Abstract
Early studies established that transcription initiates within a ~50 bp DNA segment capable of nucleating the assembly of RNA Polymerase II and associated general transcription factors necessary for transcriptional initiation; this region is called a core promoter. Subsequent analyses identified a series of conserved DNA sequence elements, present in various combinations or not at all, in core promoters. Recent genome-wide analyses have provided further insights into the complexity of core promoter architecture and function. Here we review recent studies that delineate the active role of core promoters in transcriptional regulation of diverse physiological systems.
Keywords: Transcription, Core promoter, RNA Pol II, non-coding RNAs
The Core Promoter: A platform for transcription initiation
Cellular differentiation and function depend on the accurate and regulated transcription of the genome, which encompasses both the 2–3% of the genome that encodes proteins and the ~90% that is transcribed into into non-coding RNA including ribosomal and tRNAs, long non-coding RNAs, miRNAs and other regulatory RNAs [1, 2]. Integral to this regulated expression is the ability to initiate transcription at precise genomic sites.
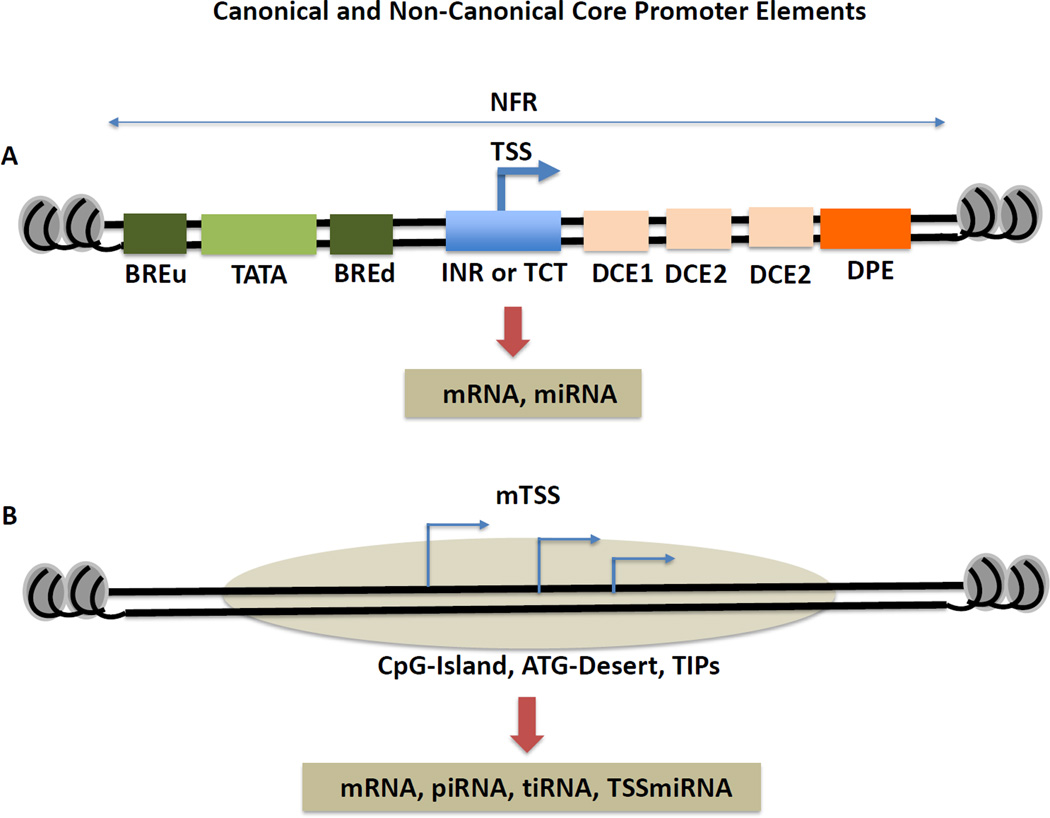
Core promoters are defined as the DNA segment of 50–100 bp within which transcription initiates [3]. Genome-wide structural analyses have identified a series of conserved DNA sequence elements that are often, but not universally, associated with of core promoters (Figure 1). The core promoter functions as a platform on which the transcription machinery assembles. Among the factors recruited to core promoters are the enzyme RNA polymerase II, which transcribes protein-coding and many non-protein coding RNAs (e.g., long non-coding RNAs, miRNAs), and the multiple general transcription factors (GTFs) and co-factors required for RNA synthesis and biogenesis. It is estimated that this transcriptional complex is well over a mega-dalton in size and can occupy over a hundred base pairs of DNA around the transcriptional start site (TSS) [3].
Figure 1.
[NN11] Canonical (A) and non-canonical (B) core promoter elements. The various established core promoter elements are shown in panel (A). Although majority of mammalian genes do not appear to have any of these elements, some protein-coding and microRNA (miRNA) promoters have canonical core promoter elements associated with them. Not all of these elements are present in all promoters and many of these elements are present in lineage-specific genes. While Inr element is utilized by TFIID, TCT element is utilized by TBP-related factor, TRF2 in ribosomal protein-coding genes. TSS is transcription start site; NFR is nucleosome free region. (B) The large majority of mammalian genes appear to lack classical core promoter elements but instead contains broad regions (100–150 bp) associated with transcription initiation. The established non-canonical elements or regions are CpG-islands, ATG-deserts and Transcription Initiation Platforms (TIPs). These are present in mRNA coding as well as various non-coding RNAs, including piwi-interacting RNA (piRNA), transcription initiation associated RNA (tiRNA) and transcription start site associated miRNA (TSSmiRNA). These promoters are frequently characterized by multiple transcription start sites (mTSS).
The assembly of the transcription machinery at the core promoter is initiated by the interaction of specific transcription factors with their cognate upstream regulatory sequences, e.g. enhancers and silencers. These specific transcription factors mediate both cell intrinsic, tissue-specific signals and extrinsic signals that modulate transcription of a given gene. However, all of these signaling events need to be integrated at the core promoter to achieve the appropriate level of transcription initiation. Although considerable effort has been expended to identify the upstream regulatory sequences that contribute to proper regulation of transcription much less attention has been focused on characterizing the structure and function of core promoters. In this review, we summarize the evidence that the core promoter functions both as a platform to integrate upstream signaling and as an active participant in regulating RNA Pol II-mediated transcription. Moreover, given that the genome-wide studies indicate that many genes lack an identifiable core promoter element, we will also discuss the implications for these recent findings.
Structure of canonical core promoter elements
Many core promoter sequences are common to a large number of protein-coding genes and have been conserved in evolution. Among these canonical core promoter elements are the TATAA box, the Initiator (Inr), Bre, DPE, DCE. Additional elements – MTE and XCP1 – have been described but occur with lower frequency. Although not formally considered a canonical core promoter element, the CCAAT box, located between −50bp and −80/−100 bp upstream of the transcription start site (TSS) has been conserved in evolution from[S1] bacteria through metazoans. However, the precise role of canonical core promoter elements in regulating transcription of all genes is unclear as not all core promoter elements are found associated with all genes. In the following sections, we’ll briefly describe the salient features of some of these elements.
Transcription initiation complex nucleators: the TATA box and the Inr
The two most common core promoter elements associated with protein-coding genes are the TATAA box and the Initiator (Inr), which occur either together or separately in the majority of eukaryotic promoters. The TATA box, TATAA, (TSS), the first core promoter element to be identified, was biochemically found to be located 20–30 bp upstream of the transcription start site and serves as the binding site for the general transcription factor TFIID [4]. However, recent studies show that the optimal spacing between the first T of the TATA box and the +1 of the TSS is 30–31 bp (5). The TATA box is found in only about 5–7% of eukaryotic promoters. Among the promoters that lack a TATA box, many contain the Inr, although the frequency of an Inr element is found as often in TATA-containing promoters (6, 7).
The Inr (consensus sequence YYA+1NT/AYY) that spans the TSS and can independently direct accurate transcription initiation [8–10]. Nearly 70% of drosophila promoters appear to have an Inr element, either alone or in combination with a TATA box; no clear estimates are available for the distribution of Inr element in mammalian promoters [6].
Although the TATA and Inr can be found together in some core promoters (7), they appear to subserve distinct functional families of genes. In general, the core promoters of tissue-specific genes are anchored by a TATA box and generally initiate transcription at a single discrete site, or at a tightly clustered locus [12]. In contrast, Inr elements are found in core promoters of ubiquitously expressed or “housekeeping” genes and initiate transcription over multiple, dispersed sites across a region of up to ~150 bp [112–14[S2][S3]]. [NN4] Thus, the promoters of immunoglobulin and globin genes contain only the TATAA box whereas actin and terminal deoxynucleotidyl transferase (TdT) promoters contain only Inr elements. Genes subject to complex regulatory patterns, such as the major histocompatibility complex (MHC) class I genes, often contain both Inr and TATAA elements [15].
Both the TATA box and the Inr provide a platform for the assembly of transcription pre-initiation complexes (PIC) by the general transcription factor, TFIID. The TATA element serves as the binding site for the TFIID component, the TATA-binding protein (TBP) to nucleate the formation of a transcription initiation complex and the subsequent recruitment of RNA Pol II [3]. Similarly, TFIID nucleates PIC assembly on Inr promoters, although the precise nature of the interaction is not known.
Downstream Promoter Element (DPE)
A variety of additional core promoter elements that modulate the activities of the TATAA and Inr elements have been identified. Among these is the Downstream Promoter Element (DPE) found in both drosophila and mammalian Inr promoters [6]. The DPE (consensus sequence: A/GGA/TCGTG), which occurs mostly but not exclusively occurs in TATA-less promoters, acts in conjunction with the Inr and is located at +28 to +32 relative to the initiating nucleotide at +1 within the Inr motif [6]. Although the DPE, like the TATA box, is a recognition site for the binding of components of the general transcription factor TFIID, it does not act independently but depends on the presence of an Inr.
TFIIB recognition element (BRE)
The requirement for core promoter element sequences upstream of the TATA box was first observed in archaeal genes through mutational analysis. Structural analysis of TBP-TFIIB-DNA as well as functional studies identified a 7 bp sequence element dubbed TFIIB recognition element (BRE). These motifs can be present on either side of the TATA box, with the upstream BRE (BREu) at −38 to −32 (consensus sequence: G/CG/CG/ACGCC) and the downstream BRE (BREd) at −23 to −17 (consensus sequence: G/ATT/AT/GT/GT/GT/G). Interestingly, BRE not only functions in activating transcription but also can repress transcription [6].
Downstream Core Element (DCE)
Another core promoter element is the DCE, which has canonical sequences at positions +6 to +11, +16 to +21, and +30 to +34 relative to the TSS (consensus sequences: CTTC, CTGT, AGC). DCE is also recognized by components of the TFIID complex [16]. Multiple start site downstream element (MED-1) and the motif ten element (MTE), which lies downstream of the transcription start site and is contacted by TFIID, have also been described to function as core promoter elements [17, 18].
TCT element
The TCT core promoter element is present in most ribosomal protein-coding genes (19, 20). Although the TCT motif encompasses the transcription start site from −2 to +6 relative to the TSS and is therefore located at the same position as the Inr motif, the TCT motif is functionally distinct from the Inr. The TCT motif cannot function in place of an Inr element and is not recognized by the TBP-containing TFIID complex (19). The distribution of TCT element beyond the ribosomal protein-coding genes remains unknown.
Although a variety of canonical core promoter elements have been identified, many genes do not have any of these core promoter elements. Similarly, the molecular mechanisms by which these canonical promoter elements are utilized in a variety of genes and recognized by the transcription machinery still remain to be fully defined.
Function of canonical core promoter elements
Why is there such heterogeneity in core promoter architecture? This heterogeneity likely reflects the diversity of regulatory mechanisms that govern gene expression. They also indicate that these core promoter elements are perhaps interchangeable or work in combination with each other. For instance, the combinatorial “mixing and matching” of the different core promoter elements may be necessary to transduce the diversity of transcriptional regulatory signaling pathways. Although the significance and distribution of mix-and-match modules in eukaryotic promoters are yet to be fully understood, different combinations of core promoter elements may support the assembly of distinct components of the transcription machinery and recruitment of RNA Pol II allowing for expression of tissue-specific and/or developmentally regulated genes [21]. The concept of “mix-and-match promoter elements” is analogous to that found in bacteria where it has been studied extensively [21].
Given the limitations of the biochemical assays used to identify and characterize canonical core promoter elements, it has been somewhat difficult to assign a clear in vivo function to each of these elements. Early in vitro and transfection studies of TATAA or Inr promoters suggested that these core promoter elements are essential for promoter function [4, 10]. However, these studies did not include analyses of complex promoters with multiple core elements; nor did they assess core promoter element function in vivo. Recent analyses of the roles of core promoter elements within the complex MHC class I promoter indicate that their functions are more nuanced. The MHC class I promoter contains both a TATAA-like element and a canonical Inr. Also within the 60 bp [DS5]promoter region is a CCAAT box and an Sp1 binding site. Each of the elements was mutated separately within the context of a full-length MHC class I transgene and introduced into mice. Surprisingly, none of these promoter elements was essential for promoter activity or transcription initiation in vivo [22]. All of the mutants supported transcription in vivo as well or better than the wild type promoter. Although none was necessary for transcription, each element had a defined role [22]. Thus, CAAT box mutations modulated constitutive expression in nonlymphoid tissues, whereas TATAA-like element mutations dysregulated transcription in lymphoid tissues. Conversely, Inr and Sp1 binding element mutations aberrantly elevated expression in both lymphoid and nonlymphoid tissues. Transgene expression correlated with RNA polymerase II binding and active histone H3K4me3 patterns while it was partly correlated with repressive H3K9me3 marks. Finally, while the wild-type, TATAA-like-mutant and CAAT-mutant promoters were activated by gamma interferon, the Sp1 and Inr mutants were repressed, implicating these elements in regulation of hormonal responses. Although the initial cell-based assays suggested an essential role for core promoter elements in transcription initiation and promoter activity [8, 23], these recent observations [22] indicate that these elements play more of a fine tuning role. It remains to be determined whether canonical core promoters are non-essential for a select few genes or this phenomenon is more wide spread in the mammalian genome.
A recent study also raises questions about the concept of a core promoter. It reports that thousands of promoters in vertebrates including those of ubiquitously expressed genes, contain at least two TSS “selection codes”. The first one is mostly utilized in oocyte and the other one in developing embryos (24). While the first code is dependent on a weak TATA-like sequence, the second code is dependent on the position of the H3K4me3-marked first nucleosome downstream of TSS. These results suggest that rather than one open promoter architecture, multiple overlapping promoter codes dictate expression of a ubiquitously expressed gene (24).
The transcriptional machinery
The complexity of core promoter architecture is further elaborated by the complexity of the transcription factors that are recruited. Decades of studies in cell-free systems revealed that there is a set of six GTFs: TFIIA, TFIIB, TFIID, TFIIE, TFIIF and TFIIH [3, 25]. These GTFs along with RNA Pol II form the transcription machinery near the TSS at the canonical core promoter elements, which predominantly includes the TATA box, Initiator (Inr) element and downstream promoter element (DPE) (Figure 1). Amongst the GTFs, TFIID is the only DNA binding protein that recognizes a canonical TATA box and initiates the nucleation of the transcriptional machinery [3]. The multi-subunit TFIID can nucleate transcription initiation via interaction of its TATA binding protein (TBP) component with the TATA box. However, TFIID also nucleates the process in many promoters that do not contain a recognizable TATA box because it can recognize other core promoter elements [6]. Apart from TFIID, which serves to nucleate transcription initiation from a variety of core promoter elements, TFIIB also makes site-specific contact via the BRE (6). Although recognition of the BRE is mediated by a helix-turn-helix motif at the C-terminus of TFIIB, this motif is missing in yeast and plants, suggesting that the BRE may not contribute to gene regulation in these organisms. It is surmised that the interactions of TFIIB with BRE help stabilize the deformations imposed on the DNA by the binding of TBP. However, BRE also functions as a repressor of basal transcription in vitro with crude nuclear extracts as well as in vivo in transfection assays (6). This repression and the TFIIB-BRE interaction were relieved when transcriptional activators were bound to distal sites, which resulted in an increased transcriptional activation (6).
Recent observations suggest that alternative initiation complexes can replace canonical TFIID [26]. For example, whereas constitutive expression of the MHC class I genes depends on the TFIID complex, γ-interferon-activated transcription bypasses the requirement for canonical TFIID, depending instead on the transcriptional co-activator class II trans-activator (CIITA). Interestingly, TFIID and CIITA target distinct families of transcription start sites [27]. Studies of the histone gene cluster also revealed that these promoters do not recruit TFIIB or TFIID, also suggesting the existence of gene-specific and/or stage specific transcriptional machinery [28]. Muscle differentiation provides an extreme example of selective usage of the transcription machinery, where the transition from myoblast to myocyte is accompanied by a loss of all TFIID components except for TAF3 [29]. The TCT core promoter element is also not recognized by TBP/TFIID. Although a direct binding of TRF2 to the TCT element has not been shown, ChIP-seq (chromatin immunoprecipitation [ChIP] combined with deep sequencing) experiments revealed the preferential localization of TRF2 at TCT versus TATA promoters, further suggesting redundancy in transcription initiation complexes (20). [DS6]
Regardless of the nature of the transcriptional machinery, the rate and/or stability of the machinery is in part dictated by the actions of transcription factors, which bind to upstream regulatory elements. Transcription factors are usually linked to the core promoter via long-range interactions that involve numerous transcriptional co-factors, general co-activators, including components of the originally described USA activity (e.g, PC4), and the more recently discovered Mediator complex [3, 30].
Lessons from genome-wide studies
While the early studies of canonical core promoters were primarily in cell free systems with isolated or cloned genes, the recent development of genome-wide methods has allowed interrogation of their role at a global level. Surprisingly, these analyses collectively indicate that most mammalian genes lack canonical core promoter elements but nevertheless recruit the transcriptional machinery. If so many genes lack a canonical core promoter element then how is transcription initiated in these genes? Recent studies clearly establish that most eukaryotic genes utilize non-canonical core promoter elements that substitute and/or function in combination with canonical core promoter elements (12, 13). The structure and function of these elements are discussed in Box 5.
Box 5.
Non-canonical core promoter elements: CpG islands and ATG deserts
The Inr-containing core promoters supporting multiple transcription start sites are often located within two broadly-defined regions: CpG islands and ATG deserts. “CpG Island” promoters generally span a 0.5–2 kbp stretch of DNA that exhibits a relatively high density of CpG dinucleotides. The CpG dinucleotide, a DNA methyltransferase substrate, is underrepresented in the genomes of many vertebrates because 5-methylcytosine can undergo deamination to form thymine, which is not repaired by DNA repair enzymes [6, 58]. CpG islands mostly remain unmethylated in all tissues and at all stages of development [58]. It has been estimated that the human genome contains approximately 30,000 of these sites, and that they are associated with approximately half of the promoters for protein-coding genes [58]. Although the precise molecular mechanisms dictated by CpG island promoters remain unclear, it is likely to be associated with the inability of stretches of CpG to assemble stable nucleosomes. Thus, CpG-island promoters have relatively low nucleosome occupancy. Accordingly, induction of expression from some CpG-island promoters does not a require SWI/SNF nucleosome remodeling complexes [59]. Interestingly, some stimuli, including serum and tumor necrosis factor-alpha, exhibit a strong bias toward activation of SWI/SNF-independent CpG-island containing genes. In contrast, induction of non-CpG-island genes requires remodeling mediated by Swi/Snf [59]. More recent observations also revealed the existence of transcriptional initiation platforms (TIPs) that are characterized by large areas of RNA Pol II and GTF recruitment at promoters, intergenic and intragenic regions [60]. TIPs show variable widths (0.4–10 kb) and correlate with high CpG content and increased tissue specificity at promoters [60].
Another class of distinct promoters with large CpG islands appears to be associated with polycomb repression-regulated genes; correspondingly, they exhibit histone H3 lysine 27 trimethylation(H3K27me3). These also have DPE, and are typically found in developmentally regulated genes [12].
ATG deserts were first discovered associated with major histocompatibility complex (MHC) class I genes (61). The ATG desert is a DNA segment that has a lower frequency of occurrence of the ATG trinucleotide than the surrounding sequences and spans a region of −1 kb both upstream and downstream of the major transcription start site. Genome-wide analysis suggests that ATG deserts are an intrinsic feature of core promoters that do not contain canonical TATAA elements. ATG desert promoters support the use of multiple, dispersed, TSS whose products all encode a single protein, thereby permitting the core promoter to serve as a platform where complex upstream regulatory signals are integrated through selective transcription start site usage [61].
Genome-wide studies also indicate that transcription is pervasive and that the transcriptional machinery not only assembles around the TSS at core promoters of protein-coding genes, but also at enhancers and “transcriptional hubs”. These latter engagements result in production of a class of non-coding RNAs called enhancer RNAs (eRNAs) [31, 32]. Until recently, it was unknown whether the eRNAs also utilize known core promoter elements or a yet unknown mechanism. However, recent studies indicate a shared architecture of transcription initiation regions between enhancers and promoters of mammalian genes (33, 34).
Another surprising finding from genome-wide studies is that the majority of mammalian promoters direct transcription initiation in both directions with opposite orientation, a phenomenon termed “divergent” transcription [35]. Divergent transcription from both promoters and enhancers might be the major source of intergenic transcription (upstream antisense RNAs and eRNAs) [35]. Although most mammalian genes [DS7]lack well-defined core promoter elements, they nevertheless recruit the transcriptional machinery. It is argued that rather than maintaining directionality, in these instances, the GTFs are recruited on both sides of a site-specific transcription factor-binding motif [35]. This model predicts that promoters with a canonical core element such as a TATA box would be “unidirectional” promoters, whereas the ones without these core elements but instead containing CpG islands would be “bidirectional” promoters [35].]. Interestingly, a recent high-resolution analysis studying nascent RNAs indicate that the divergent TSS pairs at both promoters and enhancers share common architecture, suggesting that same mechanisms of transcription initiation apply to both promoters and enhancers (33, 34).
Chromatin immunoprecipitation coupled with deep sequencing (ChIP-seq) of RNA Pol II indicate an unprecedented and genome-wide view of the recruitment of the transcriptional machinery in a given cell [12, 13, 36]. Interestingly, genome-wide studies of RNA Pol II recruitment in various human cells, including embryonic stem cells and differentiated cells, estimate that many if not most promoters are associated with “pre-loaded” or paused RNA Pol II [37, 38]. Given that most of the genome is transcribed, this is not unexpected. However, earlier biochemical quantitation showed that there are approximately 320,000 molecules of RNA Pol II in a human cell (HeLa), of which only 180,000 are engaged and perhaps an even lesser number (~65,000) engaged in active transcription initiation [39]. A far lower number (~30,000) of GTFs appear to be engaged in active complexes [39]. The discrepancy between the genome-wide studies suggesting that RNA Pol II is pre-loaded in most if not all promoters and the biochemical estimate that only a fraction of promoters have Pol II engaged in active initiation will have to be resolved in the future.
Chromatin marks and core promoters
Apart from the presence of classical or non-classical core elements, histone modifications also indicate promoter activity. Even though the molecular decoding of histone marks such as H3K4me3 and H3K27me3 is far from being fully worked out, genome-wide studies have provided important insights as to how some of these marks might function [40]. While the H3K4me3 mark is associated with active promoters around TSS, H3K27me3 is associated with repressed promoters [41]. Interestingly, in embryonic stem (ES) cells with pluripotency, often times both marks are present in promoters; these are referred to as “bivalent” promoters, and can either be activated or repressed along a particular lineage depending on the developmental cue [40].
Although H3K4me3 is generally present downstream of the TSS in active genes, RNA Pol II binding is necessary at these promoters for transcription. Across different classes of genes in vertebrates, H3K4me3 distribution is almost identical with the span of CpG islands [12]. However, ubiquitously expressed genes generally have short CpG islands, and the H3K4me3 mark and CpG island typically only overlap at the 5’-end of these genes [(12). On the other hand, developmentally regulated genes in vertebrates have features that are associated with repression, including multiple large CpG islands and exhibiting both H3K27me3 and H3K4me3 marks [12]. The large CpG islands often extend beyond the promoter region into the gene body and overlap with H3K4me3 marks,. It is noteworthy that even for the most studied epigenetic marks, only a correlation between active or repressed transcription is known [12].
Core promoters of non-coding RNAs
Micro-RNA core promoters
Because protein-coding genes constitute only a small fraction of the total transcriptome, a majority of the transcripts arise from non-coding RNAs. Thus, it is worthwhile to compare the structure and function of the core promoters of non-coding RNAs with those of the protein-coding genes.
MicroRNAs (miRNAs) are derived from large primary miRNAs (pri-miRNA) that are successively processed into first −70-nt precursors (pre-miRNA) and then their mature forms [42]. By combining nucleosome mapping with chromatin signatures, the core promoters [NN8]of 175 human miRNAs were identified [43]. 85% of miRNA promoters corresponding to previously annotated genes [NN9]exhibited a CpG island[DS10], while 51% of novel promoters contained a CpG island. Allowing for 50-bp ambiguity, 19% of the miRNA promoters had a TATA element, 21% had BRE, 47% had an INR, 7% had MTE, and surprisingly 87% exhibited DPE [43]. Although these studies suggested that the miRNA core promoters are rather similar to the mRNA core promoters, they also revealed a significant fraction of miRNAs use their own (novel) transcription initiation regions, including both miRNAs located at intergenic regions and others that are embedded within introns of known coding genes. Surprisingly, in addition to RNA Pol II, RNA Pol III was associated with a number of these miRNA promoters [43]. However, it remains to be determined whether differences in miRNA expression correlate with their transcription by RNA Pol II or RNA Pol III.
Miscellaneous non-coding RNA transcription
FANTOM4 project identified tiny RNAs of 18nt average length that map within the core promoter region (−60 to +120 nt of TSS) of humans, chicken and drosophila genes [44]. These are called transcription initiation RNA or tiRNAs, which originate from the same strand as the TSS and are generally associated with high G/C containing promoters that have regions of bound RNA Pol II and are highly transcribed [45]. Although the significance of these small RNAs is still unclear, tiRNAs may be abortive transcripts resulting from stalled or poised RNA Pol II. Alternatively, they may arise by virtue of RNA Pol II ‘backtracking’. RNA Pol II arrests +20 to +32 from the TSS on certain promoters then backtracks to approximately +12, therefore the exposed ‘backtracked’ transcript matches well to the observed position and length of tiRNAs [44, 45], tiRNAs could be TFIIS cleavage products [45]. Finally, although unlikely, it is possible that tiRNAs derive from uncapped and very short or cleaved transcripts that initiate downstream of the major TSS [45].
Another class of small non-canonical RNAs called the transcription start site-microRNAs or TSS-miRNAs was recently discovered, which originates from RNA Pol II core promoters and is bound by Argonaute-2 (Ago-2) [46]. The precise function of these small RNAs is still unclear. However, similar to miRNAs located within introns, the linked expression of TSS-miRNAs and the corresponding downstream mRNA suggests co-regulation of transcriptional pathways and shared core promoter elements [46].
PIWI-interacting RNAs (piRNA) are a germline-specific class of small noncoding RNAs that are generated from protein-coding genes by RNA Pol II and are believed to provide protection to the genome against various transposable elements [47–49]. Most primary piRNAs are generated by RNA Pol II-dependent expression of long, single-stranded precursor RNAs that are subsequently processing in specialized cytoplasmic foci [47, 50]. piRNAs can originate either from uni-strand clusters or dual-strand clusters. While transcription from dual-strand clusters appears to be specific to Drosophila, uni-strand clusters in both Drosophila and mice exhibit hallmarks of canonical Pol II transcription such as a defined Pol II peak around the TSS, an enrichment of the active histone mark H3K4me2 at their putative promoters and the expression of 5’ methyl-guanosine-capped and terminated RNAs [47, 50]. Whether the transcriptional units associated with piRNAs bear any novel core promoter features or exhibit core promoter features of protein-coding genes remain unclear at present.
Concluding remarks
Core promoter elements were first defined in cloned genes using cell free systems to study transcription initiation. However, genome-wide studies led to the discovery that the majority of mammalian genes lack a well-defined core promoter element but instead contain “promoter regions” with characteristic epigenetic marks (both histone chromatin and DNA marks). Therefore, the importance of canonical core promoter elements needs to be reevaluated. Given that RNA Pol II appears to be recruited to large stretches of the genome without identifiable core promoter elements, are core-promoter elements necessary for the recruitment of the transcriptional machinery? In addition, an in vivo study has challenged the notion that core promoter elements direct accurate transcription initiation or even that they are individually necessary. Perhaps canonical core promoter elements fine-tune physiological responses for a select few genes in a developmental fashion and work in concert with epigenetic marks and other regulatory elements such as enhancers. Although the number of these genes might be limited, they nevertheless could represent key regulatory genes essential for development and differentiation. Moreover, given the heterogeneity in core promoter architecture, it is also likely that the eukaryotic genome utilizes many of these elements in a “mix-and-match” fashion, thereby increasing the regulatory capacity of transcription initiation. Given that majority of the promoters lack canonical core promoter elements, the importance of the non-canonical promoter elements has recently been brought into light. However, the precise biochemical mechanisms that govern transcription initiation from genes with canonical versus non-canonical core promoters are still being worked out. Despite the recent high- resolution analysis of the genome and determination of the distribution of core promoter architecture, how far these principles are followed by an individual gene or a select group of genes remain to be determined also. Finally, whether the architecture and functional utilization of core promoter elements by protein-coding versus non-coding transcripts are exactly also need to be evaluated. While answers to these exciting questions are still being worked out, it is clear that the mammalian genome has taken advantage of multiple regulatory strategies to initiate and regulate transcription of both protein-coding and non-coding units.
Box 1.
Canonical Core Promoter Elements
TATA Box: ~−25 relative to the transcription start site (TSS); consensus sequence -- TATAAA or TATATA
Inr: Spans the TSS; consensus--YYANT/AYY
DPE: ~+30 relative to TSS; consensus--A/GGA/TCGTG
BREu: ~−35 relative to TSS; consensus--G/CG/CG/ACGCC
BREd: ~−20 relative to TSS; consensus--G/ATT/AT/GT/GT/GT/G
DCE: ~+9, +18, +32 relative to TSS; consensus—CTTC, CTGT, AGC
MTE: ~+23 relative to TSS; consensus--CG/CAA/GCG/CAACG
Non-Canonical Promoter Elements
CpG Island: Spans TSS; 0.5–2 kb DNA with high density of CpG dinucleotides
ATG Desert: Spans −1 kb both upstream and downstream of TSS; a segment of DNA with lower frequency of ATG trinucleotide than the surrounding sequences
TIPS: Overlapping TSS; highly variable 0.4–10 kb with high CpG content
Box 2.
As mentioned earlier, one of the more surprising findings from these genome-wide studies is the pervasive nature of transcription of the mammalian genome. What might be the reason for this? There could be several advantages of pervasive transcription initiation. First, broad expression of the genome is likely advantageous over an investment in fail-safe transcription initiation control from well-defined promoters [1]. Moreover, widespread transcription initiation at the genome-wide level may give greater opportunities for regulation. Second, allowing large genomic regions to be completely silenced may result in the formation of too tightly compacted chromatin domains, which would otherwise be hard to reopen [1]. Third, pervasive transcription may aid the three-dimensional nuclear topology by shaping chromosomes into active and repressive territories. Fourth, pervasive transcription invariably will result in accumulation of unstable transcripts, whose levels are easily modified, offering the ability of added regulation at the level of RNA turnover [1]. Finally, pervasive transcription might provide the genome with evolutionary advantages and raw material for natural selection at the level of RNA [1].
Box 3.
Packing eukaryotic genomes into high-order chromatin structures is critical for controlling most, if not all, processes derived from DNA [51, 52]. Thus, nucleosomes provide a first line of defense to prevent inappropriate RNA polymerase initiation. The minimal repeating unit of chromatin is the nucleosome, comprised of roughly 147 base pairs wrapped around a histone octamer core [51, 52]. Elegant mapping studies by Widom and colleagues found that nucleosome occupancy is relatively low at many enhancers, promoters, and transcription termination sites [53]. These are called nucleosome free regions (NFR) and reflect both intrinsic properties of the DNA sequence and active remodeling by chromatin modifiers. Nucleosome formation depends on the bendability of the DNA; CpG and AT rich regions are inherently stiff. Thus, the presence of either CpG islands or stretches of AT give rise to NFRs. Arrays of highly positioned nucleosomes surround the TSS, with positioning generally decreasing with distance from the TSS. These arrays are generated by the binding of specific transcription factors and paused RNA Pol II [51]. Thus, nonrandom mechanisms promote the proper distribution of nucleosomes, which eventually allows for correct control of transcription initiation [51].
Box 4.
Non-coding RNAs transcribed by RNA Pol III
Apart from being involved in miRNA transcription [43, 54], RNA Pol III is responsible for ~10% of all nuclear transcription and makes short noncoding RNAs, including tRNA and 5S rRNA [55, 56]. Surprisingly, RNA Pol II was found to be associated with majority of the genomic loci that are bound by RNA Pol III [56]. Another study found that only a fraction of the in silico–predicted RNA Pol III loci are actually occupied in vivo and many occupied RNA Pol III genes reside within an annotated RNA Pol II promoter (300–900 bp upstream of the TSS) [57]. Besides being associated with RNA Pol II promoters, occupied RNA Pol III genes overlap with enhancer-like chromatin. Finally, RNA Pol III occupancy coincides with the levels of nearby RNA Pol II, active chromatin and CpG content. Taken together, these results suggest that active chromatin gates RNA Pol III accessibility to the genome and RNA Pol III transcription units might function in regulating the RNA Pol II promoter activity [56, 57]. [S12]
Highlights.
In vivo experiments raise questions about the importance of core promoter elements in transcription
Genome-wide studies and pervasive transcription of the genome suggest most genes lack defined core promoter elements thus raising questions about the role of core promoter elements
Do non-coding RNAs utilize core promoter elements in the same fashion as protein coding genes?
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jensen TH, Jacquier A, Libri D. Dealing with pervasive transcription. Mol Cell. 2013 Nov 21;52(4):473–484. doi: 10.1016/j.molcel.2013.10.032. [DOI] [PubMed] [Google Scholar]
- 2.Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012 Dec 14;338(6113):1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 3.Roeder RG. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci. 1996 Sep;21(9):327–335. [PubMed] [Google Scholar]
- 4.Mathis DJ, Chambon P. The SV40 early region TATA box is required for accurate in vitro initiation of transcription. Nature. 1981 Mar 26;290(5804):310–315. doi: 10.1038/290310a0. [DOI] [PubMed] [Google Scholar]
- 5.Ponjavic J, Lenhard B, Kai C, Kawai J, Carninci P, Hayashizaki Y, Sandelin A. Transcriptional and structural impact of TATA-initiation site spacing in mammalian core promoters. Genome Biol. 2006;7(8):R78. doi: 10.1186/gb-2006-7-8-r78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smale ST, Kadonaga JT. The RNA polymerase II core promoter. Annu Rev Biochem. 2003;72:449–479. doi: 10.1146/annurev.biochem.72.121801.161520. [DOI] [PubMed] [Google Scholar]
- 7.Frith MC, Valen E, Krogh A, Hayashizaki Y, Carninci P, Sandelin A. A code for transcription initiation in mammalian genomes. Genome Res. 2008 Jan;18(1):1–12. doi: 10.1101/gr.6831208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smale ST, Baltimore D. The "initiator" as a transcription control element. Cell. 1989 Apr 7;57(1):103–113. doi: 10.1016/0092-8674(89)90176-1. [DOI] [PubMed] [Google Scholar]
- 9.Smale ST, Schmidt MC, Berk AJ, Baltimore D. Transcriptional activation by Sp1 as directed through TATA or initiator: specific requirement for mammalian transcription factor IID. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4509–4513. doi: 10.1073/pnas.87.12.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smale ST, Jain A, Kaufmann J, Emami KH, Lo K, Garraway IP. The initiator element: a paradigm for core promoter heterogeneity within metazoan protein-coding genes. Cold Spring Harb Symp Quant Biol. 1998;63:21–31. doi: 10.1101/sqb.1998.63.21. [DOI] [PubMed] [Google Scholar]
- 11.Garraway IP, Semple K, Smale ST. Transcription of the lymphocytespecific terminal deoxynucleotidyltransferase gene requires a specific core promoter structure. Proc Natl Acad Sci U S A. 1996 Apr 30;93(9):4336–4341. doi: 10.1073/pnas.93.9.4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lenhard B, Sandelin A, Carninci P. Metazoan promoters: emerging characteristics and insights into transcriptional regulation. Nat Rev Genet. 2012 Mar 6;13(4):233–245. doi: 10.1038/nrg3163. [DOI] [PubMed] [Google Scholar]
- 13.Sandelin A, Carninci P, Lenhard B, Ponjavic J, Hayashizaki Y, Hume DA. Mammalian RNA polymerase II core promoters: insights from genome-wide studies. Nat Rev Genet. 2007 Jun;8(6):424–436. doi: 10.1038/nrg2026. [DOI] [PubMed] [Google Scholar]
- 14.Gershenzon NI, Ioshikhes IP. Synergy of human Pol II core promoter elements revealed by statistical sequence analysis. Bioinformatics. 2005 Apr 15;21(8):1295–1300. doi: 10.1093/bioinformatics/bti172. [DOI] [PubMed] [Google Scholar]
- 15.Lee N, Iyer SS, Mu J, Weissman JD, Ohali A, Howcroft TK, Lewis BA, Singer DS. Three novel downstream promoter elements regulate MHC class I promoter activity in mammalian cells. PLoS One. 2010 Dec 13;5(12):e15278. doi: 10.1371/journal.pone.0015278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewis BA, Kim TK, Orkin SH. A downstream element in the human beta-globin promoter: evidence of extended sequence-specific transcription factor IID contacts. Proc Natl Acad Sci U S A. 2000 Jun 20;97(13):7172–7177. doi: 10.1073/pnas.120181197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ince TA, Scotto KW. A conserved downstream element defines a new class of RNA polymerase II promoters. J Biol Chem. 1995 Dec 22;270(51):30249–30252. doi: 10.1074/jbc.270.51.30249. [DOI] [PubMed] [Google Scholar]
- 18.Juven-Gershon T, Hsu JY, Theisen JW, Kadonaga JT. The RNA polymerase II core promoter - the gateway to transcription. Curr Opin Cell Biol. 2008 Jun;20(3):253–259. doi: 10.1016/j.ceb.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parry TJ, Theisen JW, Hsu JY, Wang YL, Corcoran DL, Eustice M, Ohler U, Kadonaga JT. The TCT motif, a key component of an RNA polymerase II transcription system for the translational machinery. Genes Dev. 2010 Sep 15;24(18):2013–2018. doi: 10.1101/gad.1951110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang YL, Duttke SH, Chen K, Johnston J, Kassavetis GA, Zeitlinger J, Kadonaga JT. TRF2, but not TBP, mediates the transcription of ribosomal protein genes. Genes Dev. 2014 Jul 15;28(14):1550–1555. doi: 10.1101/gad.245662.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Decker KB, Hinton DM. Transcription regulation at the core: similarities among bacterial, archaeal, and eukaryotic RNA polymerases. Annu Rev Microbiol. 2013;67:113–139. doi: 10.1146/annurev-micro-092412-155756. [DOI] [PubMed] [Google Scholar]
- 22.Barbash ZS, Weissman JD, Campbell JA, Jr, Mu J, Singer DS. Major histocompatibility complex class I core promoter elements are not essential for transcription in vivo. Mol Cell Biol. 2013 Nov;33(22):4395–4407. doi: 10.1128/MCB.00553-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Novina CD, Roy AL. Core promoters and transcriptional control. Trends Genet. 1996 Sep;12(9):351–355. [PubMed] [Google Scholar]
- 24.Haberle V, Li N, Hadzhiev Y, Plessy C, Previti C, Nepal C, Gehrig J, Dong X, Akalin A, Suzuki AM, van IJcken WF, Armant O, Ferg M, Strähle U, Carninci P, Müller F, Lenhard B. Two independent transcription initiation codes overlap on vertebrate core promoters. Nature. 2014 Mar 20;507(7492):381–385. doi: 10.1038/nature12974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sims RJ3rd, Belotserkovskaya R, Reinberg D. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 2004 Oct 15;18(20):2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- 26.Müller F, Tora L. Chromatin and DNA sequences in defining promoters for transcription initiation. Biochim Biophys Acta. 2014 Mar;1839(3):118–128. doi: 10.1016/j.bbagrm.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 27.Howcroft TK, Raval A, Weissman JD, Gegonne A, Singer DS. Distinct transcriptional pathways regulate basal and activated major histocompatibility complex class I expression. Mol Cell Biol. 2003 May;23(10):3377–3391. doi: 10.1128/MCB.23.10.3377-3391.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guglielmi B, La Rochelle N, Tjian R. Gene-specific transcriptional mechanisms at the histone gene cluster revealed by single-cell imaging. Mol Cell. 2013 Aug 22;51(4):480–492. doi: 10.1016/j.molcel.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deato MD, Tjian R. Switching of the core transcription machinery during myogenesis. Genes Dev. 2007 Sep 1;21(17):2137–2149. doi: 10.1101/gad.1583407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malik S, Roeder RG. Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends Biochem Sci. 2005;30:256–263. doi: 10.1016/j.tibs.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 31.Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, Markenscoff-Papadimitriou E, Kuhl D, Bito H, Worley PF, Kreiman G, Greenberg ME. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010 May 13;465(7295):182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, Muller H, Ragoussis J, Wei CL, Natoli G. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 2010 May 11;8(5):e1000384. doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Core LJ, Martins AL, Danko CG, Waters CT, Siepel A, Lis JT. Analysis of nascent RNA identifies a unified architecture of initiation regions at mammalian promoters and enhancers. Nat Genet. 2014 Dec;46(12):1311–1320. doi: 10.1038/ng.3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andersson R, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014 Mar 27;507(7493):455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu X, Sharp PA. Divergent transcription: a driving force for new gene origination? Cell. 2013 Nov 21;155(5):990–996. doi: 10.1016/j.cell.2013.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Valen E, Sandelin A. Genomic and chromatin signals underlying transcription start-site selection. Trends Genet. 2011 Nov;27(11):475–485. doi: 10.1016/j.tig.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 37.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Min IM, Waterfall JJ, Core LJ, Munroe RJ, Schimenti J, Lis JT. Regulating RNA polymerase pausing and transcription elongation in embryonic stem cells. Genes Dev. 2011;25:742–754. doi: 10.1101/gad.2005511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kimura H, Tao Y, Roeder RG, Cook PR. Quantitation of RNA polymerase II and its transcription factors in an HeLa cell: little soluble holoenzyme but significant amounts of polymerases attached to the nuclear substructure. Mol Cell Biol. 1999 Aug;19(8):5383–5392. doi: 10.1128/mcb.19.8.5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Voigt P, Tee WW, Reinberg D. A double take on bivalent promoters. Genes Dev. 2013 Jun 15;27(12):1318–1338. doi: 10.1101/gad.219626.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shilatifard A. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem. 2012;81:65–95. doi: 10.1146/annurev-biochem-051710-134100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009 Jan 23;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ozsolak F, Poling LL, Wang Z, Liu H, Liu XS, Roeder RG, Zhang X, Song JS, Fisher DE. Chromatin structure analyses identify miRNA promoters. Genes Dev. 2008 Nov 15;22(22):3172–3183. doi: 10.1101/gad.1706508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taft RJ, Glazov EA, Cloonan N, Simons C, Stephen S, Faulkner GJ, Lassmann T, Forrest AR, Grimmond SM, Schroder K, Irvine K, Arakawa T, Nakamura M, Kubosaki A, Hayashida K, Kawazu C, Murata M, Nishiyori H, Fukuda S, Kawai J, Daub CO, Hume DA, Suzuki H, Orlando V, Carninci P, Hayashizaki Y, Mattick JS. Tiny RNAs associated with transcription start sites in animals. Nat Genet. 2009 May;41(5):572–578. doi: 10.1038/ng.312. [DOI] [PubMed] [Google Scholar]
- 45.Pal M, McKean D, Luse DS. Promoter clearance by RNA polymerase II is an extended, multistep process strongly affected by sequence. Mol Cell Biol. 2001 Sep;21(17):5815–5825. doi: 10.1128/MCB.21.17.5815-5825.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zamudio JR, Kelly TJ, Sharp PA. Argonaute-bound small RNAs from promoter-proximal RNA polymerase II. Cell. 2014 Feb 27;156(5):920–934. doi: 10.1016/j.cell.2014.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sapetschnig A, Miska EA. Getting a grip on piRNA cluster transcription. Cell. 2014 Jun 5;157(6):1253–1254. doi: 10.1016/j.cell.2014.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mohn F, Sienski G, Handler D, Brennecke J. The rhino-deadlock-cutoff complex licenses noncanonical transcription of dual-strand piRNA clusters in Drosophila. Cell. 2014 Jun 5;157(6):1364–1379. doi: 10.1016/j.cell.2014.04.031. [DOI] [PubMed] [Google Scholar]
- 49.Zhang Z, Wang J, Schultz N, Zhang F, Parhad SS, Tu S, Vreven T, Zamore PD, Weng Z, Theurkauf WE. The HP1 homolog rhino anchors a nuclear complex that suppresses piRNA precursor splicing. Cell. 2014 Jun 5;157(6):1353–1363. doi: 10.1016/j.cell.2014.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Han BW, Zamore PD. piRNAs. Curr Biol. 2014 Aug 18;24(16):R730–R733. doi: 10.1016/j.cub.2014.07.037. [DOI] [PubMed] [Google Scholar]
- 51.Sadeh R, Allis CD. Genome-wide "re"-modeling of nucleosome positions. Cell. 2011 Oct 14;147(2):263–266. doi: 10.1016/j.cell.2011.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Struhl K, Segal E. Determinants of nucleosome positioning. Nat Struct Mol Biol. 2013 Mar;20(3):267–273. doi: 10.1038/nsmb.2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Segal E, Widom J. What controls nucleosome positions? Trends Genet. 2009 Aug;25(8):335–343. doi: 10.1016/j.tig.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol. 2006 Dec;13(12):1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 55.Dieci G, Fiorino G, Castelnuovo M, Teichmann M, Pagano A. The expanding RNA polymerase III transcriptome. Trends Genet. 2007 Dec;23(12):614–622. doi: 10.1016/j.tig.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 56.Barski A, Chepelev I, Liko D, Cuddapah S, Fleming AB, Birch J, Cui K, White RJ, Zhao K. Pol II and its associated epigenetic marks are present at Pol III-transcribed noncoding RNA genes. Nat Struct Mol Biol. 2010 May;17(5):629–634. doi: 10.1038/nsmb.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oler AJ, Alla RK, Roberts DN, Wong A, Hollenhorst PC, Chandler KJ, Cassiday PA, Nelson CA, Hagedorn CH, Graves BJ, Cairns BR. Human RNA polymerase III transcriptomes and relationships to Pol II promoter chromatin and enhancer-binding factors. Nat Struct Mol Biol. 2010 May;17(5):620–628. doi: 10.1038/nsmb.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011 May 15;25(10):1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ramirez-Carrozzi VR, Braas D, Bhatt DM, Cheng CS, Hong C, Doty KR, Black JC, Hoffmann A, Carey M, Smale ST. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell. 2009 Jul 10;138(1):114–128. doi: 10.1016/j.cell.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Koch F, Fenouil R, Gut M, Cauchy P, Albert TK, Zacarias-Cabeza J, Spicuglia S, de la Chapelle AL, Heidemann M, Hintermair C, Eick D, Gut I, Ferrier P, Andrau JC. Transcription initiation platforms and GTF recruitment at tissue-specific enhancers and promoters. Nat Struct Mol Biol. 2011 Jul 17;18(8):956–963. doi: 10.1038/nsmb.2085. [DOI] [PubMed] [Google Scholar]
- 61.Lee MP, Howcroft K, Kotekar A, Yang HH, Buetow KH, Singer DS. ATG deserts define a novel core promoter subclass. Genome Res. 2005 Sep;15(9):1189–1197. doi: 10.1101/gr.3873705. [DOI] [PMC free article] [PubMed] [Google Scholar]