Abstract
In the latent infection of Kaposi's sarcoma-associated herpesvirus (KSHV), its 160-kb circularized episomal DNA is replicated and maintained in the host nucleus. KSHV latency-associated nuclear antigen (LANA) is a key factor for maintaining viral latency. LANA binds to the terminal repeat (TR) DNA of the viral genome, leading to its localization to specific dot structures in the nucleus. In such an infected cell, the expression of the viral genes is restricted by a mechanism that is still unclear. Here, we found that LANA interacts with SUV39H1 histone methyltransferase, a key component of heterochromatin formation, as determined by use of a DNA pull-down assay with a biotinylated DNA fragment that contained a LANA-specific binding sequence and a maltose-binding protein pull-down assay. The diffuse localization of LANA on the chromosomes of uninfected cells changed to a punctate one with the introduction of a bacterial artificial chromosome containing most of the TR region, and SUV39H1 clearly colocalized with the LANA-associated dots. Thus, the LANA foci in KSHV-infected cells seemed to include SUV39H1 as well as heterochromatin protein 1. Furthermore, a chromatin immunoprecipitation assay revealed that the TR and the open reading frame (ORF) K1 and ORF50/RTA genes, but not the ORF73/LANA gene, lay within the heterochromatin during KSHV latency. Taken together, these observations indicate that LANA recruits heterochromatin components to the viral genome, which may lead to the establishment of viral latency and govern the transcription program.
Kaposi's sarcoma-associated herpesvirus (KSHV), also called human herpesvirus 8, was discovered in Kaposi's sarcoma (KS) lesions (8) and is strongly associated with multicentric Castleman's disease and primary effusion lymphoma (PEL), which are found predominantly in AIDS patients (6). KSHV belongs to the gammaherpesviruses, which have two distinct replication systems, lytic and latent. For lytic replication, the immediate-early gene product RTA (31), which functions as a strong transactivator that upregulates many viral genes, must be expressed. In PEL cells, KSHV is mainly in the latent state, during which RTA is not expressed (18). Thus, it is likely that KSHV has regulatory machinery that enables it to maintain the latent phase as a default state.
The latency-associated nuclear antigen (LANA) is the 1,162-amino-acid (aa) product of the KSHV open reading frame 73 (ORF73) gene, which is expressed during latency (20, 37). LANA has an acidic amino acid repeat region in the middle and a DNA-binding/dimerization domain at its C terminus (42). During latency, LANA binds a DNA sequence in the terminal repeat (TR) region of the viral genome, which consists of repeated sequences made up of 801-bp units and contains an origin of replication (OriP) (2, 3, 13). Most cell lines derived from PELs carry a viral genome with a long TR sequence of more than 20 kb (17). Given LANA's functional similarity to EBNA1, which is encoded by another gammaherpesvirus, Epstein-Barr virus, it is likely to have a key role in the replication of the KSHV genome. Thus, LANA and its binding to the TR are very important for the virus to maintain its latency in infected cells.
The pattern of cellular gene expression that defines cell fate is due to an epigenetic effect(s) of the chromatin structure (27). In PEL cells, although a few lytic genes are expressed spontaneously, most viral genes are silenced (38) via a mechanism that is still unclear. One clue is that LANA forms specific dot structures that are associated with the KSHV genome in the host heterochromatin region, where most of the genes are inactive (43).
Heterochromatin protein 1 (HP-1) binds to a methylated lysine residue at the tail of histone H3, and its homodimerization contributes to the formation of heterochromatin (4, 22). A human homologue of Drosophila su(var)3-9, named SUV39H1, was found to be a histone H3-specific methyltransferase. A protein-protein interaction between HP-1 and SUV39H1 results in expansion of the heterochromatin region (1).
In this study, we sought to determine how the viral transcription program is regulated in latency. To do this, we focused on the interaction between LANA and SUV39H1, a key factor that methylates histone H3, which recruits HP-1 to initiate the formation of heterochromatin. A maltose-binding protein (MBP) pull-down assay and a DNA pull-down assay revealed an interaction between LANA and SUV39H1, and when a bacterial artificial chromosome (BAC) of the TR was introduced into LANA-expressing 293 and Vero cells, LANA changed from being diffusely located in the chromatin region to forming dot structures in the heterochromatin that colocalized with SUV39H1 and HP-1α. A transient reporter assay with a TR-containing reporter indicated a repressive effect by LANA that was enhanced by SUV39H1. Analysis of the chromatin in the TR in a cross-linked chromatin immunoprecipitation (ChIP) assay revealed that HP-1, methylated histone H3, and LANA had accumulated on the TR. The same analysis for the ORF K1 gene, which lies close to the TR (at the left end of the KSHV genome) and encodes a membrane protein containing an immunoreceptor tyrosine-based activation motif to switch signaling, found that it was associated with the anti-HP-1 antibody and the anti-methylated histone H3 antibody in the ChIP assay. The switch from KSHV latency to lytic reactivation is regulated by the ORF50 gene product, RTA, whose DNA was also precipitated by the anti-HP-1 antibody. In contrast, the active gene locus of the latent transcript that contains LANA, v-CYC, and v-FLIP was not precipitated. Thus, these results were consistent with the expression profile of each gene during latency. Taken together, these data indicated that the interaction of the viral genome with heterochromatin was mediated by LANA along with heterochromatin factors to regulate the KSHV latent transcription program.
MATERIALS AND METHODS
Cells.
Human embryonic kidney cell line 293 was cultured in Dulbecco's modified Eagle's medium (DMEM) (Nissui, Tokyo, Japan) with 10% heat-inactivated fetal bovine serum. 293LANA cells, which were obtained by introducing the retroviral vector pHyTc-LANA into 293 cells and in which LANA was constitutively expressed, were cultured under the same conditions, except with the addition of 0.2 mg of hygromycin B (Nacalai Tesque, Kyoto, Japan) per ml. Several clones carrying ORF73/LANA cDNA and the hygromycin resistance gene were established as stable LANA-expressing cell lines. By using the same procedures, Vero cells expressing GFP-LANA (Vero/gfpLANA) were also created and cultured in DMEM containing 0.2 mg of hygromycin B per ml.
A KSHV-infected PEL cell line, BC3, was cultured in RPMI 1640 (Nissui) with 20% FBS. All cells were maintained in a 5% CO2 atmosphere.
Plasmids.
The mammalian expression vector for KSHV ORF73/LANA, pTriEX-LANA, was constructed by using the following procedure. The coding region of ORF73 was obtained by digesting phage L54 (AIDS Research and Reference Regent Program, National Institutes of Health [NIH], Rockville, Md.) with SacI. This fragment was inserted at the SacI site of pTriEX1.1 (Novagen, Madison, Wis.), resulting in pTriEX-LANA. To add the V5 epitope and histidine hexamer tag at the C terminus, the termination codon of LANA was modified and linked to the tag sequence from the pMT/V5-His A vector (Invitrogen, Carlsbad, Calif.). Truncated LANA mutants, LNΔN (aa 497 to 1162) and D1 (aa 922 to 1162), were created by PCR. PstI digestion of pTriEX-LANA (full-length) deleted the region from aa 468 to the end, and then the construct was self-ligated to generate pTriEX-LANA-N1, which encoded the LANA N terminus (aa 1 to 467) linked to the herpes simplex virus tag and the histidine hexamer of the pTriEX vector. The shorter fragment was constructed as an enhanced green fluorescent protein (EGFP) fusion gene by using pEGFP (Clontech, Palo Alto, Calif.). The polypeptides encoded by pEGFP-N and pEGFP-DE represented aa 1 to 274 and aa 275 to 467, respectively.
For the reporter plasmids, cosmid clone Z6 (NIH AIDS Research and Reference Reagent Program) was modified. Digestion of Z6 with BglII removed an approximately 32-kbp fragment from the KSHV long unique region to insert a reporter gene cassette [KSHV RTA promoter-luciferase-simian virus 40 (SV40) poly(A) signal] from the pGL3-RTA promoter-FL (40) (pTR-RpLuc-009). The digestion of pTR-RpLuc-009 with NotI deleted most of the TRs (pTR-RpLuc-903), and the resultant pTR-RpLuc-903 contained only one TR unit adjacent to the left end of the KSHV long unique region.
HP-1α (National Center for Biotechnology Information [NCBI] accession no. NM012117) was cloned by the reverse transcription-PCR technique. Total RNA was obtained from cultured 293 cells. For the reverse transcription, SuperScript II reverse transcriptase (Invitrogen) was used with random primers, according to the manufacturer's protocol. Ten percent of the cDNA product was subjected to PCR with the Expand high-fidelity PCR system (Roche Diagnostics, Mannheim, Germany) with the specific primers pHP1alpha-S-EcoRI and pHP1alpha-AS-EcoRI, whose sequences are described below. The human homolog of Drosophila Su(var)3-9, SUV39H1 (NCIB accession no. AF019968), and its mutant were cloned into pFLAGCMV5c (Sigma-Aldrich, St. Louis, Mo.) and the bacterial expression vector pMal-c2 (New England Biolabs Inc., Beverly, Mass.). Truncated SUV39H1 mutants tagged with MBP were created by PCR, digested with the appropriate restriction enzymes, and inserted into the pMal-c2 vector (39-ΔN, aa113 to 412; 39-SET, aa 250 to 412; 39-Chromo, aa 1 to 112). The primers used for plasmid construction were as follows: pHP1alpha-S-EcoRI, 5′-CGCGAATTCCATGGGAAAGAAAACCAAGCG-3′; pHP1alpha-AS-EcoRI, 5′-CGCGAATTCCTTTAGCTCTTTGCTGTTTC-3′; pSUV39-S-BamH1, 5′-CGCGGATCCAAGATGGCGGAAAATTTAAAAGGCTG-3′; pSUV39-AS-BamHI, 5′-GGCGGATCCAGAAGAGGTATTTGCGGCAGG-3′; and pSUV39-750-S-Bam, 5′-GCAGGATCCACGGATGATGGGCGTGGC-3′.
All fragments cloned by RT-PCR were confirmed by sequencing analysis with an ABI PRISM 310 Genetic Analyzer (PE Applied Biosystems, Foster City, Calif.).
BAC-TR construction and transfection.
The full-length TR obtained from BC-3 cells was inserted into the BAC vector pBeloBAC11 (Research Genetics, Inc., Huntsville, Ala.), described below. To add a drug selection marker for mammalian cells, the SV40 promoter-driven neomycin resistance gene of pEGFP-N1 (Clontech), from which a multicloning site (MCS) had been deleted by digestion with BglII and BamHI followed by religation, was inserted into the modified SphI site of pBeloBAC11. Extrachromosomal DNA was obtained from BC-3 cells by the Hirt extraction method (16). Following BamHI digestion, the resulting fragments were inserted into pBeloBAC11 (at the BamHI site) and isolated by colony hybridization. Thus, the BAC clone containing the KSHV fragment from nucleotide (nt) 134729 to 1883 was established and named BAC-TR. During the cloning process, the BAC construct lost a large portion of the GFP gene for an unknown reason, but the Neor gene was expressed in 293 cells. The KSHV fragment in BAC-TR was deleted with BamHI to generate the control vector BAC-neor. The persistence of the episomal form of this artificial and partial KSHV DNA in the 293LANA cells was confirmed by Southern blotting analysis with DpnI digestion, in which newly replicated DNA is not cut (data not shown). BAC-TR was stably maintained in 293LANA cells for more than 4 weeks under G418 selection. Vero/gfpLANA cells carrying either BAC-TR or BAC-neor were also established by using the same procedures.
IFA.
Cells were washed with phosphate-buffered saline (PBS) (140 mM NaCl, 2.7 mM KCl, 6.5 mM Na2HPO4, 1.5 mM KH2PO4), spotted on a 24-well glass slide, and then fixed with 2% paraformaldehyde-PBS for 30 min. After fixation, the cells were washed and incubated with 0.5% Triton X-100-PBS for 15 min. The plate was washed with PBS three times. A mouse monoclonal antibody against HP-1α was purchased (Euromedex, Mundolsheim, France). It was diluted 1:200 for the indirect immunofluorescence assay (IFA). An anti-LANA antiserum from an AIDS-KS patient was used at a 1:100 dilution with PBS containing 0.1% bovine serum albumin, 0.2% Tween 20, and 0.05% sodium azide. By fluorescence microscopic analysis, we did not observe any nonspecific reaction of this antiserum with cell lines that were not infected with KSHV (data not shown). As secondary antibodies, we used fluorescein isothiocyanate-conjugated rabbit anti-human immunoglobulin antibodies (Dako, Copenhagen, Denmark) and Alexa 546-conjugated goat anti-mouse immunoglobulin antibodies (Molecular Probes, Eugene, Oreg.). For the transient transfection of cells with pFLAG-SUV39H1, cells were grown on an eight-chamber culture slide (Becton Dickinson Labware, Bedford, Mass.) and incubated with the Superfect reagent (Qiagen, Hilden, Germany) for 2 h. After 24 h in culture, the cells were fixed with acetone-methanol (1:1) and reacted with suitable antibodies for each experiment. For the double staining of LANA and SUV39H1, the combination of rabbit anti-LANA polyclonal antibodies, which were a kind gift from T. Sata (National Institute of Infectious Diseases, Tokyo, Japan) (19), and a mouse anti-FLAG antibody (Sigma) was used. The fluorescent signal was observed with an LSM 510 laser confocal microscope (Carl Zeiss Microimaging Inc., Thornwood, N.Y.).
Transfection.
All transfection experiments were performed with the Superfect transfection reagent (Qiagen) according to the manufacturer's protocol. For the luciferase assay, transfection was performed in a 12-well plate. One day before the transfection, 293 cells (10,000 cells/well) were seeded onto the plate. The total amount of transfected plasmid was kept constant by adding empty plasmid. After incubation with the plasmid-reagent solution for 2 h, the cells were cultured in complete DMEM for 36 h and then assayed as described below.
Luciferase assay.
A transient reporter assay was performed with a luciferase kit (Promega). Transfected cells were harvested 36 h after transfection. The cells were washed with PBS and lysed with lysis buffer (Promega). After centrifugation at 13,000 × g for 3 min at 4°C, 10 μl of the supernatant was reacted with 50 μl of the luciferase substrate (Promega). The luciferase activity was measured immediately with a Lumat LB 9501 luminometer (EG & G Berthold, Wildbad, Germany). The transfection efficiency of each well could not be normalized by using a reference plasmid such as a β-galactosidase expression vector, because LANA influenced the promoters of such vectors, including the cytomegalovirus (CMV) immediate-early promoter, the SV40 promoter, and the Rous sarcoma virus promoter (data not shown). To overcome this issue, the experiment was performed in triplicate and at least three independent assays were performed. The protein concentration in the lysate was measured by using the Bio-Rad (Hercules, Calif.) protein assay kit to normalize the effects on cell viability.
Production of recombinant protein in E. coli.
The pMal-c2-SUV39H1 plasmid was introduced into Escherichia coli Origami B DE3 pLysS (Novagen). Expression of the MBP fusion proteins was induced for 3 h at 30°C with 1 mM isopropyl-β-d-thiogalactopyranoside (Nacalai Tesque). The cells were pelleted, resuspended in PBS, containing 2 mM phenylmethylsulfonyl fluoride (Nacalai Tesque), and sonicated. The lysate was cleared by centrifugation and then loaded onto a column filled with amylose resin (New England Biolabs Inc.). The bound protein was eluted with PBS containing 10 mM maltose and then concentrated with a Centricon-30 (Millipore, Bedford, Mass.). The protein concentration was measured with the Bio-Rad protein assay kit and verified by staining sodium dodecyl sulfate (SDS)-polyacrylamide gels with Coomassie blue. The other MBP-tagged proteins were purified by using the same procedures.
Pull-down assay with bacterial recombinant proteins.
The nuclear extract (NE) of infected cells was prepared as described elsewhere (44). Briefly, the cells (BC-3) were harvested by centrifugation, washed twice with PBS, resuspended in NE A buffer (10 mM HEPES [pH 7.9], 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol [DTT], and protease inhibitor cocktail [Sigma-Aldrich]), and allowed to incubate on ice for 10 min. After centrifugation (1,000 × g, 5 min, 4°C), the cells were lysed in NE A buffer plus 0.05% NP-40 and homogenized in a Dounce homogenizer (20 strokes) to isolate the nuclei. The nuclei were collected and resuspended in NE C buffer (5 mM HEPES [pH 7.9], 1.5 mM MgCl2, 0.1 mM EDTA, 300 mM NaCl, 20% [vol/vol] glycerol, 0.5 mM DTT, and proteinase inhibitor cocktail [Sigma-Aldrich]). The solution was rocked at 4°C for 30 min and spun at 15,000 × g for 10 min. The clear lysate was stored as NE at −80°C until use. The NE was diluted with dilution buffer (5 mM HEPES [pH 7.9], 1.5 mM MgCl2, 0.1 mM EDTA, 0.1% NP-40) to adjust the NaCl concentration to 150 mM. Before binding with the recombinant protein of interest, the NE was precleared with amylose resin (New England Biolabs Inc.) to minimize nonspecific interactions with the beads. After preincubation for 1 h at 4°C, the supernatant was subjected to a pull-down assay with fresh beads and each recombinant protein (MBP-SUV39H1 and the control MBP-LacZ α domain). After an overnight incubation at 4°C, the beads were washed five times with washing buffer (5 mM HEPES [pH 7.9], 150 mM NaCl, 1.5 mM MgCl2, 0.1 mM EDTA, 0.1% NP-40). The washed matrix was then resuspended in SDS-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer for Western blotting.
Proteins were subjected to Western blotting with an anti-LANA antibody (Advanced Biotechnologies Inc., Columbia, Md.), an anti-V5 antibody (Invitrogen), an anti-GFP antibody (Medical and Biological Laboratory, Nagoya, Japan), and an anti-MBP antibody (New England Biolabs Inc.).
Biotinylated DNA pull-down assay.
Four copies of the LANA-binding sequence (LBS) (see Fig. 3A) were tandemly arranged and inserted into the XbaI site of the pBlueScript-SKII(−) vector (Stratagene, La Jolla, Calif.), to generate pBS-4XLBS. Biotinylated primers (T7 and T3) were used in a PCR with pBS-4XLBS or empty vector (pBlueScript). The amplified DNA fragments (270 and 166 bp) were purified with a Wizard PCR cleanup system (Promega). Each 5 pmol of biotinylated DNA was incubated with NE from 293 cells that were transfected with pFLAG-SUV39H1 and TriEXLANA (wild type) or pTriEXLANA ΔN (LNΔN). The binding reaction was carried out in a 1.5-ml siliconized tube (Sarstedt, Numbrecht, Germany) with NE C buffer containing 100 mM NaCl, 0.05 mg of poly(dI-dC) (Amersham Bioscience) per ml, 0.2 mg of sheared salmon sperm DNA (Nacalai) per ml, and 0.1 mg of bovine serum albumin (Nacalai) per ml. To precipitate DNA-binding proteins, 10 μl of streptavidin-Sepharose (Amersham Biosciences, Uppsala, Sweden), which was preincubated with the binding buffer to eliminate nonspecific binding, was added to the mixture. After an overnight incubation with gentle rotation at 4°C, the precipitated matrix was washed five times with NE C buffer containing 100 mM NaCl and 0.2 mg of sheared salmon sperm DNA per ml. The washed matrix was boiled in SDS-PAGE sample buffer for Western analysis with a mouse anti-V5 monoclonal antibody (Invitrogen) and a mouse anti-FLAG monoclonal antibody (Sigma-Aldrich).
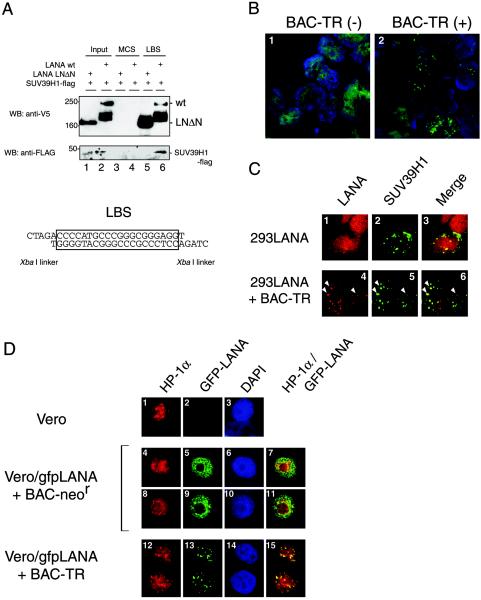
FIG. 3.
Triangular interaction among LANA, TR DNA, and SUV39H1. (A) Biotinylated-DNA pull-down assay. 293 cells were transfected with expression vectors for LANA (wild type [wt]) or its mutant (LNΔN), which lacked the SUV39H1-interacting site, and pCMV5-flagSUV39H1. Biotinylated DNA of either the LBS or the MCS of pBS-SKII (Stratagene) as a control DNA fragment was incubated with NE from the transfectants and bound to streptavidin-Sepharose (Amersham Bioscience). Precipitates were analyzed on Western blots (WB) probed with a mouse anti-V5 monoclonal antibody and a mouse anti-FLAG monoclonal antibody. The DNA sequence of the LBS is shown. (B) IFA of 293LANA cells with or without the BAC-TR construct. The focal concentration of LANA was detected in 293LANA cells carrying BAC-TR. (C) Colocalization of LANA with SUV39H1 in 293 cells bearing the artificial KSHV DNA (BAC-TR). 293LANA cells were transiently transfected with pCMV5-FLAG-SUV39H1 with (panels 1 to 3) or without (panels 4 to 6) BAC-TR. (D) Association between LANA and heterochromatin in Vero cells stably expressing GFP-LANA with BAC-TR or the control BAC construct (BAC-neor). Parental Vero cell (panels 1 to 3), Vero/gfpLANA cells with the empty BAC (panels 4 to 11), and Vero/gfpLANA cells with BAC-TR (panels 12 to 15) were stained with an anti-HP-1α antibody (panels 1, 4, 8, and 12). The IFA was performed as described in Materials and Methods.
Generation of a monoclonal antibody against LANA.
Procedures to establish hybridoma cell lines were the same as previously described (33). Briefly, the purified GST-LANA (aa 1 to 270) was administered to female BALB/c mice with Freund's complete adjuvant. The antibody titer was boosted 3 weeks later with Freund's incomplete adjuvant and reboosted with the same antigen without adjuvant 1 month after the second injection. The spleen was removed 3 days after the last booster injection. The splenic cells were fused with the SP2 cells by use of polyethylene glycol and cultured in 96-well microplates with hypoxanthine-aminopterin-thymidine medium to screen for the secretion of antibody. A clone, A23-9, was obtained by this procedure, and the antibody from this hybridoma was used for the Western blotting shown in Fig. 2B and for the IFA shown in Fig. 4A.
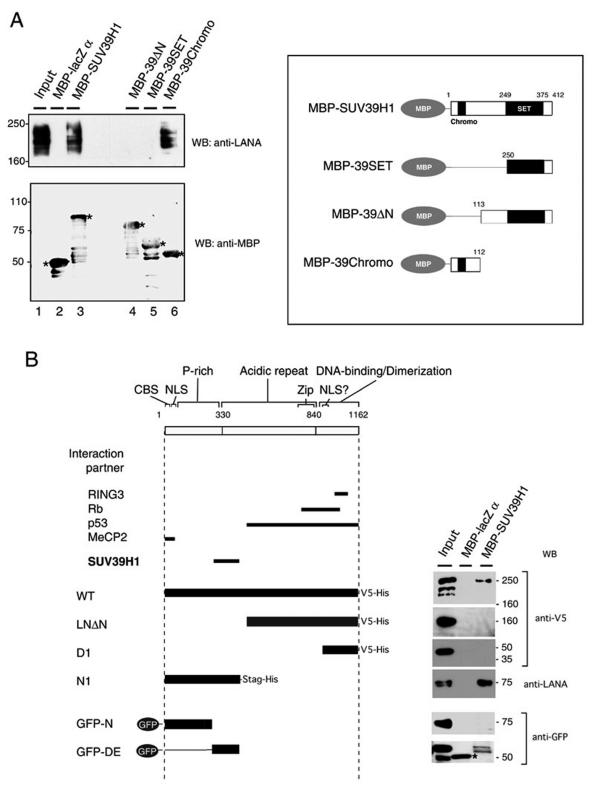
FIG. 2.
LANA interacts with SUV39H1. (A) Pull-down assay with recombinant MBP-SUV39H1 and its mutants. The constructs are shown on the right. NE from BC3 cells was incubated with 4 μg of MBP-LacZ (lane 2) and with MBP-fused SUV39H1 (lane 3) and its mutants (39-ΔN [aa 113 to 412] [lane 4], 39-SET [aa 250 to 412] [lane 5], and 39-Chromo [aa 1 to 112] [lane 6]). Each precipitate was probed with an anti-LANA antibody (Advanced Biotechnologies Inc.) and an anti-MBP antibody (New England Biolabs Inc.). Asterisks indicate the MBP fusion proteins. WB, Western blotting. (B) Mapping the site of LANA that was required for the interaction with SUV39H1. Truncated LANA mutants (left panel) were expressed in 293 cells and subjected to the pull-down assay described for panel A (1 μg of recombinant proteins was used for each experiment). Each precipitate was probed with an anti-V5 antibody (Invitrogen) to detect wild-type protein (WT), LNΔN, and D1; with a mouse anti-LANA antibody (generated in our laboratory) to detect LANA; and with rabbit polyclonal anti-GFP antibodies (Medical and Biological Laboratory) to detect GFP-N1 and GFP-NE. The asterisk shows a nonspecific band seen in the MBP-LacZ α precipitation.
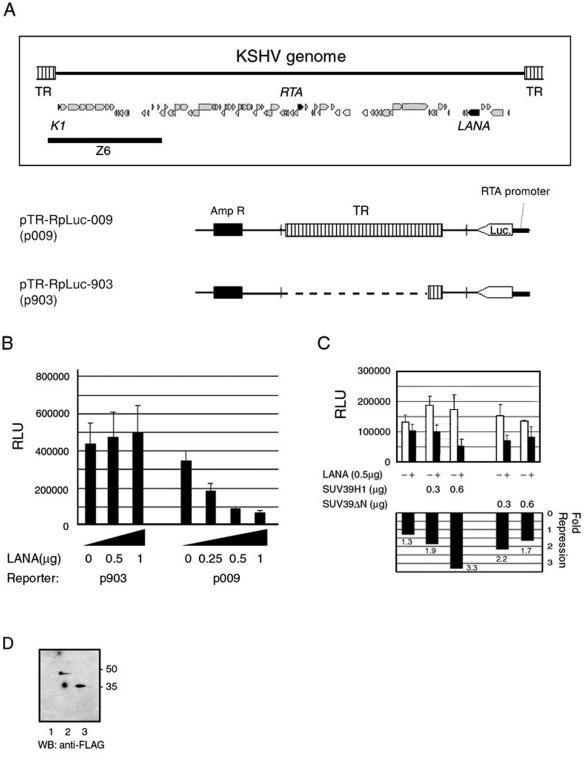
FIG. 4.
Repression of gene expression by LANA in a reporter plasmid harboring the TR. (A) Schematic representation of the reporter plasmids used in the transient-transfection assay and a representative structure of the KSHV genome and its genes (top). Part of the TR-containing fragment from cosmid Z6 (EcoRI-BglII) was linked to the RTA-promoter luciferase cassette. Z6, cloning region in cosmid Z6 (NIH AIDS Research and Reference Reagent Program); Amp R, ampicillin resistance gene; Luc, luciferase gene. (B) Transient assay for transcriptional regulation by LANA of a TR-bearing plasmid. The transfection and luciferase assay were performed as described in Materials and Methods. 293 cells were transfected with the effector plasmid (total of 2 μg/well in a 12-well plate) and the reporter plasmid (0.2 μg/well). (C) SUV39H1 enhances the repression by LANA. 293 cells were cotransfected transiently with pTriEX-LANA with pFLAG-SUV39H1 (0.3 and 0.6 μg/well) or pFLAG-SUV39H1ΔN (SUV39ΔN) (0.3 and 0.6 μg/well). The luciferase activities were normalized to the protein concentration and are shown in the upper graph, in which the open bars indicate empty vector (pTriEX1.1) (0.5 μg/well) and the closed bars indicate pTriEX-LANA (0.5 μg/well). Transfection efficiency could not be normalized to a reference plasmid such as a β-galactosidase expression vector due to LANA's modulation of some promoters. The fold repression by LANA was calculated as follows and is shown in the lower graph: [(pTriEXLANA + X)/(pTriEX + X)]−1, where the designations in parentheses show the luciferase units (relative light units [RLU]) for each combination and X is pFLAGCMV1, pFLAG-SUV39H1, or pFLAG-SUV39H1ΔN (SUV39ΔN). The assays shown in panels B and C were performed in triplicate for each set of transfections, and the mean value and standard deviation were calculated. (D) Western blotting (WB) of 293 cells transfected with SUV39H1 and its mutant expression vector. The expression of each construct (0.6 μg/well) transfected with pFLAGCMV1 (lane 1), pFLAG-SUV39H1 (lane 2), and pFLAG-SUV39ΔN (lane 3) in 293 cells was analyzed by Western blotting with an anti-FLAG antibody (Sigma Aldrich).
ChIP.
Procedures described elsewhere (23) were modified. BC-3 cells (108 cells for Western blotting and 107 cells for PCR to analyze the precipitants) were washed with PBS and cross-linked in vivo in PBS containing 1% formaldehyde at 37°C for 10 min. The cross-linked cells were washed in ice-cold PBS twice and resuspended in buffer A (10 mM Tris-HCl [pH 7.5], 10 mM NaCl, 3 mM MgCl2, 0.2% Triton X-100, 1 mM DTT, 0.5 mM EDTA, 0.2 mM phenylmethylsulfonyl fluoride, and proteinase inhibitor cocktail [Sigma-Aldrich]), and the nuclei were successfully released from the cells by homogenizing them in a Dounce homogenizer. The nuclei were pelleted by low-speed centrifugation and resuspended in buffer B (10 mM Tris-HCl [pH 7.5], 10 mM NaCl, 3 mM MgCl2, 1 mM CaCl2). To a 400-μl aliquot of the nucleus solution, 50 U of micrococcal endonuclease (Takara Shuzo, Kyoto, Japan) was added, and the reaction mixture was incubated at 37°C for 10 min. After the incubation, a 1/20 volume of 200 μM EGTA was added to terminate the reaction, and the solution was spun at 10,000 × g for 5 min at 4°C. The supernatant was collected, and the pellet was resuspended in 400 μl of LS buffer (10 mM Tris-HCl, 10 mM NaCl, 1 mM EDTA) and subjected to a second centrifugation. The supernatant was collected again and the pellet was resuspended in 200 μl of HS buffer (50 mM Tris-HCl [pH 7.6], 500 mM NaCl, 5 mM EDTA, 0.5% sodium deoxycholate, 1% NP-40, and proteinase inhibitor cocktail [Sigma-Aldrich]). The supernatants were combined and subjected to immunoprecipitation with 1 μg of a rat monoclonal anti-LANA antibody (Advanced Biotechnologies Inc.) and 10 μl of protein G-Sepharose (Amersham Biosciences), which was preincubated in a binding buffer containing 0.2 mg of salmon sperm DNA per ml and 0.05 mg of poly(dI-dC) per ml for 30 min, in a 1.5-ml siliconized tube (Sarstedt). After an overnight incubation at 4°C, the precipitated matrix was washed five times with radioimmunoprecipitation assay buffer (50 mM Tris-HCl [pH 7.6], 150 mM NaCl, 1 mM EDTA, 0.1% SDS, 0.5% deoxycholate, and 1% NP-40), four times with LiCl buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA, 0.1% SDS, 0.5% deoxycholate, 1% NP-40, and 150 mM LiCl), and once with TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA). Rat immunoglobulin (Dako) was used as a negative control.
Western blotting for HP-1 isoforms was performed with 10 to 20% gradient gels (Daiichi Pure Chemicals Co., Ltd., Tokyo, Japan) to separate HP-1 from the immunoglobulin light chain used for immunoprecipitation. The procedures for Western blotting are described above.
To detect the DNA fragment in the precipitates by PCR, TE with 1% SDS and 0.2 M NaCl was added to the washed matrix. After incubation at 65°C for 8 h, proteinase K (Roche Diagnostics) was added to the matrix suspension, and the suspension was incubated at 37°C overnight. The eluted solution was subjected to phenol-CHCl3 extraction and ethanol precipitation with yeast tRNA as the carrier. One-sixth of the purified precipitated DNA was subjected to PCR with the FailSafe PCR system (Epicentre, Madison, Wis.), with some optimization to amplify the TR region. One-third of the PCR products was analyzed by electrophoresis in a 2% agarose gel (Takara Shuzo). TR R3 and TR F4 generated a 200-bp fragment within the KSHV TR. The LANA/v-CYC/v-FLIP promoter region [LT(3)S and LT(3)AS for nt 127868 to 128106], the ORF73/LANA N-terminal region [ORF73chip(4)S and ORF73chip(4)AS for nt 126492 to 126720], the K1 locus (K1-3S and K1-3AS for nt 46 to 273), the RTA promoter region [Rpchip(1)S and Rpchip(1)AS for nt 71192 to 71425], and the first exon of ORF50/RTA [ORF50chip(2)S and ORF50chip(2)AS for nt 71456 to 71673] were amplified in the assay. The positions of the nucleotides were identified by using the KSHV genome sequence under NCBI accession no. U75698. The primers used in the reaction were TR R3 (5′-CCTGTCCCCGCGCGGGCCCG-3′), TR F4 (5′-GGCGCCCCCTTCCCTCGCTGC-3′), LT(3)S (5′-CGCAGCTGCCTCCAAATGATACAC-3′), LT(3)AS (5′-CGAGAGGCACTCGGCGTCGTCCAC-3′), ORF73chip(4)S (5′-CGTCTACCCTGCGTAGCCTG-3′), ORF73chip(4)AS (5′-CCAGGATCCCTCAGACGGGGATG-3′), K1-3S (5′-CTGGCGGCCCTTGTGTAAACC-3′), K1-3AS (5′-GCCGAGTATTGTTGCAATACC-3′), Rpchip(1)S (5′-GGGAGGCCAGCGTATTCAGGAC-3′), Rpchip(1)AS (5′-GTAGCTGGGTCCTATGGGGGTTGG-3′), ORF50chip(2)S (5′-TACCAGGCAGCTACCGGCGACTC-3′), and ORF50chip(2)AS (5′-CCCACCTACACCATTGTAAACACGC-3′).
Nucleotide sequence accession number.
The sequence of the 200-bp fragment within the KSHV TR generated by TR R3 and TR F4 was deposited in the NCBI under accession no. U75699.
RESULTS
LANA interacts with heterochromatin in latently infected PEL cells.
Transcription is often regulated by the nucleosome structure, which is composed of DNA and histone and nonhistone chromatin proteins. Given that there is less transcription during KSHV latency in infected cells, such as BC-3 cells, we tested whether the viral genome, which persists as a closed circular double-stranded DNA in the nucleus, exploits the host chromatin regulatory machinery to suppress viral transcription.
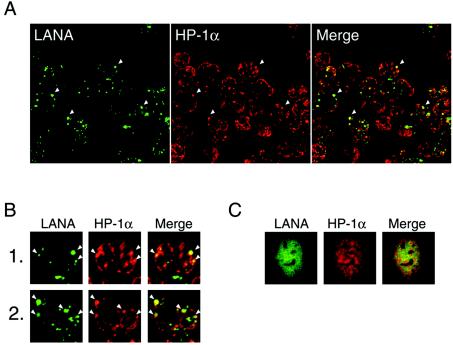
We first performed an IFA to verify whether one of the major components of heterochromatin, HP-1α, was involved in KSHV latency, because one report showed that it interacted with LANA (26) and another showed it did not (21). LANA can be used as a marker for the viral genome, because it binds to the TR (3, 13). The antibody staining LANA was highly colocalized with HP-1α in the infected cells (Fig. 1A and B). This suggests that heterochromatin factor HP-1 associates with the viral genome. On the other hand, in 293 cells coexpressing LANA and DsRed-HP-1α, LANA did not appear to localize to the focal and dense region of HP-1α (Fig. 1C), as reported elsewhere (21). These results indicated that LANA colocalized with HP-1 in the presence of the viral genome and suggested that the recruited HP-1 could be involved in the heterochromatin formation of the viral genome.
FIG. 1.
LANA colocalizes with HP-1 in BC-3 cells. (A and B) Paraformaldehyde-fixed BC-3 cells were reacted with a mouse monoclonal antibody against HP-1α (red) and serum from an AIDS-KS patient that was used to detect LANA (green). (B) A higher magnification of the image (×400). Arrowheads show representative colocalization of these proteins. (C) LANA does not colocalize with HP-1α in the absence of the KSHV genome. 293 cells in which LANA and DsRed-HP-1α were transiently coexpressed were fixed and reacted with a mouse monoclonal antibody against LANA (generated in our laboratory).
Interaction between LANA and SUV39H1 histone methyltransferase.
We next tested whether there was an association between LANA and SUV39H1, a histone H3-specific methyltransferase that is a key regulator of heterochromatin components. SUV39H1 methylates histone H3 on a key lysine residue (K9). HP-1 recognizes the methylated K9 and is recruited to histone H3, leading to heterochromatin formation (29). Although it has been recently reported that LANA interacts with HP-1α directly (26), it was important to verify whether the histone methylase itself was involved in the interaction and/or associated with LANA, because this step occurs prior to the recruitment of HP-1 and HP-1 did not appear to simply colocalize with LANA in the absence of the viral DNA (Fig. 1C).
To this end, we performed a pull-down assay with MBP-tagged recombinant SUV39H1 (Fig. 2A, lanes 1 to 3). We observed that MBP-SUV39H1 precipitated LANA from the BC-3 NE, whereas MBP-LacZ α domain (∼50 kDa), the negative control, did not.
To determine the SUV39H1 domain required for this interaction, several truncated forms of SUV39H1 (39-ΔN, 39-SET, and 39-Chromo) were generated. Of these, 39-Chromo (aa 1 to 112) was required for the interaction with LANA, whereas the C-terminal catalytic domain was not. (Fig. 2A, lanes 4 to 6).
To map the LANA domain that interacts with SUV39H1, several truncated mutants of LANA were constructed, as shown in Fig. 2B, and expressed in 293 cells. The mutants were then subjected to a pull-down assay with MBP-SUV39H1 or a control protein. An anti-V5 mouse monoclonal antibody was used to detect wild-type LANA, LNΔN (aa 497 to 1162), and D1 (aa 922 to 1162). A mouse anti-LANA monoclonal antibody (generated in our laboratory) was used to detect N1 (aa 1 to 467), and an anti-GFP rabbit polyclonal antibody was used to detect GFP-N (aa 1 to 274) and GFP-DE (aa 275 to 467). We observed that wild-type LANA conjugated with the V5His tag and the N-terminal domain N1 were pulled down by MBP-SUV39H1 but that LANA lacking the N terminus (LNΔN) and the DNA-binding domain (D1) were not (Fig. 2B). Under the same conditions, a GFP-tagged aspartic acid and glutamic acid repeat region of LANA (GFP-DE) was precipitated by MBP-SUV39H1, whereas only a nonspecific reaction with rabbit anti-GFP antibodies was observed in the precipitate with the MBP-LacZ α domain. The N-terminal peptide (GFP-N), which contained the chromosome-binding site, nuclear localization signal, and proline-rich domain (34), was not precipitated. Taking these findings together, we concluded that the DE repeat region (aa 275 to 467) of LANA was required to bind the SUV39H1 methyltransferase.
LANA recruits SUV39H1 to the viral genome.
For the reciprocal test for an interaction between LANA and SUV39H1 and to examine whether LANA binding to the LBS could recruit SUV39H1 to the TR, we performed a biotinylated-DNA pull-down assay. This experiment was also designed to show in vivo interactions between LANA and SUV39H1. We could not perform a direct immunoprecipitation followed by Western blotting analysis because there was no available antibody against SUV39H1 and because its apparent molecular mass on SDS-PAGE (∼50 kDa) overlapped with that of the immunoglobulin heavy chains.
Biotinylated-4XLBS was incubated with NE from 293 cells that had been transiently transfected with LANA-V5His and SUV39H1-flag. After the incubation, the protein-DNA complex was pulled down with streptavidin beads and subjected to SDS-PAGE. We found that the control DNA fragment [the MCS of the cloning plasmid pBlueScript-SKII(−) (Stratagene)] did not bind with either LANA or its mutant (LNΔN) (Fig. 3A, lanes 3 and 4) but that the LBS effectively precipitated these proteins (Fig. 3A, lanes 5 and 6). Under these conditions, the participation of SUV39H1-flag in the interaction between wild-type LANA and the DNA was observed by Western blotting with an anti-FLAG antibody (Fig. 3A, lane 6). Because LNΔN lacked the binding site for SUV39H1, as shown in Fig. 2B, SUV39H1-flag was not precipitated in this case (Fig. 3A, lane 5).
To observe this tertiary interaction in vivo, a BAC clone containing the whole TR region of the viral genome was transfected into 293LANA cells, which were then selected in G418-containing medium. The surviving cells were designated 293LANA/BAC-TR, and the LANA staining pattern changed from a diffuse to a dotted one (Fig. 3B). This was probably because LANA came to recognize the presence of BAC-TR (Fig. 3B, panel 2) (2). The presence of the stable episomal form of BAC-TR DNA was confirmed by Southern blotting a few weeks after the establishment of the cell clone (data not shown).
Using the 293LANA/BAC-TR and the 293LANA cells, we examined the localization of SUV39H1 by confocal fluorescence microscopy. In the absence of BAC-TR, LANA (red) was dispersed over the chromatin region, and the FLAG-tagged SUV39H1 (green) showed a punctate pattern, which overlapped LANA only because LANA was so broadly distributed (Fig. 3C, panels 1 to 3). On the other hand, the LANA staining pattern changed to dots in the nucleus when BAC-TR was introduced, and the LANA immunofluorescence was highly colocalized with SUV39H1 (Fig. 3C, panels 4 to 6). These observations indicated that the interaction between LANA and heterochromatin depended on the presence of the TR in vivo.
293 cells were useful for the assay described above because of the high efficiency of transfection that could be attained. However, it is difficult to observe the heterochromatin and nuclear structure in these cells. To overcome this difficulty, Vero cells stably expressing a GFP-fused LANA were obtained, and either BAC-TR or the control BAC construct (BAC-neor) was transfected into them, resulting in the establishment of Vero/gfpLANA+BAC-neor and Vero/gfpLANA+BAC-TR (Fig. 3D, panels 4 to 11 and 12 to 15, respectively). BAC-neor was retained in the cells, probably due to integration of the neomycin resistance gene. With these cells, an immunofluorescence assay with the mouse monoclonal antibody against HP-1α was performed. In Vero/gfpLANA+BAC-neor cells, a diffuse pattern of GFP-LANA was observed in the chromatin region (Fig. 3D, panels 5 and 9), in which the endogenous HP-1α localization was disturbed (Fig. 3D, panels 4 and 8) and excluded from its original site observed in the parental Vero cells (Fig. 3D, panel 1), in which a dense and focal deposition of HP-1α was observed along with a weak and diffuse background. Thus, the typical colocalization of LANA and HP-1 was not observed (Fig. 3D, panels 7 and 11), in agreement with Fig. 1C. In the presence of the artificial KSHV genome, LANA localized to the heterochromatin region, where it was colocalized with HP-1α (Fig. 3D, panels 12 to 15). Taken together, these observations indicated that LANA's binding with the TR caused it to interact with SUV39H1 in vivo and, finally, that the viral genome was associated with heterochromatin during latency.
LANA represses the expression of a luciferase reporter bearing the TR, and the repression is enhanced by SUV39H1.
To test the effect of heterochromatin formation mediated by LANA on the transcription of the viral genome, we designed a transient reporter assay. Eight copies of TR from the cosmid clone Z6 were linked to a luciferase reporter gene regulated by the KSHV RTA promoter (Fig. 4A). The RTA promoter is one of the strongest promoters in the KSHV genome and regulates RTA expression, which launches the reactivation of KSHV (9, 31, 45). 293 cells were transfected with the reporter plasmid and either the LANA expression vector or empty vector (pTriEX-LANA or pTriEX1.1). Thirty-six hours later, the cells were harvested and the luciferase activity was measured. pTR-RpLuc-009 (p009) contained the TR, while most of this region was removed in pTR-RpLuc-903 (p903). As shown in Fig. 4B, the luciferase activity of p009 was decreased by LANA in a dose-dependent manner, although that of p903 was not affected. In the reporter plasmid, the RTA promoter was located far away (about 3 kbp) from the LBSs, suggesting that LANA could repress transcription over a long distance and that the repression is dependent on TR.
We next used the reporter assay to test the enhancement of the transcriptional repression by SUV39H1. It was observed that coexpression of LANA with SUV39H1 further enhanced the repression in dose-dependent manner (Fig. 4C). The results of the pull-down assay (Fig. 2A) had indicated that the amino-terminal domain of SUV39H1 was required for its interaction with LANA. The FLAG-tagged SUV39H1 lacking its amino terminus (SUV39ΔN), which encoded aa 113 to 412, was examined in the reporter assay. As expected, the deletion of the domain that was important for the interaction abolished the enhancement of the transcriptional repression by LANA (Fig. 4C). Comparable expression levels of SUV39H1 and SUV39ΔN were confirmed by Western blotting when each expression vector (0.6 μg/well) was transfected into 293 cells (Fig. 4D). Taken together, these results suggested that the repression effect of LANA on transcription should be attributed to its recruitment of SUV39H1, which methylates histone H3, leading to HP-1 accumulation.
Chromatin within the TR associates with heterochromatin components in BC-3 cells.
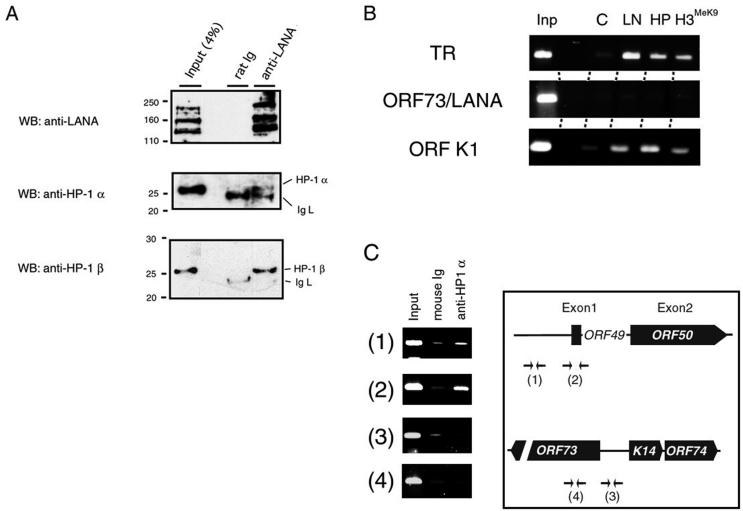
To confirm the heterochromatin association with LANA in KSHV-infected cells, in vivo cross-linked chromatin was subjected to the immunoprecipitation assay. We first tested whether HP-1 was involved in a LANA-associated complex. As shown in Fig. 5A, both HP-1α and -β were precipitated with the anti-LANA antibody but not by control rat immunoglobulin. This result showed that the complex containing LANA associated with heterochromatin factors, such as HP-1.
FIG. 5.
ChIP. (A) ChIP and Western blotting (WB) with BC-3 cells. In vivo cross-linked chromatin was digested with micrococcal nuclease (DNA length of < 1kb) and immunoprecipitated with 1 μg of a rat anti-LANA antibody or control rat immunoglobulin (Dako). The precipitated matrix was analyzed by Western blotting with an anti-LANA antibody, an anti-HP-1α antibody (Euromedex), and an anti-HP-1β antibody (Euromedex). IgL, light chain of immunoglobulin. (B) Chromatin states of the TR and the K1 and LANA promoters. PCR was performed with primers for the TR, the K1 gene, and the promoter of the latent transcript (LANA) after ChIP. The amplified products were separated by electrophoresis in a 2% agarose gel and stained with ethidium bromide (Inp, 5% of input; C, control rat immunoglobulin; LN, rat anti-LANA monoclonal antibody [Advanced Biotechnologies Inc.]; HP, mouse anti-HP-1α monoclonal antibody [Upstate]; H3MeK9, mouse anti-methylated histone H3 monoclonal antibody [Upstate]). (C) The KSHV ORF50/RTA region was specifically precipitated by the anti-HP-1α antibody in the ChIP assay. Precipitated DNA was reacted in the PCR with the primers for the ORF50/RTA locus (panels 1 and 2) or the ORF73/LANA locus (panels 3 and 4), and the results of 2% agarose-TBE electrophoresis are shown. A schematic presentation of KSHV ORF50/RTA and ORF73/LANA is also shown. Arrows indicated the priming site of each PCR primer.
Next, the immunoprecipitate was used as a template for PCR with primers designed to amplify the TR, LANA promoter, and K1 promoter. Almost no amplification was observed in any of the samples immunoprecipitated with rat immunoglobulin (Fig. 5B, lane C). Amplified TR and K1 promoter DNAs were observed in the immunoprecipitate brought down with the anti-HP-1 antibody and the anti-methylated histone H3 antibody at levels comparable to those brought down by the anti-LANA antibody (Fig. 5B). This result indicated that the heterochromatin factors were accumulated at the TR region and the K1 promoter during KSHV latency. The K1 gene, which is close to the TR region and is not transcribed during latency, encodes a type 1 transmembrane glycoprotein that is an immunoreceptor with a tyrosine-based activation motif (24). This finding is also consistent with the recent report by Cotter and Robertson that LANA also binds to a sequence that partially overlaps the K1 ORF (10).
In contrast, these antibodies did not precipitate the active promoter in latency for three genes, LANA, v-CYC, and v-FLIP, whose transcripts are regulated by alternative splicing and an internal ribosome entry site (15, 28, 41) (Fig. 5B). This was also consistent with the expression profiles of these genes, because the association of a gene with HP-1 and methylated histone H3 indicates that it is in a suppressed state. In conclusion, the interaction of LANA with the TR seemed to elicit heterochromatin formation on the viral genome, by recruiting a histone methyltransferase, SUV39H1, which results in the association of HP-1 and methylated histone H3 within the TR region, leading to the suppression of lytic gene expression, such as that of K1.
Finally, the RTA gene locus (ORF50) was analyzed in a ChIP assay to verify the association of heterochromatin components with RTA expression and latent-lytic switching. During the early onset of the lytic phase, expression of the viral transactivator RTA is induced by an unknown trigger to disrupt the latency. ORF50/RTA is highly suppressed in cells latently infected with KSHV (18), and phorbol esters and sodium butyrate can induce RTA expression (38). The results of the ChIP assay indicated specific binding of HP-1 to the ORF50 locus (Fig. 5C). This finding suggested that the switching from latency to the lytic cycle of KSHV could be regulated by the epigenetic state of the viral genome.
DISCUSSION
KSHV LANA is thought to be an essential protein for viral latency. In KSHV-infected cells, LANA marks the viral genome, and its localization to the heterochromatin area (Fig. 1) led us to speculate that the viral gene expression during latency is regulated by certain heterochromatin proteins; the regulation mechanisms include position effects and the insulation of genes from the effects of heterochromatin. However, colocalization of LANA and HP-1α was not perfect, probably due to a distinct localization of HP-1 isoforms (HP-1β and -γ). Despite their sequence conservation, HP-1 isoforms differ in their localization and mitotic distribution (30).
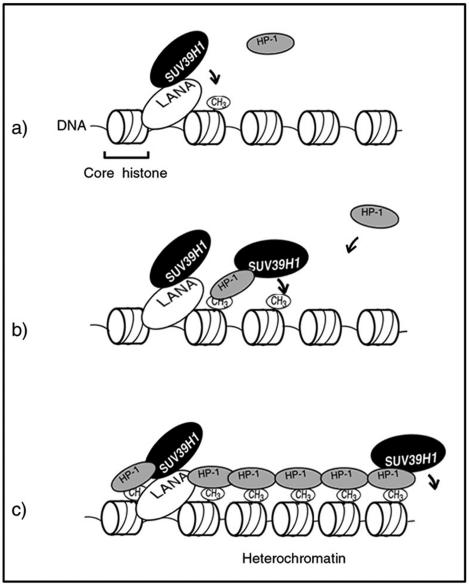
We showed that TR-bound LANA interacted with SUV39H1 to suppress transcription of the viral genome. Mapping experiments showed that the DE-rich domain in the N terminus of LANA was an SUV39H1-interacting domain (Fig. 2B). Many cellular nuclear proteins, such as RING3, Rb, p53, and MeCP2, have been reported to be interaction partners of LANA (12, 21, 35, 36). The domains required for each of these interactions are shown in Fig. 2B. In addition, it was notable that the domain of LANA that interacted with SUV39H1 was distinct from that reported to interact with HP-1α directly (26). However, it was still unclear whether LANA just tethered the viral genome to the host heterochromatin or whether it built the heterochromatin structure on the viral genome for the transcriptional regulation. Our findings confirmed that the colocalization of LANA with HP-1 observed in BC-3 cells (Fig. 1A and B) was a result of their interaction with SUV39H1, since this methyltransferase catalyzes the methylation of lysine residue K9 on histone H3, leading to heterochromatin formation in the TR region (Fig. 5), and because the HP-1 recruitment to histone H3 K9 is methylation specific and continuous (Fig. 6 [our working hypothesis]). Once this structure is made, then the structure spreads via the interaction between HP-1 and SUV39H1. While the heterochromatin state is stably inherited during cell divisions in general (14), our model explains how KSHV latency is retained in the infected cells without viral lytic gene expression.
FIG. 6.
Working hypothesis for viral transcriptional control by LANA in latency. (a) First, LANA binds with the TR DNA sequence in the KSHV genome and recruits the SUV39H1 histone methyltransferase. SUV39H1 methylates lysine K9 in the amino terminus of histone H3, where HP-1 is specifically recruited. (b) HP-1 then interacts with SUV39H1, resulting in the methylation of neighboring histones. (c) Finally, the heterochromatin architecture spreads over the viral genome. Some genes might be suppressed if their regulatory elements lie in the heterochromatin region.
The relocation of LANA in uninfected cells by the exogenous expression of heterochromatin factors is controversial. One report described an interaction of LANA with MeCP2 (21), which binds CpG-methylated DNA and always localizes to heterochromatin (25, 32). MeCP2 physically interacts with the chromosome-binding site adjacent to the nuclear localization signal in the N terminus of LANA and relocates LANA to the heterochromatin dots in the absence of the viral genome (21). In that report, it was also shown that LANA and exogenous human HP-1α were not colocalized in mouse NIH 3T3 cells. We did not observe their colocalization in 293 cells in the absence of the KSHV genome either (Fig. 1C). On the other hand, Lim et al. reported that LANA was relocated to the nuclear dots by exogenous HP-1 in 293T cells (26).
The tandem-repeated TR caused the focal concentration of LANA in non-KSHV-infected cells (Fig. 3B), which was also seen in LANA-expressing BJAB cells (Burkitt lymphoma KSHV- and EBV-negative cells) with artificial KSHV episomes (2). Although the colocalization of LANA and SUV39H1 was not observed in non-KSHV-infected cells (Fig. 3C, panels 1 to 3), SUV39H1 was highly colocalized with LANA in the presence of BAC-TR (Fig. 3C, panels 4 to 6). We have shown that the LANA bound with TR DNA (LBS) could interact with SUV39H1 (Fig. 3A), indicating that SUV39H1 was recruited by LANA to the TR. Thus, a structural change in LANA as a result of its binding to TR might be important for the complex formation.
LANA has another binding site in the K1 locus in vitro (11), and we confirmed by ChIP assay that LANA and heterochromatin factors bound to the K1 gene locus in vivo (Fig. 5B). The K1 promoter region, which is located at the left end of the KSHV genome, was recently analyzed (7). It showed relatively high activity in various cell lines in a transfection study, but its expression was observed in only a few PEL cells during latency, probably due to spontaneous viral reactivation. This difference could be explained by the recruitment of heterochromatin by LANA to the viral genome in the infected cells. On the other hand, LANA's promoter region did not contain heterochromatin factors such as HP-1 and methylated histone H3. This explains why the LANA promoter is active in latency, even if other viral promoters are not.
Here we showed that the recruitment of SUV39H1 and then HP-1 to the KSHV genome by LANA could result in the formation of heterochromatin, which governs the latent transcription program. The RTA promoter was blocked by the HP-1 recruitment (Fig. 5C), which is heritable and stable.
However, some latent genes (LANA, vCYC, vFLIP, vIRF2, and vIRF3/LANA2) can be expressed under these circumstances. There may be several elements that can insulate these latent genes from the effects of heterochromatin. For example, some CTCF motifs are found in the DNA sequence of the KSHV genome. CTCF is a DNA-binding factor that is responsible for enhancer-blocking activity of boundaries at the chicken β-globin locus (5′HS4 and 3′HS) to shield the locus from regulatory effects of neighboring genes (5, 39).
The next questions will be how the heterochromatin status of the viral genome changes upon viral reactivation and what distinguishes promoters that are active in latency from those that are not. The answers to such questions will lead to a total understanding of the viral transcription and replication program in latency and reactivation.
Acknowledgments
We thank Y. B. Jiang, M. Kinoshita, and D. Hayami for technical support. We also thank T. Sata for the rabbit anti-LANA polyclonal antibodies.
This work was supported by PRESTO, Japanese Science and Technology Corporation (JST).
REFERENCES
- 1.Aagaard, L., G. Laible, P. Selenko, M. Schmid, R. Dorn, G. Schotta, S. Kuhfittig, A. Wolf, A. Lebersorger, P. B. Singh, G. Reuter, and T. Jenuwein. 1999. Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3-9 encode centromere-associated proteins which complex with the heterochromatin component M31. EMBO J. 18:1923-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ballestas, M. E., P. A. Chatis, and K. M. Kaye. 1999. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284:641-644. [DOI] [PubMed] [Google Scholar]
- 3.Ballestas, M. E., and K. M. Kaye. 2001. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 mediates episome persistence through cis-acting terminal repeat (TR) sequence and specifically binds TR DNA. J. Virol. 75:3250-3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bannister, A. J., P. Zegerman, J. F. Partridge, E. A. Miska, J. O. Thomas, R. C. Allshire, and T. Kouzarides. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410:120-124. [DOI] [PubMed] [Google Scholar]
- 5.Bell, A. C., A. G. West, and G. Felsenfeld. 1999. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98:387-396. [DOI] [PubMed] [Google Scholar]
- 6.Boshoff, C., and R. A. Weiss. 2001. Epidemiology and pathogenesis of Kaposi's sarcoma-associated herpesvirus. Philos. Trans. R Soc. London B 356:517-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bowser, B. S., S. M. DeWire, and B. Damania. 2002. Transcriptional regulation of the K1 gene product of Kaposi's sarcoma-associated herpesvirus. J. Virol. 76:12574-12583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 9.Chen, J., K. Ueda, S. Sakakibara, T. Okuno, C. Parravicini, M. Corbellino, and K. Yamanishi. 2001. Activation of latent Kaposi's sarcoma-associated herpesvirus by demethylation of the promoter of the lytic transactivator. Proc. Natl. Acad. Sci. USA 98:4119-4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cotter, M. A., II, and E. S. Robertson. 1999. The latency-associated nuclear antigen tethers the Kaposi's sarcoma-associated herpesvirus genome to host chromosomes in body cavity-based lymphoma cells. Virology 264:254-264. [DOI] [PubMed] [Google Scholar]
- 11.Cotter, M. A., II, C. Subramanian, and E. S. Robertson. 2001. The Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen binds to specific sequences at the left end of the viral genome through its carboxy-terminus. Virology 291:241-259. [DOI] [PubMed] [Google Scholar]
- 12.Friborg, J., Jr., W. Kong, M. O. Hottiger, and G. J. Nabel. 1999. p53 inhibition by the LANA protein of KSHV protects against cell death. Nature 402:889-894. [DOI] [PubMed] [Google Scholar]
- 13.Garber, A. C., M. A. Shu, J. Hu, and R. Renne. 2001. DNA binding and modulation of gene expression by the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:7882-7892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grewal, S. I., and D. Moazed. 2003. Heterochromatin and epigenetic control of gene expression. Science 301:798-802. [DOI] [PubMed] [Google Scholar]
- 15.Grundhoff, A., and D. Ganem. 2001. Mechanisms governing expression of the v-FLIP gene of Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:1857-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365-369. [DOI] [PubMed] [Google Scholar]
- 17.Judde, J. G., V. Lacoste, J. Briere, E. Kassa-Kelembho, E. Clyti, P. Couppie, C. Buchrieser, M. Tulliez, J. Morvan, and A. Gessain. 2000. Monoclonality or oligoclonality of human herpesvirus 8 terminal repeat sequences in Kaposi's sarcoma and other diseases. J. Natl. Cancer Inst. 92:729-736. [DOI] [PubMed] [Google Scholar]
- 18.Katano, H., Y. Sato, H. Itoh, and T. Sata. 2001. Expression of human herpesvirus 8 (HHV-8)-encoded immediate early protein, open reading frame 50, in HHV-8-associated diseases. J. Hum. Virol. 4:96-102. [PubMed] [Google Scholar]
- 19.Katano, H., Y. Sato, T. Kurata, S. Mori, and T. Sata. 1999. High expression of HHV-8-encoded ORF73 protein in spindle-shaped cells of Kaposi's sarcoma. Am. J. Pathol. 155:47-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kedes, D. H., M. Lagunoff, R. Renne, and D. Ganem. 1997. Identification of the gene encoding the major latency-associated nuclear antigen of the Kaposi's sarcoma-associated herpesvirus. J. Clin. Investig. 100:2606-2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krithivas, A., M. Fujimuro, M. Weidner, D. B. Young, and S. D. Hayward. 2002. Protein interactions targeting the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus to cell chromosomes. J. Virol. 76:11596-11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lachner, M., D. O'Carroll, S. Rea, K. Mechtler, and T. Jenuwein. 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410:116-120. [DOI] [PubMed] [Google Scholar]
- 23.Ladenburger, E. M., C. Keller, and R. Knippers. 2002. Identification of a binding region for human origin recognition complex proteins 1 and 2 that coincides with an origin of DNA replication. Mol. Cell. Biol. 22:1036-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, H., J. Guo, M. Li, J. K. Choi, M. DeMaria, M. Rosenzweig, and J. U. Jung. 1998. Identification of an immunoreceptor tyrosine-based activation motif of K1 transforming protein of Kaposi's sarcoma-associated herpesvirus. Mol. Cell. Biol. 18:5219-5228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lewis, J. D., R. R. Meehan, W. J. Henzel, I. Maurer-Fogy, P. Jeppesen, F. Klein, and A. Bird. 1992. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell 69:905-914. [DOI] [PubMed] [Google Scholar]
- 26.Lim, C., D. Lee, T. Seo, C. Choi, and J. Choe. 2003. Latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus functionally interacts with heterochromatin protein 1. J. Biol. Chem. 278:7397-7405. [DOI] [PubMed] [Google Scholar]
- 27.Litt, M. D., M. Simpson, M. Gaszner, C. D. Allis, and G. Felsenfeld. 2001. Correlation between histone lysine methylation and developmental changes at the chicken beta-globin locus. Science 293:2453-2455. [DOI] [PubMed] [Google Scholar]
- 28.Low, W., M. Harries, H. Ye, M. Q. Du, C. Boshoff, and M. Collins. 2001. Internal ribosome entry site regulates translation of Kaposi's sarcoma-associated herpesvirus FLICE inhibitory protein. J. Virol. 75:2938-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Melcher, M., M. Schmid, L. Aagaard, P. Selenko, G. Laible, and T. Jenuwein. 2000. Structure-function analysis of SUV39H1 reveals a dominant role in heterochromatin organization, chromosome segregation, and mitotic progression. Mol. Cell. Biol. 20:3728-3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Minc, E., Y. Allory, H. J. Worman, J. C. Courvalin, and B. Buendia. 1999. Localization and phosphorylation of HP1 proteins during the cell cycle in mammalian cells. Chromosoma 108:220-234. [DOI] [PubMed] [Google Scholar]
- 31.Nakamura, H., M. Lu, Y. Gwack, J. Souvlis, S. L. Zeichner, and J. U. Jung. 2003. Global changes in Kaposi's sarcoma-associated virus gene expression patterns following expression of a tetracycline-inducible Rta transactivator. J. Virol. 77:4205-4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nan, X., P. Tate, E. Li, and A. Bird. 1996. DNA methylation specifies chromosomal localization of MeCP2. Mol. Cell. Biol. 16:414-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okuno, T., Y. B. Jiang, K. Ueda, K. Nishimura, T. Tamura, and K. Yamanishi. 2002. Activation of human herpesvirus 8 open reading frame K5 independent of ORF50 expression. Virus Res. 90:77-89. [DOI] [PubMed] [Google Scholar]
- 34.Piolot, T., M. Tramier, M. Coppey, J. C. Nicolas, and V. Marechal. 2001. Close but distinct regions of human herpesvirus 8 latency-associated nuclear antigen 1 are responsible for nuclear targeting and binding to human mitotic chromosomes. J. Virol. 75:3948-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Platt, G. M., G. R. Simpson, S. Mittnacht, and T. F. Schulz. 1999. Latent nuclear antigen of Kaposi's sarcoma-associated herpesvirus interacts with RING3, a homolog of the Drosophila female sterile homeotic (fsh) gene. J. Virol. 73:9789-9795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Radkov, S. A., P. Kellam, and C. Boshoff. 2000. The latent nuclear antigen of Kaposi sarcoma-associated herpesvirus targets the retinoblastoma-E2F pathway and with the oncogene Hras transforms primary rat cells. Nat. Med. 6:1121-1127. [DOI] [PubMed] [Google Scholar]
- 37.Rainbow, L., G. M. Platt, G. R. Simpson, R. Sarid, S. J. Gao, H. Stoiber, C. S. Herrington, P. S. Moore, and T. F. Schulz. 1997. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J. Virol. 71:5915-5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Renne, R., W. Zhong, B. Herndier, M. McGrath, N. Abbey, D. Kedes, and D. Ganem. 1996. Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat. Med. 2:342-346. [DOI] [PubMed] [Google Scholar]
- 39.Saitoh, N., A. C. Bell, F. Recillas-Targa, A. G. West, M. Simpson, M. Pikaart, and G. Felsenfeld. 2000. Structural and functional conservation at the boundaries of the chicken beta-globin domain. EMBO J. 19:2315-2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sakakibara, S., K. Ueda, J. Chen, T. Okuno, and K. Yamanishi. 2001. Octamer-binding sequence is a key element for the autoregulation of Kaposi's sarcoma-associated herpesvirus ORF50/Lyta gene expression. J. Virol. 75:6894-6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sarid, R., J. S. Wiezorek, P. S. Moore, and Y. Chang. 1999. Characterization and cell cycle regulation of the major Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) latent genes and their promoter. J. Virol. 73:1438-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwam, D. R., R. L. Luciano, S. S. Mahajan, L. Wong, and A. C. Wilson. 2000. Carboxy terminus of human herpesvirus 8 latency-associated nuclear antigen mediates dimerization, transcriptional repression, and targeting to nuclear bodies. J. Virol. 74:8532-8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Szekely, L., C. Kiss, K. Mattsson, E. Kashuba, K. Pokrovskaja, A. Juhasz, P. Holmvall, and G. Klein. 1999. Human herpesvirus-8-encoded LNA-1 accumulates in heterochromatin-associated nuclear bodies. J. Gen. Virol. 80:2889-2900. [DOI] [PubMed] [Google Scholar]
- 44.Ueda, K., K. Ishikawa, K. Nishimura, S. Sakakibara, E. Do, and K. Yamanishi. 2002. Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) replication and transcription factor activates the K9 (vIRF) gene through two distinct cis elements by a non-DNA-binding mechanism. J. Virol. 76:12044-12054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.West, J. T., and C. Wood. 2003. The role of Kaposi's sarcoma-associated herpesvirus/human herpesvirus-8 regulator of transcription activation (RTA) in control of gene expression. Oncogene 22:5150-5163. [DOI] [PubMed] [Google Scholar]