Abstract
The MHC class I-chain-related proteins (MICs) and the UL16-binding proteins (ULBPs) are inducible stress response molecules that work as activators of a specific receptor, NKG2D, which is expressed on effector cells, such as NK cells and subsets of T cells. In this study, we sought to explore the biological significance of NKG2D ligands in human neoplasms by comprehensively examining the immunohistochemical expression profile of NKG2D ligands in a variety of human epithelial neoplasms. Following careful validation of the immunohistochemical specificity and availability of anti-human ULBP antibodies for formalin-fixed paraffin-embedded (FFPE) materials, the expression of NKG2D ligands was analyzed in FFPE tissue microarrays comprising 22 types of epithelial neoplastic tissue with their non-neoplastic counterpart from various organs. Hierarchical cluster analysis demonstrated a positive relationship among ULBP2/6, ULBP3, ULBP1, and ULBP5, whose expression patterns were similar across all of the neoplastic tissues examined. In contrast, MICA/B, as well as ULBP4, did not appear to be related to any other ligand. These expression profiles of NKG2D ligands in human neoplasms based on well-validated specific antibodies, followed by hierarchical cluster analysis, should help to clarify some functional aspects of these molecules in cancer biology, and also provide a path to the development of novel tumor-type-specific treatment strategies.
Keywords: NKG2D ligands, immunohistochemistry, epithelial neoplasms
Introduction
It is widely accepted that the natural history of human neoplasms is influenced by a variety of microenvironmental factors, such as blood supply, hypoxia, and immunological surveillance. Natural killer (NK) cells are one of the key players in the immunological response to neoplastic cells, and their function is regulated by a delicate balance of signals initiated from a variety of activating and inhibitory receptors on NK cells. The activating receptor NKG2D (natural-killer group 2, member D) belongs to the family of C-type lectin-like type II transmembrane proteins and is expressed by a range of effector cells, such as NK cells, NKT cells, γδT cells (Wu et al. 1999; Jamieson et al. 2002) and CD8+ T cells (Ehrlich et al. 2005). One of the characteristics of the NKG2D system is that there are multiple ligands for the receptor. The NKG2D receptor ligands are distant homologs of major histocompatibility complex (MHC) class I molecules (Bauer et al. 1999; Cerwenka et al. 2000; Diefenbach et al. 2000), and include two families in humans: the MHC class I-chain-related proteins (MIC) A and B (Bauer et al. 1999), and the UL16-binding proteins (ULBPs) 1-6 (Cosman et al. 2001; Radosavljevic et al. 2002; Chalupny et al. 2003; Bacon et al. 2004; Eagle et al. 2009b). The amino acid sequences and domain structures of NKG2D ligands are variable. MICA and MICB consist of α1, α2 and α3 domains (Bauer et al. 2000; Bahram et al. 1994), whereas each ULBP consists of only two Ig-like domains (α1 and α2). Moreover, two members (ULBPs 4, -5) of the ULBP family are anchored to the cell membrane by a transmembrane region, whereas other members are linked to the cell surface via glycosyl-phosphatidylinositol anchors (Bacon et al. 2004; Eagle et al. 2009b). Although NKG2D, as a single receptor, combines with these several distinct ligands, it is still unclear why multiple ligands exist for this one invariant receptor.
The cellular expression of these ligands can be up-regulated in response to a variety of stimuli, such as viral infection, tissue ischemia, heat shock and malignant transformation (Groh et al. 1996; Gasser et al. 2005; Groh et al. 2001). In humans, the expression of NKG2D ligands is known to be rare in normal tissues, but frequent in both primary tumors and tumor-derived cell lines (Groh et al. 1996; Raffaghello et al. 2004; Pende et al. 2002; Coudert et al. 2006). Many reports have already described the expression of MICA/B in a broad range of normal and tumor tissues in humans, including various carcinomas (breast, lung, colon, kidney, ovary and prostate), leukemias, gliomas, neuroblastomas and melanomas (Groh et al. 1999; Vetter et al. 2002; Friese et al. 2003; Salih et al. 2003; Watson et al. 2006; Castriconi et al. 2007). Similarly, it has been reported that ULBPs are expressed in several types of human tumors, and that up-regulation of NKG2D ligands in cancer is associated with patient survival (McGilvray et al. 2010; McGilvray et al. 2009). Moreover, recent studies have strongly suggested that the expression levels of these ligands are associated with enhanced antibody-dependent cell-mediated cytotoxicity (ADCC) activity (Inagaki et al. 2009), which is one of the key mechanisms responsible for the antitumor effect of antibody therapeutics. This topic is of considerable interest because the potential to manipulate NKG2D ligand expression could offer promise in the treatment of tumors. However, as comprehensive details of NKG2D ligand expression patterns in human tissues are still largely lacking, the significance of NKG2D ligands in the pathobiological behavior of human neoplasms remains speculative. In previous immunohistochemical studies, the specificity of the antibodies employed was unclear, and both the analyzed neoplasms and ligands were considerably limited.
This study identified the specificity of the antibodies using transfected cells, and clarified differences in ligand expression between non-neoplastic and neoplastic tissues in the same individual. This is the first reported study to have clarified the expression patterns of eight NKG2D ligands during malignant transformation using six validated and specific antibodies.
Materials & Methods
Plasmid Construction
To construct vectors for FLAG-ULBP overexpression, the following cDNA clones were used: ULBP1 (clone AK292519; NITE Biological Resource Center (NBRC)), ULBP2 (clone MGC:21383; American Type Culture Collection (ATCC)), ULBP3 (clone AK315275; NBRC) and ULBP4 (clone MGC:125309; ATCC). ULBP5 and ULBP6 were cloned by PCR from cDNA derived from HEK-293 cells and HeLa cells, respectively. The amplification was performed using KOD plus version 2 DNA polymerase (Toyobo; Osaka, Japan). The primer sequences for ULBP5 and ULBP6 were 5’-TGCTGTCCC CTGCGATCCAA-3’ and 5’-TCAAGATATGGAGACCTGTAGTGGC-3’, and 5’-GTCCCCAGCCCTCCTGGT-3’ and 5’-TCAGATGCCAGGGAGGATGAAG-3’, respectively. The amplified products were cloned into the pGEM T-easy vector (Promega; Madison, WI). No substantive mutation was found in the sequences of the isolated ULBP5 and ULBP6 cDNAs. Each of the ULBP1-6 clones was amplified using gene-specific primers (Table 1), and ligated into the pFLAG-CMV-3 vector (Sigma-Aldrich; St. Louis, MO).
Table 1.
Primer Sequences for Cloning of FLAG-tagged UL16-binding proteins (ULBPs).
| Forward (5′ to 3′) | Reverse (5′ to 3′) | |
|---|---|---|
| ULBP1 | cccAAGCTTgggGGATGGGTCGACA | tgctctagagcaTCATCTGCCAGCTAGAATGA |
| ULBP2 | cccAAGCTTgggGACCCTCACTCTCTT | tgctctagagcaTCAGATGCCAGGGAGGAT |
| ULBP3 | cccAAGCTTgggGACGCTCACTCTCTC | tgctctagagcaTCAGATGCCAGGGAGGAT |
| ULBP4 | cccAAGCTTgggCACTCTCTTTGCTTCA | tgctctagagcaCTAAGACGTCCTCAAGGGCCA |
| ULBP5 | cccAAGCTTgggGACCCTCACTCTCTT | tgctctagagcaTCAAGATATGGAGACCTGTAG |
| ULBP6 | cccAAGCTTgggAGGCGAGACGACC | tgctctagagcaTCAGATGCCAGGGAGGAT |
Transfection
COS7 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen; Carlsbad, CA) containing 10% FCS, at 37C in a humidified 5% CO2 atmosphere. For transient transfection, cells were seeded on 6-well plates and grown to 40%–50% confluence. Transfection was performed using FugeneHD (Roche Diagnostics; Indianapolis, IN) in accordance with the manufacturer’s protocol. After incubation for 48 hr, the transfected cells were scraped off the plates, and the expression of each ULBP was checked using some of the cells by western blot analysis with rabbit anti-FLAG antibody (Sigma-Aldrich). Collected cell pellets not lysed for western blotting were fixed in 10% formalin and embedded in paraffin to prepare FFPE cell blocks. NKG2D ligand-transfected and non-transfected COS7 cells were mixed at a ratio of 1:9 in the cell block.
Antibodies and Immunohistochemical Validation using FFPE-transfected Cell Blocks
Antibodies against NKG2D ligands were obtained from the following sources: anti-MICA/B monoclonal antibody (mAb) from Biolegend (San Diego, CA); anti-ULBP1 and anti-ULBP3 goat polyclonal antibodies (pAbs) from R&D Systems (Minneapolis, MN); anti-ULBP2 rabbit pAb from Novus Biologicals (Littleton, CO); and anti-ULBP4 goat pAb from Santa Cruz Biotechnology (Dallas, TX). To obtain a specific anti-ULBP5 antibody capable of discriminating ULBP5 from the very similar proteins ULBP2 and ULBP6, a rabbit pAb was generated in-house against a peptide corresponding to part of the cytoplasmic region of ULBP5 (Eagle et al. 2009a). This synthetic peptide (CNNGAARYSEPLQVSIS; Hokudo, Sapporo, Japan) was conjugated to keyhole limpet hemocyanin for immunizations and to bovine serum albumin for ELISA screening. We performed the immunostaining with negative control antibodies (mouse IgG, goat IgG, and rabbit IgG; Santa Cruz Biotechnology) to confirm the non-specific binding of immunoglobulins to tissue.
Immunohistochemical analysis of ULBPs was performed using FFPE cell blocks. Fresh, 5-μm-thick sections were placed on coated glass slides, deparaffinized, and rehydrated. The deparaffinized sections were then heat-treated with antigen retrieval solution (Target Retrieval Solution, pH 9.0; Dako; Glostrup, Denmark) at 95C for 20 min using the Dako PT Link system. After blocking of endogenous peroxidase using Dako Peroxidase-Blocking Solution, the sections were incubated for 30 min at room temperature with specific antibodies: anti-MICA/B (dilution 1:50), anti-ULBP1 (1:100), anti-ULBP2 (1:500), anti-ULBP3 (1:50), anti-ULBP4 (1:50), and anti-ULBP5 (1:50). Detection was then performed using a standard polymer method in accordance with the manufacturer’s instructions (EnVision Flex system for mouse mAb and rabbit pAb, Dako; and SimpleStain system for goat pAb, Nichirei; Tokyo, Japan). These immunohistochemical reactions were performed using an automated immunostaining system (Autostainer Plus, Dako).
Immunohistochemical Analysis using Tissue Microarray (TMA)
Tumors and adjacent non-neoplastic tissues were retrieved from surgical specimens in the pathology files of Hokkaido University Hospital covering the period from 1997 to 2005. The tissue specimens were fixed in 10% neutral-buffered formalin for 24 to 48 hr, and then embedded in paraffin wax. TMA was prepared using the following procedure. Specifically, each hematoxylin and eosin-stained sections from FFPE tissue blocks were evaluated to locate representative areas for further analysis. Needle core samples (2.0 mm) were cut out from the corresponding areas of the block and then placed at pre-specified coordinates in recipient paraffin array blocks using a manual tissue microarrayer (Sakura Finetek Japan, Tokyo). Thus, array blocks, each containing between 37 and 42 cores, were constructed, covering a total of 123 FFPE tumor tissue samples derived from 22 types of primary epithelial neoplasms (n=5 for each case, except for thyroid follicular carcinoma [n=3]) and their non-neoplastic counterpart with normal morphology (Table 2). Characteristics of the patients and tumors are shown in Supplementary Table 1.
Table 2.
Comparison of NKG2D Ligand Expression between Non-neoplastic and Neoplastic Tissues.
| ULBP1 |
ULBP2/6 |
ULBP3 |
ULBP4 |
ULBP5 |
MICA/B |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tissue type | Positivity rate (%) | p | Positivity rate (%) | p | Positivity rate (%) | p | Positivity rate (%) | p | Positivity rate (%) | p | Positivity rate (%) | p | |
| Lung | Alveolar epithelium | 20 | 0.524 | 0 | 1.000 | 20 | 1.000 | 0 | 1.000 | 0 | 0.167 | 20 | 1.000 |
| Adenocarcinoma | 60 | 20 | 40 | 20 | 60 | 20 | |||||||
| Lung | Bronchial epithelium | 100 | 0.143 | 33 | 1.000 | 33 | 0.375 | 0 | - | 67 | 0.107 | 0 | - |
| Squamous cell carcinoma | 20 | 20 | 0 | 0 | 0 | 0 | |||||||
| Esophagus | Squamous epithelium | 0 | 0.444 | 0 | 0.524 | 0 | 1.000 | 0 | 0.206 | 0 | 0.048* | 0 | 1.000 |
| Squamous cell carcinoma | 20 | 20 | 20 | 40 | 0 | 20 | |||||||
| Stomach | Mucosal epithelium | 60 | 1.000 | 40 | 1.000 | 80 | 1.000 | 40 | 0.524 | 20 | 0.524 | 40 | 0.524 |
| Adenocarcinoma | 80 | 60 | 80 | 80 | 60 | 60 | |||||||
| Colon | Mucosal epithelium | 60 | 0.444 | 40 | 0.524 | 60 | 0.444 | 20 | 0.206 | 20 | 0.206 | 100 | - |
| Adenocarcinoma | 100 | 80 | 100 | 80 | 80 | 100 | |||||||
| Liver | Hepatocyte | 100 | - | 60 | 1.000 | 80 | 1.000 | 100 | 0.444 | 80 | 1.000 | 40 | 1.000 |
| Hepatocellular carcinoma | 100 | 60 | 40 | 60 | 100 | 40 | |||||||
| Liver | Intrahepatic bile duct epithelium | 60 | 1.000 | 0 | 0.167 | 60 | 1.000 | 0 | - | 0 | 1.000 | 0 | 0.206 |
| Intrahepatic cholangiocarcinoma | 80 | 60 | 60 | 0 | 20 | 40 | |||||||
| Bile duct | Bile duct epithelium | 80 | 0.524 | 40 | 1.000 | 40 | 1.000 | 40 | 1.000 | 80 | 0.524 | 80 | 0.206 |
| Adenocarcinoma | 40 | 40 | 60 | 40 | 40 | 20 | |||||||
| Pancreas | Pancreatic ducts | 100 | 0.008** | 20 | 1.000 | 60 | 0.524 | 60 | 0.167 | 60 | 0.524 | 20 | 1.000 |
| Adenocarcinoma | 0 | 0 | 20 | 0 | 20 | 0 | |||||||
| Prostate | Glandular epithelium | 100 | 1.000 | 40 | 1.000 | 0 | 1.000 | 60 | 0.524 | 0 | - | 0 | 1.000 |
| Adenocarcinoma | 80 | 20 | 20 | 20 | 0 | 0 | |||||||
| Kidney | Renal tubular epithelium | 100 | 0.048** | 100 | 0.048** | 100 | 0.008** | 100 | 0.008** | 75 | 0.206 | 50 | 0.286 |
| Renal cell carcinoma | 20 | 20 | 0 | 0 | 20 | 0 | |||||||
| Renal pelvis | Urothelial epithelium | 50 | 1.000 | 50 | 0.524 | 100 | - | 100 | 1.000 | 75 | 1.000 | 75 | 0.206 |
| Urothelial carcinoma | 60 | 80 | 100 | 80 | 60 | 20 | |||||||
| Breast | Ductal epithelium | 50 | 1.000 | 25 | 0.206 | 50 | 1.000 | 25 | 0.524 | 75 | 0.524 | 25 | 1.000 |
| Ductal carcinoma | 60 | 80 | 60 | 60 | 40 | 40 | |||||||
| Uterine cervix | Squamous epithelium | 0 | 1.000 | 0 | 0.444 | 0 | 0.008* | 0 | 0.048* | 0 | 0.444 | 0 | 0.286 |
| Squamous cell carcinoma | 20 | 40 | 100 | 80 | 40 | 20 | |||||||
| Uterine corpus | Endometrial glands | 33 | 0.679 | 0 | 0.018* | 67 | 0.375 | 67 | 1.000 | 67 | 0.375 | 33 | 1.000 |
| Adenocarcinoma | 60 | 100 | 100 | 80 | 100 | 20 | |||||||
| Thyroid | Follicular epithelium | 0 | 0.048* | 20 | 0.460 | 0 | 0.016* | 0 | 0.048* | 0 | 0.008* | 0 | 0.365 |
| Papillary carcinoma | 80 | 60 | 80 | 80 | 100 | 60 | |||||||
| Thyroid | Follicular epithelium | 0 | 0.100 | 0 | 0.100 | 0 | 0.400 | 0 | 0.400 | 0 | 0.100 | 0 | 1.000 |
| Follicular carcinoma | 100 | 100 | 67 | 0 | 100 | 33 | |||||||
| Tongue | Squamous epithelium | 0 | 0.048* | 0 | 0.206 | 0 | - | 0 | 0.095 | 0 | 0.206 | 0 | 1.000 |
| Squamous cell carcinoma | 0 | 0 | 0 | 60 | 0 | 0 | |||||||
| Larynx | Squamous epithelium | 0 | 0.206 | 0 | 0.444 | 0 | 0.524 | 0 | 0.048* | 0 | 0.008* | 0 | 0.444 |
| Squamous cell carcinoma | 20 | 0 | 0 | 40 | 60 | 20 | |||||||
| Skin | Squamous epithelium | 0 | 1.000 | 0 | 1.000 | 0 | - | 20 | 1.000 | 0 | 1.000 | 20 | 1.000 |
| Squamous cell carcinoma | 0 | 0 | 0 | 40 | 20 | 20 | |||||||
The significance of differences in expression of each NKG2D ligand between neoplastic and non-neoplastic tissues was determined using Fisher’s exact test (p<0.05).
Positive correlation, ** Negative correlation. ULBP, UL16-binding protein; MIC, MHC class I-chain-related proteins.
Immunohistochemical analysis of MICA/B and ULBPs was performed using TMA materials as described above. The intensity of staining was assessed according to a semi-quantitative system as no (score 0), low (score 1) or high (score 2) expression (Fig. 1A–1C). The judgment of Score 2 was based on the previous study of Henriksen and others (2007), which examined the use of the Allred Score for semi-quantitative scoring of cytoplasmic staining. An Intensity Score (IS) for the Allred Score of 2 (intermediate) or 3 (strong) was considered equivalent to Score 2 in the present study, and Score 1 in the present study was assumed to represent a positive reaction with lower intensity.
Figure 1.
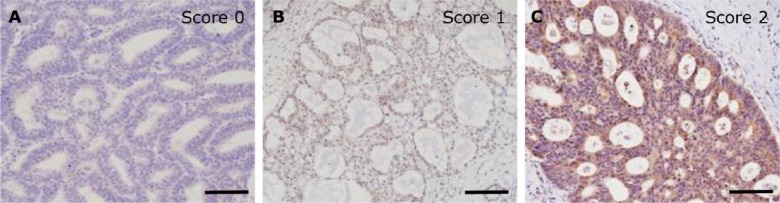
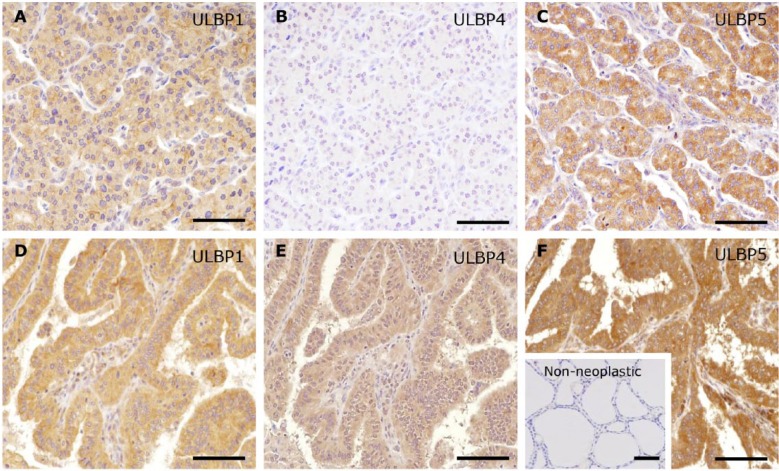
Representative immunohistochemical patterns of NKG2D ligands. The intensity of staining, as the proportion of positive cases among all cases, was assessed using a semi-quantitative system as: no (A; score 0 in MICA/B), low (B, score 1 in ULBP3) or high (C; score 2 in ULBP5) by two observers. Scores of 0 and 1 were defined as negative, and a score of 2 was defined as positive. Scale, 100 μm.
Each assessment was performed independently by two observers and, in a few cases where there was discrepancy between the observers, a joint review was performed using a double-headed microscope, and a consensus was reached. In each case, scores of 0 and 1 were defined as negative, and a score of 2 was defined as positive.
Statistical Analysis
Hierarchical clustering analysis was performed based on the proportion of positive cases (positivity rate) for each tissue type in order to analyze inter-ligand or inter-tissue relationships. Significance was established at p <0.05. All statistical analyses were performed using the SPSS software package (SPSS Inc; Chicago, IL). Fisher’s exact test was used to determine the significance of differences in ligand expression between neoplastic and non-neoplastic tissues based on the scoring results (Score 0–2).
Results
Validation of Specific Antibodies against NKG2D Ligands
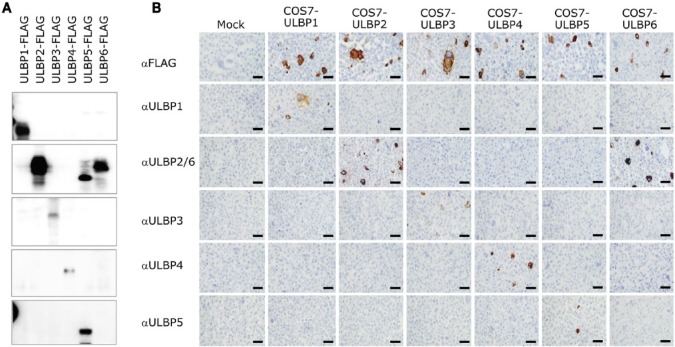
For antibody validation, several commercial antibodies were screened using western blotting with cell lysates prepared from each respective ULBP transfectant. Immunohistochemical specificity and applicability on the FFPE cell block of each transfectant were also checked. As shown in Figure 2, the pAbs against ULBP1, ULBP3, ULBP4 and ULBP5 used in this study all showed a specific reaction in western blotting, and were applicable to the FFPE cell blocks. The pAb against ULBP2 was shown to be cross-reactive with both ULBP5 and ULBP6 in western blotting (Fig. 2A), with only ULBP6 showing cross-reactivity with FFPE immunohistochemistry (Fig. 2B). Therefore, this pAb was evaluated as a dual antibody against ULBP2/6 in subsequent immunohistochemistry experiments. We performed additional western blot analysis using lysate prepared from HeLa cells, which are known to express NKG2D ligands, and this revealed a band at the expected position. In addition, we confirmed that the expression was changed by cellular stress (using phorbol myristate acetate and cobalt chloride) (Supplementary Fig. S1). The MICA/B mAb (clone 6D4) showed no cross-reaction with any of the ULBP transfectants and was specifically applicable to FFPE immunohistochemistry (data not shown).
Figure 2.
Validation of antibodies using ULBP-transfected COS7 cells. We confirmed the specificity of all antibodies against UL16-binding proteins (ULBP) using western blotting with cell lysates (A) and immunohistochemistry with a FFPE cell block (B) prepared from each ULBP transfectant, respectively. The anti-ULBP2 antibody cross-reacted with ULBP5 and ULBP6 in western blotting but only with ULBP6 in FFPE. Therefore, this antibody was used to mark ULBP2 and ULBP6. Scale, 50 μm.
Immunohistochemical Distribution and Expression Profile of NKG2D Ligands in Non-neoplastic Tissues
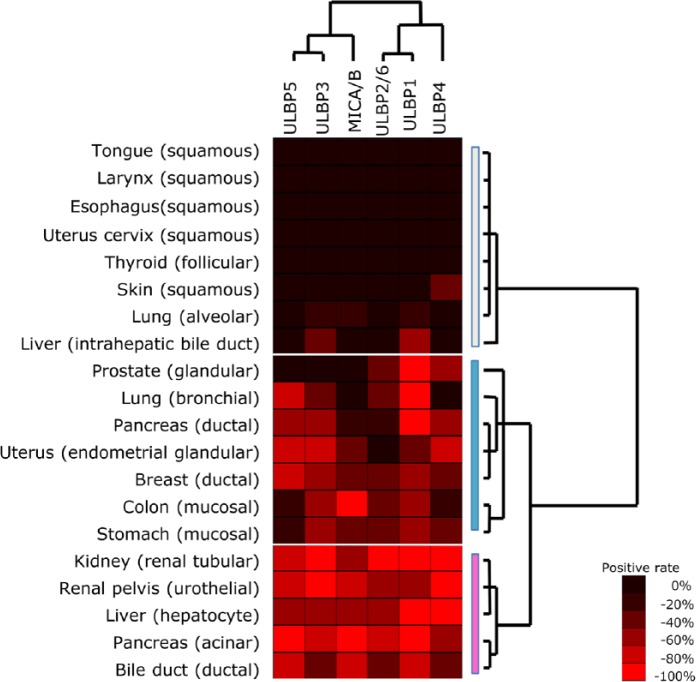
Immunohistochemistry for NKG2D ligands consistently demonstrated a predominantly diffuse cytoplasmic and partial membranous staining pattern, as reported previously (Groh et al. 1999; McGilvray et al. 2010; McGilvray et al. 2009; Eagle et al. 2009a; Eagle et al. 2009b). Among non-neoplastic tissues, there were several patterns of NKG2D ligand expression. As shown by the heatmap in Figure 3, the positivity rate for each tissue type varied widely between 20% and 80% depending on ligand species. Generally, squamous epithelium of organs such as the tongue, larynx, esophagus and skin expressed NKG2D ligands less frequently than glandular epithelium of organs such as the endometrium, breast, gastrointestinal tract, and prostate (Fig.4).
Figure 3.
Expression profiles for NKG2D ligands in non-neoplastic epithelial tissues. Hierarchical cluster analysis based on the expression profiles of NKG2D ligands demonstrated two distinct ligand-based clusters and three distinct tissue-based clusters: white, N-null type; blue, N-variable type; pink, N-complete type (right side).
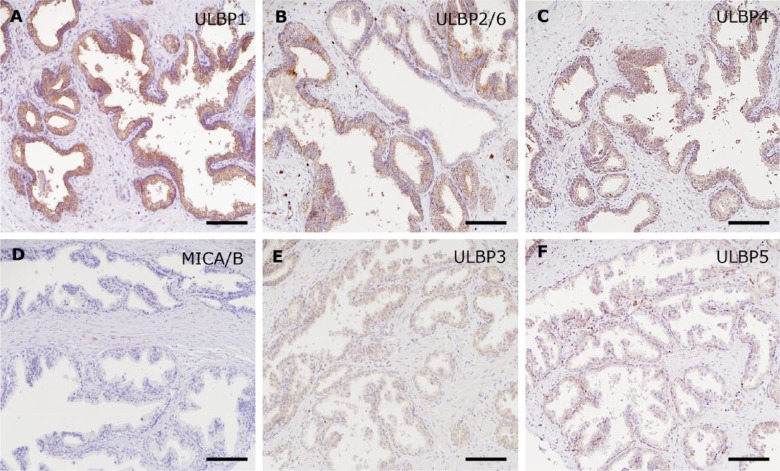
Figure 4.
Diverse expression of NKG2D ligands in non-neoplastic prostate tissue. The upper panels show immunohistochemistry for ULBP1 (A), ULBP2/6 (B), and ULBP4 (C) as positive, and the lower panels for MICA/B (D), ULBP3 (E), and ULBP5 (F) as negative. Scale, 100 μm.
To obtain immunohistochemical expression profiles for NKG2D ligands in non-neoplastic tissues, hierarchical cluster analysis was performed based on the positivity rate for each non-neoplastic tissue type (Fig. 3). According to tissue-based cluster analysis, non-neoplastic tissues were divided into three clusters: 1) N-null type, which was effectively negative for all ligands; 2) N-variable type, which showed diverse expression among the ligands; and 3) N-complete type, which showed a high positivity rate for almost all of the ligands (N-; normal). The N-variable type showed a closer relationship to the N-complete type than did the N-null type (dendrogram on the right side in Fig. 3). The N-null type typically included all squamous epithelia, as well as the pulmonary alveolar epithelium. The N-variable type included mainly mucosal glandular epithelium. The N-complete type included epithelial cells with specific functions, such as urothelial epithelium, hepatocytes, and pancreatic acinar cells. Ligand-based cluster analysis was then performed, and this demonstrated two distinct clusters: ULBP5-ULBP3-MICA/B and ULBP2/6-ULBP1-ULBP4 (Fig. 3, upper side).
Immunohistochemical Expression Profile of NKG2D Ligands in Neoplastic Tissues
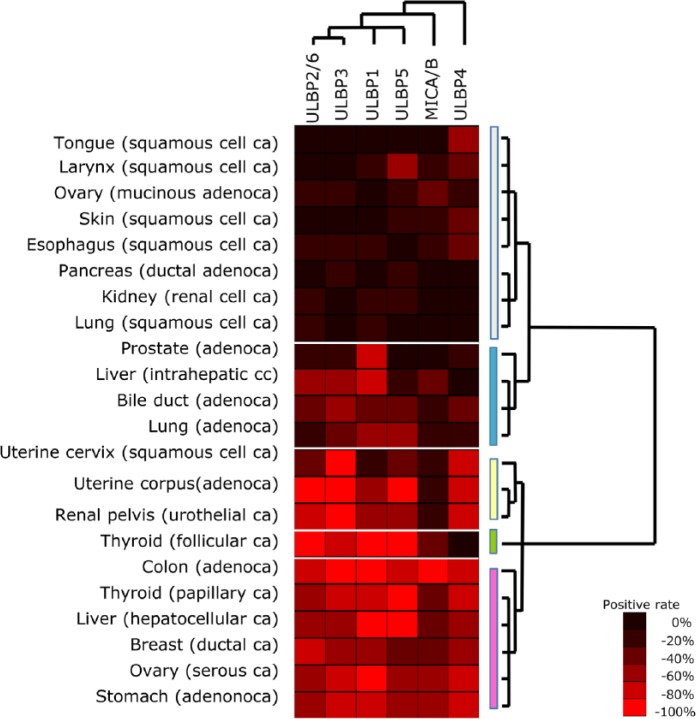
Unlike with non-neoplastic tissues, the neoplastic tissues were divided into five clusters by tissue-based cluster analysis (Fig. 5): T-null type, T-variable A, B, and C types, and T-complete type (T-; tumor). Squamous cell carcinomas of the tongue, skin, esophagus, and lung were categorized as T-null type. The T-variable A type, which showed an expression pattern similar to that of the T-null type and thus belonged to the same parental cluster, included pulmonary adenocarcinoma and intrahepatic cholangiocarcinoma. The T-variable B and C types, derived from the same parental clusters as the T-complete type, lacked MICA/B and ULBP4 expression, respectively (Figs. 5 and 6). Among squamous cell carcinomas, only that of the uterine cervix was categorized into the T-variable B type cluster, showing an increased positivity rate for several NKG2D ligands such as ULBP3 and ULBP4, unlike squamous cell carcinomas of other organs. Interestingly, some tumors showed different expression patterns of NKG2D ligands according to histological type, even though they arose from the same tissues and organs, such as carcinomas of the thyroid and ovary (Fig. 7). Ligand-based cluster analysis demonstrated a relatively close relationship among ULBP2/6, ULBP3, ULBP1, and ULBP5, whose expression patterns were similar across the neoplastic tissues examined. On the other hand, MICA/B and ULBP4 were unlike the other ligands and also each other, showing a unique expression pattern (dendrogram at the top of Fig. 5).
Figure 5.
Expression profiles of NKG2D ligands in neoplastic epithelial tissues. Hierarchical cluster analysis based on the expression profiles of NKG2D ligands demonstrated five distinct tissue-based clusters: white, T-null type; blue, T-variable A type; yellow, T-variable B type; green, T-variable C type; pink, T-complete type. A dendrogram constructed from the expression profiles obtained in the ligand-based cluster analysis (top) was similar to the phylogenetic tree based on the DNA promoter sequences of the ligands reported previously by Eagle et al. (2006).
Figure 6.
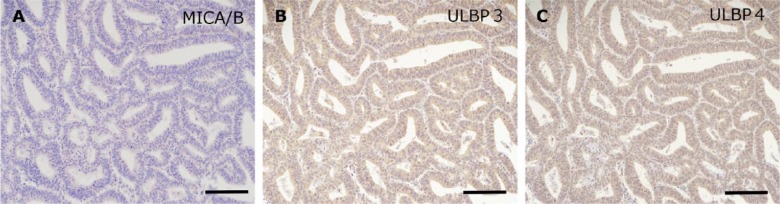
Variable expression pattern of NKG2D ligands in uterine endometrial carcinoma. Uterine endometrial carcinoma was placed in the T-variable B group (Figure 5). Only MICA/B showed a negative pattern (A), whereas the other ligands, including ULBP3 (B) and ULBP4 (C), were positive. Scale, 100 μm.
Figure 7.
Differential expression of NKG2D ligands between the two histological types of thyroid carcinoma. Thyroid follicular carcinoma was divided into the T-variable C type (A–C; B was Score 0), and papillary carcinoma into the T-complete type (D–F). (F) Inset shows non-neoplastic follicular epithelium. Scale, 50 μm.
Comparison of NKG2D Ligand Distribution between Neoplastic and Non-neoplastic Tissues in Human Organs
To evaluate the biological significance of NKG2D ligands in human tumors, differences in the immunohistochemical scores for each ligand in a specific tissue type were analyzed between neoplastic tissues and their non-neoplastic counterparts with normal morphology. Some types of neoplastic tissues had lower expression scores than their non-neoplastic counterpart with normal morphology. Pancreatic adenocarcinoma showed significantly lower scores for ULBP1 than did non-neoplastic pancreatic ducts. Scores for ULBP1, ULBP2/6, ULBP3 and ULBP4 were significantly lower in clear cell renal cell carcinoma than in renal tubules. In contrast, several tumor types showed significantly higher expression scores than their non-neoplastic counterparts with normal morphology. For instance, in the uterine cervix, squamous cell carcinoma showed higher expression scores than non-neoplastic squamous epithelium (ULBP3 and ULBP4). Endometrioid adenocarcinoma showed higher expression scores than endometrial glands (ULBP2/6), and squamous cell carcinoma of the larynx showed higher expression scores than laryngeal squamous epithelium (ULBP4 and ULBP5). For MICA/B, there was no significant difference in expression between neoplastic and non-neoplastic tissues.
Discussion
In this study, we attempted to obtain an accurate overall picture of the expression patterns of NKG2D ligands in a variety of human tissues, both neoplastic and non-neoplastic, by employing well-validated specific antibodies. A critical issue affecting the reliabilityof immunohistochemistry is the specificity and applicability of the antibodies used. Because of this point, the results of previous immunohistochemical studies of human NKG2D ligands need to be validated. In fact, our pilot study using western blotting revealed that several commercially available antibodies against ULBP2 cross-reacted with ULBP5 and ULBP6 (data not shown), and existing commercially available antibodies against ULBP5 were not applicable for FFPE immunohistochemistry. Therefore, we first validated the specificity and FFPE applicability of the antibodies very carefully, including commercially available antibodies and an antibody we had raised ourselves. As a result, we succeeded in distinguishing six different NKG2D ligands reliably on the basis of FFPE tissue immunohistochemistry: ULBP1, ULBP2/6, ULBP3, ULBP4, ULBP5, and MICA/B.
As described above, previous reports have indicated that there was almost no expression of NKG2D ligands in normal tissues, whereas the present study demonstrated diverse expression of NKG2D in non-neoplastic tissues with an apparently normal histology. This difference may be attributable to the fact that, in the present study, clinical samples were obtained from cancer patients, but not from healthy individuals, and tumor-associated changes, such as inflammation or immunological reaction, may have occurred in the otherwise apparently normal cells and tissues from these patients. Therefore, in order to minimize the impact of this potential limitation on our evaluation of normal tissues, we focused on differences in the expression levels of each NKG2D ligand between neoplastic and corresponding non-neoplastic lesions in the same individual.
Tissue-based cluster analysis divided non-neoplastic tissues into the N-null type, which consisted mainly of common stratified squamous epithelium (tongue, larynx, esophagus, uterine cervix, and skin) and simple flattened epithelium (alveolar epithelium of the lung); the N-variable and N-complete types consisted mainly of non-squamous epithelia, such as ductal and glandular epithelial cells. Accordingly, it appears that, in general, NKG2D ligands are of less importance for maintaining the function of squamous epithelial cells unless they are subjected to severe pathological disturbance. Although most squamous epithelia showed a low level of expression, characteristic expression of ULBP4 was observed in the skin, in agreement with a recent study (Chalupny et al. 2003). In contrast, in ductal or glandular epithelial cells, at least one of the NKG2D ligands was frequently expressed, though any significant relationship was not found with specific ligands. This implies that local NK cell-mediated immunological defense may differ between squamous- and glandular epithelium-lined tissues, and may be variably regulated among tissues and organs. In addition, groups of ligands shared a similar expression pattern in non-neoplastic tissues, and cluster analysis revealed two distinct clusters: the ULBP5-ULBP3-MICA/B cluster and the ULBP2/6-ULBP1-ULBP4 cluster (Fig. 3). Thus, it seems possible to assume that ligand expression in normal cells might be regulated in a ligand-set manner, at least in some tissues. More interestingly, on the basis of a comparative analysis of the gene promoters of the six NKG2D ligands reported by Eagle et al. (2006), ligands were divisible into three groups using specific antibodies: ULBP2/6 and ULBP5; ULBP1 and ULBP3; and MICA/B and ULBP4. In fact, each of the two clusters presented in this study includes either one of the ligands in each group.
With regard to neoplastic tissues, recent studies have revealed that MICs and ULBPs are variably expressed, and often co-expressed in ovarian (McGilvray et al. 2010) and colorectal (McGilvary et al. 2009) cancers, and that their expression is associated with prognosis. However, whether there are many correlations of ligand expression pattern among various tissues and organs remains an interesting question. In the present study, ligand-based cluster analysis of neoplastic tissues indicated that the dendrogram (based on immunohistochemical expression profiles) was closely similar to the phylogenetic tree (constructed on the basis of the ligand promoter DNA sequences; Fig. 5, inset shows the phylogenic tree presented by Eagle et al. (2009)). This similarity between these two molecular features—the protein expression profile in neoplastic tissues and DNA phylogeny in normal cells—may imply that genetic control of NKG2D ligand expression is basically conserved even during cellular stress resulting from malignant transformation, without marked epigenetic alteration.
Histologically, most squamous cell carcinomas, especially those of the upper aerodigestive tract (tongue, larynx, esophagus and lung) and skin, were included in the T-null type. The only exception was squamous cell carcinoma of the uterine cervix, because it showed a relatively high rate of ligand positivity (T-variable B). Textor et al. (2008) also investigated the differential expression of NKG2D ligands in cervical carcinogenesis, and demonstrated an increased rate of positivity for MICA in 20% of cervical carcinomas, being similar to the rate we observed in the present study. Close association with HPV infection as a carcinogenetic factor may be one of the explanations for this unique characteristic of squamous cell carcinoma of the uterine cervix. On the other hand, tissue-based cluster analysis of neoplastic tissues demonstrated that null-type squamous cell carcinomas, except for those of the lung, specifically expressed ULBP4, unlike other tissues in the null-type cluster. This expression pattern of ULBP4 is similar to that of a murine homologue H60c (Takada et al. 2008), as reported by Whang et al. (2009). However, no expression of ULBP4 was observed in squamous cell carcinoma of the lung. This difference may be attributable to a non-squamous origin of squamous cell carcinoma in the lung; i.e., de novo squamous carcinogenesis from bronchial columnar epithelium.
In this study, using validated and specific antibodies, we analyzed the immunohistochemical expression of NKG2D ligands in neoplastic lesions and their normal counterpart tissues. The results indicated that epithelial neoplasms show a characteristic pattern of NKG2D ligand expression, suggesting that expression of the ligand proteins may be controlled by promoter-dependent transcriptional regulation. The data presented should serve as a useful reference for other investigators in future studies of NKG2D ligand functions. For instance, neoplasms expressing NKG2D ligands might be potential targets for antibody-based therapy, and NKG2D ligand expression might be useful as a surrogate marker reflecting ADCC activity. Our present study has attempted to clarify why multiple NKG2D ligands exist, and also the patterns of expression of NKG2D ligands in various neoplasms, as we considered that these molecules could have potential therapeutic applications as direct targets and modulators of ADCC activity in a variety of neoplasms through antibody treatment. In addition, these IHC evaluations may yield some predictive markers. We also considered that future studies focusing on correlations with clinical factors might lead to the identification of prognostic markers.
Supplementary Material
Acknowledgments
We thank Ms Kyoko Fujii for her excellent technical assistance in the antibody validation.
Footnotes
Supplementary material for this article is available on the Journal of Histochemistry & Cytochemistry Web site at http://jhc.sagepub.com/supplemental.
Declaration of Conflicting Interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: This study was supported in part by a Grant-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan (Research Project No. 24590441).
References
- Bacon L, Eagle RA, Meyer M, Easom N, Young NT, Trowsdale J. (2004). Two human ULBP/RAET1 molecules with transmembrane regions are ligands for NKG2D. J Immunol 173:1078-1084. [DOI] [PubMed] [Google Scholar]
- Bahram S, Bresnahan M, Geraghty DE, Spies T. (1994). A second lineage of mammalian major histocompatibility complex class I genes. Proc Natl Acad Sci U S A 91:6259-6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer S, Groh V, Wu J, Steinle A, Phillips JH, Lanier LL, Spies T. (1999). Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science 285:727-729. [DOI] [PubMed] [Google Scholar]
- Castriconi R, Dondero A, Negri F, Bellora F, Nozza P, Carnemolla B, Raso A, Moretta L, Bottino C. (2007). Both CD133(+) and CD133(-) medulloblastoma cell lines express ligands for triggering NK receptors and are susceptible to NK-mediated cytotoxicity. Eur J Immunol 37:3190-196. [DOI] [PubMed] [Google Scholar]
- Cerwenka A, Bakker AB, McClanahan T, Wagner J, Wu J, Phillips JH, Lanier LL. (2000). Retinoic acid early inducible genes define a ligand family for the activating NKG2D receptor in mice. Immunity 12:721-727. [DOI] [PubMed] [Google Scholar]
- Chalupny N, Sutherland C, Lawrence W, Rein-Weston A, Cosman D. (2003). ULBP4 is a novel ligand for human NKG2D. Biochem Biophys Res Commun 305:129-135. [DOI] [PubMed] [Google Scholar]
- Cosman D, Müllberg J, Sutherland C, Chin W, Armitage R, Fanslow W, Kubin M, Chalupny N. (2001). ULBPs novel MHC class I-related molecules, bind to CMV glycoprotein UL16 and stimulate NK cytotoxicity through the NKG2D receptor. Immunity 14:123-133. [DOI] [PubMed] [Google Scholar]
- Coudert JD, Held W. (2006). The role of the NKG2D receptor for tumor immunity. Semin Cancer Biol 16:333-343. [DOI] [PubMed] [Google Scholar]
- Diefenbach A, Jamieson AM, Liu SD, Shastri N, Raulet DH. (2000). Ligands for the murine NKG2D receptor: expression by tumor cells and activation of NK cells and macrophages. Nat Immunol 1:119-126. [DOI] [PubMed] [Google Scholar]
- Eagle RA, Traherne J, Ashiru O, Wills M, Trowsdale J. (2006). Regulation of NKG2D ligand gene expression. Hum Immunol 67:159-169. [DOI] [PubMed] [Google Scholar]
- Eagle RA, Flack G, Warford A, Martínez-Borra J, Jafferji I, Traheme JA, Ohashi M, Boyle LH, Barrow AD, Caillat-Zucman S, Young NT, Trowsdale J. (2009a). Cellular expression, trafficking, and function of two isoforms of human ULBP5/RAET1G. PLoS One 4:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eagle RA, Traherne JA, Hair TR, Jaferji I, Trowsdale J. (2009b). ULBP6/RAET1L is an additional human NKG2D ligand. Eur J Immunol 39:3207-3016. [DOI] [PubMed] [Google Scholar]
- Ehrlich LI, Ogasawara K, Hamerman JA, Takaki R, Zin-goni A, Allison JP, Lanier LL. (2005). Engagement of NKG2D by cognate ligand or antibody alone is insufficient to mediate costimulation of human and mouse CD8+ T cells. J Immunol 174:1922-1931. [DOI] [PubMed] [Google Scholar]
- Friese MA, Platten M, Lutz SZ, Naumann U, Aulwurm S, Bischof F, Buhring HJ, Rammensee HG, Steinle A, Weller M. (2003). MICA/NKG2D-mediated immunogene therapy of experimental gliomas. Cancer Res 63:8996-9006. [PubMed] [Google Scholar]
- Gasser S, Orsulic S, Brown EJ, Raulet DH. (2005). The DNA damage pathway regulates innate immune system ligands of the NKG2D receptor. Nature 436:1186-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groh V, Bahram S, Bauer S, Herman A, Beauchamp M, Spies T. (1996). Cell stress- regulated human major histocompatibility complex class I gene expressed in gastrointestinal epithelium. Proc Natl Acad Sci U S A 93:12445-12450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groh V, Rhinehart R, Secrist H, Bauer S, Grabstein KH, Spies T. (1999). Broad tumor-associated expression and recognition by tumor-derived gamma delta T cells of MICA and MICB. Proc Natl Acad Sci U S A 96:6879-6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groh V, Rhinehart R, Randolph-Habecker J, Topp MS, Riddell SR, Spies T. (2001). Costimulation of CD8alphabeta Tcells by NKG2D via engagement by MIC induced on virus-infected cells. Nat Immunol 2:255-260. [DOI] [PubMed] [Google Scholar]
- Henriksen KL, Rasmussen BB, Lykkesfeldt AE, Moller S, Ejlersten B, et al. (2007). Semi-quantitative scoring of potentially predictive markers for endocrine treatment of breast cancer : a comparison between whole sections and tissue microarrays. J Clin Pathol 60:397-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki A, Ishida T, Yano H, Ishii T, Kusumoto S, Ito A, RI M, Mori F, Dind J, Komatsu H, Iida S, Ueda R. (2009). Expression of the ULBP ligands for NKG2D by B-NHL cells plays an important role in determining their susceptibility to rituximab-induced ADCC. Int J Cancer 125:212-221. [DOI] [PubMed] [Google Scholar]
- Jamieson AM, Diefenbach A, McMahon CW, Xiong N, Carlyle JR, Raulet DH. (2002). The role of the NKG2D immunoreceptor in immune cell activation and natural killing. Immunity 17:19-29. [DOI] [PubMed] [Google Scholar]
- McGilvray RW, Eagle RA, Rolland P, Jafferji I, Trowsdale J, Durrant LG. (2010). ULBP2 and RAET1E NKG2D ligands are independent predictors of poor prognosis in ovarian cancer patients. Int J Cancer 127:1412-1420. [DOI] [PubMed] [Google Scholar]
- McGilvray RW, Eagle RA, Watson NF, Al-Attar A, Ball G, Jafferiji I, Trowsdalw J, Durrant LG. (2009). NKG2D ligand expression in human colorectal cancer reveals associations with prognosis and evidence for immunoediting. Clin Cancer Res 15:6993-7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pende D, Rivera P, Marcenaro S, Chang CC, Biassoni R, Conte R, Kubin M, Cosman D, Ferrone S, Moretta L, Moretta A. (2002). Major histocompatibility complex class I-related chain A and UL16-binding protein expression on tumor cell lines of different histotypes: analysis of tumor susceptibility to NKG2D-dependent natural killer cell cytotoxicity. Cancer Res 62:6178-6186. [PubMed] [Google Scholar]
- Radosavljevic M, Cuillerier B, Wilson M, Clément O, Wicker S, Gilfillan S, Beck S, Trowsdale J, Bahram S. (2002). A cluster of ten novel MHC class I related genes on human chromosome 6q24.2-q25.3. Genomics 79:114-123. [DOI] [PubMed] [Google Scholar]
- Raffaghello L, Prigione I, Airoldi I, Camoriano M, Levreri I, Gambini C, Pende D, Steinle A, Ferrone S, Pistoia V.(2004). Downregulation and/or release of NKG2D ligands as immune evasion strategy of human neuroblastoma. Neoplasia 6:558-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salih HR, Antropius H, Gieseke F, Lutz SZ, Kanz L, Rammensee HG, Steinie A. (2003). Functional expression and release of ligands for the activating immunoreceptor NKG2D in leukemia. Blood 102:1389-1396. [DOI] [PubMed] [Google Scholar]
- Takada A, Yoshida S, Kajikawa M, Miyatake Y, Tomaru U, Sakai M, Chiba H, Maenaka K, Kohda D, Fugo K, Kasahara M.(2008). Two novel NKG2D ligands of the mouse H60 family with differential expression patterns and binding affinities to NKG2D. J Immunol 180:1678-1685. [DOI] [PubMed] [Google Scholar]
- Textor S, Dürst M, Jansen L, Accardi R, Tommasino M, Trunk Mj, Porgador A, Watzl C, Gissmann L, Cerwenka A. (2008). Activating NK cell receptor ligands are differentially expressed during progression to cervical cancer. Int J Cancer 123:2343-2353. [DOI] [PubMed] [Google Scholar]
- Vetter CS, Groh V, thor Straten P, Spies T, Brocker EB, Becker JC. (2002). Expression of stress-induced MHC class I related chain molecules on human melanoma. J Invest Dermatol 118:600-605. [DOI] [PubMed] [Google Scholar]
- Watson NF, Spendlove I, Madjd Z, McGilvray R, Green AR, Ellis IO, Scholefield JH, Durrant LG. (2006). Expression of the stress-related MHC class I chain-related protein MICA is an indicator of good prognosis in colorectal cancer patients. Int J Cancer 118:1445-1452. [DOI] [PubMed] [Google Scholar]
- Whang MI, Guerra N, Raulet DH. (2009). Costimulation of dendritic epidermal gammadelta T cells by a new NKG2D ligand expressed specifically in the skin. J Immunol 182:4557-4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Song Y, Bakker AB, Bauer S, Spies T, Lanier LL, Phillips JH. (1999). An activating immunoreceptor complex formed by NKG2D and DAP10. Science 285:730–732. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.