensitive cdc7 mutations of Saccharomyces cerevisiae are suppressed by the DBF4 gene, which is required for the G1/S cell cycle transition. Genetics 131, 21–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kumagai H., Sato N., Yamada M., Mahony D., Seghezzi W., Lees E., Arai K., Masai H. (1999) A novel growth- and cell cycle-regulated protein, ASK, activates human Cdc7-related kinase and is essential for G1/S transition in mammalian cells. Mol. Cell. Biol. 19, 5083–5095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lepke M., Pütter V., Staib C., Kneissl M., Berger C., Hoehn K., Nanda I., Schmid M., Grummt F. (1999) Identification, characterization and chromosomal localization of the cognate human and murine DBF4 genes. Mol. Gen. Genet. 262, 220–229 [DOI] [PubMed] [Google Scholar]
- 66. Nakamura T., Kishida M., Shimoda C. (2000) The Schizosaccharomyces pombe spo6+ gene encoding a nuclear protein with sequence similarity to budding yeast Dbf4 is required for meiotic second division and sporulation. Genes Cells 5, 463–479 [DOI] [PubMed] [Google Scholar]
- 67. Masai H., Arai K. (2002) Cdc7 kinase complex: a key regulator in the initiation of DNA replication. J. Cell. Physiol. 190, 287–296 [DOI] [PubMed] [Google Scholar]
- 68. Sclafani R. A. (2000) Cdc7p-Dbf4p becomes famous in the cell cycle. J. Cell Sci. 113, 2111–2117 [DOI] [PubMed] [Google Scholar]
- 69. Cheng L., Collyer T., Hardy C. F. (1999) Cell cycle regulation of DNA replication initiator factor Dbf4p. Mol. Cell. Biol. 19, 4270–4278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ferreira M. F., Santocanale C., Drury L. S., Diffley J. F. (2000) Dbf4p, an essential S phase-promoting factor, is targeted for degradation by the anaphase-promoting complex. Mol. Cell. Biol. 20, 242–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Yoon H. J., Loo S., Campbell J. L. (1993) Regulation of Saccharomyces cerevisiae CDC7 function during the cell cycle. Mol. Biol. Cell 4, 195–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Bruck I., Kaplan D. (2009) Dbf4-Cdc7 phosphorylation of Mcm2 is required for cell growth. J. Biol. Chem. 284, 28823–28831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Montagnoli A., Valsasina B., Brotherton D., Troiani S., Rainoldi S., Tenca P., Molinari A., Santocanale C. (2006) Identification of Mcm2 phosphorylation sites by S-phase-regulating kinases. J. Biol. Chem. 281, 10281–10290 [DOI] [PubMed] [Google Scholar]
- 74. Jares P., Blow J. J. (2000) Xenopus cdc7 function is dependent on licensing but not on XORC, XCdc6, or CDK activity and is required for XCdc45 loading. Genes Dev. 14, 1528–1540 [PMC free article] [PubMed] [Google Scholar]
- 75. Roberts B. T., Ying C. Y., Gautier J., Maller J. L. (1999) DNA replication in vertebrates requires a homolog of the Cdc7 protein kinase. Proc. Natl. Acad. Sci. U.S.A. 96, 2800–2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Walter J., Newport J. (2000) Initiation of eukaryotic DNA replication: origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase α. Mol. Cell 5, 617–627 [DOI] [PubMed] [Google Scholar]
- 77. Walter J. C. (2000) Evidence for sequential action of cdc7 and cdk2 protein kinases during initiation of DNA replication in Xenopus egg extracts. J. Biol. Chem. 275, 39773–39778 [DOI] [PubMed] [Google Scholar]
- 78. Kim J. M., Nakao K., Nakamura K., Saito I., Katsuki M., Arai K., Masai H. (2002) Inactivation of Cdc7 kinase in mouse ES cells results in S-phase arrest and p53-dependent cell death. EMBO J. 21, 2168–2179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Peter A., Schöttler P., Werner M., Beinert N., Dowe G., Burkert P., Mourkioti F., Dentzer L., He Y., Deak P., Benos P. V., Gatt M. K., Murphy L., Harris D., Barrell B., Ferraz C., Vidal S., Brun C., Demaille J., Cadieu E., Dreano S., Gloux S., Lelaure V., Mottier S., Galibert F., Borkova D., Miñana B., Kafatos F. C., Bolshakov S., Sidén-Kiamos I., Papagiannakis G., Spanos L., Louis C., Madueño E., de Pablos B., Modolell J., Bucheton A., Callister D., Campbell L., Henderson N. S., McMillan P. J., Salles C., Tait E., Valenti P., Saunders R. D., Billaud A., Pachter L., Klapper R., Janning W., Glover D. M., Ashburner M., Bellen H. J., Jackle H., Schafer U. (2002) Mapping and identification of essential gene functions on the X chromosome of Drosophila. EMBO Rep. 3, 34–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Calvi B. R., Lilly M. A., Spradling A. C. (1998) Cell cycle control of chorion gene amplification. Genes Dev. 12, 734–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Theodosiou N. A., Xu T. (1998) Use of FLP/FRT system to study Drosophila development. Methods 14, 355–365 [DOI] [PubMed] [Google Scholar]
- 82. Perrimon N. (1998) Creating mosaics in Drosophila. Int. J. Dev. Biol. 42, 243–247 [PubMed] [Google Scholar]
- 83. Zhang H., Tower J. (2004) Sequence requirements for function of the Drosophila chorion gene locus ACE3 replicator and ori-β origin elements. Development 131, 2089–2099 [DOI] [PubMed] [Google Scholar]
- 84. Marston A. L. (2009) Meiosis: DDK is not just for replication. Curr. Biol. 19, R74–R76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Bunt S. M., Monk A. C., Siddall N. A., Johnston N. L., Hime G. R. (2012) GAL4 enhancer traps that can be used to drive gene expression in developing Drosophila spermatocytes. Genesis 50, 914–920 [DOI] [PubMed] [Google Scholar]
- 86. Tower J. (2004) Developmental gene amplification and origin regulation. Annu. Rev. Genet. 38, 273–304 [DOI] [PubMed] [Google Scholar]
- 87. Montagnoli A., Bosotti R., Villa F., Rialland M., Brotherton D., Mercurio C., Berthelsen J., Santocanale C. (2002) Drf1, a novel regulatory subunit for human Cdc7 kinase. EMBO J. 21, 3171–3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Yanow S. K., Gold D. A., Yoo H. Y., Dunphy W. G. (2003) Xenopus Drf1, a regulator of Cdc7, displays checkpoint-dependent accumulation on chromatin during an S-phase arrest. J. Biol. Chem. 278, 41083–41092 [DOI] [PubMed] [Google Scholar]
- 89. Yoshizawa-Sugata N., Ishii A., Taniyama C., Matsui E., Arai K., Masai H. (2005) A second human Dbf4/ASK-related protein, Drf1/ASKL1, is required for efficient progression of S and M phases. J. Biol. Chem. 280, 13062–13070 [DOI] [PubMed] [Google Scholar]
- 90. Takahashi T. S., Walter J. C. (2005) Cdc7-Drf1 is a developmentally regulated protein kinase required for the initiation of vertebrate DNA replication. Genes Dev. 19, 2295–2300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Silva T., Bradley R. H., Gao Y., Coue M. (2006) Xenopus CDC7/DRF1 complex is required for the initiation of DNA replication. J. Biol. Chem. 281, 11569–11576 [DOI] [PubMed] [Google Scholar]
- 92. Shi N., Xie W. B., Chen S. Y. (2012) Cell division cycle 7 is a novel regulator of transforming growth factor-beta-induced smooth muscle cell differentiation. J. Biol. Chem. 287, 6860–6867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Lake C. M., Teeter K., Page S. L., Nielsen R., Hawley R. S. (2007) A genetic analysis of the Drosophila mcm5 gene defines a domain specifically required for meiotic recombination. Genetics 176, 2151–2163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Su T. T., Feger G., O'Farrell P. H. (1996) Drosophila MCM protein complexes. Mol. Biol. Cell 7, 319–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Ramer M. D., Suman E. S., Richter H., Stanger K., Spranger M., Bieberstein N., Duncker B. P. (2013) Dbf4 and Cdc7 proteins promote DNA replication through interactions with distinct Mcm2-7 protein subunits. J. Biol. Chem. 288, 14926–14935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Randell J. C., Fan A., Chan C., Francis L. I., Heller R. C., Galani K., Bell S. P. (2010) Mec1 is one of multiple kinases that prime the Mcm2-7 helicase for phosphorylation by Cdc7. Mol. Cell 40, 353–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Francis L. I., Randell J. C., Takara T. J., Uchima L., Bell S. P. (2009) Incorporation into the prereplicative complex activates the Mcm2-7 helicase for Cdc7-Dbf4 phosphorylation. Genes Dev. 23, 643–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Hong A., Narbonne-Reveau K., Riesgo-Escovar J., Fu H., Aladjem M. I., Lilly M. A. (2007) The cyclin-dependent kinase inhibitor Dacapo promotes replication licensing during Drosophila endocycles. EMBO J. 26, 2071–2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Su T. T., O'Farrell P. H. (1998) Chromosome association of minichromosome maintenance proteins in Drosophila endoreplication cycles. J. Cell Biol. 140, 451–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Sato N., Sato M., Nakayama M., Saitoh R., Arai K., Masai H. (2003) Cell cycle regulation of chromatin binding and nuclear localization of human Cdc7-ASK kinase complex. Genes Cells 8, 451–463 [DOI] [PubMed] [Google Scholar]
- 101. Hughes S., Jenkins V., Dar M. J., Engelman A., Cherepanov P. (2010) Transcriptional co-activator LEDGF interacts with Cdc7-activator of S-phase kinase (ASK) and stimulates its enzymatic activity. J. Biol. Chem. 285, 541–554 [DOI] [PMC free article] [PubMed] [Google Scholar]